Abstract
Ras is a key signal transduction protein in the cell. Mutants of Gly12 and Gln61 impair GTPase activity and are found prominently in cancers. In wild type Ras-GTP, an allosteric switch promotes disorder to order transition in switch II, placing Gln61 in the active site. We show that the “on” and “off” conformations of the allosteric switch can also be attained in RasG12V and RasQ61L. Although both mutants have similarly impaired active sites in the on state, RasQ61L stabilizes an anti-catalytic conformation of switch II in the off state of the allosteric switch when bound to Raf. This translates into more potent activation of the MAPK pathway involving Ras, Raf kinase, MEK, and ERK (Ras/Raf/MEK/ERK) in cells transfected with RasQ61L relative to RasG12V. This differential is not observed in the Raf-independent pathway involving Ras, phosphoinositide 3-kinase (PI3K), and Akt (Ras/PI3K/Akt). Using a combination of structural analysis, hydrolysis rates, and experiments in NIH-3T3 cells, we link the allosteric switch to the control of signaling in the Ras/Raf/MEK/ERK pathway, supporting a GTPase-activating protein-independent model for duration of the Ras-Raf complex.
Keywords: Allosteric Regulation, Crystal Structure, ERK, G Proteins, Protein Structure, Raf, Ras
Introduction
Ras GTPase is a central protein in signal transduction pathways in the cell that mediate the control of cell proliferation as well as many other functions (1, 2). It is isoprenylated at the C terminus through which it is anchored to the cell membrane (3). When bound to GTP, Ras propagates its signal by interacting with effector proteins such as Raf (4) and phosphoinositide 3-kinase (PI3K) (5) among others (6, 7). Once GTP is hydrolyzed to GDP on Ras, complexes with effectors are no longer favored, and signaling is turned off. The signal is thus directly connected to the level of Ras-GTP, which in turn is kept in check by the opposing actions of guanine nucleotide exchange factors that catalyze the loading of GTP (8), and of GTPase-activating proteins (GAPs)2 that increase the intrinsically slow GTPase activity of Ras for timely depletion of Ras-GTP (9).
We have recently discovered a mechanism through which the active site of Ras can be modulated by ligand binding at an allosteric site near the Ras-membrane interface, suggesting a GAP-independent mechanism through which hydrolysis rates could be increased (10). The binding of calcium and a negatively charged ligand, mimicked in our Ras-GppNHp crystal structure by an acetate molecule, promote a network of H-bonding interactions that connect the allosteric site to switch II, resulting in a disorder to order transition that positions key residues for catalysis (10). Thus, an “on” state of the allosteric switch implies increased hydrolysis rates associated with signaling being turned off. When the allosteric switch is “off,” GTP hydrolysis is impaired and signaling remains on.
The active site residues are situated primarily in the so-called switch I, switch II, and the phosphate-binding loop (P-loop) composed of residues 30–40, 60–76, and 10–17, respectively (11). In particular, mutants of residues Gly12 in the P-loop and Gln61 in switch II are often highly oncogenic and have been the focus of numerous structural biology (12–15), biochemical (16, 17), and cell biology studies (18, 19), with RasG12V and RasQ61L receiving great attention for being highly transforming mutants commonly found in human cancers (20, 21). These residues are near the γ-phosphate and oxygen atom that bridges the β- and γ-phosphate groups in GTP. The dissociative-like reaction mechanism observed in the presence and absence of GAP (22, 23) requires accumulation of negative charge at the β-γ-bridging oxygen atom of GTP in the transition state of the hydrolysis reaction (24). Stabilization of this charge in the GAP-catalyzed reaction is aided by the arginine finger (Arg789), which GAP inserts in the active site, and the ordering of both switch I and switch II at the Ras/RasGAP interface (9). The Ras-GppNHp crystal form in which wild type and most mutant Ras proteins have been studied until recently have symmetry of the space group P3221, and the active site has a conformation similar to that found in the Ras-GppNHp-RasGAP complex (25). The structures of RasG12V and RasQ61L in this crystal form reveal active sites that are very similar in conformation to the wild type structure and give insight into how these mutants interfere with the GAP-catalyzed reaction (13). These structures do not account for an effect on the active site configured by the allosteric switch and the resulting consequences for intrinsic hydrolysis in the Ras-Raf complex.
All known effector protein-binding sites on Ras overlap with that for GAPs and most interact with micromolar affinities similar to that for the Ras/RasGAP interaction (5, 26–28). The striking exception is the effector Raf, which mediates signaling through the Ras/Raf/MEK/ERK signal transduction cascade linked directly to the control of cell proliferation (2). The Raf-binding site overlaps with that for GAP through switch I but not switch II (4), yet its affinity to Ras is 3.5 nm (29), 3 orders of magnitude greater than the 2 μm Ras affinity for p120GAP (27, 28). Thus, although effectors such as PI3K could be easily displaced by GAP, we propose that the timing of the Ras/Raf interaction is controlled by intrinsic hydrolysis where switch I is modulated by the binding of Raf, and switch II is positioned for catalysis by the allosteric switch (10). In this case, switch I residue Tyr32 is situated in a position similar to that of the GAP arginine finger, where it interacts with a water molecule (WAT189) that bridges it to the γ-phosphate of GTP (PDB code 3K8Y). Gln61 is positioned by the allosteric switch to interact with this same water molecule, which could receive a proton and develop a partial positive charge near the β-γ-bridging oxygen atom of GTP during catalysis. We propose that WAT189, aided by Tyr32 and Gln61, is a critical element in stabilizing the transition state of the reaction catalyzed by Ras in the absence of GAP when the allosteric switch is in the on state (10). The details of the mechanism proposed based on this active site conformation were derived from our structure of wild type Ras-GppNHp obtained from a crystal of symmetry R32, where switch I and active site water molecules are as found in the Raps-GppNHp-RafRBD complex (30). Our published structure of RasQ61L-GppNHp in this crystal form has a switch II that closes over the nucleotide with Leu61 at the center of a hydrophobic cluster that isolates the γ-phosphate from bulk solvent (31). Based on this structure, we predicted that the hydrolysis rate in RasQ61L is severely impaired by Raf. We suggested then that it is in the presence of Raf that this mutant exerts its potently transforming phenotype, because Raf stabilizes a switch I conformation that is critical to the hydrophobic cluster.
