Abstract
Tight junctions are multiprotein complexes that form the fundamental physiologic and anatomic barrier between epithelial and endothelial cells, yet little information is available about their molecular organization. To begin to understand how the transmembrane proteins of the tight junction are organized into multiprotein complexes, we used blue native-PAGE (BN-PAGE) and cross-linking techniques to identify complexes extracted from MDCK II cells and mouse liver. In nonionic detergent extracts from MDCK II cells, the tight junction integral membrane protein claudin-2 was preferentially isolated as a homodimer, whereas claudin-4 was monomeric. Analysis of the interactions between chimeras of claudin-2 and -4 are consistent with the transmembrane domains of claudin-2 being responsible for dimerization, and mutational analysis followed by cross-linking indicated that the second transmembrane domains were arranged in close proximity in homodimers. BN-PAGE of mouse liver membrane identified a relatively discrete high molecular weight complex containing at least claudin-1, claudin-2, and occludin; the difference in the protein complex sizes between cultured cells and tissues may reflect differences in tight junction protein or lipid composition or post-translational modifications. Our results suggest that BN-PAGE may be a useful tool in understanding tight junction structure.
Keywords: Cell Junctions, Epithelial Cell, Membrane Proteins, Protein Cross-linking, Protein-Protein Interactions, Tight Junction, Claudin, Occludin
Introduction
Tight junctions are multiprotein complexes that form the physiologic barrier to the paracellular movement of water, ions, and solutes between epithelial and endothelial cells. This structure encircles the apical end of the lateral membrane of epithelial cells and can be visualized by transmission electron microscopic analysis as a series of close membrane appositions. However, the fine structure of tight junctions is best revealed by freeze fracture electron microscopy, where the sites of membrane apposition appear as a network of anastomosing transmembrane protein strands that interact between adjacent cells. In unfixed tissue, these continuous strands appear as rows of 10 nm particles (reviewed in Ref. 1), suggesting that the transmembrane protein components of the tight junction are organized into large repeating protein complexes. Defining the nature of these complexes is critical to understanding how the selective barrier properties of the tight junction are created, regulated, and altered in disease.
Although several tight junction transmembrane proteins have been identified, including the members of the Marvel domain-containing family (occludin (2), tricellulin (3), and Marvel D3 (4, 5)) and the JAM IgG family (6), only members of the claudin family (7) are able to recapitulate the tight junction strands seen in freeze fracture EM when expressed in tight junction-null cells. It is presumed that these strands represent claudin oligomers, with other proteins like occludin recruited to the strands by an interaction with claudins (8). Despite considerable effort, however, only a few studies have been able to show biochemical evidence for claudin oligomers (9–13), and in general, these interactions have been demonstrated using overexpressed proteins in fibroblastic cells.
Blue native-PAGE (BN-PAGE)3 has been successfully used to identify membrane protein-protein interactions and complexes (14). Mild neutral detergents are used for initial solubilization, and the anionic dye Coomassie Blue G-250 is used instead of SDS to introduce a negative charge to the protein complex for electrophoretic separation. Although this method has been used most extensively to analyze components of mitochondrial protein complexes (15), it has also been used to study aquaporin oligomerization (16–18), cadherin interactions (19), and as a tool for proteomic analysis of multiprotein plasma membrane complexes (20, 21).
In this study, we use BN-PAGE to demonstrate that in MDCK II cells a homodimer is the favored oligomeric unit for claudin-2 (cldn2) but that in the same cell line both claudin-4 (cldn4) and occludin are monomers. Formation of the cldn2 dimer is not dependent on extracellular domains or on the cytoplasmic carboxyl-terminal domain, suggesting that dimerization is mediated by one or more of the transmembrane domains. Consistent with this idea, we found that dimerization does not require cell surface localization and thus does not represent trans-interactions between cells. Additionally, we demonstrate that in mouse liver, cldn2, along with at least cldn1 and occludin, is present in a high molecular weight protein complex.
EXPERIMENTAL PROCEDURES
Antibodies, Constructs, and Cell Lines
Antibodies were purchased from Zymed Laboratories Inc./Invitrogen, except the M2 anti-FLAG monoclonal antibody (Sigma), the ZO-1 rat monoclonal R40.76 (a gift from Bruce Stevenson, (22)) and VSV-g monoclonal antibody (a gift from Thomas Kreis). Tet-Off MDCK II cells and inducible cell lines expressing cldn2, cldn4, and chimeras have been described previously (23–25). Amino-terminally His6- and FLAG-tagged cldn2 constructs were generated using PCR and cloned into pTRE (Clontech) and pcDNA 3.1 (Clontech); primers used were for His6 tag, 5′-cgaattcaccatggcgtactaccatcaccatcaccatcacaccggtggtatggcctcccttggcgt-3′, and for FLAG tag, 5′-gaattcgatctacccatggactacaaagacgatgacgacaagatggcctcccttggcgt-3′. Cldn2 mutants were generated using the Stratagene QuikChange site-directed mutagenesis kit; the cldn2 A3 mutants were made by replacing the highly conserved 49GLW51 in the first extracellular domain with AAA using the primers 5′-attgtgacggcggttggcttttccaaggccgccgccatggagtgtgcgacacacagcacaggc-3′ and 5′-gcctgtgctgtgtgtcgcacactccatggcggcggccttggaaaagccaaccgccgtcacaat-3′ (all QuikChange primers are sense, followed by antisense sequences). The cldn2 cysteine to serine mutants were created first by separately replacing perimembrane cysteines in the second (TM2Cys → Ser) and fourth (TM4 Cys → Ser) transmembrane domains with serines. C104S was created using primers 5′-ctctgtggtgggcatgagaagcaccgtgtt-3′ and 5′-aacacggtgcttctcatgcccaccacagag-3′, and subsequently C104S,C108S was made using 5′-agatgcaccgtgttcagccaggattc-tcgag-3′ and 5′-ctcgagaatcctggctgaacacggtgcatct-3′. C182S,C185S was made using 5′-agccggagtcatccttagcttttccagctcgcccc-3′ and 5′-ggggcgagctggaaaagctaaggatgactccggct-3′. Stable cell lines were generated in MDCK II Tet-Off cells by cloning mutant claudin cDNAs into pTRE and co-transfection with pSV Zeo with Lipofectamine (Invitrogen); stable cell clones were selected using 1 mg/ml Zeocin; at least duplicate cell lines expressing each transgene were isolated. HEK 293T cells were transiently transfected with pcDNA 3.1 His- and FLAG-tagged claudins using FuGENE (Roche Applied Science); cells were collected 2–3 days after transfection.
