Abstract
The innate immune response to Listeria monocytogenes depends on phagosomal bacterial degradation by macrophages. Here, we describe the role of LIMP-2, a lysosomal type III transmembrane glycoprotein and scavenger-like protein, in Listeria phagocytosis. LIMP-2-deficient mice display a macrophage-related defect in Listeria innate immunity. They produce less acute phase pro-inflammatory cytokines/chemokines, MCP-1, TNF-α, and IL-6 but normal levels of IL-12, IL-10, and IFN-γ and a 25-fold increase in susceptibility to Listeria infection. This macrophage defect results in a low listericidal potential, poor response to TNF-α activation signals, impaired phago-lysosome transformation into antigen-processing compartments, and uncontrolled LM cytosolic growth that fails to induce normal levels of acute phase pro-inflammatory cytokines. LIMP-2 transfection of CHO cells confirmed that LIMP-2 participates in the degradation of Listeria within phagosomes, controls the late endosomal/lysosomal fusion machinery, and is linked to the activation of Rab5a. Therefore, the role of LIMP-2 appears to be connected to the TNF-α-dependent and early activation of Listeria macrophages through internal signals linking the regulation of late trafficking events with the onset of the innate Listeria immune response.
Keywords: Bacteria, Cellular Immune Response, Immunology, Innate Immunity, Lysosomal Glycoproteins, Macrophage
Introduction
Infection with a sublethal dose of Listeria monocytogenes (LM)4 triggers an innate immune response in which MØs play a central role. In fact, LM replicates intracellularly, and the number of bacteria is limited by activated MØs, which prevent the dissemination of LM into the bloodstream. Recent studies suggest that the onset of LM innate immune response in MØs involves at least three intracellular stages (1–3). The first stage corresponds with LM internalization by the Toll-like receptor (TLR) recognition system and several scavenger receptors (4–9). Next, the phagosomal stage promotes the degradation of LM via the action of oxidative pathways, such as phox and inducible NOS enzymatic complexes (10–13), and nonoxidative microbicidal components, such as endosomal-lysosomal proteins (11, 14, 15). Lastly, a newly described cytosolic surveillance system mediated by NOD-like receptors senses degraded phagosomal bacteria (2, 3) and induces the production of MØ-derived pro-inflammatory cytokines/chemokines MCP-1, TNF-α, IL-6, and IL-12. The culmination of these intracellular responses is the transformation of LM-primed MØs into powerful microbicidal cells that promote the clearance of LM and contribute to adaptive immunity as antigen-presenting cells. LM-primed MØs respond to different signals to acquire features of microbicidal cells and antigen-presenting cells. TNF-α is produced by LM-primed MØs and controls the early signals that grant these cells the ability to kill intracellular bacteria. IFN-γ is produced by NK and T cells and regulates the late signals that transform MØs into listericidal cells and activated antigen-presenting cells with high MHC class II expression (10, 12, 13). Although the overall picture of LM-driven innate immunity is well described (1, 10, 12, 13), the intracellular molecules connecting the phagosomal and cytosolic stages are currently unknown. Moreover, the trafficking components involved in the innate immune response that promotes the transformation of the less bactericidal phagosomes into fully competent microbicidal and MHC class II antigen-processing compartments (MIICs) remain elusive.
Our group has been analyzing the role of MØs in LM-driven innate immunity and in deciphering the phagosomal, nonoxidative components for several years. We have previously reported the direct participation of the soluble lysosomal protease cathepsin-D (Ctsd) in LM degradation in the phagosome (14–16). Similarly, lysosomal proteins and regulatory factors, cathepsin-L, Rab7, LAMP-1, and LIMP-2, participate in other phagocytic systems (5, 7, 17–22).
Here, we have investigated the possibility that two major components of phago-lysosomal membranes, LAMP-1 and LIMP-2 (19), were involved in the innate immune response to LM by linking the phagosomal and cytosolic stages. We based our hypothesis on two observations: (i) different MIIC and phagocytic vesicles contain LAMP-1 and LIMP-2 as characteristic markers (11, 17, 19, 20, 23–26), and (ii) both proteins appear as key regulators of late trafficking events (19, 27–29). Therefore, they were good candidates for the molecules that connect the late trafficking processes with innate immunity to LM.
We infected LIMP-2- and LAMP-1-deficient mice (LIMP-2−/− and LAMP-1−/−, respectively) with LM and found that only the LIMP-2−/− mice were highly susceptible to LM infection and displayed defective MØ activation. This MØ defect affected the amount of LM able to escape to the cytosol, the production of early acute phase pro-inflammatory cytokines, and the ability of LM phagosomes to interact with MIIC vesicles. To confirm a role for LIMP-2 in phagocytic late trafficking events such as phagosome-lysosome fusion or bacterial proteolysis, we used CHO cells that overexpressed LIMP-2. Our results suggest that in concert with active Rab5a, LIMP-2 regulated the phagosomal fusion machinery of the late endosomes-lysosomes and the cytosolic levels of LM.
EXPERIMENTAL PROCEDURES
Bacteria
The L. monocytogenes strains used were: 10403S wild type (LMwt); hly-deficient mutant DPL-2161 (LMhly−) (D. A. Portnoy, UCLA-Berkeley); and DHL-L1039, a GFP-Listeria variant (D. E. Higgins, Boston College). Heat-killed LM (HKLM) was prepared as reported previously (14, 30, 31).
Cells, Reagents, and Mice
CHO cells were acquired from ATCC. We previously described the use of LIMP-2-deficient (LIMP-2−/−) (29), LAMP-1-deficient (LAMP-1−/−) (19), and Ctsd-deficient mice (Ctsd−/−) (15). The animals were used in accordance with the regulations of the University of Kiel and the Hospital Universitario Marqués de Valdecilla Instituto de Formación e Investigación Marqués de Valdecilla animal use and welfare board. BM-DM and mouse embryonic fibroblast culture conditions and requirements, as well as in vivo and in vitro infections and treatments, were described previously (15).
Transfections, FACS Analysis, Phagoburst, and Cytokine Measurements
CHO cells were transfected as described previously (15) with the following cDNAs subcloned into the peGFP-C1 vector: LAMP-1 (provided by S. Meresse, Centre d'Immunologie Marseille-Luminy, Marseille, France); murine cathepsin-D (provided by M. Dustin, New York University); LIMP-2 (provided by I. V. Sandoval, Centro de Biología Molecular Severo Ochoa, Madrid, Spain); and cathepsin-L, which was subcloned from the pK14-cathepsin-L vector using the SmaI site. hRab5a, Q79L (AQ), or S34N (AS) cDNAs were subcloned into the pcDNA3.1 hygromycin vector using the HindIII/BamHI sites. Transfected cells were FACS-sorted twice (FACSCalibur; BD Biosciences), and the brightest 5% of cells was collected. Thereafter, we analyzed 100,000 cells/transfectant cell line by FACS using the CellQuest software program (BD Biosciences) for data acquisition and analysis. The mean of arbitrary fluorescent units (mean fluorescent intensity) represents the percentage of fluorescent intensity per cell line as described previously (31). We also used FACS analysis for evaluation of the cell surface phenotype of BM-DM (100,000 cells) using FITC-labeled monoclonal antibodies (BD, Biosciences) against the following MØ activation markers: CD11b, Gr1, CD11c, IAb, F4/80, and CD54. BM-DM oxidative burst was examined using LM as the stimulus (10 min at 37 °C) and the Phagoburst kit (Orpegen Pharma, Heidelberg, Germany). FACS was used to examine the levels of pro-inflammatory cytokines in the sera of LM-infected mice or supernatants of infected BM-DM cultures using the CBA kit (BD Biosciences) that simultaneously analyzes IFN-γ, TNF-α, MCP-1, IL-6, IL-10, and IL-12.