In this work, we provide evidence that our interpretation of the role of Raf in relation to the RasQ61L mutant is functionally relevant in cells. We use structural analysis and in vitro hydrolysis experiments to show that the G12V and Q61L mutations perturb the on state active site in similar ways but that in the presence of Raf the accessibility to a catalytically competent, albeit impaired, conformation is greatly diminished in RasQ61L but unaffected in RasG12V under conditions lacking an activator of the allosteric switch. We then show that RasG12V and RasQ61L exhibit distinct behavior in terms of MEK and ERK but not Akt phosphorylation in signaling involving the Raf/MEK/ERK and PI3K/Akt pathways in NIH-3T3 cells. Structural features of switch I, switch II, and the allosteric switch in the two mutants correlate with their behavior in vivo. Taken together, the structural biology, kinetics, and cell biology experiments presented here point to the Ras-Raf complex as timing the signal through the Ras/Raf/MEK/ERK pathway, modulated by the Ras allosteric switch.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization
C-terminally truncated H-Ras, consisting of residues 1–166, cloned into the pET21 expression vector was used for generating the RasG12V mutant with the QuikChange II kit from Stratagene, following the manufacturer's instructions. RasQ61L had been obtained previously in our laboratory (31). Expression, purification, and nucleotide exchange for loading the GTP analog GppNHp in both RasG12V and RasQ61L were as described previously for the wild type protein (25).
Crystals of RasG12V-GppNHp with the allosteric switch in either the on or off states were obtained by the hanging drop vapor diffusion method in 24-well plates with drops containing 3 μl of protein and 3 μl of reservoir solutions. In both cases, crystals grew in about 2 weeks at 18 °C. The protein was prepared in stabilization buffer (20 mm Hepes, pH 7.5, 50 mm NaCl, 20 mm MgCl2, and 10 mm DTT) at 12 and 14.4 mg/ml for the on and off states, respectively. For crystals of the on state, the reservoir solution consisted of 500 ml of 200 mm calcium acetate and 20% PEG 3350 (Hampton Research PEG Ion Screen number 28). Crystals were transferred to a solution containing stabilization buffer plus 15% glycerol, 20% PEG 3350, and 200 mm calcium acetate before flash freezing for data collection. For crystals in the off state, the reservoir solution contained 400 μl of 200 mm calcium chloride, 20% PEG 3350 (Hampton Research PEG Ion Screen number 7), 120–160 μl of stabilization buffer, and 0–30 μl of 50% PEG 3350. Prior to flash freezing, crystals were placed in a solution consisting of 80% PEG Ion Screen number 7 (Hampton Research) and 20% PEG 400.
Crystals of RasQ61L-GppNHp with the allosteric switch in the on state were obtained by the sitting drop vapor diffusion method in 96-well plates (3-well Intelli-PlateTM, ARI) with drops containing 1 μl of protein and 1 μl of reservoir solutions. The protein was at a concentration of 12 mg/ml in stabilization buffer. The reservoir solution contained 365 mm calcium acetate and 24% PEG 3350 in stabilization buffer. Crystal trays were set using a Phoenix crystallization robot (ARI). Crystals grew in about 1 week at 18 °C and were transferred to a buffer containing 80% reservoir solution and 20% PEG 400 before flash freezing for data collection.
Synchrotron data were collected for the three structures at a temperature of 100 K and x-ray wavelength of 1.0 Å at the Southeast Regional Collaborative Access Team (SER-CAT) beamline, Advanced Photon Source (APS), Argonne, IL. Data were processed with HKL2000 (32) and structures refined with PHENIX (33) and Coot (34). The wild type structures with the allosteric switch in the on state (PDB code 3K8Y) and off state (PDB code 2RGE) were used in molecular replacement for phasing the corresponding structures of the mutants. The RasG12V on, RasG12V off, and RasQ61L on structures had 96.99, 96.27, and 98.20% dihedral angles in favored regions of the Ramachandran plot, respectively, and there were no outliers. Data collection and refinement statistics are in Table 1.
TABLE 1.
Diffraction data collection and structure refinement statistics
A single crystal was used for each structure. The values in parentheses are for the highest resolution shell. NA means not applicable.
