Abstract
Three specific β(1,3)glucan synthase (GS) inhibitor families, papulacandins, acidic terpenoids, and echinocandins, have been analyzed in Schizosaccharomyces pombe wild-type and papulacandin-resistant cells and GS activities. Papulacandin and enfumafungin produced similar in vivo effects, different from that of echinocandins. Also, papulacandin was the strongest in vitro GS inhibitor (IC50 103–104-fold lower than with enfumafungin or pneumocandin), but caspofungin was by far the most efficient antifungal because of the following. 1) It was the only drug that affected resistant cells (minimal inhibitory concentration close to that of the wild type). 2) It was a strong inhibitor of wild-type GS (IC50 close to that of papulacandin). 3) It was the best inhibitor of mutant GS. Moreover, caspofungin showed a special effect for two GS inhibition activities, of high and low affinity, separated by 2 log orders, with no increase in inhibition. pbr1-8 and pbr1-6 resistances are due to single substitutions in the essential Bgs4 GS, located close to the resistance hot spot 1 region described in Saccharomyces and Candida Fks mutants. Bgs4pbr1-8 contains the E700V change, four residues N-terminal from hot spot 1 defining a larger resistance hot spot 1-1 of 13 amino acids. Bgs4pbr1-6 contains the W760S substitution, defining a new resistance hot spot 1-2. We observed spontaneous revertants of the spherical pbr1-6 phenotype and found that an additional A914V change is involved in the recovery of the wild-type cell shape, but it maintains the resistance phenotype. A better understanding of the mechanism of action of the antifungals available should help to improve their activity and to identify new antifungal targets.
Keywords: Antibiotics, Cell Wall, Enzyme Inhibitors, Membrane Enzymes, Yeast, Antifungals, Caspofungin, Fks Proteins, Glucan, Glucan Synthase
Introduction
The number of antifungal families available and their use in therapy is very limited (1–4). Recently, a new family of specific fungal cell wall synthesis inhibitors has emerged as an alternative antifungal therapy and is gaining increasing relevance yearly (5–7).
The cell wall is a structure external to the plasma membrane and is present in all fungal cells. Its integrity is crucial, and it constitutes the exoskeleton that confers mechanical strength and osmotic resistance to fungal cells (8–10). In mammalian cells, the cell wall is absent, and consequently, drugs that interfere with its synthesis are attractive as potential antifungal agents. β(1,3)glucan is a major contributor to the framework of the cell wall. There are several families of antifungal drugs whose mode of action is not well known, although they clearly interfere with β(1,3)glucan synthesis by inhibiting the β(1,3)glucan synthase (GS)6 enzyme. These inhibitors include echinocandins (lipopeptides), papulacandins (glycolipids), and acidic terpenoids such as enfumafungin (2, 11). To date, only the echinocandins caspofungin, micafungin, and anidulafungin have been approved (in years 2002, 2005, and 2006, respectively) for treatment of invasive fungal infections (1, 3, 5, 7, 12).
The fission yeast Schizosaccharomyces pombe provides an appealing model for studies addressing cell wall synthesis and morphogenesis. The S. pombe cell wall has no detectable chitin (10), but it contains three different essential β-glucans as follows: a branched β(1,3)glucan, which is the major contributor to the cell wall structure; a minor linear β(1,3)glucan, concentrated in the primary septum, with minor amounts in the cell wall; and a minor branched β(1,6)glucan (13, 14). S. pombe contains four essential putative GS catalytic subunits, Bgs1 to Bgs4. Bgs1 is responsible for the synthesis of the linear β(1,3)glucan and primary septum. Bgs2 is essential for spore wall maturation, and Bgs3 is essential, although its function remains unknown. Bgs4 is the only subunit that has been shown to form part of the GS enzyme. It is responsible for the major part of cell wall β(1,3)glucan synthesis and in vitro GS activity, and it is essential for the maintenance of cell integrity during cell growth and separation (14–17). The different essential functions of Bgs proteins in cell morphogenesis make them good targets for the study of antifungal drugs that specifically inhibit β(1,3)glucan synthesis.
The S. pombe Bgs family is homologous to fungal Fks and plant CalS proteins, considered to be putative GS catalytic subunits (8, 18). Fungal resistance to GS inhibitors is clearly associated with mutations in conserved short regions (hot spots) of the Fks proteins, indicating that this mechanism is well conserved in fungi (12, 19, 20). In addition, intrinsic echinocandin-resistant fungi contain natural substitutions in the conserved Fks region that are determinants of their resistance (19, 21–23). Most of the mutants resistant to GS inhibitors have been isolated as resistant to echinocandins. Only some Saccharomyces cerevisiae and S. pombe mutants have been selected as resistant to papulacandin (24, 25), in each case defining a single complementation group called pbr1. However, although the S. cerevisiae pbr1-1 mutation has been associated with FKS1, the S. pombe pbr1+ gene remains unknown.