Preparation of MDCK Cell Membranes
Tet-Off MDCK II cells were plated in duplicate 150 mm dishes and cultured for 8d; cells were washed and scraped in cold PBS; plasma membranes were enriched by a modification of the method of Boone et al. (26). Briefly, cells were homogenized after incubation in 10 mm Tris, pH 7.5, 1 mm MgCl2 supplemented with protease inhibitors (Halt protease inhibitor mixture, Thermo Scientific), washed with 10 mm Tris, 1 mm MgCl2, 250 mm sucrose, and the pellets resuspended in 50% sucrose in 10 mm Tris, pH 7.4, 1 mm MgCl2. Aliquots of 0.8 ml were transferred to 2-ml ultracentrifuge tubes and overlaid with 0.8 ml of 42.9% sucrose and 0.5 ml of 20% sucrose. Sucrose step gradients were centrifuged at 100,00 × g for 60 min in a Beckman TLS-55 rotor; the band at the 20/42.9% interface was collected and stored on ice overnight. The sample was diluted 10-fold with 10 mm Tris, pH 7.5, centrifuged for 10 min at 15,000 × g, and the resulting pellet extracted twice by incubation on ice for 20 min with 4 m urea in 10 mm Tris, pH 7.5. The insoluble material after each extraction and two subsequent washes with 10 mm Tris, pH 7.5, was collected by centrifugation at 100,000 × g for 10 min. The pellets were resuspended in 1% dodecyl maltoside (DDM, Anatrace, Maumee, OH) in 10 mm Tris, pH 7.5, and 1× native-PAGE sample buffer (Invitrogen) and incubated on ice for 15 min. Insoluble material was removed by centrifugation at 100,000 × g for 10 min in a TLA100 rotor (Beckman Coulter), and 0.1 volume native-PAGE 5% G-250 sample additive (Invitrogen) was added.
Preparation of MDCK and HEK 293T Cells Expressing Transgenes
Cells were washed with PBS or HBSS and resuspended in 1% DDM, 20 mm Tris, pH 7.5, 50 mm NaCl, incubated on ice for 20 min, and insoluble material was removed by centrifugation at 15,000 × g for 20 min, and the supernatants were prepared for BN-PAGE. When tagged claudins were purified by affinity chromatography, cells were resuspended in 1% DDM, 20 mm Hepes, pH 7.4, 50 mm NaCl and clarified as above before incubation with FLAG affinity resin (Sigma) or Talon resin (Clontech) according to the manufacturers' directions. Tagged claudins were eluted from affinity resins using either FLAG peptide (Sigma) or 250 mm imidazole as appropriate.
Cross-linking with Photo-activated Cross-linkers
His-tagged cldn2 was eluted from Talon resin and incubated with 2 mm NHS-diazirine (SDA), NHS-LC-diazirine or Sulfo-NHS-SS-diazirine (S-SDAD) (Thermo Scientific) for 30 min. Samples were quenched with 0.1 m Tris, pH 7.5, dialyzed against four changes of 20 mm Hepes, pH 7.4, 50 mm NaCl, 0.3% DDM in Slide-a-Lyzer mini dialysis cups (Thermo Scientific), and 15 μl transferred to glass slides; drops were constrained within circles drawn with a wax pencil. Cross-linking was performed by exposing drops to UV light at 350 nm for the indicated times. Equal volumes of SDS sample buffer were added to terminate the cross-linking, and samples were subjected to SDS-PAGE, transferred, and immunoblotted as described below.
Cross-linking with Glutaraldehyde
His-tagged cldn2 and cldn4 were eluted from Talon resin and incubated with 100 μm glutaraldehyde for the indicated times (27); SDS was added to one sample before addition of glutaraldehyde to disrupt oligomeric organization. Reactions were quenched by the addition of 0.1 m Tris, pH 7.5, and samples were processed for SDS-PAGE and immunoblotting as described below.
Cysteine Cross-linking
MDCK II cells induced to express VSV-g-tagged cldn2 with and without mutation of perimembrane cysteines to serines were treated overnight with the palmitoylation inhibitor, 2-bromopalmitate, as described previously (28), washed twice with Dulbecco's PBS, and incubated with 2 mg/ml dithiobismaleimidoethane (DTME, Thermo Scientific) for 30 min at 4 °C. Cross-linker was quenched with 2 mm cysteine in Dulbecco's PBS, and cells were extracted with SDS sample buffer lacking reducing agents. Samples subjected to SDS-PAGE and immunoblotted as described below.
Preparation of a High Molecular Weight Complex from Mouse Liver
Mouse liver membranes were prepared with a simplified method based on that of Goodenough and co-workers (29). One mouse liver was minced with razor blades and incubated with 1 mm NaHCO3 supplemented with protease inhibitors for 10 min on ice and homogenized using a glass/Teflon homogenizer. The resulting homogenate was filtered through four layers of cotton gauze and centrifuged at 2000 × g for 15 min. The supernatant was discarded and the soft pellet made 50% sucrose by addition of 3 volumes of 67% sucrose in 1 mm NaHCO3. Aliquots of 0.8 ml were placed at the bottom of each of four 2-ml ultracentrifuge tubes and overlaid with 0.8 ml of 42.9% sucrose and 0.5 ml of 20% sucrose. Samples were centrifuged in a Beckman Coulter TLS-55 rotor at 100,00 × g for 60 min, the upper interfaces collected, combined, and diluted 10-fold with 1 mm NaHCO3, and the diluted material centrifuged at 4000 × g for 30 min. The resulting pellet was resuspended in 200 μl of 1% dodecyl maltoside in 10 mm Tris, pH 7.5, 50 mm NaCl and incubated on ice for 20 min. Resuspended pellets were centrifuged at 100,000 × g for 10 min to remove insoluble material, and the resulting supernatant prepared for BN-PAGE. Samples were electrophoresed on duplicate gels; one was stored at 4 °C, and the second was transferred and immunoblotted for cldn1 as described previously. The position of the immunoreactive band was determined and the corresponding region excised from the duplicate BN gel; the excised band was forced three times through a 30-gauge needle and electroeluted overnight in 25 mm Tricine, 3.75 mm imidazole, 5 mm aminocaproic acid, pH 7.0, for 16 h at 100 V using an Elutrap (Schleicher & Schüll). Following brief voltage reversal (200 V, 20 s), the electroeluted material was removed, precipitated with 20% trichloroacetic acid, washed three times with cold acetone, and air-dried. The pellet was resuspended in 4× SDS/Laemmli sample buffer, incubated at 37 °C for 10 min, and briefly sonicated. Aliquots were electrophoresed in SDS-polyacrylamide gels, transferred to nitrocellulose, and probed with cldn1, cldn2, and occludin antibodies.