| RasG12V-GppNHp allosteric switch on | RasG12V-GppNHp allosteric switch off | RasQ61L-GppNHp allosteric switch on | |
|---|---|---|---|
| Data collection | |||
| Space group | R32 | R32 | R32 |
| Cell dimensions | |||
| a, b, c | 88.52, 88.52, 134.39 ; | 87.88, 87.88, 133.06 Å | 87.99, 87.99, 135.15 Å |
| α, β, γ | 90, 90, 120° | 90, 90, 120° | 90, 90, 120° |
| Resolution | 50-1.24 Å (1.29 to 1.24 Å) | 50-1.84 Å (1.91 to 1.84 Å) | 50-1.32 Å (1.35 to 1.32 Å) |
| Rsym or Rmerge | 0.056 (0.853) | 0.131 (0.643) | 0.103 (0.859) |
| I/sI | 54 (2) | 18 (2) | 56 (3.2) |
| Completeness | 90.7% (82.7%) | 94.6% (81.3%) | 93.6% (82.5%) |
| Redundancy | 9.5 (7.1) | 10.1 (7.1) | 9.7 (7.4) |
| Refinement | |||
| Resolution | 26.6 to 1.30 Å | 26.4 to 1.84 Å | 30.89 to 1.32 Å |
| No. of reflections | 47,378 | 16,527 | 44,574 |
| Rwork/Rfree | 19.6/21.0 | 15.8/19.8 | 18.2/19.2 |
| No. of atoms | |||
| Protein | 1330 | 1294 | 1334 |
| GppNHp | 32 | 32 | 32 |
| Acetate/DTT | 4/0 | 0/16 | 4/0 |
| Calcium/magnesium | 3/2 | 1/3 | 3/2 |
| Water | 180 | 148 | 172 |
| B-factors | |||
| Protein | 18.21 | 18.16 | 18.52 |
| GppNHp | 13.26 | 11.80 | 12.04 |
| Acetate/DTT | 24.51/NA | NA/48.46 | 23.16/NA |
| Calcium/magnesium | 21.93/12.18 | 13.64/25.70 | 24.51/12.10 |
| Water | 27.92 | 28.73 | 27.68 |
| Root mean square deviations | |||
| Bond lengths | 0.007 Å | 0.008 Å | 0.007 Å |
| Bond angles | 1.13° | 1.22° | 1.20° |
Measurement of Hydrolysis Rates
The methods used for the hydrolysis experiments presented here involved the exchange of GDP for [γ-32P]GTP in the purified mutant Ras proteins and were as described in detail for wild type Ras and RasY32F (10). The c-Raf construct used to test the effects of Raf on the hydrolysis rates of RasG12V and RasQ61L includes both the Ras binding domain and cysteine-rich Ras-binding domain (residues 52–184) fused at the N terminus to residues 1–56 of the immunoglobulin-binding domain of streptococcal protein G (GB1). This construct was expressed, purified, stored, and used as described previously (10).
The rates of hydrolysis are given in Table 2 as 1/t in units of min−1 and represent the mean value of duplicate experiments. The standard deviations from the mean are given in each case. Because the rates of intrinsic hydrolysis for RasG12V and RasQ61L are too slow for the reaction to go to completion in the time frame where single turnover conditions are unaffected, mutant hydrolysis experiments were run side by side with wild type Ras hydrolysis, and the final count for mutant hydrolysis was obtained by counting the total radioactivity of a given aliquot, without charcoal treatment, normalized by 100% wild type Ras hydrolysis counts.
TABLE 2.
Hydrolysis rates for Ras, RasG12V, and RasQ61L in the absence and presence of Raf
Rates given are averages of two independent experiments. NA means not applicable.
| Ras | Raf_RBD_CRD | 1/t0 | Standard deviation | Rate WT/Rate measured |
|---|---|---|---|---|
| min−1 | ||||
| Wild type | No | 0.01565 | 0.00171 | 1 |
| Yes | 0.01600 | 0.00180 | 1 | |
| G12V | No | 0.00137 | 0.00033 | 11 |
| Yes | 0.00136 | 0.00037 | 11 | |
| Q61L | No | 0.00061 | 0.00002 | 26 |
| Yes | Not measurablea | NA | NA |
a The radioactivity counts measured for RasQ61L in the presence of Raf do not change with time in the first 40 min where one would expect a linear response and are within the background level obtained in control experiments where Ras is replaced by Raf in our protocol. This background noise level would translate into an apparent hydrolysis rate of 0.0002 min−1 and defines an approximate lower limit of detection.
In Vivo Experiments with NIH-3T3 Cells
All tissue culture reagents were from Invitrogen. Human recombinant PDGF-BB was purchased from PeproTech (Rocky Hill, NJ). Antibodies against total ERK1/2 and pan-Ras and phospho-specific antibodies against Akt Ser(P)473, ERK Thr(P)202/Tyr(P)204, and MEK Ser(P)217/Ser(P)221 were from Cell Signaling Technology (Beverly, MA); antibodies against Akt-1/2 N terminus were from Santa Cruz Biotechnology (Santa Cruz, CA). LY294002 was purchased from Calbiochem. Unless otherwise noted, all other reagents were from Sigma.
NIH-3T3 fibroblasts (American Type Culture Collection, Manassas, VA) were cultured at 37 °C, 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and the antibiotics penicillin and streptomycin. NIH-3T3 cells were serially infected with retrovirus bearing empty pBM-IRES-Puro vector or vector carrying the indicated full-length H-Ras variants and selected using puromycin prior to each experiment, as described previously (35, 36).
Cells were serum-starved for 4 h prior to stimulation. Detergent lysates were prepared for quantitative immunoblotting, and immunoblots were performed using enhanced chemiluminescence, as described previously (36, 37). Blots comparing lysates prepared on the same day were performed in parallel and exposed at the same time. The Fluor S-Max system (Bio-Rad), which gives a linear response with respect to light output, was used, and band intensity was quantified using local background subtraction. The data were first normalized by an appropriate loading control and then further normalized to evaluate the consistency of relative trends across independent experiments, according to the procedures described in detail previously (36).
RESULTS
The allosteric switch, consisting of a shift in Helix3/Loop7 that triggers ordering of switch II in the on state, can be modulated in wild type Ras-GppNHp crystals by the binding of calcium acetate from the crystallization mother liquor (PDB code 3K8Y) (10). The off state, with a disordered switch II, is obtained by growing crystals in conditions containing calcium chloride instead (PDB code 2RGE) (31). This modulation can similarly be achieved in the RasG12V and RasQ61L mutants by growing crystals with symmetry R32 in the presence of calcium chloride to yield the off state and in the presence of calcium acetate to promote the on state of the allosteric switch. We have already published the crystal structure of RasQ61L-GppNHp in which the allosteric switch is off (PDB code 2RGD). Here, we present the crystal structures of RasG12V-GppNHp in the on state (1.30 Å resolution) and in the off state (1.84 Å resolution), as well as the structure of RasQ61L-GppNHp in the on state (1.32 Å resolution). These structures, together with the previously published ones mentioned above, provide a comprehensive picture of how the oncogenic mutants impair the active site and a model through which the Q61L mutation obstructs the equilibrium between the on and off states of the allosteric switch in Ras.