In light of the above data, we were prompted to study and compare the mechanism of action of the following three antifungal families: papulacandins, acidic terpenoids, and echinocandins. Using S. pombe as a model, we examined the in vivo and in vitro effect of the antifungals on cells and the GS of wild-type and resistant mutants. Our data point to important differences among the antifungal families in both cells and GS activity. Caspofungin was overall the best inhibitor of cells and GS tested, not only of the wild-type but also resistant cells. Although S. pombe vegetative cells contain three essential Bgs subunits, the antifungal resistance is exclusively associated with Bgs4, suggesting that Bgs1 and Bgs3 are intrinsic resistant subunits. Papulacandin selection afforded two new amino acid substitutions, expanding the resistance hot spot 1 to 13 residues and defining a new resistance hot spot 1-2. These new sites, which are important for resistance and interaction with antifungals, should help to understand the mechanism of action of antifungals and the resistance mechanism of the Bgs/Fks proteins.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
The S. pombe strains used were isogenic to wild-type strain h− 972. The pbr1-1, pbr1-2, pbr1-3, pbr1-6, and pbr1-8 mutants were obtained by ethyl methanesulfonate mutagenesis (15–30% survival) and selection in the presence of 20 μg/ml papulacandin B (25). The five mutants were backcrossed three times with the wild-type strain. In all the cases, tetrad analysis revealed a 2R:2S segregation, indicating the monogenic trait or the resistance phenotype. The dominant/recessive analysis was performed in stable heterozygous diploid strains, using the mat2P-B102 mutation (26). All the papulacandin resistance mutations proved to be recessive. Complementation analysis was carried out in diploid strains and showed that the five resistance mutations are alleles of the same gene, which was named pbr1 (from papulacandin B resistant).
The standard complete yeast growth medium (YES), selective medium (EMM) supplemented with the appropriate amino acids, and sporulation medium (SPA) (26, 27) have been described previously. Cell growth was monitored by measuring the A600 of early log phase cell cultures in a Coleman Junior II spectrophotometer (A600 0.1 = 1 × 107 cells/ml). The general procedures for yeast and bacterial culture and genetic manipulations were carried out as described previously (27, 28).
Plasmids and DNA Techniques
The multicopy pAL-bgs4+ (S. cerevisiae LEU2 selection) and integrative single copy pJK-bgs4+ plasmids (S. pombe leu1+ selection) have been described elsewhere (15). All the bgs4+ plasmids contain an 8.84-kb PstI-NheI bgs4+ fragment. The integrative phis3-bgs4+ plasmid (his3+ selection) contains the Bluescript KS+ backbone, the 2.3-kb Eco47III-DraI his3+ sequence cloned into EcoRV (KpnI-SacI orientation), and the bgs4+ sequence from pAL-bgs4+ cloned into PstI-NotI. p81X-bgs4+ is pJR2-81XL (LEU2 selection and 81X version of the thiamine-repressible nmt1+ promoter) (29) containing the bgs4+ ORF (15).
pAL-bgs4pbr1-8 contained bgs4pbr1-8 obtained from the pbr1-8 strain by plasmid gap repair. pAL-bgs4+ was digested with SacI (removing the bgs4+ ORF, 680 bp of promoter, and 188 bp of terminator) and transferred into strain pbr1-8, and repaired pAL-bgs4pbr1-8 plasmids were recovered from transformants. Integrative pJK-bgs4pbr1-8 and phis3-bgs4pbr1-8 and regulatable p81X-bgs4pbr1-8 contained bgs4pbr1-8 from pAL-bgs4pbr1-8. The 8.8-kb region of bgs4pbr1-8 contained a single base substitution, A2099T, resulting in the E700V amino acid change of Bgs4, and bgs4pbr1-6 contained the single G2279C base substitution, resulting in the W760S amino acid change of Bgs4 (see under “Results”).
bgs4pbr1-8 gene deletion was performed in a pbr1-8/pbr1-8 diploid by removing the entire ORF of a bgs4pbr1-8 copy, as described for bgs4+ gene deletion (15). Bgs4 is essential, and therefore haploid bgs4pbr1-8Δ deletion strains were maintained viable with a plasmid expressing bgs4+.
Antifungal Drugs and Resistance Assays
The antifungals were generous gifts from Ciba-Geigy/Novartis (papulacandin B), Tokyo Jozo (aculeacin A) and Merck Sharp and Dohme (enfumafungin, pneumocandin B0, and caspofungin). The antifungals were kept at −20 °C in stock solution (10 mg/ml in DMSO, except caspofungin, which is highly soluble in water) and assayed at the final concentrations specified in the text and figures.
For micro-culture assays of large numbers of samples, late log phase cultures grown in YES medium were diluted to a cell density of 2 × 106 cells/ml in 200 μl of YES medium containing increasing concentrations of antifungal (0, 1, 2, 5, 10, 20, 50, and 100 μg/ml) or an equivalent volume of solvent. The cell cultures were incubated in an orbital roller at 28 °C, and turbidity was analyzed after 24 and 48 h of incubation, affording values ranging from 0 (−, no turbidity, wild-type cells in the presence of a lethal concentration of antifungal) to 100 (+++, total turbidity, wild-type cells in the absence of antifungal). The MIC was determined as the minimal concentration of antifungal that produced complete cell growth inhibition after 24 h of incubation. The values were calculated from at least three independent experiments.