BN-PAGE, SDS-PAGE, Immunoblotting, and Immunofluorescence
BN electrophoresis was performed using native-PAGE Novex BisTris gels (Invitrogen) according to the manufacturer's directions; the protocols are based on techniques developed by Schagger and von Jagow (15). Gels were transferred for 60 min at 30 V to PVDF membrane (Immobilon P, Millipore) in Tris-glycine buffer containing 20% methanol and 0.1% SDS. Membranes were fixed in 8% acetic acid for 15 min, air-dried, destained in 100% methanol for 20 s, washed with H2O, and incubated in blocking buffer (10% NFDM in PBS) for 60 min. Immunoblotting for BN-polyacrylamide gels was performed as described previously (30), except HRP-coupled secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used, and detection was performed using SuperSignal West chemiluminescent substrate (Thermo Scientific). SDS-PAGE, immunoblotting, IR detection, and immunofluorescence were all performed as described previously (30). Recombinant aquaporin Z was extracted from bacterial pellets with 1% DDM, purified on Talon resin, dialyzed, electrophoresed on BN-PAGE, and stained with Coomassie Brilliant Blue; aquaporin Z plasmid construct was generously provided by Dr. Michael Wiener, University of Virginia.
RESULTS
Some Claudins Form Discrete Oligomers in MDCK Cells
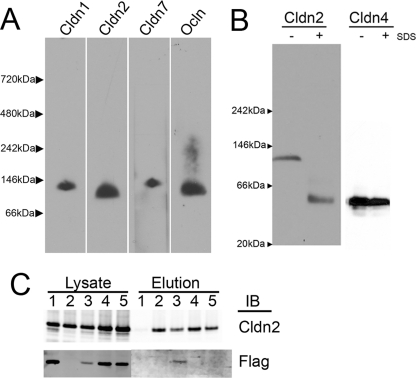
To analyze the oligomeric organization of endogenous claudins in cultured epithelial cells (MDCK II), we used sucrose density fractionation to enrich tight junction-containing membranes and extracted these fractions with 4 m urea to remove peripheral membrane proteins. Protein pellets were dissolved in the nonionic detergent DDM, and samples were subjected to BN-PAGE. Immunoblot analysis of BN-polyacrylamide gels transferred to PVDF membranes revealed that cldn1, -2, -7 and occludin were easily detectable (Fig. 1A). When compared with soluble molecular weight markers, cldn2 migrated at ∼112 kDa. There are considerable pitfalls in molecular weight estimation using BN-PAGE (31) especially for small membrane proteins (32), partially because unit of mass includes the protein, variable amounts of bound Coomassie dye, and an unknown amount of lipid and partly because electrophoresis of soluble protein markers variably deviates from that of membrane proteins of known molecular weight in BN-PAGE. We applied a correction factor to obtain the protein alone based on an estimate of dye binding of 0.56 g/g protein (32) (observed range is about 0.35–0.6 g/g (31)); after this correction, the approximate molecular mass for the protein component of the complex was ∼62 kDa. This size is consistent with either a dimer or trimer of cldn2. Similar analysis suggested cldn1 and cldn7 were either dimers or trimers, whereas endogenous occludin (correction: 130 × 0.56 = 65 kDa) appeared to migrate as a monomer. These relative molecular weight assumptions were substantiated by comparison with that of aquaporin Z electrophoresed in parallel in BN-PAGE. Aquaporin Z (monomer = 25 kDa) is known from crystallographic studies to assemble as a tetramer (33); its apparent molecular mass in BN-PAGE was 185 kDa; correction for the bound Coomassie dye (185 × 0.56 = 103 kDa) resulted in a calculated molecular mass consistent with a protein tetramer containing four 25-kDa monomers (supplemental Fig. 1). The various claudins had slightly different electrophoretic mobility; the reason for this is unclear but may be result from differences in protein molecular weights, isoelectric points, and associated lipids or due to difference in protein interactions (homo- or hetero-oligomerization). The extraction and BN-PAGE was repeated using different nonionic detergents with similar results, although use of digitonin resulted in slightly higher molecular weight complexes (data not shown), consistent with other reports that digitonin removes less associated lipid from membrane protein complexes than does DDM (34). Despite several attempts, we were unable to detect endogenous cldn4 after BN-PAGE from wild-type MDCK II cells.
FIGURE 1.
Claudins 1, 2, and 7 form discrete oligomers in MDCK cells. A, sucrose gradient-enriched plasma membrane fractions from MDCK II cells were extracted with 1% DDM, electrophoresed in BN-PAGE, transferred to PVDF membrane, and immunoblotted for cldn1, -2, and -7 and occludin (ocln); molecular weights determined by comparison with native mark molecular weight markers (Invitrogen) reveal that claudins but not occludin migrate consistent with being either dimers or trimers. B, stably transfected MDCK II cells were induced to express cldn2 or cldn4, extracted with 1% DDM without (−) or with (+) the addition of 1% SDS, and electrophoresed in BN-PAGE. SDS-treated samples reveal position of monomer; DDM-extracted cldn2 but not cldn4 migrates as a dimer. C, HEK cells were transfected with FLAG-cldn 2 (lane 1), His-cldn2 (lane 2), co-transfected with both His- and FLAG-cldn2 (lane 3), separately transfected with His- and FLAG-cldn2, and then co-plated (lane 4); all cells were extracted with 1% DDM, and supernatants from separately transfected cells were mixed after extraction (lane 5). Aliquots of DDM extracts were electrophoresed in SDS-PAGE and immunoblotted (IB) for cldn2 (upper left) and FLAG epitope (lower left). Remaining lysates were applied to Talon resin, washed, and eluted with imidazole; eluates were subjected to SDS-PAGE and immunoblotted with cldn2 (upper right) and FLAG antibodies (lower right). FLAG-cldn2 is retained on Talon resin when co-transfected with His-cldn2, consistent with homodimer formation during biosynthesis or trafficking but not via intercellular interactions.
To determine whether induced cldn2 in stably transfected MDCK cells showed the same oligomeric organization as the endogenous cldn2, we next performed BN-PAGE on DDM-extracted MDCK Tet-Off cells induced to express mouse cldn2 by removal of doxycyline. To distinguish cross-linking of subunits within a complex versus freely diffusing uncomplexed proteins, duplicate samples were first treated with 1% SDS to denature and thus dissociate oligomers (Fig. 1B, left panel). When induced mouse cldn2 was extracted in DDM, it migrated at approximately the same molecular mass as endogenous canine cldn2, whereas SDS-treated cldn2 migrated at ∼55 kDa, and when corrected for bound detergent (55 × 0.56 = 31 kDa) approximates a monomer, suggesting that the 112-kDa band most likely represents a cldn2 dimer and not trimer. Because of the high level of transgene induction, the endogenous cldn2 was not detected at the exposure times employed (data not shown). In contrast to the results seen with cldn2, BN-PAGE analysis of induced human cldn4 resulted in identical migration behavior for DDM- and SDS-treated samples (Fig. 1B, right panel), suggesting that in MDCK cells, cldn4 exists as a monomer.