RasG12V, Gln61 and Bridging Water Molecule Perturbed by Steric Hindrance
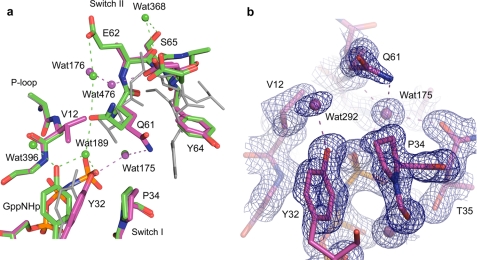
The crystal structure of RasG12V-GppNHp in the presence of calcium acetate has all of the features of the on state of the allosteric switch except in the local surroundings of the active site, where critical elements are perturbed. A bound calcium acetate at the remote allosteric site positions Loop7 and Helix3 with Arg97 at the center of an H-bonding network such that Val103 from Loop7 and several residues in Helix3 move away from switch II as described for the wild type structure (Fig. 1) (10). This allows the placement of the C-terminal portion of switch II such that Arg68 is at the center of a second H-bonding network that modulates the conformation of the N-terminal portion of the switch, including catalytic residue Gln61. The result is that the main chain conformation of the entire switch II in RasG12V-GppNHp is the same as in the wild type structure (Fig. 2a). The side chain of Gln61, however, cannot adopt the conformation seen in the wild type due to steric hindrance with the Val12 side chain. Instead it is rotated about χ2 to interact with the nucleophilic water molecule WAT175, as observed in the structures solved from crystals of symmetry P3221 (PDB code 1CTQ) and in the Ras-RasGAP complex (PDB code 1WQ1). Interestingly, the side chain of Glu62 is disordered in RasG12V, whereas in the wild type it is highly ordered and interacts through water molecule WAT176 with Gln61 to help position it near the bridging water molecule WAT189 (10). The side chain of Val12 does not allow placement of WAT189 between the hydroxyl group of Tyr32 and the O1γ atom of the GppNHp γ-phosphate (Fig. 2b). Instead, Tyr32 makes a direct H-bond with O1γ of the γ-phosphate.
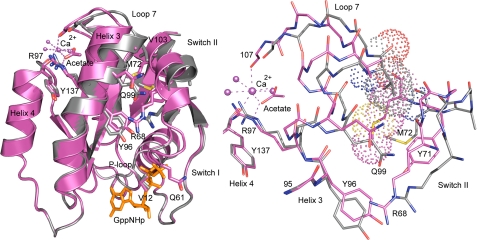
FIGURE 1.
Superimposed structures of RasG12V with the allosteric switch in the on (magenta) and off (gray) conformations. Left panel, ribbon diagram of the structures showing the shift in Helix3/Loop7 and switch II. Arg68 and Arg97 are shown explicitly in stick form as is the calcium acetate bound in the allosteric site with coordinating water molecules and other key residues involved in activation of the allosteric switch. The GTP analog, GppNHp, is shown in orange. Right panel, interface between Helix3 and switch II with van der Waal's surfaces showing the clash that would occur between switch II residue Met72 in the on state (magenta) and the Loop7 residue Val103 in the off state (gray). The binding of calcium acetate shifts Helix3/Loop7, creating room for proper placement of switch II to complete the Ras active site. All figures in this article were created with PyMOL (Delano Scientific).
FIGURE 2.
Active site of RasG12V in the on state. a, RasG12V (magenta) superimposed on the wild type protein (green) (PDB code 3K8Y). The off state in RasQ61L with the ordered switch II conformation (PDB code 2RGD) is shown in gray for comparison. Water molecules and dashed lines indicating H-bonds are colored as their respective protein models. b, 2Fo − Fc electron density map contoured at 1.2 s shown for active site protein residues and water molecules within 6 Å of the O1γ atom of GppNHp.
In the structure of RasG12V-GppNHp from crystals grown in calcium chloride, the allosteric site is “empty” and Helix3/Loop7 is as in the wild type Ras-GppNHp with the allosteric switch in the off state. The N-terminal portion of switch II, from residues 61 to 66, is disordered as in its wild type counterpart. The switch I and P-loop residues are the same in the on and off states, including Tyr32, which interacts directly with the O1γ atom of the γ-phosphate. As in the wild type, the nucleophilic water molecule is in place, despite the disordered switch II, including Gln61, for which there is no electron density.
We had previously measured the effect of a Raf construct containing its two Ras binding domains (Raf_RBD_CRD) on the in vitro hydrolysis rate of wild type Ras. We found the rate to be unchanged in the presence of the effector protein (10). This result was reproduced and is shown in Table 2. This makes sense in a context in which the ordering of switch II with placement of Gln61 in the active site is the rate-determining step in the reaction. Because Raf binds at switch I, but not switch II, the rate should not be expected to change. We show here that, as is the case for wild type, Raf_RBD_CRD has no effect on the intrinsic hydrolysis rate of the RasG12V mutant. Table 2 shows that there is a 11-fold decrease in hydrolysis rate of RasG12V relative to wild type whether or not Raf is present. All hydrolysis rates presented in Table 2 were obtained in the absence of any component that could activate the allosteric switch, as was the case for previously published hydrolysis rates measured by others for Ras (16). The slow rates thus reflect the state in which the Ras allosteric switch is off.