Infection Kinetics
Infection assays were performed as described (30) and indices of intracellular growth corresponded to the ratio of CFU at 8 h divided by CFU at 0 h. Intraphagosomal viability was estimated from intact phagosomal fractions as previously reported (11, 15). CFU from controls were expressed as 100% of viable LM ± S.D. of three different experiments. Conventional fluorescence microscopy (Nikon Eclipse E400 microscope) was used for visualizing transfectants grown on glass coverslips and fixed with paraformaldehyde. Confocal fluorescence microscopy (63× Zeiss microscope) was used for GFP-LM infection of cells and co-localization with biotinylate anti-IAb antibodies followed by streptavidin-phycoerythrin. In some experiments, BM-DM were pretreated with IFN-γ (10 ng/ml) or TNF-α (10 ng/ml) for 24 h before LM infection.
[35S]LM Binding and Degradation Experiments
Binding assays were performed using nontransfected CHO cells and CHO transfectants as described previously (31). In brief, the cells were cultured to confluency (4 × 105 cells/well) on polyvinyl 96-well plates (Falcon; Becton Dickinson), fixed with 1% paraformaldehyde to prevent internalization of the bacteria, and blocked with PBS, 10% FCS to block nonspecific binding sites. [35S]LM or [35S]HKLM (500,000 cpm/well) were added to the cells and incubated on ice for 60 min to allow adherence. After washing, the wells were cut out, and the radioactivity associated with adherent cells was measured in a β-counter. For degradation experiments, [35S]HKLM (500,000 cpm/well) were internalized by cells cultured on regular cultured and treated 96-well plates (4 × 105 cells/well), centrifuged to synchronize the infection, and incubated for 20 min to allow for bacterial uptake. Next, supernatant and cell lysates were collected after 60 min. The proteins were precipitated from cell lysates and supernatants with 10% TCA, and cell-associated and supernatant radioactivity was measured in all of the samples following a protocol described previously (14). Individual data points were performed in triplicate, and the data correspond to the means ± S.D.
Phagosome Isolation and Comparative Analysis of Phagosomal and Cytosolic Bacteria
CHO cells, mouse embryonic fibroblasts, or BM-DM were infected with LM for 20 min (20:1 bacteria:cells for CHO and mouse embryonic fibroblasts cells and 10:1 for BM-DM), and phagosomal fractions or LMPs were isolated from thawed post-nuclear supernatants (PNS). A 100-μl aliquot of PNS was applied to a 8.8–20-40% discontinuous sucrose gradient and centrifuged. Phagosomes or LMPs were recovered from the 8.8–20% interface and the lower 20% (15). The purity of the organelles was monitored by biochemical analysis, as previously reported, to assess contamination with other cell components (11, 14–16). For phagosomal protein separation, 30 μg of purified phagosomal proteins were separated on 10% SDS-PAGE gels, blotted on a membrane, and analyzed for several markers with the following antibodies: 4F11 for Rab5a, goat anti-ASMase (kindly provided by O. Utermöhlen), and rabbit anti-cathepsin D. The percentage of stable/unstable MHC II molecules in the phagosomes and the whole cells were estimated after cell labeling with 10 μCi of [35S]Met/Cys translabel and dividing the phagosomal or cell preparations into nonboiled or boiled samples as reported previously (32, 33). To estimate the percentages of phagosomal and cytosolic bacteria, we used a previously reported procedure (15, 16). In brief, LM infection was carried out for 0 and 8 h. Phagosomes were isolated from an aliquot of PNS. An additional PNS aliquot was used to estimate the percentage of total internalized LM expressed as CFU. The percentage of cytosolic bacteria was calculated as (PNS − phagosomal numbers/PNS) × 100 (15, 16).
Assays for Phagosome-Endosome or Phagosome-Lysosome Fusion Transfers
For phagosome-early endosome fusion transfer, we incubated BM-DM with streptavidin-HRP (500 μg/ml) for 5 min followed by infection with biotinylated LM for 20 min (14). For phagosome-late endosome fusion transfer, BM-DM cells were incubated for 5 min with streptavidin-HRP and chased for 15 min in DMEM before infection for 20 min with biotinylated LM. For phagosome-lysosome fusion, BM-DM cells were treated as above with streptavidin-HRP for 5 min and chased for 16 h in DMEM, 1% FCS to allow for labeling of the lysosomal compartment before infection with biotinylated LM for 20 min (14). The results are expressed as percentages of total PNS before phagosome isolation.
Statistical Analysis
For statistical analysis, the Student's t test was applied. A p < 0.05 was considered statistically significant.
RESULTS
LIMP-2−/− Mice Are More Susceptible to LM
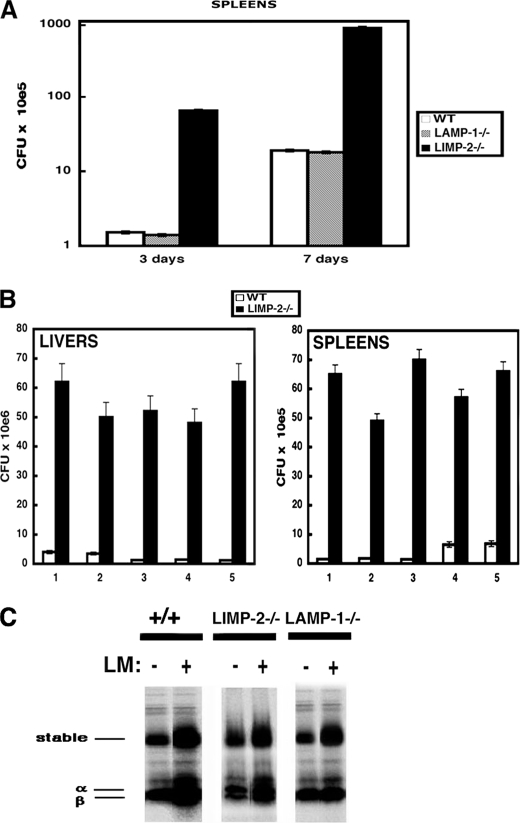
In listeriosis, control of the infection is performed in the spleen by resident MØs as early as 3 days post-infection (dpi). However, complete bacterial clearance is not achieved until 7 dpi. To determine the involvement of LAMP-1 or LIMP-2 in the immune response to LM, we compared the bacterial LM burden of spleens from infected LIMP-2−/− and LAMP-1−/− mice at two time points, 3 and 7 dpi, which allowed for the evaluation of the innate and adaptive immune responses. As shown in Fig. 1A, LIMP-2−/− mice had 25- and 100-fold increases in susceptibility to LM infection at 3 and 7 dpi, respectively, compared with both the WT and LAMP-1−/− mice. These data suggest that LAMP-1 is not required for immunity to LM. To evaluate in detail the impact of the innate immune response to LM and the role of LIMP-2, we examined early listeriosis at 3 dpi following the bacterial burden in both the liver and spleen of infected LIMP-2−/− mice after intraperitoneal injection of 5 × 104 CFU, which is LD50 for WT mice. Infection periods beyond 3 days would lead to the induction of adaptive immune responses to LM, which begins between 3 and 5 dpi (10, 12, 13). We performed five different experiments with five mice/genotype (Fig. 1B; 1–5; p < 0.01). LIMP-2−/− mice had an average bacterial burden of 8 × 108 CFU/spleen and 7 × 109 CFU/liver compared with 3.2 × 107 CFU in the spleen and 3 × 108 CFU in liver of WT mice. This increase in bacteria load translated into a ∼25-fold increase in susceptibility to LM infection for the LIMP-2−/− mice compared with the WT animals.
FIGURE 1.