Enzyme Preparation and β(1,3)Glucan Synthase Assay
Cell extracts and GS assays were performed essentially as described previously (30). Cell extracts were obtained from early log phase cells grown in YES medium at 28 °C. The cells were washed with buffer A (50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 1 mm β-mercaptoethanol), suspended in 100 μl of buffer A containing 50 μm GTPγS to preserve enzyme activity, and broken with glass beads. Membrane enzyme extracts were resuspended in the same buffer containing 33% glycerol and 50 μm GTPγS and stored at −80 °C. All GS assays (150 μm GTPγS and 15–25 μg of protein) were carried out in duplicate, and the values were calculated from at least three independent cell cultures.
Other Procedures
The fractionation of cell wall polysaccharides, enzymatic lysis of cell suspensions (31), and fluorescence microscopy of cell walls stained with Calcofluor White (30) have been described previously.
RESULTS
pbr1 Mutants Present Increasing Cell Wall-related Phenotypes
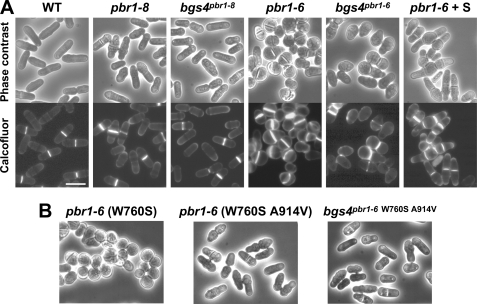
The isolation of S. pombe mutants resistant to papulacandin B, which defined a single complementation group designated pbr1 (for papulacandin B resistant), has been reported previously (25). pbr1-6 cells show a special spherical morphology (Fig. 1), which has been associated with a single recessive gene called sph1-1 (24). This mutant was characterized as being defective in cell wall galactomannan, which could account for a cell integrity defect responsible for the spherical phenotype. In fact, osmotic stabilization with sorbitol suppressed that phenotype (Fig. 1A). However, the morphology of the other pbr1 alleles was that of both normal and lemon-shaped cells (Fig. 1A) (data not shown). This led us to consider the possibility that both phenotypes might be related, pbr1-6 being the result of a more aggravated phenotype. Accordingly, the cell wall-related phenotypes of pbr1-6 and pbr1-8 (the latter being representative of the other mutant alleles) were examined (Table 1).
FIGURE 1.
Morphology of pbr1-8 and pbr1-6 cells. A, normal and lemon-shaped morphology of pbr1-8 and bgs4Δ bgs4pbr1-8 cells and rounded morphology of pbr1-6 and bgs4Δ bgs4pbr1-6 cells. Osmotic stabilization with sorbitol (S) suppresses the pbr1-6 rounded morphology, after which a multiseptated phenotype appears. Phase contrast and Calcofluor white-stained (50 μg/ml) micrographs of early log phase cells grown on YES medium at 28 °C. Bar, 10 μm. B, spherical morphology of original pbr1-6 strain (W760S substitution) and elongated cell shape of a spontaneous pbr1-6 morphological revertant (W760S and A914V changes) and of a bgs4Δ bgs4pbr1-6 (W760S A914V) strain. The pbr1-6 resistance is not affected with the A914V suppressor mutation (Table 3).
TABLE 1.
Increasing cell wall defects of S. pombe pbr1-8 and pbr1-6 strains
Values are the means ± S.D. calculated from at least three independent experiments.
| Strain | Alkali extractiona% residual cell wall | Lysis of cell suspensionsb |
Incorporation of [14C]glucosec |
Cell wall hexosesd |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Novozyme | Zymolyase | Cell wall | α-Glucan | β-Glucan | Galactomannan | Glc | Man | Gal | ||
| % | % | % | ||||||||
| WT | 12.7 ± 1.6 | 13.4 ± 3.3 | 71.7 ± 3.0 | 25.4 ± 0.3 | 7.6 ± 0.4 | 14.0 ± 0.2 | 3.8 ± 0.3 | 88.8 ± 0.1 | 7.4 ± 0.2 | 3.7 ± 0.2 |
| pbr1-8 | 10.9 ± 0.6 | 50.1 ± 6.5 | 60.9 ± 2.6 | 27.7 ± 0.7 | 9.2 ± 0.5 | 15.1 ± 0.5 | 3.4 ± 0.1 | 91.3 ± 1.0 | 5.5 ± 0.6 | 3.2 ± 0.3 |
| pbr1-6 | 1.8 ± 0.2 | 69.6 ± 4.1 | 44.1 ± 3.9 | 28.2 ± 1.0 | 11.7 ± 0.8 | 15.2 ± 0.9 | 1.3 ± 0.2 | 97.4 ± 0.6 | 1.8 ± 0.3 | 0.7 ± 0.2 |
a Mild alkali extraction of the cell wall (6% NaOH, room temperature, 5 min) is shown. Standard alkali extraction removes the entire cell wall.
b Percentage of A600 decrease after 6 h in the presence of Novozyme (10 mg/ml) or Zymolyase (2 mg/ml). Novozyme 234 complex degrades the entire cell wall, whereas Zymolyase 100T degrades all the cell wall except α-glucan.
c Percentage incorporation of [14C]glucose into the cell wall polysaccharides (cpm incorporated per fraction × 100/total cpm incorporated).
d Percentage of cell wall hexoses analyzed by gas-liquid chromatography.