To test if the cldn2 complex visualized by BN-PAGE likely represented a homodimer or a complex containing cldn2 and a different protein, HEK cells were transiently transfected with His6-tagged cldn2 and FLAG-tagged cldn2 separately or together (Fig. 1C). FLAG- and His-tagged cldn2 migrated identically to wild-type cldn2 in BN-PAGE when expressed either transiently or stably (data not shown). All transfected HEK cells showed approximately equal levels of cldn2 expression (Fig. 1C, lysates, top left); untransfected HEK cells do not express endogenous claudins (data not shown). The FLAG antibody did not recognize His-tagged cldn2 (Fig. 1C, lane 2) in cells transfected with His-tagged cldn2 alone (lysates, bottom left). DDM extract of transfected cells was purified on metal affinity resin; only extracts from cells transfected with His-tagged cldn2 were specifically eluted with imidazole (Fig. 1C, upper right panel). Only when FLAG-tagged cldn 2 was co-transfected with His-tagged cldn2 did it specifically elute from metal affinity resin (Fig. 1C, lane 3, lower right panel), suggesting that the FLAG-tagged protein was retained on the resin via an interaction with the His-tagged protein (lane 3). In contrast, there was no apparent interaction between His- and FLAG-tagged cldn2 when separately transfected cells were co-cultured (Fig. 1C, lane 4) or supernatants of separately transfected cells were mixed before incubation with affinity resin (lane 5). These results suggest that dimerization is not due to transcellular interactions between proteins expressed in apposing cells or as an artifactual interaction formed during isolation. Because FLAG-His dimers depend on co-transfection, it is possible that dimers form during synthesis or trafficking.
Chemical Cross-linking of cldn2 Results in Formation of Dimers but Not Higher Molecular Weight Forms
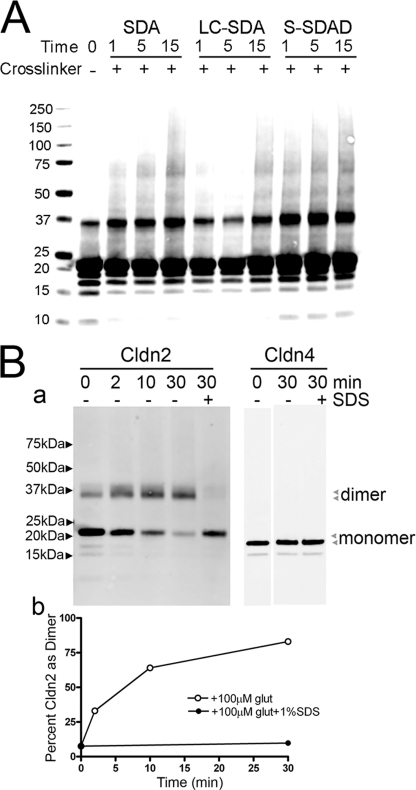
Chemical cross-linking is a commonly used method to define protein-protein interactions within oligomers. We used two separate approaches to determine whether chemical cross-linking could confirm the existence of cldn2 dimers in DDM extracts and perhaps identify other higher molecular weight complexes. First, we used a series of photoactivatable succinimidyl ester diazirine (SDA) cross-linkers to probe for cross-linking between His-tagged cldn2, which had been induced in MDCK II cells, extracted in DDM, and purified by metal affinity resin chromatography (Fig. 2A). As reported previously, we variably observed a low background of SDS-resistant dimer even in the absence of cross-linking (12). Incubation of purified cldn2 with SDA, NHS-LC-diazirine, or S-SDAD resulted in increased levels of dimeric cldn2, with only very minor appearance of higher molecular weight bands. Of note, although in particular after S-SDAD, as much as 15–20% of cldn2 appears as dimer, most cldn2 migrates as monomer; the reason for this limited cross-linking is unclear. There was little time-dependent increase in UV cross-linking; it seems likely that the rate-limiting step is the first amine-dependent cross-linking and that the UV-dependent cross-linking is relatively efficient and rapid.
FIGURE 2.
Chemical cross-linking of cldn2 results in formation of dimers but not higher molecular weight forms. A, His-tagged cldn2 was extracted from induced MDCK II cells with 1% DDM and purified on Talon resin and cross-linked using photoactivated cross-linkers, electrophoresed using SDS-PAGE, and immunoblotted with cldn2 antibody. Some SDS-resistant cldn2 dimer is evident in the absence of cross-linker (−); addition of three cross-linkers of different lengths all resulted in an increase in dimer formation with only minor increases in higher molecular weight bands. Water-insoluble SDA and NHS-LC-diazirine showed time-dependent increases in cross-linking (1, 5, and 15 min), whereas water-soluble S-SDAD showed maximal cross-linking at shortest period (1 min) of UV exposure; in no case was cross-linked cld2 more than 20% of monomeric cldn2. B, His-tagged cldn2 and cldn4 were purified as described above and cross-linked by incubation with glutaraldehyde for 0–30 min; 1% SDS was added to some samples to disrupt oligomerization. Panel a, cross-linking to dimer is evident for cldn2 (left panel) but not cldn4 (right panel) in the absence (−) but not the presence (+) of SDS; the positions of monomers and dimers for cldn2 and cldn4 are indicated by arrowheads. Panel b, conversion to dimer is quantified by densitometry of the immunoblots in (panel a).
Glutaraldehyde-dependent cross-linking has often been used to obtain information about subunit number or proximity, and although highly efficient, its lack of chemical specificity can at times produce oligomers of questionable biologic relevance (35). However, despite this lack of specificity, the efficiency of glutaraldehyde cross-linking suggested that it might provide a useful approach to identify potential higher order cldn2 oligomers from DDM-extracted MDCK cells (Fig. 2B, panel a, top left panel). Time-dependent glutaraldehyde cross-linking of purified cldn2 resulted in conversion of most cldn2 to a dimer with no increase in any specific higher molecular weight species; preincubation of DDM-extracted purified cldn2 with 1% SDS prior to cross-linking prevented cross-linking to a dimer. There was, however, loss of some fraction of the immunoreactive cldn2 signal after extended incubation in glutaraldehyde, consistent with nonspecific cross-linking to nonclaudin protein contaminants, to insoluble aggregates, or to sizes exceeding resolution on the gel. In contrast to glutaraldehyde-dimerized cldn2, purified DDM-extracted cldn4 inducibly expressed in MDCK cells was not cross-linked by glutaraldehyde (Fig. 2B, top right panel). The conversion of cldn2 monomer to dimer is presented graphically in Fig. 2B, panel b, bottom panel, supporting a kinetic plateau of cross-linking at a dimer. Together these cross-linking studies support a dimer as the fundamental oligomeric state of cldn2.