LIMP-2−/− but not LAMP-1−/− mice were highly permissive to LM infection and displayed an impaired LM innate immunity. WT mice (white bars), LAMP-1−/− (gray bars), or LIMP-2−/− littermates (black bars) were infected with 5 × 104 LM for different times. Next, CFU were quantified in homogenized organs. The experiments were performed at least three times. A, CFU analysis of spleen from LM infected LIMP-2−/−, LAMP-1−/−, and wild-type mice at 3 and 7 dpi. The results are expressed as the CFU × 105 ± S.D. B, columns 1–5 represent different experiments of five mice/genotype in each experiment. CFU analysis of liver and spleen of LM infected LIMP-2−/− and WT mice at 3 dpi. The results are expressed as the CFU × 106 ± S.D. in liver and as CFU × 105 ± S.D. in spleen of triplicate samples. C, spleen from wild-type mice (labeled as +/+), LAMP-1−/−, or LIMP-2−/− infected, or not, with 5 × 104 LM for 7 days. Homogenates were incubated with 10 μCi of 35S-translabel for 2 h. Next, the spleen cells were incubated with 2% dialyzed FCS-DMEM-Met/Cys-free for 14 h. The cells were lysed and immunoprecipitated with Y3P antibody to capture the newly synthesized MHC II molecules. The immunoprecipitates were run under SDS-PAGE, and the gels were exposed to x-ray films.
LIMP-2 Participates in Early Innate Immune Responses to LM
To consolidate the role of LIMP-2 in the innate immune response to LM, we examined the level of pro-inflammatory cytokines in the sera of LM-infected LIMP-2−/− and WT mice at 3 dpi. To differentiate between a possible early or late role for LIMP-2 in the innate immune response to LM, we evaluated the production of early pro-inflammatory cytokines, such as TNF-α, MCP-1, IL-6, and IL-12, and late pro-inflammatory cytokines, such as IFN-γ and IL-10. After LM infection, the serum concentrations of IFN-γ and IL-10 were similar in the LIMP-2−/− and WT mice. LIMP-2−/− mice infected with LM had a 10-fold reduced concentration of acute phase pro-inflammatory cytokines, TNF-α, MCP-1, and IL-6 compared with the levels seen in WT mice (Table 1). LAMP-1−/− mice and WT control animals had comparable concentrations of all pro-inflammatory cytokines (Table 1). These data suggested that LIMP-2 regulates the early innate immune response to LM. Moreover, we discarded a role for LIMP-2 in the adaptive immune response following the increase on the levels of newly formed stable MHC II αβ dimers in the spleen after LM infection that correlated with the onset of LM-specific immunity and can be detected 4–5 dpi (10, 12, 13). LIMP-2−/−, LAMP-1−/−, and WT mice showed similar levels of stable αβ dimers in the spleen of 7-dpi LM-infected mice (Fig. 1C).
TABLE 1.
Innate immune parameters elicited by LM infection
| LIMP-2−/− | Wild type | |
|---|---|---|
| Sera cytokine concentrationa | ||
| TNF-α | 309.5 ± 11 | 3.500 ± 35 |
| MCP-1 | 612.5 ± 13 | 2.803 ± 21 |
| IL-6 | 809.5 ± 12 | 4.500 ± 45 |
| IL-12 | 34.1 ± 5 | 31.2 ± 8 |
| IL-10 | 5.1 ± 1 | 6.2 ± 2 |
| IFN-γ | 177.3 ± 12 | 163.45 ± 13 |
| BM-DM cytokine productionb | ||
| TNF-α/LMwt | 816.8 ± 13 | 5101.2 ± 21 |
| MCP-1/LMwt | 942.1 ± 25 | 4619.8 ± 16 |
| IL-6/LMwt | 2909 ± 11 | 6502 ± 14 |
| IL-10/LMwt | 189 ± 5 | 193 ± 2 |
| TNF-α/LMhly− | 140 ± 11 | 150 ± 15 |
| MCP-1/LMhly− | 153 ± 15 | 157 ± 16 |
| IL-6/LMhly− | 86 ± 2 | 81 ± 4 |
| TNF-α/LMHKLM | 310 ± 11 | 321 ± 15 |
| MCP-1/LMHKLM | 123 ± 15 | 133 ± 16 |
| IL-6/LMHKLM | 156 ± 2 | 131 ± 4 |
| TNF-α/LPS | 3004 ± 35 | 3400 ± 31 |
| MCP-1/LPS | 3012 ± 21 | 3500 ± 24 |
| IL-6/LPS | 2204 ± 20 | 2550 ± 31 |
a LIMP-2−/− or wild type mice (n = 5) were infected with 1× LD50 intraperitoneally for 3 days. The mice were sacrificed, and sera were collected for cytokine measurement by FACS analysis. The results are expressed as concentrations in pg/ml extrapolated from an internal standard curve. LAMP-1−/− concentration of cytokines was as follows: 3050 ± 17 for TNF-α, 2500 ± 20 for MCP-1, 4280 ± 35 for IL-6, 30 ± 5 for IL-12, 5.6 ± 2 for IL-10, and 165 ± 11 for IFN-γ.
b BM-DM were infected with wild type LMwt, LLO-deficient mutant LMhly−, or heat-killed bacteria, LMHKLM (ratio 10:1 of bacteria: cell) for 1 h or treated with LPS from E. coli (1 ng/ml) and supernatants collected after 24 h. Cytokine measurement was performed as above.
LIMP-2−/− Bone Marrow-derived MØ Showed a Defect in Endogenous Activation Signals
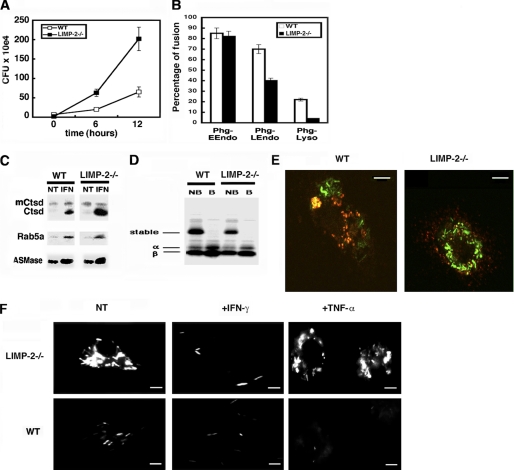
The high susceptibility to LM infection in LIMP-2−/− mice might reflect a defect in endogenous signals that activate MØ such as bacterial infections. We examined BM-DM from LIMP-2−/− and WT mice after in vitro infection with pathogenic LM (LMwt) and observed an increase in LM intracellular growth rates compared with WT BM-DM (Fig. 2A), reflecting reduced microbicidal function. These results also correlated with reduced production of MCP-1, TNF-α, and IL-6 but normal levels of IL-10, after infection with pathogenic LMwt (Table 1). Therefore, the activation of LIMP-2−/− BM-DM may be compromised to some extent, although they displayed a competent oxidative burst capacity (Table 2). Evaluation of cell surface markers characteristic for activated MØ, such as F4/80, MHC II molecules (IAb), and CD54, demonstrated that their expression was reduced on the LIMP-2−/− BM-DM compared with WT BM-DM. Other cell surface markers common to resting and activated MØ as well as dendritic cells or granulocytes, such as CD11b, CD11c, and Gr-1, were expressed at similar levels on the LIMP-2−/− and WT BM-DM (Table 2).
FIGURE 2.