The fluorochrome Calcofluor White specifically binds to growing poles, with especially high affinity for the septum (14). pbr1-8 and pbr1-6 cells showed increasing Calcofluor staining, which was more intense in pbr1-6 cells, where the compound stained the whole cell wall and the septa appeared thicker and more strongly stained (Fig. 1A). Cell wall analyses also revealed increasing phenotypes as follows: from the wild-type to pbr1-8 and to pbr1-6 (Table 1). With mild alkali extraction of the cell walls, a gradual increase in susceptibility was observed. Lysis of cell suspensions with Novozyme also elicited increasing susceptibility. By contrast, the mutant cells exhibited increasing resistance to Zymolyase degradation. Novozyme degrades the cell wall, suggesting an increasing defect in cell wall structure and integrity. However, Zymolyase does not contain α-glucanases and hence resistance to degradation indicates a progressive increase in cell wall α-glucan levels. This was confirmed by cell wall fractionation, which revealed a gradual increase in total cell wall, α-glucan, and β-glucan and a gradual decrease in galactomannan. Similarly, the amount of cell wall hexoses changed as follows: glucose increased whereas mannose and galactose decreased (Table 1).
The above findings indicate that both pbr1-8 and pbr1-6 mutants show similar cell wall-related phenotypes, more aggravated in pbr1-6. However, although the mutants were selected as being resistant to a GS inhibitor, the kinetic parameters of in vitro mutant GS activities (specific activity, Km and Vmax) were not altered either in the absence (24, 25) or in the presence of papulacandin (50 μg/ml, data not shown).
Differential in Vivo Effect of Three Families of Specific GS Inhibitors on Wild-type and Resistant Stains
The pbr1 mutants were obtained as being resistant to papulacandin B and also showed resistance to aculeacin A. However, pbr1-6 was again different because it showed normal sensitivity to aculeacin A. Because of the different behavior of the pbr1-6 and pbr1-8 mutants with aculeacin A and the fact that only a few antifungals have been analyzed, antifungals representative of the different families of specific GS inhibitors (2) were selected to study their in vivo and in vitro effects on wild-type and resistant strains and GS activities as follows: papulacandin B (papulacandins), enfumafungin (acidic terpenoids), aculeacin A, pneumocandin B0, and caspofungin (echinocandins) (Fig. 2).
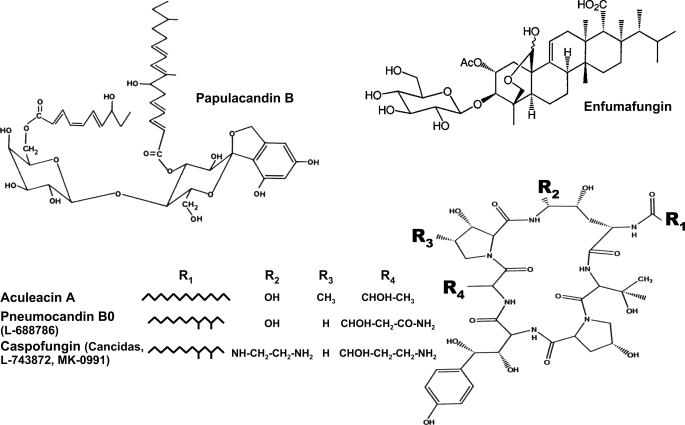
FIGURE 2.
Chemical structures of the three families of specific β(1,3)glucan synthase inhibitors. Structures of the inhibitors representative of each family used in this work are as follows: papulacandin B (papulacandins), enfumafungin (acidic terpenoids), aculeacin A, pneumocandin B0, and caspofungin (echinocandins).
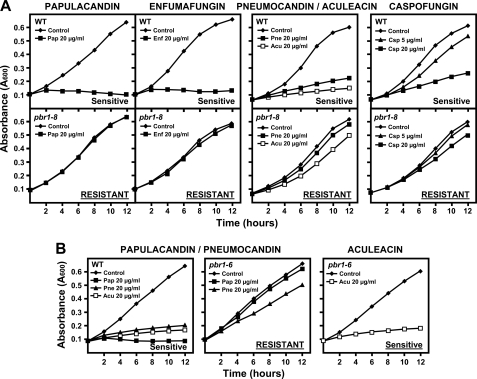
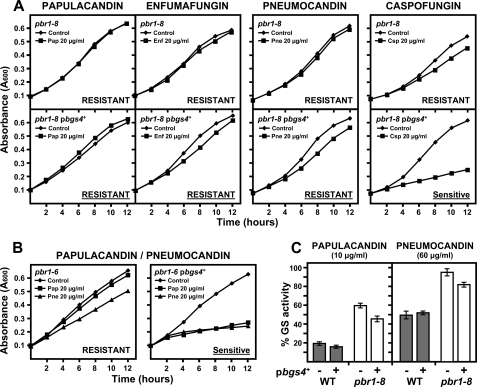
First, a different in vivo inhibitory capacity was observed, depending on the antifungal family. In wild-type cells, papulacandin and enfumafungin produced a dramatic and complete arrest of cell growth, whereas the echinocandins permitted residual cell growth even at higher concentrations (Fig. 3A). This was accompanied by a different lytic effect. Papulacandin and enfumafungin led to rapid and total cell lysis, and the echinocandins produced lysed cells accompanied by swollen, round, and aberrant cells (data not shown). Similar two types of antifungal effects were also observed with the pbr1-8 and pbr1-6 resistances as follows: complete resistance and an unaltered morphology with papulacandin and enfumafungin, and weaker resistance and cells becoming round and aggregated with the echinocandins (Fig. 3, A and B) or even no resistance at all, as in the case of pbr1-6 strain with aculeacin (Fig. 3B).