Dimerization of cldn2 Is Not Mediated through Its Extracellular Domains or Its Cytoplasmic Carboxyl-terminal Domain
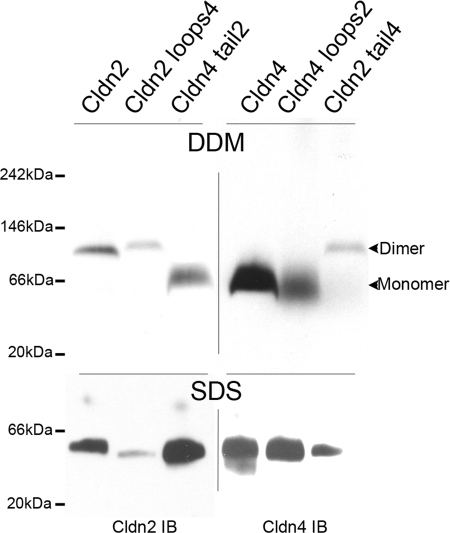
Because cldn2 forms dimers and cldn4 does not, we used previously generated MDCK II cell lines inducibly expressing chimeras between cldn2 and cldn4 (24, 25) to determine the region of cldn2 that was responsible for dimer formation. A diagram of the previously published chimeras is included as supplemental Fig. 2. MDCK cells expressing wild-type cldn2, cldn2 with the extracellular domains of cldn4, and cldn4 with the cytoplasmic carboxyl-terminal domain of cldn2 were extracted with DDM, electrophoresed using BN-PAGE, and immunoblotted with cldn2 antibody (Fig. 3, top left panel). We observed that wild-type cldn2 and cldn2 with the extracellular domain of cldn4 both migrate as dimers, whereas cldn4 with the tail of cldn2 migrates at a position smaller than the 66-kDa molecular mass marker, in the size range consistent with a monomer. Consistent with the lack of a role for the extracellular domains in cross-linking, cldn4 and cldn4 with the extracellular domains of cldn2 appear to be monomers, whereas cldn2 with the extracellular domain of cldn4 migrates as a dimer (Fig. 3, top right panel). When 1% SDS is added, the electrophoretic mobility is increased and similar for all transgenes (Fig. 3, bottom panels). These results suggested that cldn2 dimerization does not require the cytoplasmic or extracellular domains and is likely mediated through interaction of one or several of its four transmembrane segments.
FIGURE 3.
Dimerization of cldn2 is not mediated through its extracellular domains or its cytoplasmic carboxyl-terminal domain. Top left panel, stably transfected MDCK II Tet-Off cells induced to express wild-type cldn2, chimeric cld2 with the extracellular domains of cldn4 (cldn2 loops4), or cld4 with the cytoplasmic carboxyl terminus of cldn2 (cldn4 tail2) were extracted with DDM, electrophoresed in BN-PAGE, and immunoblotted for cldn2. cldn2 and cldn2 loops4 migrate as dimers, whereas cld4 tail 2 migrates in a broad band consistent with monomer. Top right panel, cells expressing wild-type cldn4, cldn4 loops 2, and cldn2 tail4 were similarly processed but immunoblotted for cldn4; cldn4 and cldn4 loops2 migrate as broad band in the range of monomers, whereas cldn2 tail4 migrates as a dimer. Bottom panels, treatment of DDM-extracted claudins and chimeras with 1% SDS results in similar electrophoretic mobility for all transgenes.
Dimers Form within a Single Cell
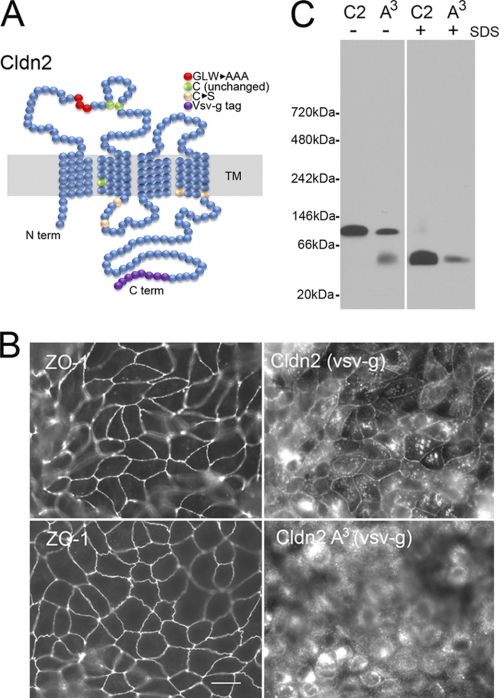
Highly conserved residues in the first extracellular domain of claudins (GLW) have been implicated in proper claudin localization and in the formation of claudin-dependent cell contacts (36, 37). To test if claudin dimer formation was dependent on intercellular contacts, we mutated GLW residues of this conserved region to alanines; the location of these mutations is shown in the diagram in Fig. 4A. Immunofluorescent analysis of induced wild-type cldn2, tagged at the carboxyl terminus with a VSV-g motif, showed that it co-localized with the tight junction cytoplasmic protein ZO-1 but was also present in intracellular vesicles as reported previously (Fig. 4B, top right) (25). Localization of cldn2 with the triple alanine mutation (A3) was almost exclusively intracellular, and it did not co-localize with ZO-1 (Fig. 4A, bottom right). When cells plated in parallel were extracted with DDM and analyzed by BN-PAGE (Fig. 4C), both wild-type cldn2 and the A3 mutant primarily migrated as dimers and were similarly converted to monomers after treatment with 1% SDS. These results were consistent with the idea that the observed dimerization does not represent transcellular interaction or that it requires localization to the plasma membrane, but instead it represents cis contacts mediated by transmembrane segments.
FIGURE 4.
Dimerization of cldn2 does not require cell surface expression and thus does not represent intercellular cross-linking. A, diagram showing the location of mutated amino acid residues; red, GLW to AAA; pale pinks, Cys to Ser mutants; purple, position of the VSV-g tag. B, immunofluorescence of VSV-g tagged cldn2 and A3 mutants demonstrates that tagged wild-type cldn2 co-localizes with ZO-1, whereas cldn2 A3 mutant is largely intracellular. Stably transfected MDCK cells were induced to express wild-type or mutant cldn2, fixed, and processed for immunofluorescence. Top panels, left, ZO-1; right, VSV-g tagged wild-type cldn 2 co-localize at the tight junction, although a considerable fraction of cldn2 is in intracellular compartments. Bottom panels, left, ZO-1 localization is unaffected by induction of mutant cldn2; right the A3 mutant of cldn2 is largely intracellular. C, BN-PAGE of VSV-g-tagged Cldn2 and Cldn2 A3 mutants reveals dimer formation is not dependent on cell surface localization or intercellular interactions. MDCK cells induced in parallel with those used for immunofluorescence (above) were extracted with 1% DDM without (−) or with (+) 1% SDS and analyzed by immunoblot after BN-PAGE. Both cldn2 and cldn2A3 migrate as dimers in the absence of SDS.