RasQ61L, Impaired On State as in RasG12V and Anti-catalytic Off State Stabilized by Raf
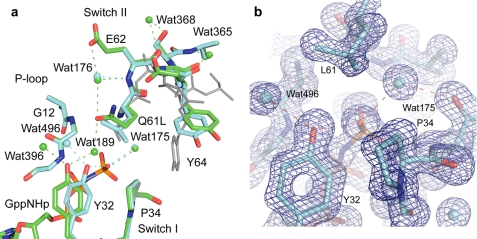
As was observed for RasG12V-GppNHp, the structure of RasQ61L-GppNHp from crystals grown in the presence of calcium acetate has the features of the on state of the allosteric switch with a bound calcium acetate at the allosteric site and the two H-bonding networks centered around Arg97 and Arg68 linking it to switch II. This structure differs from wild type only in the details of the active site, which is perturbed by the presence of Leu rather than Gln at position 61 (Fig. 3). The main chain of switch II is ordered, with a break in the electron density at Cα of Ser65. The side chains of Glu62, Glu63, Ser65, and Ala66 are disordered in this structure in contrast to the wild type where these side chains are ordered. Leu61 adopts a conformation where its Cβ and Cγ atoms overlap with those of Gln61 in the wild type (Fig. 3a). Interestingly, its Cδ1 atom roughly overlaps with the Cδ of Gln61 in the conformation where it interacts with the nucleophilic water molecule WAT175 as in the Ras-RasGAP complex, and its Cδ2 atom overlaps with the Cδ of Gln61 in the conformation where it interacts with the bridging water molecule WAT189 as it is positioned by the allosteric switch in the presence of calcium acetate. In this fashion, the branched side chain of Leu61 roughly overlaps the position of Gln61 in both conformations. The side chain of Tyr64 is rotated a few degrees about χ1 toward Leu61 in this structure, bringing one edge of the aromatic ring within van der Waal's contact of the aliphatic Leu61 side chain. The active site includes the nucleophilic water molecule WAT175 but no bridging water molecule WAT189 (Fig. 3b) due to steric hindrance with Leu61. Thus, both Val12 and Leu61 do not allow the presence of the bridging water molecule WAT189 in the active site. Not surprisingly, in the RasQ61L-GppNHp structure Tyr32 also makes a direct H-bond with the O1γ atom of the γ-phosphate. However, the position of Tyr32 in this structure is different from that in the G12V mutant (shown in Fig. 4 for the superimposed structures in the off state where switch I in all structures have the same conformation as in their on state counterparts). In RasG12V, the presence of the bulky Val12 side chain pushes Tyr32 toward Pro34, and its position is such as to minimize steric hindrance with these two side chains.
FIGURE 3.
Active site of RasQ61L in the on state. a, RasQ61L (cyan) superimposed on the wild type protein (green) (PDB code 3K8Y). The off state in RasQ61L with the ordered switch II conformation (PDB code 2RGD) is shown in gray for comparison. Water molecules and dashed lines indicating H-bonds are colored as their respective protein models. b, 2Fo − Fc electron density map contoured at 1.2 s shown for active site protein residues and water molecules within 6 Å of the O1γ atom of GppNHp.
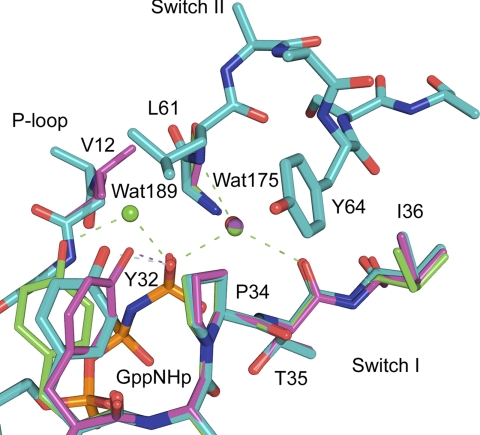
FIGURE 4.
Allosteric switch in the off state in Ras. Superimposed active sites of RasG12V (magenta), RasQ61L (cyan) (PDB code 2RGD), and wild type Ras (green) (PDB code 2RGE) are shown. The nucleophilic and bridging water molecules are shown, WAT175 and WAT189, respectively. Water molecules and dashed lines indicating H-bonds are colored as their respective protein models.
The RasQ61L-GppNHp structure from crystals grown in the presence of calcium chloride has an empty allosteric site. Helix3/Loop7 and the C-terminal portion of switch II are in the same conformation as in the wild type and RasG12V structures with the allosteric switch in the off state. Unlike the other two structures, however, the N-terminal portion of switch II is well ordered and in a conformation where switch II extends over switch I, and the Leu61 residue is at the center of a hydrophobic cluster that stabilizes this structure (Fig. 4) (31).
As for RasG12V, we measured the hydrolysis rates of RasQ61L in the absence and presence of Raf_RBD_CRD (Table 2). In the absence of the effector, the rate is 26-fold slower than wild type, only slightly slower than the rate we measured for RasG12V. For the purposes of correlations with the cell biology experiments below, RasG12V and RasQ61L can both be considered to be 1 order of magnitude slower than wild type in the absence of Raf. However, in the presence of Raf_RBD_CRD hydrolysis is so slow as to not be measurable in our experiments. It is clear that in the presence of Raf, RasQ61L does not appreciably hydrolyze GTP in vitro. We correlate this result with the anti-catalytic conformation that is stabilized under conditions where the allosteric switch is off.