FIGURE 3.
Differential effect of papulacandin B and enfumafungin and of pneumocandin B0, aculeacin A, and caspofungin on the cell growth of the wild-type, pbr1-8, and pbr1-6 strains. Early log phase cells were grown in YES medium at 28 °C either in the absence or in the presence of the indicated antifungal concentrations. Cell growth was monitored at the indicated times. A, papulacandin and enfumafungin produce total cell growth arrest in wild-type cells, whereas pbr1-8 cells are completely resistant. However, echinocandins permit a residual growth of wild-type cells and produce slow cell growth in pbr1-8 cells. B, pbr1-6 cells are resistant to only some antifungals, very resistant to papulacandin, partially resistant to pneumocandin, and sensitive to aculeacin. pbr1-6 cells are also resistant to enfumafungin and caspofungin (see Tables 2 and 3). Pap, papulacandin; Enf, enfumafungin; Pne, pneumocandin; Acu, aculeacin; Csp, caspofungin.
The MIC of the three antifungal families was similar in the wild-type strain (5–10 μg/ml, Table 2) and again defined two antifungal groups in the resistant strains as follows: highly resistant with papulacandin and enfumafungin and less resistant with the echinocandins (Table 2). Like aculeacin, the other echinocandins also elicited differences between the resistant strains, pbr1-8 exhibiting higher resistance than pbr1-6 (Table 2).
TABLE 2.
In vivo MIC and in vitro IC50 of wild-type, pbr1-8, and pbr1-6 strains
| Strain | MIC |
|||
|---|---|---|---|---|
| Papulacandin | Enfumafungin | Pneumocandin | Caspofungin | |
| μg/ml | ||||
| WT | 5 | 10 | 5 | 10 |
| pbr1-8 | >100 | >100 | >100 | 50 |
| pbr1-6 | >100 | >100 | 50 | 30 |
| IC50 | ||||
| μg/ml | ||||
| WT | 0.02 | 40 | 120 | 0.3 |
| pbr1-8 | >250 | >250 | >250 | 250 |
| pbr1-6 | >250 | >250 | >250 | 150 |
Differential in Vitro Effect of Three Families of Specific GS Inhibitors on Wild-type and Resistant GS Activities
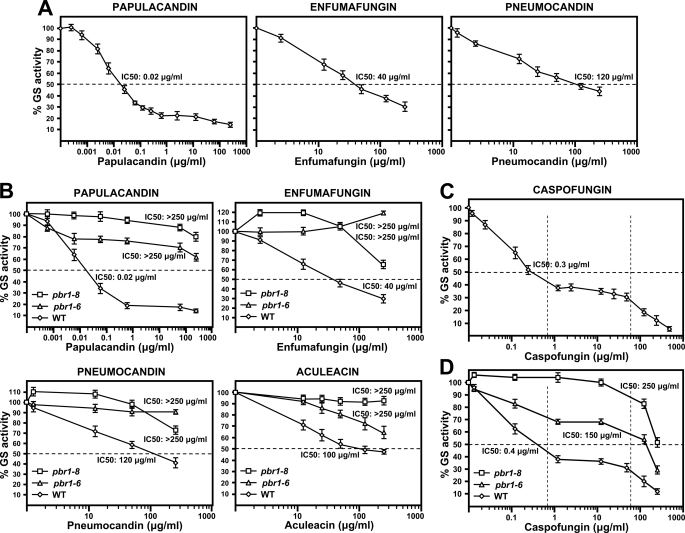
Because of the different effects observed in vivo with the antifungals analyzed and to the fact that all these antifungals are specific GS inhibitors, the effect of the three inhibitor families on the in vitro GS activity of the wild-type strain was analyzed (Fig. 4A). Enfumafungin and pneumocandin showed similar IC50 (half-maximal inhibitory concentration) values. Surprisingly, papulacandin showed a very strong GS inhibitory activity, with an IC50 103–104-fold lower than that of enfumafungin and pneumocandin (Fig. 4A and Table 2).
FIGURE 4.
Differential inhibitory effect of papulacandin B, enfumafungin, pneumocandin B0, aculeacin A, and caspofungin on the in vitro GS activity of wild-type and pbr1 cell extracts. A, inhibitory effect on the wild-type GS activity. Papulacandin shows an inhibitory capacity 103–104-fold higher than the other antifungals, except caspofungin (see Table 2). B, inhibitory effect on the GS activity of pbr1 cell extracts. pbr1-6 and pbr1-8 GS are highly resistant, with IC50 values above 250 μg/ml. The pbr1-6 strain was characterized in vivo as aculeacin A-sensitive, but its in vitro GS activity was partially resistant. C, inhibitory effect of caspofungin on the wild-type GS activity. Caspofungin exhibits a very high inhibitory capacity, 102–103-fold higher than other antifungals, except papulacandin (see Table 2). Caspofungin shows a dual inhibitory effect, at low and high concentrations (1st and 3rd sections marked by dotted lines), separated by a plateau of a 100-fold drug increase with no increase in inhibition (2nd section marked by dotted lines). D, resistance of pbr1-6 and pbr1-8 GS activities to caspofungin. pbr1-8 GS is totally resistant, and pbr1-6 is partially resistant to the low concentration inhibitory effect, but both GS are sensitive to the high concentration inhibitory effect. Caspofungin is the only antifungal able to produce a noticeable inhibition of both mutant GS, with IC50 values below 250 μg/ml. Results are shown as percentages of residual GS activity. Error bars show standard deviations.