The activation of LIMP-2−/− BM-DM was impaired, and they failed to control cytosolic LM growth. A, BM-DM from WT or LIMP-2−/− littermates were infected with LM for 0, 6, or 12 h. The results are expressed as CFU × 104 ± S.D. of triplicate samples (“Experimental Procedures”). B, BM-DM were incubated with streptavidin-HRP to label early endosomes, late endosomes, or lysosomes and next infected with biotinylated LM to isolate phagosomes according to the protocol described under “Experimental Procedures.” The HRP enzymatic activity was measured by the absorbance at A450 nm. The results are expressed as the percentages of total internalized HRP activity and reflect the percentage of fusion of phagosomes with either early, late endosomes or lysosomes. C, LM phagosomes from BM-DM pretreated with mIFN-γ before infection (IFN lanes) or nontreated (NT lanes) were isolated as described under “Experimental Procedures,” and 30 μg of phagosomal proteins were loaded per lane. Western blots indicate the inactive and membrane-bound Ctsd forms (mCtsd) and the active and soluble Ctsd bands (Ctsd), developed with a rabbit anti-Ctsd antibody. The Rab5a bands were developed with a mouse monoclonal anti-Rab5a antibody and the ASMase bands with a goat anti-mouse ASMase antibody. D, LM phagosomes from BM-DM labeled with 10 μCi of 35S-translabel, as in Fig. 4C, were isolated as under “Experimental Procedures,” and 30 μg of phagosomal proteins were boiled (B lanes) or nonboiled (NB lanes) before loading. The labels show stable αβ dimers and unstable α and β chains as reported (32, 33). E, WT or LIMP-2−/− BM-DM were infected with GFP-LM for 1 h, and MIIC compartments were labeled with biotinylated anti-IAb antibody followed by streptavidin-phycoerythrin. Co-localization images appearing as yellow fluorescence were analyzed by confocal microscopy. The scale bars correspond with 5 μm. F, different BM-DM pretreated with IFN-γ (+IFN-γ samples), TNF-α (+TNF-α samples), or untreated (NT samples) were infected with GFP-LM (ratio 10:1, bacteria:cell) for 12 h, and fluorescent images were analyzed by conventional fluorescent microscopy. The scale bars correspond with 6 μm. CFU values at 12 h were 10 × 104 (WT and untreated images), 2 × 102 (WT and +IFN-γ images), 3 × 102 (WT and +TNF-α images), 65 × 104 (LIMP-2−/− and untreated images), 13 × 102 (LIMP-2−/− and +IFN-γ images), and 56 × 104 (LIMP-2−/− and TNF-α images).
TABLE 2.
Activation parameters and markers elicited by LM infection
| BM-DM activation response | LIMP-2−/− | Wild type |
|---|---|---|
| Oxybursta | 77.5 | 78.2 |
| Control isotypeb | 0.18 | 0.18 |
| CD11b | 22.3 | 21.1 |
| CD11c | 8.5 | 10 |
| Gr1 | 0.33 | 0.33 |
| IAb | 27.5 | 57.2 |
| F4/80 | 2.1 | 23 |
| CD54 | 67.4 | 89.9 |
a Oxidative burst capacity of BM-DM using the Phagoburst fluorescent reagent after 10 min of incubation with LM as stimulus and according to the instructions of the supplier. The results correspond with the fluorescent intensity mean of 100,000 cells using the CellQuest software program of data acquisition and analysis.
b BM-DM were also surface-labeled with specific monoclonal antibodies phycoerythrin-labeled. The percentages reflect the amount of positive cells for a given marker or stimulus.
The significantly higher rate of intracellular LM growth in the LIMP-2−/− BM-DM compared with the WT BM-DM indicated uncontrolled cytosolic growth of the pathogen. To analyze the subcellular compartment of LM growth in LIMP-2−/− BM-DM, we directly evaluated the percentage of phagosomal and cytosolic LM. We reasoned that the first 8 h post-infection would allow us to examine the phagocytic and cytosolic stages of LM growth, because LM remains in the phagosome for 30–90 min post-infection, depending on the cell type (34), before escaping to the cytosol. Therefore, we calculated the percentage of phagosomal and cytosolic bacteria at 0 and 8 h post-infection using a previously described procedure (16). As controls, we also examined the intraphagosomal growth of LM in Ctsd−/− and Ctsd+/+ BM-DM, in which we have previously shown enhanced intraphagosomal viability of LM (16). The percentage of phagosomal LM at 0 h in the Ctsd−/− and LIMP-2−/− BM-DM was lower than in WT cells (Table 3). These results suggest a lower listericidal potential in Ctsd−/−- and LIMP-2−/−-derived MØ compared with WT cells (Ctsd+/+ or LIMP-2+/+ mice). The intracellular distribution of LM at 8 h displayed clear differences between LIMP-2−/− and Ctsd−/− BM-DM. In Ctsd−/− BM-DM, LM was found mainly within the phagosomes. However, it was primarily found in the cytosol in the LIMP-2−/− BM-DM (Table 3). Therefore, LM preferentially grows within the phagosomes of Ctsd−/− BM-DM and in the cytosol in the LIMP-2−/− BM-DM. Exclusively cytosolic LM triggers the cytosolic surveillance system of NOD-2 receptors responsible for the production of the acute phase pro-inflammatory cytokines, such as MCP-1, TNF-α, and IL-6. In fact, hly-deficient mutants of LM (LMhly−), which are trapped in the phagosome, do not induce these cytokines in BM-DM (1–3). LIMP-2−/− and WT BM-DM infected with LMhly− produced only background levels of MCP-1, TNF-α, or IL-6 and infected with heat-killed LM (LMHKLM) low levels (Table 1). However, LIMP-2−/− BM-DM infected with LMwt showed a 10-fold reduced production of the acute phase pro-inflammatory cytokines compared with WT. Exogenous signals such as the one mediated by LPS through activation of TLR2 or TLR4 were not impaired in LIMP-2−/− BM-DM (Table 1). These data indicated that LM-primed BM-DM from LIMP-2−/− mice showed a defect in endogenous signals, which had an impact on their intracellular activation.
TABLE 3.
Subcellular compartments for LM growth in different BM-DM genotypes
| BM-DM genotypesa | Phagosomal |
Cytosolic |
||
|---|---|---|---|---|
| 0 h | 8 h | 0 h | 8 h | |
| % | % | |||
| Ctsd+/+ | 32 | 2 | 68 | 98 |
| Ctsd−/− | 12 | 70 | 88 | 30 |
| LIMP-2−/− | 16 | 20 | 84 | 80 |
| LIMP-2+/+ | 33 | 1 | 67 | 99 |
a BM-DM from LIMP-2−/− or WT mice were infected with LM at a 10: 1 (cell: bacteria) ratio as described under “Experimental Procedures.” Phagosomal and cytosolic fractions were purified from PNS (30 μg) and solubilized, and viable LM were quantified as CFU to calculate the percentages of phagosomal and cytosolic bacteria as previously reported (15, 16). The results are expressed as percentages of total CFU internalized in PNS as described under “Experimental Procedures.” CFU values for PNS at 0 h were: 6.5 ± 0.03 for Ctsd+/+, 32.5 ± 0.1 for Ctsd−/−, 63.7 ± 0.01 for LIMP-2−/−, and 7.0 ± 0.03 for LIMP-2+/+.
LIMP-2−/− BM-DM Showed Impaired Transformation of LM Phagosomes into Late Endosomal and Antigen Processing Competent MIIC
A defect in the MØ bactericidal ability may correspond with impaired late intracellular trafficking events. In this regard, LM phagosomes from LIMP-2−/− BM-DM exhibited normal early endosome fusion, whereas the percentage of interactions with late endosomes or lysosomes was reduced (Fig. 2B). LM phagosomes from LIMP-2−/− and WT BM-DM had also similar levels of endocytic markers such as Rab5a, inactive/immature Ctsd forms, iCtsd, or ASMase (25, 35) (Fig. 2C). LIMP-2−/− BM-DM also displayed a defect to transform LM phagosomes into MIIC because we detected low percentages of peptide-loaded MHC II molecules or stable αβ-MHC II dimers (32, 33) in LM phagosomes compared with WT BM-DM (Fig. 2D). Moreover, we performed co-localization experiments of GFP-LM with anti-MHC-class II antibodies and showed co-localization in WT BM-DM (Fig. 2E and supplemental Fig. S1). In contrast, we observed low levels of co-localization in LIMP-2−/− BM-DM (Fig. 2E and supplemental Fig. S1A). Therefore, the defect of LIMP-2−/− BM-DM to respond to endogenous activation signals and produce acute phase pro-inflammatory cytokines (Table 1) may correlate with impaired late trafficking (Fig. 2, B–E).