Targeted Cysteine Cross-linking Reveals Proximity of the Second Transmembrane Domains
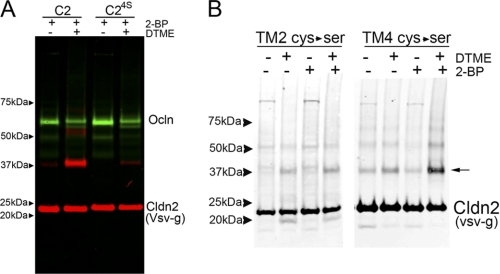
Claudins, like tetraspanins (38), are palmitoylated on perimembrane cysteines located at the cytoplasmic ends of the transmembrane segments (28). Using cldn2 sequences mutated at various perimembrane cysteines allowed us to use cross-linking to ask whether these specific cysteine positions were in close proximity within the dimer. To prevent palmitoylation and leave the cysteines free for cross-linking, cells were preincubated with 2-bromopalmitate overnight (39). Vsv-g-tagged cldn2 isolated from MDCK II cells preincubated in 2-bromopalmitate was minimally cross-linked to dimer in the absence of cysteine cross-linker (Fig. 5A, red signal), but dimer formation was significantly increased after incubation with the cysteine cross-linker DTME. The specificity of the role of the perimembrane cysteines in mediating this cross-linking is shown by the significant reduction of dimer formation in cells expressing cldn2 with mutations of four of the perimembrane cysteines to serines (C24S). Occludin (Fig. 5A, green signal) is not palmitoylated and not cross-linked to a specific product after DTME treatment; however, the decrease in occludin signal in the DTME-treated samples does suggest the formation of some occludin adducts that are not detected as specific bands. DTME cross-links cysteines at a distance of 13.3 Å. Cross-linking with a cysteine cross-linker with shorter spacer arms (bis(maleimido)ethane, 8 Å) resulted in a similar level of cross-linking (data not shown).
FIGURE 5.
Cysteine cross-linking suggests proximity of the second transmembrane domains. A, cldn2 can be cross-linked with disulfide cross-linkers by perimembrane cysteines. MDCK cells induced to express wild-type cldn2 (C2) and cldn2 with perimembrane cysteines mutated to serines (C24S) were incubated overnight in 2-bromopalmitate to inhibit palmitoylation and then incubated without (−) and with (+) the cysteine cross-linker DTME. Cell extracts were subjected to SDS-PAGE and immunoblotted for VSV-g cldn2 (red) and occludin (green); wild-type cldn2 was cross-linked with much greater efficiency than cldn24S; cldn24S migrates as a dimer in BN-PAGE (data not shown). B, mutational analysis suggests that the second transmembrane domains may be in close proximity. MDCK cells stably transfected and induced to express VSV-g-tagged cldn2 with perimembrane cysteines mutated to serines after the second (TM2 Cys → Ser) or fourth (TM4 Cys → Ser) were incubated overnight without (−) or with (+) 2-bromopalmitate and then treated (+) with cysteine cross-linker. The only sample where cross-linking to dimer (arrow) was clearly dependent on preincubation with 2-bromopalmitate and DTME was TM4 (Cys → Ser), suggesting a critical role for perimembrane cysteines following the second transmembrane domain.
The perimembrane cysteines in cldn2 are located at the carboxyl-terminal ends of the second and fourth transmembrane domains; this location is conserved among claudins (28). To determine the relative contributions of the cysteines proximal to the second versus fourth transmembrane segments to the cysteine-dependent cross-linking, we separately mutated these cysteines to serine and performed similar cross-linking experiments as described above. Some background dimer formation was detectable in all cell lines after treatment with the cross-linking agent, perhaps due to less than complete palmitoylation of perimembrane cysteines, to incomplete oxidation of the extracellular cysteines, or to the unmutated cysteine within TM2 (see diagram in Fig. 4A). However, only in the cells expressing cldn2 where the perimembrane cysteines at the end of the fourth transmembrane domain were mutated to serines was there an appreciable increase in specific cross-linking to dimer (Fig. 5B). These data suggest that perimembrane cysteines at the end of the second, but not the fourth, transmembrane domains can participate in DTME-mediated cross-linking and thus suggest that the second transmembrane domains of two cldn2 molecules are likely to be in close proximity.
BN-PAGE of Purified Mouse Liver Membranes Reveals That Claudins and Occludin Are Components of a High Molecular Weight Complex
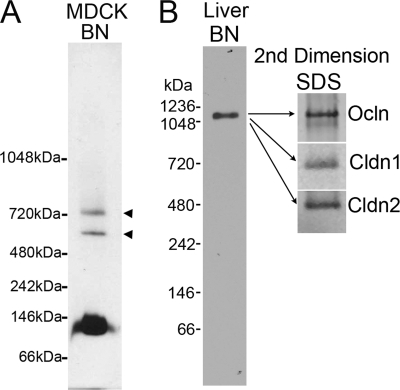
Although most cldn2 detectable by BN-PAGE of MDCK cells appears as a dimer, longer exposure of the immunoblots revealed some higher molecular weight cldn2-containing bands (Fig. 6A). Freeze fracture EM of tight junctions reveals that claudins can exist in 10 nm particle complexes, which would represent protein complexes significantly larger than dimers. We speculated that larger complexes might be more easily detected in native tissues, like liver, where the tight junction network is more developed and cell turnover rates are slower than in MDCK cells. Using a simplified protocol based on biochemical enrichment of murine hepatocyte tight junctions (29), we used sucrose density gradients to enrich junction fractions before extraction with DDM, electrophoresis in BN-PAGE, and immunoblotting. Anti-cldn1 antibody was used on tissue blots because it gave a robust signal compared with cldn2 polyclonal antibodies. The observed size of the immunoreactive complex from liver was ∼1100 kDa. Correcting for bound Coomassie dye (×0.56) suggests a complex of ∼600 kDa for the protein fraction (Fig. 6B). Of note, Coomassie binding should be limited to the surface of this large complex, and thus the actual protein content may be higher (31). When the corresponding area was excised from a duplicate gel, electroeluted, precipitated, and electrophoresed in SDS-PAGE, the complex was found to contain at least cldn1, cldn2, and occludin; the presence or absence of other tight junction proteins has not yet been tested. This is the first evidence for a large relatively discrete biochemical complex containing tight junction proteins. With further purification and characterization, we expect it may provide a source for identifying new tight junction components as well as allowing analysis of protein stoichiometry.
FIGURE 6.