Accordingly, the pbr1-6 and pbr1-8 mutants were analyzed for their in vitro GS resistance to the three antifungals (Fig. 4B and Table 2), and the results were compared with the data previously reported for aculeacin (25). Both mutant GS showed high resistance to all the antifungals, with an IC50 above 250 μg/ml even in the case of pbr1-6 GS with aculeacin. However, some differences were observed between both mutant GS (Fig. 4B). 1) pbr1-8 GS showed higher resistance to the inhibitors than pbr1-6 GS, which at least correlates with the lower resistance to the echinocandins of the pbr1-6 cells. 2) Enfumafungin and pneumocandin promoted a specific activation of pbr1-8 (110–120%) but not of pbr1-6 GS activity. 3) pbr1-8 GS resistance to papulacandin remained above that of pbr1-6 across the range analyzed, and with enfumafungin and pneumocandin, it decreased faster, with lower resistance observed at high drug concentrations. These results show the following: 1) each antifungal family produces a different effect on wild-type and mutant GS activities; 2) each mutant GS exhibits a different resistance pattern, depending on the inhibitor analyzed.
Novel and Special Effect of Caspofungin on in Vitro Wild-type and Resistant GS Activities
Because of its clinical relevance, the effect of the echinocandin derivative caspofungin on wild-type GS activity was analyzed (Fig. 4C and Table 2). Caspofungin proved to be a potent GS inhibitor, with an interesting novel effect on its activity. First, the compound showed strong GS inhibitory activity, with an IC50 102–103-fold lower than that of the antifungals tested, except papulacandin (IC50 still 10-fold lower). Second, caspofungin promoted two GS inhibition levels as follows: a high affinity inhibition, with a gradual decrease in GS levels down to 40% activity, and a low affinity inhibition, with a new decrease in GS activity until complete inhibition. Both inhibition ranges were separated by a plateau of 2 orders of magnitude in which increasing caspofungin did not produce any additional inhibitory effect (Fig. 4C).
As observed with other antifungals, pbr1-8 GS was more resistant to caspofungin than pbr1-6 GS and showed a constant activation (110%) over a broad concentration range of inhibitors (Fig. 4D). pbr1-8 GS proved to be resistant to the high affinity inhibition but sensitive to the low affinity inhibition. pbr1-6 GS showed lower resistance, with partial inhibition in the high affinity inhibition range, remaining constant during the plateau of 2 log orders of caspofungin. Increasing concentrations of caspofungin elicited a complete inhibition, similar to the wild-type inhibition. This indicates that pbr1-8 and pbr1-6 GS are only resistant to the high affinity inhibition effect of caspofungin. In addition, caspofungin was a potent inhibitor of mutant GS because it was the only drug that produced measurable IC50 values, which were lower in pbr1-6 than in pbr1-8 GS (150 and 250 μg/ml, respectively; Fig. 4D and Table 2).
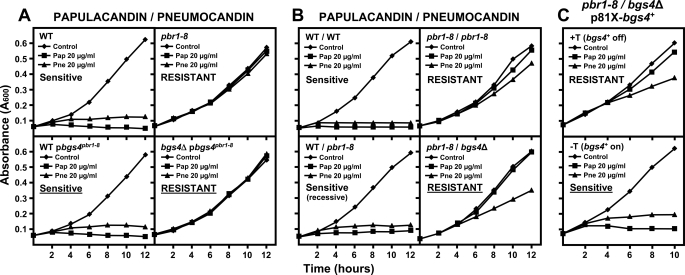
bgs4+ Is Unable to Suppress the pbr1-8 Resistance Phenotype
Previously, pbr1-8 has been genetically linked to cwg1-1. Both are GS-related genes, suggesting that pbr1+ and cwg1+ could be the same gene. pbr1-8 resistance is recessive, but cloning of the pbr1+ gene has been unsuccessful, as is the case of cwg1+ (25, 32). Cloning from a cosmid clone of the bgs4+ gene of the essential GS subunit revealed that bgs4+ is allelic to cwg1+ (15).
The above results also suggested that pbr1+ and bgs4+ could be the same gene. The fact that in S. cerevisiae and other fungi the resistance to GS antifungals is due to mutations in the Fks GS homologues (12, 19, 25, 33–36) supports the idea that specific bgs4 mutations could be responsible for the resistance phenotypes. To test this notion, the resistance phenotypes of pbr1-8 and pbr1-6 mutants expressing bgs4+ (own promoter, multicopy, and single copy integrative plasmids) were analyzed. pbr1-8 and pbr1-6 resistances are recessive (Fig. 6B and Table 3) (25). However, bgs4+ was unable to suppress the papulacandin, enfumafungin, and pneumocandin resistances of pbr1-8, although it did suppress the resistance to caspofungin (Fig. 5A and Table 3). This was not the case with pbr1-6, in which suppression of all the resistances by bgs4+ was observed (Fig. 5B and Table 3), as would be expected if pbr1+ and bgs4+ were the same gene. A similar absence of pbr1-8 suppression by bgs4+ was observed in the in vitro GS resistance of pbr1-8 cells expressing bgs4+ (Fig. 5C). This effect was not due to a defective pbgs4+ because no changes were found in the 8.8-kb bgs4+ sequence, and pbgs4+ was able to suppress the phenotype of other bgs4 mutants (15).