LIMP-2−/− BM-DM Display Impaired Responses to Early Exogenous Activation Signals but Normal Response to Late Signals
Activation of MØ is also modulated by two exogenous responses: an early activating signal modulated by TNF-α and a late activating signal controlled by IFN-γ. To analyze the participation of LIMP-2 in both exogenous activating signals, we used GFP-conjugated LM and examined cytosolic LM after 12 h post-infection as described previously (5, 6, 22). LIMP-2−/− BM-DM displayed a higher level of cytosolic GFP-LM fluorescence than WT BM-DM (Fig. 2E). IFN-γ pretreatment of LIMP-2−/− and WT BM-DM resulted in a clearly reduced GFP-LM fluorescence signal, and this reduction was comparable between the LIMP-2−/− and WT BM-DM (Fig. 2F). However, pretreatment of BM-DM with TNF-α induced a reduced GFP-LM fluorescence signal only in WT BM-DM but not in LIMP-2−/− BM-DM (Fig. 2, F and G). Replication index (RI) values for these cells confirmed LM intracellular killing after TNF-α and IFN-γ treatment in WT BM-DM as well as in LIMP-2−/− BM-DM with IFN-γ treatment (Fig. 2E). LM phagosomes from IFN-γ-treated LIMP-2−/− and WT BM-DM also displayed high levels of Rab5a, mCtsd, and ASMase as expected (Fig. 2C) (11, 25). Therefore, LIMP-2−/− BM-DM could respond normally to exogenous signals such as IFN-γ.
LIMP-2 Regulates LM Intracellular Growth in LM Permissive Cells
To evaluate the participation of LIMP-2 as a regulator of LM cytosolic growth without the interference of MØ bactericidal mechanisms, we used LM-permissive cells. WT or LIMP-2-deficient mouse embryonic fibroblasts (Limp2+/+ and Limp2−/−, in supplemental Fig. S2A) were infected for different periods with LM. Limp2−/− cells showed exaggerated LM intracellular growth with rates 20-fold higher as compared with WT cells. We also included Ctsd−/− mouse embryonic fibroblasts, previously reported to show increased LM intracellular growth within the phagosomes (15). We also studied LM distribution between phagosomes and cytosol at two different time points: 0 and 6 h. Limp2−/− cells showed a clear LM distribution and uncontrolled growth in the cytosol, compared with the phagosomal growth in Ctsd−/− cells (supplemental Fig. S2B). To verify these results, we infected WT, Limp2−/−, and Ctsd−/− mouse embryonic fibroblasts with GFP-LM and labeled the cytoskeleton with TRITC-phalloidin. Co-localization of GFP-LM with TRITC-phalloidin was pronounced in Limp2−/− mouse embryonic fibroblasts, WT cells showed some co-localization, but GFP-LM in Ctsd−/− cells hardly co-localized with TRITC-phalloidin as expected, because bacteria grows within the phagosomes (15) (lower images in supplemental Fig. S2B). These results indicate that LIMP-2 influences the phagosomal permeability of LM phagosomes. In the absence of LIMP-2, the membrane permeability changes and allows a higher number of LM to escape to the cytosol.
LIMP-2 may also be involved in late endocytic trafficking events in endocytosis and phagocytosis requiring active Rab5a (28). It also functions as the sorting receptor for β-glucocerebrosidase (29, 36) or acts as the invasive receptor for the enterovirus 71 (37). To discard a role of LIMP-2 in bacterial surface adherence and to evaluate the molecular mechanism to control cytosolic LM growth, we transfected LM permissive CHO cells with the GFP-tagged cDNAs of LIMP-2, LAMP-1, and Ctsd. Transfection efficiency was verified by immunoblot analysis of whole cell lysates using anti-GFP antibodies for LAMP-1 and LIMP-2 and a specific rabbit anti-Ctsd antibody (supplemental Fig. S3A, left panel). An equivalent fluorescent intensity among transfectants was further confirmed by FACS analysis (supplemental Fig. S3A, right panel). We evaluated the role of LAMP-1, Ctsd, or LIMP-2 in LM recognition using a described previously [35S]LM binding assay with CHO cells to directly examine binding of bacteria to the cell surface (30, 31). Control cells and different CHO transfectants displayed similar binding abilities (supplemental Fig. S3B), demonstrating that none of these lysosomal components participated in LM cell surface recognition. We also observed that LIMP-2 and Ctsd transfectants compromised the intracellular growth of LM (supplemental Fig. S3B), whereas LAMP-1 transfectants displayed similar LM growth as CHO or peGFP-mock transfected cells.
LIMP-2 and Active Rab5a Regulate Late Trafficking Events Involved in LM Bacterial Proteolysis
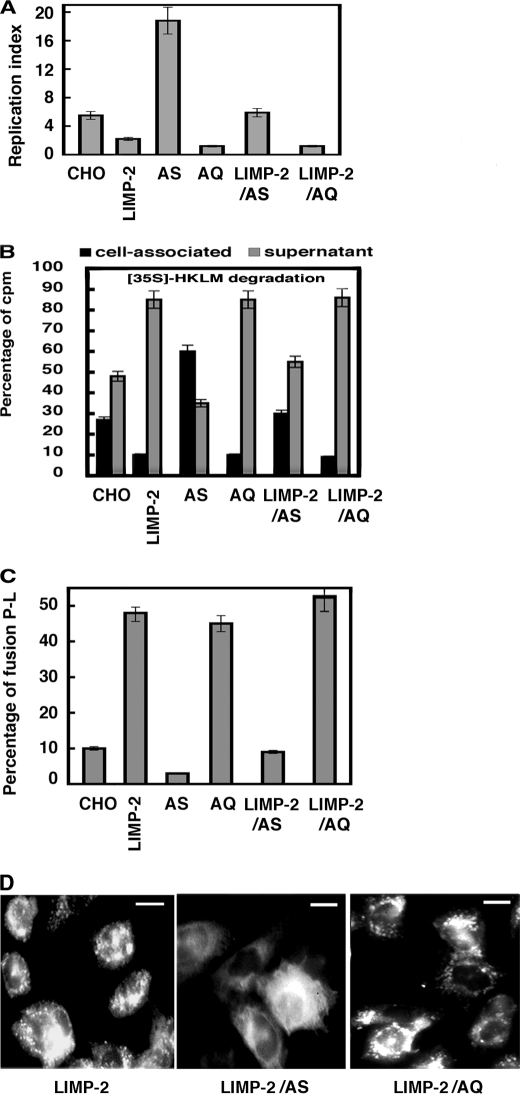
We examined the possibility that Rab5a controls the involvement of LIMP-2 in different phagocytic events in LM infection using CHO co-transfectants of LIMP-2 with Rab5a inactive (Rab5a: S34N named here as AS) or Rab5a active forms (Rab5a: Q79L named here as AQ). LM killing was analyzed by estimating the pathogen RI that measures the rate of LM intracellular growth (Fig. 3A) (16). LIMP-2 and AQ transfectants displayed RI values that were 2.5-fold lower than control cells, indicating high LM killing efficiencies. AS transfectants showed RI values that were 3.5-fold higher than controls reflecting low LM killing abilities, as expected (14). LIMP-2/AQ co-transfectants had RI values at the same range as compared with LIMP-2 and AQ transfectants. However, LIMP-2/AS co-transfectants reverted the high killing efficiencies of LIMP-2 transfectants to RI values of control cells, indicating that Rab5a activation might be required for the negative LIMP-2 effect on LM intracellular growth.
FIGURE 3.