RasQ61L Is More Potent than RasG12V as an Activator of ERK in NIH-3T3 Cells
To circumvent the limitations of measuring intrinsic hydrolysis in vitro with the allosteric switch in the off state and to test the potency of RasQ61L compared with RasG12V in pathways with and without Raf, we turned to experiments in NIH-3T3 mouse fibroblasts. Based on our structural biology analysis combined with measurements of hydrolysis rates in vitro, the state in which the allosteric switch is off appears to be stabilized in the RasQ61L mutant in the presence of Raf_RBD_CRD, relative to the off states in either the wild type or RasG12V. This means that in vivo, the allosteric switch, which we propose is involved in the timing of the Ras-Raf complex and thus the signaling through the Ras/Raf/MEK/ERK pathway, may be more easily activated to the on state in the RasG12V mutant than in RasQ61L. If this is the case, we would expect higher levels of MEK and ERK phosphorylation in cells transfected with RasQ61L than with RasG12V even though they have similarly perturbed active sites in the on state and similar in vitro hydrolysis rates in the absence of Raf. RasG12V would in turn show higher levels of phosphorylated MEK and ERK than in wild type due to its perturbed active site. In a pathway such as the Ras/PI3K/Akt where Raf does not play a role, and where PI3K interacts with Ras at both switch I and switch II precluding modulation by the allosteric switch, transfection with either RasG12V or RasQ61L should result in similar levels of Akt phosphorylation, as neither residue 12 nor 61 is involved in affinity hot spots for the Ras/PI3K interaction (5).
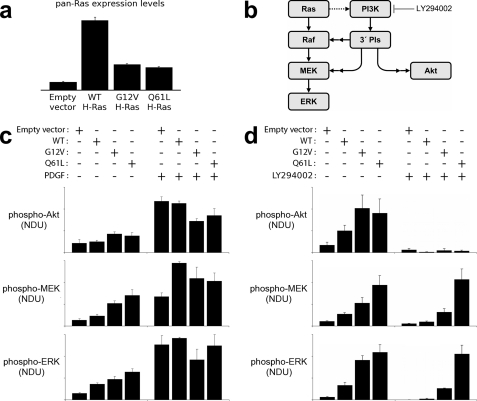
Fig. 5 shows the results of stable expression of the two transforming mutants and wild type H-Ras, alongside an empty vector control, in NIH-3T3 cells. The increased expression of Ras above endogenous levels in these cells was quantified by pan-Ras immunoblot and found to be approximately the same for RasG12V and RasQ61L but significantly higher for wild type H-Ras (Fig. 5a). Fig. 5b summarizes the anticipated effects of these mutants on the activation of Ras/Raf/MEK/ERK and Ras/PI3K/Akt pathways in these cells, including cross-talk of the PI3K-dependent signaling to MEK/ERK as characterized previously (36, 37). As expected, basal levels of Akt phosphorylation were moderately higher in RasG12V- and RasQ61L-expressing cells relative to those with empty vector or wild type Ras; interestingly, saturating the PI3K/Akt pathway by stimulation with platelet-derived growth factor (PDGF) was somewhat attenuated by the expression of constitutively active Ras mutants (Fig. 5c). The reason for this reduction may involve differential activation of PI3K by PDGF receptors and Ras mutants as postulated previously (35). The important result here is that there was no significant difference between the two mutants in terms of their effects on Akt phosphorylation. In contrast, a 5-fold range of basal MEK phosphorylation levels was observed in this panel of Ras-expressing cells, with low signal intensity for the empty vector and progressively higher signal readout with overexpression of wild type, RasG12V, and RasQ61L. As expected, ERK phosphorylation levels followed the same qualitative trends seen for MEK (Fig. 5c). To decouple the contributions of Ras/Raf and Ras/PI3K interactions in activating MEK and ERK phosphorylation, we reassessed the basal signaling states in the presence or absence of LY294002, an inhibitor of PI3K catalytic activity. In accord with the hypothesis that RasQ61L has an abnormally long lived interaction with Raf in particular, blocking the PI3K-dependent contribution dramatically widened the difference between RasQ61L- and RasG12V-expressing cells in their basal MEK and ERK phosphorylation (Fig. 5d; p = 0.026 and p = 0.012, respectively (Student's t test)). Indeed, in RasQ61L-transfected cells the inhibition of PI3K resulted in no significant change in MEK or ERK phosphorylation, suggesting that the Ras/Raf signaling alone is sufficient to saturate the rate of MEK phosphorylation under these conditions. This is not the case for cells transfected with RasG12V. The exciting result here is that activation of the Ras/Raf/MEK/ERK cascade is clearly greater in cells expressing RasQ61L relative to those expressing RasG12V, as expected from our structural biology and in vitro hydrolysis experiments.
FIGURE 5.
Quantitative comparison of basal and growth factor-stimulated signal transduction in mouse fibroblasts expressing endogenous Ras or supplemented with expression of wild type, RasG12V, or RasQ61L variants. a, expression of wild type (WT) H-Ras or mutant H-Ras in NIH 3T3 fibroblasts was assessed by immunoblotting with pan-Ras antibodies, normalized by total ERK1/2 as a loading control (averages of two independent experiments). Control cells show endogenous Ras expression. b, ERK signaling network in mouse fibroblasts is composed of the canonical Ras-dependent pathway and a parallel PI3K-dependent pathway. The dotted arrow signifies that excessive levels of active Ras are required to mobilize PI3K signaling in these cells, and double arrows signify that additional signaling proteins, not depicted, are involved. c, phosphorylation of Akt1/2/3 (phospho-Akt), MEK1/2 (phospho-MEK), and ERK1/2 (phospho-ERK) in unstimulated and PDGF-BB-stimulated (1 nm, 15 min) NIH-3T3 fibroblasts were measured by quantitative immunoblotting alongside total Akt and total ERK as loading controls. The bar graphs show phosphorylation levels, expressed in normalized densitometry units (NDU) as described under “Experimental Procedures,” reported as mean ± S.E. (n = 4–5). d, same as c, except here the effect of PI3K inhibition (100 μm LY294002) on the unstimulated phosphorylation levels is assessed alongside controls with DMSO vehicle only (n = 3–6).