FIGURE 6.
bgs4 is responsible for at least part of the resistance phenotype of pbr1-8 strain. Cells were grown and monitored as in Fig 3. A, bgs4 cloned from the pbr1-8 strain confers antifungal resistance to a bgs4Δ strain. The bgs4pbr1-8 resistance is recessive in a wild-type (bgs4+) strain. B, pbr1-8 resistance is recessive in heterozygosis (WT/pbr1-8). However, pbr1-8 confers resistance to pbr1-8/bgs4Δ diploid strains. C, pbr1-8/bgs4Δ resistance phenotype can be suppressed by bgs4+ expression. The expression of p81X-bgs4+ suppresses the pbr1-8/bgs4Δ resistance when the 81X promoter is induced (− thiamine, −T) and maintains the pbr1-8/bgs4Δ resistance when the 81X promoter is repressed (+ thiamine, +T).
TABLE 3.
bgs4+ plasmids cannot suppress the recessive resistance mutation of the pbr1-8 strain but are able to suppress that of the pbr1-6 strain
The (pJK-bgs4+) is a single copy integrative plasmid; pAL-bgs4+ is a multicopy plasmid. The pbr1-6 (W760S A914V) strain contains the spontaneous A914V substitution that suppresses the spherical morphology but maintains the resistance phenotype of pbr1-6 (W760S) strain. − indicates no growth; +++ indicates total growth.
| Strain | Papulacandin (20 μg/ml) | Enfumafungin (20 μg/ml) | Echinocandin (20 μg/ml) | Caspofungin (20 μg/ml) |
|---|---|---|---|---|
| WT | − | − | − | ± |
| WT (pJK-bgs4pbr1-8) | ± | ± | ± | ± |
| WT pAL-bgs4pbr1-8 | ± | ± | ± | ± |
| pbr1-8 | +++ | +++ | +++ | +++ |
| pbr1-8 (pJK-bgs4+) | +++ | +++ | ++ | + |
| pbr1-8 pAL-bgs4+ | +++ | +++ | ++ | + |
| WT/WT | − | − | − | − |
| pbr1-8/pbr1-8 | +++ | +++ | +++ | +++ |
| WT/pbr1-8 | ± | ± | ± | − |
| WT/WT (pJK-bgs4pbr1-8) | − | − | ± | − |
| WT/WT pAL-bgs4pbr1-8 | ± | − | ± | − |
| pbr1-8/pbr1-8 (pJK-bgs4+) | +++ | +++ | +++ | + |
| pbr1-8/pbr1-8 pAL-bgs4+ | +++ | +++ | +++ | + |
| pbr1-6 (W760S) | +++ | +++ | +++ | ++ |
| pbr1-6 (W760S A914V) | +++ | +++ | +++ | ++ |
| pbr1-6/pbr1-6 | +++ | +++ | +++ | +++ |
| WT/pbr1-6 | − | − | − | − |
| WT (pJK-bgs4pbr1-6) | ± | ± | ± | − |
| WT pAL-bgs4pbr1-6 | ± | ± | ± | − |
| pbr1-6 (pJK-bgs4+) | + | ± | ± | ± |
| pbr1-6 pAL-bgs4+ | + | ± | ± | ± |
FIGURE 5.
Resistance phenotypes of pbr1-6, but not those of pbr1-8, can be suppressed by bgs4+ expression. A, bgs4+ expression cannot suppress the recessive resistances to papulacandin, enfumafungin, and pneumocandin of pbr1-8. bgs4+ is only able to suppress the caspofungin resistance of pbr1-8. Cells were grown and monitored as in Fig. 3. B, recessive pbr1-6 resistances to GS antifungals are suppressed by bgs4+ expression (see also Table 3). C, bgs4+ is unable to suppress the in vitro pbr1-8 GS resistance. Extracts of cells transformed with empty or bgs4+ multicopy plasmids were assayed for in vitro GS activity in the presence of 10 μg/ml papulacandin or 60 μg/ml pneumocandin (concentrations that produce high inhibition of the wild-type GS) as in Fig. 4.
To study whether pbr1-8 resistance was related to bgs4+, bgs4 from the pbr1-8 strain (bgs4pbr1-8) was cloned and analyzed. pbgs4pbr1-8 did not confer resistance to a wild-type strain, as expected for a recessive mutation, but it did provide resistance to a bgs4Δ strain (Fig. 6A). Similarly, a heterozygous WT/pbr1-8 diploid was sensitive, but bgs4Δ/pbr1-8 showed resistance to the GS inhibitors (Fig. 6B and Table 5). Contrary to the incapacity of bgs4+ to suppress the haploid pbr1-8 resistance, bgs4+ was able to suppress the bgs4Δ/pbr1-8 resistance. When a regulatable promoter was used, the bgs4Δ/pbr1-8 strain showed resistance when bgs4+ expression was repressed, and the sensitive phenotype when bgs4+ was induced (Fig. 6C). This suggested that bgs4pbr1-8 is responsible for at least part of the resistance of pbr1-8.