Linked actions of Rab5a activation and LIMP-2 degrade LM and promote lysosome fusion. LIMP-2 transfectants and active Rab5a: Q79L (AQ) or inactive Rab5a: S34N (AS) co-transfectants were infected with LM, and different trafficking parameters were analyzed. A, LM replication indices at 6 h post-infection in CHO control cells, LIMP-2, LIMP-2/AQ, and LIMP-2/AS co-transfectants. The results are expressed as RI ± S.D. of triplicate samples. B, degradation of [35S]HKLM (500,000 cpm/sample) added to 4 × 105 CHO cells/well in 96-well plates, centrifuged to synchronize infection, and internalized for 20 min. The cells were washed and cell-associated, and supernatant radioactivities were recorded after a 60-min incubation (14). The results are expressed as percentages of cell-associated (black bars) or supernatant (gray bars) compared with internalized radioactivity ± S.D. of triplicate samples. C, percentages of phagosome-lysosome fusion using HRP-loaded lysosomes transfer fusion assays (14). The results correspond to percentages ± S.D. of triplicate experiments. D, conventional fluorescent patterns of LIMP-2 transfectants, LIMP-2/AQ, and LIMP-2/AS co-transfectants. Scale bars, 4 μm.
We also examined directly the role of LIMP-2 and Rab5a in bacterial proteolysis of radiolabeled and HKLM ([35S]HKLM in Fig. 3B). This assay measures two ratios of radioactivity that reflect bacterial proteolysis (30): (i) the percentage of cell-associated radioactivity (Fig. 3B, black bars) and (ii) the percentage of radioactivity in the supernatant (Fig. 3B, gray bars). LIMP-2 transfectants had a clear increase in bacterial proteolysis similar to AQ transfectants and LIMP-2/AQ co-transfectants reflected by a decrease in cell-associated radioactivity compared with control cells. AS transfectants showed a defect in bacterial proteolysis, as expected (14). LIMP-2/AS co-transfectants reverted the increase in bacterial proteolysis to control cells values (Fig. 3B). Therefore, active Rab5a seems to regulate the high bacterial proteolytic capacity of LIMP-2 transfectants.
Because bacterial proteolysis is linked to the transformation of phagosomes into phago-lysosomes, we also monitored phagosome-lysosome fusion by the transfer of the lysosomal content to LM phagocytic vesicles (LMP) as described previously (16). LMP from LIMP-2, AQ transfectants, and LIMP-2/AQ co-transfectants showed a higher percentage of lysosomal fusion events than control cells (48, 45, and 50%, respectively; Fig. 3C). LIMP-2/AS co-transfectants reverted the high percentages of LMP-lysosome fusion values seen in the LIMP-2 transfectants to control CHO values (9–10%). Rab5a activation appears required for LIMP-2-mediated enhancement of phagosome/lysosome fusion. LIMP-2-mediated endosome-lysosome fusion has been linked to an enlarged endosomal compartment regulated by active Rab5a (28). We also observed enlarged endocytic vesicles in LIMP-2 transfectants, which were absent in LIMP-2/AS co-transfectants but unchanged in the LIMP-2/AQ co-transfected cells (Fig. 3D), suggesting a general role for LIMP-2 in late trafficking events both in endocytosis and phagocytosis. These results suggest a concerted involvement of LIMP-2 and active Rab5a in several phagocytic parameters, such as LM intracellular growth, bacterial proteolysis, and phagosome-lysosome fusion events. We examined the phagocytic rates of different CHO transfectants and co-transfectants at 60 min to analyze late trafficking events. Only LIMP-2 transfectants and LIMP-2/AQ co-transfectants showed high phagocytic rates, whereas AQ transfectants showed normal ratios (supplemental Table S1). LIMP-2/AS co-transfectants reverted the high phagocytic rates promoted by LIMP-2. These data suggest that LIMP-2 acts as a late regulator of phagocytosis, whereas active Rab5a regulates early fusion events (30). Therefore, it seems that Rab5a precedes the LIMP-2 involvement in phagocytosis.
DISCUSSION
In this study, we have investigated the role of LAMP-1 and LIMP-2 in bacterial immunity. We present evidence for the specific role of LIMP-2/SCARB2 in the innate immune response to L. monocytogenes and in phagocytosis, whereas LAMP-1 seems to play no important role. Studies with LM-infected LIMP-2−/− BM-DM and LIMP-2−/− mice indicated that LIMP-2 participates in two processes in the activation of LM-primed MØ. First, LIMP-2 tightly controls the number of cytosolic LM and the induction of acute phase pro-inflammatory cytokines such as MCP-1, TNF-α, and IL-6. Therefore, it appears that low cytosolic LM numbers modulate the normal production of these cytokines in BM-DM. However, the production of late pro-inflammatory cytokines, such as IFN-γ and IL-10, was not regulated by LIMP-2. LIMP-2 also participates in late trafficking events transforming LM phagosomes into microbicidal and antigen processing compartments without affecting early intracellular processes. In fact, LIMP-2−/− BM-DM display impaired listericidal function and low expression of cell surface markers characteristic of activated MØ (i.e. F4/80 and IAb), albeit a normal oxidative burst capacity. They showed impaired interactions of LM phagosomes with late endosomes and lysosomes. Moreover, we observed decreased recruitment of peptide-loaded MHC II molecules in LIMP-2−/− BM-DM and hardly any co-localization of LM with MHC-II molecules compared with the WT cells, indicating impaired interactions with late MIIC vesicles. However, we also observed normal levels of several other endosomal proteins in the LM phagosomes from LIMP-2−/− BM-DM, such as Rab5a, mCtsd, and ASMase, suggesting normal interactions with endosomes. LIMP-2 appears to require active Rab5a to regulate all of these late trafficking processes. Nevertheless, Rab5a seems to precede the role of LIMP-2 in phagocytosis as previously suggested in MØ with inhibited Rab5a expression, which presented reduced LIMP-2 translocation to LM phagosomes (11). In fact, experiments in LIMP-2-transfected CHO cells and co-transfection with Rab5a-inactive forms abrogated LIMP-2 effects in different LM phagocytic parameters such as LM killing, phagosome-lysosome fusion, and late phagocytic rates.
The proposed linkage of LAMP-1 and Rab7 in different phagocytic systems (19, 20) and the recent demonstration that ASMase functions in the interaction between LM phagosomes and endosomes (25) suggest the possibility that late endocytic vesicles are subdivided into different compartments that are targeted by different trafficking regulators (18, 19, 21).
The specific intracellular action of LIMP-2/SCARB2 in LM phagocytosis and the lack of involvement in bacterial cell surface adherence or TLR2/TLR4 signaling correlate with the role assigned to LIMP-2 in endocytosis participating in late trafficking events (27–29, 38) and also correlate with TLR2/TLR4-independent killing of LM by activated MØ (39–41). In fact, LIMP-2−/− BM-DM showed a failure in listericidal abilities and acute phase cytokines production, whereas they displayed normal LPS responses and oxidative burst capacity. This intracellular specificity of LIMP-2/SCARB2 for late endocytic vesicles appears dependent on the presence of a coiled-coil domain in the luminal region and a preceding N-terminal transmembrane segment (36, 38). These LIMP-2/SCARB2 domains are absent in other CD36 family members such as CLA-1/SCARB1 or CD36, which have been shown to participate in bacterial recognition and uptake (7–9). The fine division of tasks among the CD36 family of scavenger-like molecules seems highlighted by the differential roles of CLA-1/SCARB1 and LIMP-2/SCARB2 in LM phagocytosis. In this regard, CLA-1/SCARB1 participates in the recognition and uptake of LM (5). However, LIMP-2/SCARB2 functions in the late phagosomal trafficking events involved in the onset of the innate immune response to LM but not in bacterial adherence (this study).