BN-PAGE reveals evidence of higher molecular weight claudin-containing complexes. A, electrophoresis of DDM-extracted sucrose gradient-enriched MDCK II membrane in BN-PAGE reveals a predominant band at 112 kDa, consistent with dimer formation. However, long exposure reveals two higher molecular weight species, migrating at ∼600 and 720 kDa (arrowheads) suggesting a small fraction of cldn2 may be present as a higher order protein complex. B, BN-PAGE of purified mouse liver membranes reveals that claudins and occludin are components of a higher molecular weight complex. Sucrose gradient-enriched mouse liver membranes were extracted with 1% DDM and subjected to BN-PAGE followed by immunoblot for cldn1, because this polyclonal antibody was more sensitive than the available cldn 2 polyclonal antibodies. The area corresponding to the immunoreactive band was excised on a duplicate gel, and the sample was electroeluted, precipitated, electrophoresed in SDS-PAGE, and immunoblotted for cldn1, cldn2, and occludin, all of which were present in the original high molecular weight band.
DISCUSSION
Two significant new findings about tight junction structure emerge from our studies. First, we demonstrate that cldn2 extracted with DDM from MDCK cells and analyzed by BN-PAGE is assembled as a homodimer. Cross-linking of chimeric cldn2/cldn4 molecules reveals that dimerization is likely mediated through the transmembrane segments. Mutational analysis followed by cysteine cross-linking suggests that the second transmembrane segments are in very close proximity, speculatively organized as a helix-helix interaction. Second, we find that BN-PAGE of junction-enriched mouse liver fractions contains a relatively discrete ∼600-kDa multiprotein complex containing at least cldn1, cldn2, and occludin. Although the dimer is clearly a fundamental structural unit of claudins, we speculate the organization of the large complex may include the 10 nm particle seen in freeze fracture electron microscopic images of tight junction strands, either as a cell-to-cell particle complex and/or including bound cytoplasmic proteins. Further characterization of this complex is warranted to understand the hierarchical assembly of the tight junction barrier.
The variable sensitivity of different membrane proteins and their associated lipids to solubilization by detergents means that isolation of membrane protein complexes is highly empirical. The complexes obtained depend on the detergent used (40) and to some extent even on the tissue source (41). In DDM extracts from cultured cells, we were only reliably able to detect dimers, suggesting this represents the most stable or fundamental claudin oligomeric unit. In contrast, the finding that transfected cldn4 behaves as a monomer was somewhat surprising, because induction of this transgene in MDCK II cells is associated with a striking increase in the number of freeze fracture strands and a correlating increase in the physiologic barrier (23). However, we have also noted that cldn4 in MDCK II cells does not localize to tight junctions but extends all along the lateral membrane, and when expressed in fibroblasts, it does not concentrate in plaques at cell-cell junctions as do cldn1 or cldn2 (data not shown). These results suggest that it does not behave as a typical adhesive claudin and we speculate may lack some ability to self-organize; this could explain its failure to form stable dimers in BN-PAGE. In support of this possibility, a recent study (42) demonstrated that cldn4 interacted with cldn8 in a split ubiquitin yeast two-hybrid assay but did not interact with itself. These authors further showed that in cultured renal collecting duct cells, cldn4 localization required cldn8 co-expression. Unlike cldn2, cldn4 and cldn16 (see below) may participate not in homodimers but in obligate heterodimers with selected other claudins. The interesting implication is that combinatorial diversity in tight junction structure might be generated if sets of claudins follow different assembly rules.
Our finding that cldn2 formed dimers based on transmembrane interactions fits much published indirect evidence suggesting claudins interact via lateral association and that strand assembly requires cooperative cis and trans interactions. Soon after the initial identification of claudins, Tsukita and co-workers (43) demonstrated by immuno-freeze fracture electron microscopy that all claudins they tested, along with occludin, were co-localized to the same tight junction strands, despite the observation that only a subset of claudins appeared to interact across cells. These results were supported by subsequent studies using FRET analysis, which also were consistent with lateral claudin interactions (9). A more direct demonstration of the importance of the transmembrane segments in claudin interactions came from the work of Hou et al. (44). As in their analysis of cldn4, they used a split ubiquitin yeast two-hybrid assay to demonstrate that cldn19 could interact both with itself and with cldn16 and further found that a mutation in the first transmembrane domain of cldn19 could abrogate its ability to interact itself and with cldn16. The transmembrane-based interaction seems likely to happen during protein synthesis or trafficking as we observed that the intracellular A3 mutants of cldn2 formed dimers, and other studies have noted heterotypic claudin association in Golgi (42). Piontek et al. (10) recently proposed a model where claudins first form cis-oligomers in the plane of the membrane and suggested that polymerization into continuous strands depended on subsequent engagement of trans interactions across the intercellular space.
Claudin dimers are too small to account for the 10-nm tight junction particles seen in freeze fracture electron microscopic images. It has been suggested that these particles may be formed from claudin hexamers, based on analogy to the connexins, similar sized tetraspanning transmembrane proteins, which organize into hexamers and form similar sized particles freeze fracture particles (1). Unlike analysis of connexins, however, it has been difficult to demonstrate the existence of higher order claudin oligomers. Studies that suggested the existence of claudin hexamers were dependent on claudin overexpression (12, 13) and may represent nonspecific aggregation. Alternatively, these studies may demonstrate labile physiologic hexamers that are only maintained during biochemical isolation when there is an unusually high claudin concentration. Defining the composition of the 10 nm particle may depend on a better understanding of essential tight junction components, including other cellular proteins and lipids that may be recruited into a claudin-based particle.
Along with trying to understand how claudins are organized into oligomers, the approach we have used is to try to define a larger, multiprotein complex to begin to define a critical set of interacting proteins in tight junction assembly. Anatomic data suggested that claudin and occludin, at a minimum, are organized into a membrane protein complex; however, the lack of unambiguous biochemical evidence for such a complex indicates that these protein interactions might be labile or very sensitive to extraction conditions. We were able to demonstrate the existence of a high molecular weight claudin- and occludin-containing protein complex from mouse liver using BN-PAGE but not from MDCK cells despite the fact that, like liver tissue, this cell line contains cldn1, cldn2, and occludin. It is possible, as suggested above, that the minimal tight junctions of rapidly dividing MDCK cell are less stable to detergent extraction or that stabilization of the interactions of these proteins depends on unknown factors, including perhaps tissue-specific differences in protein or lipid composition, stoichiometry, or post-translational modifications. We plan to examine complex formation in Caco-2 and T84 cells after 3 weeks in culture, because at this time these cells have more elaborate tight junction organization than MDCK cells. Another possibility is that extraction of liver but not MDCK cells maintains association of cytoplasmic scaffolding proteins that might be critical in stabilizing the oligomeric organization. Further purification and characterization of the BN-PAGE purified tight junction protein complex may provide insight into these factors and should allow us to further characterize the molecular composition and organization of tight junctions.
Supplementary Material
Acknowledgments
We thank Alan Fanning, Laurel Rogers, and Jennifer Holmes of University of North Carolina, Chapel Hill, for valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 DK45134.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- BN-PAGE
- blue native-PAGE
- MDCK
- Madin-Darby canine kidney cell
- DDM
- dodecyl maltoside
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- SDA
- succinimidyl ester diazirine
- SDAD
- sulfo-NHS-SS-diazirine.