DISCUSSION
The intrinsic hydrolysis rate measured in vitro for Ras has fueled the notion that Ras is too slow to turn off signaling in cells and that it is only in the presence of GAPs that hydrolysis rates are biologically relevant. Our discovery of an allosteric switch has led us to reconsider the functional relevance of intrinsic hydrolysis in Ras and to study the effects of oncogenic mutants from this new perspective.
Although it is possible to control the state of the allosteric switch in the crystal by the presence or absence of calcium acetate, this has not been the case in solution, most likely because in the crystals the allosteric binding site is stabilized by a crystal contact interaction between Arg135 and the C-terminal carboxyl group of a symmetry-related molecule, mimicking a salt bridge between Arg135 and a membrane phospholipid (10, 38). We are therefore limited to measuring the hydrolysis rates for wild type Ras and its mutants in the state in which the allosteric switch is off. In solution, there is an equilibrium between conformational states, and we must think of the off state as one in which the catalytically competent conformation is accessed only rarely. This leads to the slow rate measured for the wild type in vitro. We attribute the decreased rates measured for both mutants in the absence of Raf primarily to the perturbations described above for the active site prevalent in the on state, with only small variation, if any, in accessibility to this state in the mutants relative to wild type. Although the presence of Raf does not appear to affect accessibility to the catalytic conformation in the wild type or in RasG12V as reflected in unchanged hydrolysis rates in vitro, it greatly diminishes accessibility in the RasQ61L mutant by stabilizing a noncatalytic conformation of switch II, resulting in lack of hydrolysis under in vitro conditions where the allosteric switch is off.
Our experiments in NIH-3T3 cells show an increase in MEK and ERK phosphorylation associated with the RasQ61L mutant compared with RasG12V as a readout of signaling through the Ras-Raf complex, whereas these two mutants result in similar levels of Akt phosphorylation in a Raf-independent pathway. In this in vivo system, with cellular components in place, we do expect binding at the allosteric site to shift equilibrium toward the on state both in the wild type and in the mutants. Our analysis here is aided by the fact that the affinity of Raf, which interacts near the mutation sites in Ras, is the same for wild type and both mutants (39, 40), eliminating the possibility that higher levels of phosphorylation of MEK and ERK for the mutants are related to higher affinity for one or both mutants in the GTP-bound form, rather than to hydrolysis rates in the Ras-Raf complex. We thus interpret the increase in MEK and ERK phosphorylation for RasG12V relative to wild type to reflect the fact that the impaired active site, lacking the bridging water molecule WAT189 and appropriate positioning of Gln61, leads to an on state in which the intrinsic hydrolysis rate is reduced. We interpret the further increase in MEK and ERK phosphorylation observed for transfection with RasQ61L versus RasG12V to be due to stabilization of the off state in the Q61L mutant in the presence of Raf, resulting in a greater energy barrier to attain the on state promoted by binding of the appropriate cellular components at the allosteric site in vivo. This is supported by the similarity of the on state active sites between the RasG12V and RasQ61L mutants, their similar intrinsic hydrolysis rates measured in vitro in the absence of Raf, the fact that Raf stabilizes an anti-catalytic conformation in RasQ61L that abrogates hydrolysis in vitro, and the similar effects of both mutants on the Raf-independent Akt phosphorylation in cells. The stabilization of the off state results in greater signaling through the Ras/Raf/MEK/ERK cascade in RasQ61L relative to RasG12V, a differential not seen in the Ras/PI3K/Akt pathway. This is a Ras-Raf complex-dependent effect connected to a substantially altered equilibrium between the on and off states of the allosteric switch. This effect is to be distinguished from the impaired active site that most likely provides the major contribution in both mutants to increased signaling through the Raf-independent pathway leading to Akt phosphorylation.
Together, our experiments provide strong support for the intrinsic hydrolysis control of the duration of the Ras/Raf interaction and of the signal propagated through the Ras/Raf/MEK/ERK pathway. In this GAP-independent model for activation of GTP hydrolysis, Raf remains actively bound to Ras until the allosteric site near the cell membrane is activated, promoting hydrolysis of GTP to GDP and dissociation of the Ras-Raf complex. This model is consistent with the high affinity of Raf for Ras-GTP and the fact that switch II is not part of the interface in the Ras-Raf complex. The role of RasGAP in modulating this pathway may be to quickly deplete available free Ras-GTP so that Raf can no longer bind. The interplay between the roles of the allosteric switch and GAPs in signaling pathways involving Ras must be further investigated in light of the results presented here.
Acknowledgments
Kathleen Davis and Susan Fetics made the RasG12V and RasQ61L mutants of the full-length protein inserted into the pBM-IRES-Puro vector for expression in NIH-3T3 cells. Stephanie Holsenbeck made the truncated RasG12V mutant in pET21a. Use of the Advanced Photon Source at the Argonne National Laboratory was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W31-109-Eng-38.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-CA096867 and R56-CA096867.
The atomic coordinates and structure factors (codes 3OIW, 3OIV, and 3OIU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GAP
- GTPase-activating protein
- GppNHp
- guanosine 5′-(β,γ-imido)triphosphate
- PDB
- Protein Data Bank
- Raf_RBD_CRD
- construct containing the two Ras binding domains of c-Raf, the Ras binding domain (RBD), and the cysteine-rich domain (CRD).