Listeriosis is characterized by the effect of two cytokines involved in MØ activation: TNF-α and IFN-γ. Although TNF-α acts as an early signal in innate immunity, IFN-γ is a late signal. In fact, IFN-γ constitutes a late pro-inflammatory cytokine, bridging the innate and adaptive immune response and facilitating the full clearance of LM from the spleen. Another difference between TNF-α and IFN-γ signaling is the endogenous and exogenous role of TNF-α to activate MØs. LM-primed MØs produce TNF-α that serves as a feedback mechanism to activate MØs exogenously. MØs show several activating states before turning into powerful bactericidal cells with antigen-presenting abilities. It has been claimed that the exogenous action of TNF-α promotes an early activating state in MØs that triggers the cytosolic microbicidal mechanisms (12, 13, 39). The exogenous effect of IFN-γ acts in MØ as a late activation signal that induces a strong microbicidal cascade affecting different subcellular compartments, phagosomes, endosomes, and cytosol (10, 12, 13).
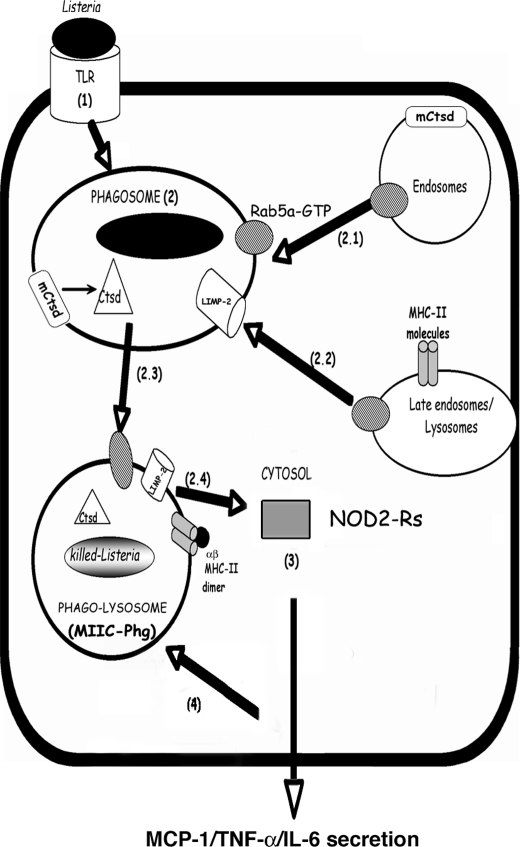
LIMP-2 participates in a second process involved in exogenous MØ activation, the early signals modulated by TNF-α. In fact, WT BM-DM responds to listericidal signals induced by TNF-α and IFN-γ. LIMP-2−/− BM-DM responds normally to IFN-γ listericidal signals but failed to respond to TNF-α listericidal signals, showing high rates of LM growth. We have compiled all of these data in a proposed model (Fig. 4) for LIMP-2 role in early activation of LM-primed MØ, connecting late trafficking events with the acute phase pro-inflammatory cytokines signaling in listeriosis.
FIGURE 4.
Model for the role of LIMP-2 in LM-primed MØ activity. Step 1, LM binds to different TLRs or scavenger receptors and is internalized into phagosomes. Step 2, the early phagosomal steps include Rab5a-GTP-mediated fusion with endosomes, the transport of membrane-bound and inactive Ctsd (mCtsd) and the activation of Ctsd into a listericidal, soluble, and active enzyme (step 2.1). LIMP-2 action on the phagosomal membrane requires Rab5a-GTP and promotes fusion with late endosomes/lysosomes, including late endocytic vesicles containing MHC II molecules (step 2.2). This overall process allows the transformation of phagosomes into phago-lysosomes with bactericidal abilities and features of MIIC (MIIC-Phg) (step 2.3). LIMP-2 regulation of this phagosomal transformation process influences the LM membrane permeability and confines the bacteria within phagosomes. Therefore, the number of cytosolic LM is limited (step 2.4). Step 3, the low numbers of cytosolic LM are sensed by NOD-2 receptors in the cytosol that activate the production of the acute phase pro-inflammatory cytokines/chemokines: MCP-1, TNF-α, and IL-6. Step 4, the release of these cytokines/chemokines such as TNF-α acts as a feedback loop that promotes the full activation of MØ, making them more efficient at degradation of LM.
The phagocytosis of LM appears to be regulated by the function of Rab5a (steps 1–3 in Fig. 4) (11, 15). Rab5a activation (step 2.1 in Fig. 4) promotes early interactions with the endosomal compartment and transport to LM phagosomes containing Ctsd-endosomal hydrolytic enzymes that, upon activation, have listericidal potential (15) (step 2 in Fig. 4). In fact, Ctsd enzymatic action inactivates listeriolysin O, which is the LM virulence factor responsible for phagosomal membrane lysis, and decreases the bacterial viability within the phagosomal lumen (16). Next, specific late endocytic vesicles transform the LM phagosomes into MIIC (steps 2.2–2.3 in Fig. 4). The action of LIMP-2 on the membrane permeability of LM phagosomes, which confines the nondegraded LM to the phagosomal compartment, contributes to bacterial proteolysis by other listericidal oxidative or nonoxidative mechanisms. LIMP-2 regulation of the transformation of phagosomes into MIIC (MIIC-Phg in Fig. 4) would also limit the number of cytosolic LM bacteria. Notably, only low numbers of cytosolic LM appear to be sensed by the cytosolic surveillance system of NOD2 receptors (Fig. 4, step 2.4), activating the production of acute phase pro-inflammatory cytokines/chemokines such as MCP-1, TNF-α, and IL-6 (Fig. 4, step 3) (2, 22). The secretion of these cytokines/chemokines such as TNF-α may act as a feedback loop that activates MØ to transform LM phagosomes into proteolytic and MIIC compartments that control the number of bacteria accessing the cytosolic compartment (Fig. 4, step 4). In conclusion, LIMP-2-mediated regulation of the late trafficking events of MØ may be due to TNF-α signaling pathways induced during the innate response to L. monocytogenes.
Supplementary Material
Acknowledgments
We thank H. Goldfine and Lee Shaw for critical review of the manuscript and English style corrections; I. V. Sandoval, M. Dustin, D. A. Portnoy, D. E. Higgins, S. Meresse, Jenny Schröder, O. Uttermöhlen, and J. P. Gorvel for reagents and mice; and the Unidad de Microscopía Avanzada in the Instituto de Formación e Investigación Marqués de Valdecilla directed by M. Lopez-Fanárraga for assistance with confocal images.
This work was supported in part by Fondo de Investigaciones Sanitarias Grants FIS-ISCIII-00/3073, PI01/3128, PI03-1009, and PI07-0289 (to E. C.-M.); grants from Ministerio de Ciencia, Investigación e Innovación (BIO2002-0628, SAF-2006-08968, and SAF2009-08695) and from Fondo de Investigaciones Sanitarias PI04-0324 (to C. A.-D.); Deutsche Forschungsgemeinschaft Grant SA683/6-1 and the Cluster of Excellence “Inflammation at Interfaces” (to P. S.); and Fondo de Investigaciones Sanitarias Contract CA06-0062 and Fundación Marqués de Valdecilla-Instituto de Formación e Investigación Marqués de Valdecilla Grant API09-SAF2009-08695 (to L. F.-P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- LM
- L. monocytogenes
- dpi
- days post-infection
- MØ
- macrophages
- LMP
- L. monocytogenes phagosomes-like vesicles
- TLR
- Toll-like receptor
- MIIC
- MHC class II antigen-processing compartment
- HKLM
- heat-killed LM
- CFU
- colony-forming unit(s)
- PNS
- post-nuclear supernatant(s)
- BM-DM
- bone marrow-derived MØ
- ASMase
- acid sphingomyelinase
- RI
- replication index
- TRITC
- tetramethylrhodamine isothiocyanate
- Ctsd
- cathepsin-D.