REFERENCES
- 1. Anderson J. M. (2001) News Physiol. Sci. 16, 126–130 [DOI] [PubMed] [Google Scholar]
- 2. Furuse M., Hirase T., Itoh M., Nagafuchi A., Yonemura S., Tsukita S., Tsukita S. (1993) J. Cell Biol. 123, 1777–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ikenouchi J., Furuse M., Furuse K., Sasaki H., Tsukita S., Tsukita S. (2005) J. Cell Biol. 171, 939–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raleigh D. R., Marchiando A. M., Zhang Y., Shen L., Sasaki H., Wang Y., Long M., Turner J. R. (2010) Mol. Biol. Cell 21, 1200–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Steed E., Rodrigues N. T., Balda M. S., Matter K. (2009) BMC Cell Biol. 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martìn-Padura I., Lostaglio S., Schneemann M., Williams L., Romano M., Fruscella P., Panzeri C., Stoppacciaro A., Ruco L., Villa A., Simmons D., Dejana E. (1998) J. Cell Biol. 142, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S. (1998) J. Cell Biol. 141, 1539–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Furuse M., Sasaki H., Fujimoto K., Tsukita S. (1998) J. Cell Biol. 143, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blasig I. E., Winkler L., Lassowski B., Mueller S. L., Zuleger N., Krause E., Krause G., Gast K., Kolbe M., Piontek J. (2006) Cell. Mol. Life Sci. 63, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piontek J., Winkler L., Wolburg H., Müller S. L., Zuleger N., Piehl C., Wiesner B., Krause G., Blasig I. E. (2008) FASEB J. 22, 146–158 [DOI] [PubMed] [Google Scholar]
- 11. Daugherty B. L., Ward C., Smith T., Ritzenthaler J. D., Koval M. (2007) J. Biol. Chem. 282, 30005–30013 [DOI] [PubMed] [Google Scholar]
- 12. Mitic L. L., Unger V. M., Anderson J. M. (2003) Protein Sci. 12, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coyne C. B., Gambling T. M., Boucher R. C., Carson J. L., Johnson L. G. (2003) Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L1166–L1178 [DOI] [PubMed] [Google Scholar]
- 14. Wittig I., Schägger H. (2009) Proteomics 9, 5214–5223 [DOI] [PubMed] [Google Scholar]
- 15. Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 16. Crane J. M., Bennett J. L., Verkman A. S. (2009) J. Biol. Chem. 284, 35850–35860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galka J. J., Baturin S. J., Manley D. M., Kehler A. J., O'Neil J. D. (2008) Biochemistry 47, 3513–3524 [DOI] [PubMed] [Google Scholar]
- 18. Sorbo J. G., Moe S. E., Ottersen O. P., Holen T. (2008) Biochemistry 47, 2631–2637 [DOI] [PubMed] [Google Scholar]
- 19. Kiss A., Troyanovsky R. B., Troyanovsky S. M. (2008) Mol. Biol. Cell 19, 4042–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Babusiak M., Man P., Petrak J., Vyoral D. (2007) Proteomics 7, 121–129 [DOI] [PubMed] [Google Scholar]
- 21. Li X., Xie C., Jin Q., Liu M., He Q., Cao R., Lin Y., Li J., Li Y., Chen P., Liang S. (2009) J. Proteome Res. 8, 3475–3486 [DOI] [PubMed] [Google Scholar]
- 22. Stevenson B. R., Siliciano J. D., Mooseker M. S., Goodenough D. A. (1986) J. Cell Biol. 103, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Itallie C., Rahner C., Anderson J. M. (2001) J. Clin. Invest. 107, 1319–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Itallie C. M., Colegio O. R., Anderson J. M. (2004) J. Membr. Biol. 199, 29–38 [DOI] [PubMed] [Google Scholar]
- 25. Colegio O. R., Van Itallie C., Rahner C., Anderson J. M. (2003) Am. J. Physiol. Cell Physiol. 284, C1346–C1354 [DOI] [PubMed] [Google Scholar]
- 26. Boone C. W., Ford L. E., Bond H. E., Stuart D. C., Lorenz D. (1969) J. Cell Biol. 41, 378–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Azem A., Tsfadia Y., Hajouj O., Shaked I., Daniel E. (2010) Biochim. Biophys. Acta 1804, 768–780 [DOI] [PubMed] [Google Scholar]
- 28. Van Itallie C. M., Gambling T. M., Carson J. L., Anderson J. M. (2005) J. Cell Sci. 118, 1427–1436 [DOI] [PubMed] [Google Scholar]
- 29. Stevenson B. R., Goodenough D. A. (1984) J. Cell Biol. 98, 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Itallie C. M., Fanning A. S., Bridges A., Anderson J. M. (2009) Mol. Biol. Cell 20, 3930–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wittig I., Beckhaus T., Wumaier Z., Karas M., Schägger H. (2010) Mol. Cell. Proteomics 9, 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H., Poolman B. (2002) J. Mol. Biol. 317, 591–600 [DOI] [PubMed] [Google Scholar]
- 33. Jiang J., Daniels B. V., Fu D. (2006) J. Biol. Chem. 281, 454–460 [DOI] [PubMed] [Google Scholar]
- 34. Reisinger V., Eichacker L. A. (2007) Proteomics 7, Suppl. 1, 6–16 [DOI] [PubMed] [Google Scholar]
- 35. Payne J. W. (1973) Biochem. J. 135, 867–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cukierman L., Meertens L., Bertaux C., Kajumo F., Dragic T. (2009) J. Virol. 83, 5477–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mrsny R. J., Brown G. T., Gerner-Smidt K., Buret A. G., Meddings J. B., Quan C., Koval M., Nusrat A. (2008) Am. J. Pathol. 172, 905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang X., Claas C., Kraeft S. K., Chen L. B., Wang Z., Kreidberg J. A., Hemler M. E. (2002) Mol. Biol. Cell 13, 767–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kovalenko O. V., Yang X., Kolesnikova T. V., Hemler M. E. (2004) Biochem. J. 377, 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reisinger V., Eichacker L. A. (2008) J. Proteomics 71, 277–283 [DOI] [PubMed] [Google Scholar]
- 41. Reifschneider N. H., Goto S., Nakamoto H., Takahashi R., Sugawa M., Dencher N. A., Krause F. (2006) J. Proteome Res. 5, 1117–1132 [DOI] [PubMed] [Google Scholar]
- 42. Hou J., Renigunta A., Yang J., Waldegger S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18010–18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Furuse M., Sasaki H., Tsukita S. (1999) J. Cell Biol. 147, 891–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hou J., Renigunta A., Konrad M., Gomes A. S., Schneeberger E. E., Paul D. L., Waldegger S., Goodenough D. A. (2008) J. Clin. Invest. 118, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.