REFERENCES
- 1. Cox A. D., Der C. J. (2003) Oncogene 22, 8999–9006 [DOI] [PubMed] [Google Scholar]
- 2. McCubrey J. A., Steelman L. S., Chappell W. H., Abrams S. L., Wong E. W., Chang F., Lehmann B., Terrian D. M., Milella M., Tafuri A., Stivala F., Libra M., Basecke J., Evangelisti C., Martelli A. M., Franklin R. A. (2007) Biochim. Biophys. Acta 1773, 1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casey P. J., Solski P. A., Der C. J., Buss J. E. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 8323–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thapar R., Williams J. G., Campbell S. L. (2004) J. Mol. Biol. 343, 1391–1408 [DOI] [PubMed] [Google Scholar]
- 5. Pacold M. E., Suire S., Perisic O., Lara-Gonzalez S., Davis C. T., Walker E. H., Hawkins P. T., Stephens L., Eccleston J. F., Williams R. L. (2000) Cell 103, 931–943 [DOI] [PubMed] [Google Scholar]
- 6. Akasaka K., Tamada M., Wang F., Kariya K., Shima F., Kikuchi A., Yamamoto M., Shirouzu M., Yokoyama S., Kataoka T. (1996) J. Biol. Chem. 271, 5353–5360 [DOI] [PubMed] [Google Scholar]
- 7. Huang L., Hofer F., Martin G. S., Kim S. H. (1998) Nat. Struct. Biol. 5, 422–426 [DOI] [PubMed] [Google Scholar]
- 8. Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J. (1998) Nature 394, 337–343 [DOI] [PubMed] [Google Scholar]
- 9. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 10. Buhrman G., Holzapfel G., Fetics S., Mattos C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4931–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milburn M. V., Tong L., deVos A. M., Brünger A., Yamaizumi Z., Nishimura S., Kim S. H. (1990) Science 247, 939–945 [DOI] [PubMed] [Google Scholar]
- 12. Franken S. M., Scheidig A. J., Krengel U., Rensland H., Lautwein A., Geyer M., Scheffzek K., Goody R. S., Kalbitzer H. R., Pai E. F. (1993) Biochemistry 32, 8411–8420 [DOI] [PubMed] [Google Scholar]
- 13. Krengel U., Schlichting I., Scherer A., Schumann R., Frech M., John J., Kabsch W., Pai E. F., Wittinghofer A. (1990) Cell 62, 539–548 [DOI] [PubMed] [Google Scholar]
- 14. Tong L. A., de Vos A. M., Milburn M. V., Jancarik J., Noguchi S., Nishimura S., Miura K., Ohtsuka E., Kim S. H. (1989) Nature 337, 90–93 [DOI] [PubMed] [Google Scholar]
- 15. Tong L. A., de Vos A. M., Milburn M. V., Kim S. H. (1991) J. Mol. Biol. 217, 503–516 [DOI] [PubMed] [Google Scholar]
- 16. John J., Schlichting I., Schiltz E., Rösch P., Wittinghofer A. (1989) J. Biol. Chem. 264, 13086–13092 [PubMed] [Google Scholar]
- 17. Neal S. E., Eccleston J. F., Hall A., Webb M. R. (1988) J. Biol. Chem. 263, 19718–19722 [PubMed] [Google Scholar]
- 18. Der C. J., Finkel T., Cooper G. M. (1986) Cell 44, 167–176 [DOI] [PubMed] [Google Scholar]
- 19. Seeburg P. H., Colby W. W., Capon D. J., Goeddel D. V., Levinson A. D. (1984) Nature 312, 71–75 [DOI] [PubMed] [Google Scholar]
- 20. Loupakis F., Ruzzo A., Cremolini C., Vincenzi B., Salvatore L., Santini D., Masi G., Stasi I., Canestrari E., Rulli E., Floriani I., Bencardino K., Galluccio N., Catalano V., Tonini G., Magnani M., Fontanini G., Basolo F., Falcone A., Graziano F. (2009) Br. J. Cancer 101, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 22. Du X., Black G. E., Lecchi P., Abramson F. P., Sprang S. R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8858–8863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Du X., Sprang S. R. (2009) Biochemistry 48, 4538–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maegley K. A., Admiraal S. J., Herschlag D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8160–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Buhrman G., de Serrano V., Mattos C. (2003) Structure 11, 747–751 [DOI] [PubMed] [Google Scholar]
- 26. Herrmann C., Horn G., Spaargaren M., Wittinghofer A. (1996) J. Biol. Chem. 271, 6794–6800 [DOI] [PubMed] [Google Scholar]
- 27. Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. (1990) Cell 63, 843–849 [DOI] [PubMed] [Google Scholar]
- 28. Vogel U. S., Dixon R. A., Schaber M. D., Diehl R. E., Marshall M. S., Scolnick E. M., Sigal I. S., Gibbs J. B. (1988) Nature 335, 90–93 [DOI] [PubMed] [Google Scholar]
- 29. Minato T., Wang J., Akasaka K., Okada T., Suzuki N., Kataoka T. (1994) J. Biol. Chem. 269, 20845–20851 [PubMed] [Google Scholar]
- 30. Nassar N., Horn G., Herrmann C., Block C., Janknecht R., Wittinghofer A. (1996) Nat. Struct. Biol. 3, 723–729 [DOI] [PubMed] [Google Scholar]
- 31. Buhrman G., Wink G., Mattos C. (2007) Structure 15, 1618–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 34. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 35. Kaur H., Park C. S., Lewis J. M., Haugh J. M. (2006) Biochem. J. 393, 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang C. C., Cirit M., Haugh J. M. (2009) Mol. Syst. Biol. 5, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cirit M., Wang C. C., Haugh J. M. (2010) J. Biol. Chem. 285, 36736–36744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gorfe A. A., Hanzal-Bayer M., Abankwa D., Hancock J. F., McCammon J. A. (2007) J. Med. Chem. 50, 674–684 [DOI] [PubMed] [Google Scholar]
- 39. Chuang E., Barnard D., Hettich L., Zhang X. F., Avruch J., Marshall M. S. (1994) Mol. Cell. Biol. 14, 5318–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herrmann C., Martin G. A., Wittinghofer A. (1995) J. Biol. Chem. 270, 2901–2905 [DOI] [PubMed] [Google Scholar]