REFERENCES
- 1. Kobayashi K. S., Chamaillard M., Ogura Y., Henegariu O., Inohara N., Nuñez G., Flavell R. A. (2005) Science 307, 731–734 [DOI] [PubMed] [Google Scholar]
- 2. Leber J. H., Crimmins G. T., Raghavan S., Meyer-Morse N. P., Cox J. S., Portnoy D. A. (2008) PLoS Pathog. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bauler L. D., Duckett C. S., O'Riordan M. X. (2008) PLoS Pathog. 4, e1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crombie R., Silverstein R. (1998) J. Biol. Chem. 273, 4855–4863 [DOI] [PubMed] [Google Scholar]
- 5. Agaisse H., Burrack L. S., Philips J. A., Rubin E. J., Perrimon N., Higgins D. E. (2005) Science 309, 1248–1251 [DOI] [PubMed] [Google Scholar]
- 6. Cheng L. W., Viala J. P., Stuurman N., Wiedemann U., Vale R. D., Portnoy D. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13646–13651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Philips J. A., Rubin E. J., Perrimon N. (2005) Science 309, 1251–1253 [DOI] [PubMed] [Google Scholar]
- 8. Vishnyakova T. G., Kurlander R., Bocharov A. V., Baranova I. N., Chen Z., Abu-Asab M. S., Tsokos M., Malide D., Basso F., Remaley A., Csako G., Eggerman T. L., Patterson A. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16888–16893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rennemeier C., Hammerschmidt S., Niemann S., Inamura S., Zähringer U., Kehrel B. E. (2007) FASEB J. 21, 3118–3132 [DOI] [PubMed] [Google Scholar]
- 10. Unanue E. R. (1997) Curr. Opin. Immunol. 9, 35–43 [DOI] [PubMed] [Google Scholar]
- 11. Prada-Delgado A., Carrasco-Marin E., Bokoch G. M., Alvarez-Dominguez C. (2001) J. Biol. Chem. 276, 19059–19065 [DOI] [PubMed] [Google Scholar]
- 12. Pamer E. G. (2004) Nat. Rev. Immunol. 4, 812–823 [DOI] [PubMed] [Google Scholar]
- 13. Zenewicz L. A., Shen H. (2007) Microbes Infect. 9, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prada-Delgado A., Carrasco-Marín E., Peña-Macarro C., Del Cerro-Vadillo E., Fresno-Escudero M., Leyva-Cobián F., Alvarez-Dominguez C. (2005) Traffic 6, 252–265 [DOI] [PubMed] [Google Scholar]
- 15. del Cerro-Vadillo E., Madrazo-Toca F., Carrasco-Marín E., Fernandez-Prieto L., Beck C., Leyva-Cobián F., Saftig P., Alvarez-Dominguez C. (2006) J. Immunol. 176, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 16. Carrasco-Marín E., Madrazo-Toca F., de los Toyos J. R., Cacho-Alonso E., Tobes R., Pareja E., Paradela A., Albar J. P., Chen W., Gomez-Lopez M. T., Alvarez-Dominguez C. (2009) Mol. Microbiol. 72, 668–682 [DOI] [PubMed] [Google Scholar]
- 17. Schaible U. E., Sturgill-Koszycki S., Schlesinger P. H., Russell D. G. (1998). J. Immunol. 160, 1290–1296 [PubMed] [Google Scholar]
- 18. Harrison R. E., Brumell J. H., Khandani A., Bucci C., Scott C. C., Jiang X., Finlay B. B., Grinstein S. (2004) Mol. Biol. Cell 15, 3146–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huynh K. K., Eskelinen E. L., Scott C. C., Malevanets A., Saftig P., Grinstein S. (2007) EMBO J. 26, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Binker M. G., Cosen-Binker L. I., Terebiznik M. R., Mallo G. V., McCaw S. E., Eskelinen E. L., Willenborg M., Brumell J. H., Saftig P., Grinstein S., Gray-Owen S. D. (2007) Cell Microbiol. 9, 2153–2166 [DOI] [PubMed] [Google Scholar]
- 21. Sun J., Deghmane A. E., Soualhine H., Hong T., Bucci C., Solodkin A., Hmama Z. (2007) J. Leukocyte Biol. 82, 1437–1445 [DOI] [PubMed] [Google Scholar]
- 22. Via L. E., Deretic D., Ulmer R. J., Hibler N. S., Huber L. A., Deretic V. (1997) J. Biol. Chem. 272, 13326–13331 [DOI] [PubMed] [Google Scholar]
- 23. Harding C. V., Geuze H. J. (1992) J. Cell Biol. 119, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. von Delwig A., Musson J. A., Shim H. K., Lee J. J., Walter N., Harding C. V., Williamson E. D., Robinson J. H. (2005) Scand. J. Immunol. 62, 243–250 [DOI] [PubMed] [Google Scholar]
- 25. Schramm M., Herz J., Haas A., Krönke M., Utermöhlen O. (2008) Cell Microbiol. 10, 1839–1853 [DOI] [PubMed] [Google Scholar]
- 26. Jutras I., Houde M., Currier N., Boulais J., Duclos S., LaBoissière S., Bonneil E., Kearney P., Thibault P., Paramithiotis E., Hugo P., Desjardins M. (2008) Mol. Cell. Proteomics 7, 697–715 [DOI] [PubMed] [Google Scholar]
- 27. Sandoval I. V., Martinez-Arca S., Valdueza J., Palacios S., Holman G. D. (2000) J. Biol. Chem. 275, 39874–39885 [DOI] [PubMed] [Google Scholar]
- 28. Kuronita T., Eskelinen E. L., Fujita H., Saftig P., Himeno M., Tanaka Y. (2002) J. Cell Sci. 115, 4117–4131 [DOI] [PubMed] [Google Scholar]
- 29. Reczek D., Schwake M., Schröder J., Hughes H., Blanz J., Jin X., Brondyk W., Van Patten S., Edmunds T., Saftig P. (2007) Cell 131, 770–783 [DOI] [PubMed] [Google Scholar]
- 30. Alvarez-Dominguez C., Roberts R., Stahl P. D. (1997) J. Cell Sci. 110, 731–743 [DOI] [PubMed] [Google Scholar]
- 31. Alvarez-Domínguez C., Vázquez-Boland J. A., Carrasco-Marín E., López-Mato P., Leyva-Cobián F. (1997) Infect. Immun. 65, 78–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carrasco-Marin E., Shimizu J., Kanagawa O., Unanue E. R. (1996) J. Immunol. 156, 450–458 [PubMed] [Google Scholar]
- 33. Forestier C., Deleuil F., Lapaque N., Moreno E., Gorvel J. P. (2000) J. Immunol. 165, 5202–5210 [DOI] [PubMed] [Google Scholar]
- 34. de Chastellier C., Berche P. (1994) Infect. Immun. 62, 543–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blum J. S., Fiani M. L., Stahl P. D. (1991) Adv Exp Med Biol. 306, 281–287 [DOI] [PubMed] [Google Scholar]
- 36. Blanz J., Groth J., Zachos C., Wehling C., Saftig P., Schwake M. (2010) Human Mol. Genet. 19, 563–572 [DOI] [PubMed] [Google Scholar]
- 37. Yamayoshi S., Yamashita Y., Li J., Hanagata N., Minowa T., Takemura T., Koike S. (2009) Nat. Med. 15, 798–801 [DOI] [PubMed] [Google Scholar]
- 38. Kuronita T., Hatano T., Furuyama A., Hirota Y., Masuyama N., Saftig P., Himeno M., Fujita H., Tanaka Y. (2005) Traffic 6, 895–906 [DOI] [PubMed] [Google Scholar]
- 39. Edelson B. T., Unanue E. R. (2002) J. Immunol. 169, 3869–3875 [DOI] [PubMed] [Google Scholar]
- 40. O'Connell R. M., Vaidya S. A., Perry A. K., Saha S. K., Dempsey P. W., Cheng G. (2005) J. Immunol. 174, 1602–1607 [DOI] [PubMed] [Google Scholar]
- 41. Herskovits A. A., Auerbuch V., Portnoy D. A. (2007) PLoS Pathog. 3, e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.