TABLE 5.
Suppression of pbr1-8 resistances by regulatable bgs4+ expression plasmid and of hemi-dosage pbr1-8/bgs4Δ diploid resistances by bgs4+ plasmids (own promoter) suggests a dosage-dependent suppression (compare with Table 3)
p81X-bgs4+ indicates multicopy plasmid with the 81X version of the thiamine repressible nmt1+ promoter. −T indicates absence of thiamine, induced conditions (on). +T indicates presence of thiamine, repressed conditions (off). pJK-bgs4+ indicates single copy integrative plasmid; pAL-bgs4+ indicates multicopy plasmid. − indicates no growth; +++ indicates total growth.
| Strain | Papulacandin (20 μg/ml) | Enfumafungin (20 μg/ml) | Echinocandin (20 μg/ml) | Caspofungin (20 μg/ml) |
|---|---|---|---|---|
| WT p81X-bgs4pbr1-8 −T (on) | − | − | − | − |
| WT p81X-bgs4pbr1-8 +T (off) | − | − | − | − |
| pbr1-8 p81X-bgs4+ −T (on) | − | ± | ± | − |
| pbr1-8 p81X-bgs4+ +T (off) | +++ | +++ | +++ | ++ |
| WT/WT p81X- bgs4pbr1-8 −T (on) | − | − | − | − |
| WT/WT p81X-bgs4pbr1-8 +T (off) | − | − | − | − |
| pbr1-8/pbr1-8 p81X-bgs4+ −T (on) | − | ± | − | ± |
| pbr1-8/pbr1-8 p81X-bgs4+ +T (off) | +++ | +++ | +++ | ++ |
| WT/bgs4Δ | − | − | − | − |
| pbr1-8/bgs4Δ | +++ | +++ | +++ | +++ |
| WT/bgs4Δ (pJK- bgs4pbr1-8) | ± | ± | ± | ± |
| WT/bgs4Δ pAL-bgs4pbr1-8 | ± | ± | − | − |
| pbr1-8/bgs4Δ (pJK-bgs4+) | ± | ± | ± | ± |
| pbr1-8/bgs4Δ pAL-bgs4+ | ± | ± | − | − |
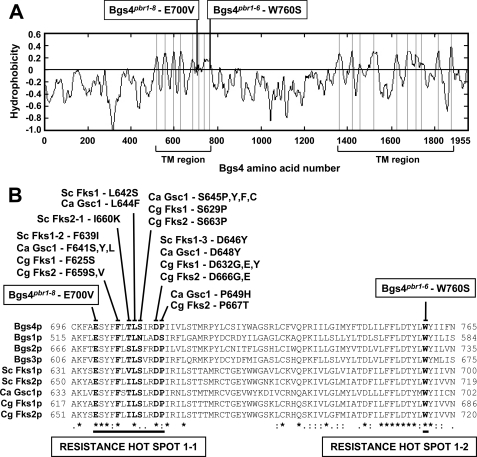
Sequencing of the bgs4 ORF and adjacent regions of the pbr1 mutant alleles revealed the substitution of only one base in the 8.8-kb DNA sequence of each mutant. pbr1-8 and the rest of the alleles, except pbr1-6, contained the same A2099T substitution, resulting in the E700V amino acid change. pbr1-6 contained the G2279C substitution, encoding the W760S amino acid change, 60 amino acids C-terminal from the pbr1-8 mutation. Both residues, Glu-700 and Trp-760, are very well conserved and are located in the predicted first transmembrane region of the Bgs/Fks/CalS protein family (Fig. 7A). Interestingly, the Bgs4pbr1-8 mutation is close to a conserved 9-amino acid resistance hot spot 1 described from the Fks sequences of Saccharomyces and Candida species, whose mutations confer resistance to echinocandins (Fig. 7B) (12, 21, 22, 37–43). The Bgs4pbr1-8 mutation permitted a larger 13-amino acid hot spot 1-1 to be defined. In addition, the Bgspbr1-6 mutation was located in a new resistance site, C-terminal from the hot spot 1-1, defining a new hot spot 1-2 of resistance to papulacandins, enfumafungin, and echinocandins (Fig. 7B).
FIGURE 7.
pbr1-6 and pbr1-8 resistances are due to single amino acid substitutions of the Bgs4 sequence. A, hydropathy profile of Bgs4. The two predicted transmembrane (TM) regions and the sites where Bgs4pbr1-8-E700V and Bgs4pbr1-6-W760S are located in the first transmembrane region are shown. B, sequence alignment of a conserved region of 70 amino acids of Bgs1, Bgs2, Bgs3, and Bgs4 from S. pombe, Fks1 and Fks2 from S. cerevisiae (Sc), Gsc1 (Fks1) from Candida albicans (Ca), and Fks1 and Fks2 from Candida glabrata (Cg). The amino acid mutations described to confer resistance to echinocandins in S. cerevisiae, C. albicans, and C. glabrata, defining a resistance hot spot 1 of 9 amino acids, are shown. The Bgs4pbr1-8 mutation is located 4 amino acids N-terminal from hot spot 1, increasing the cluster to a 13-amino acid hot spot 1-1 of resistance to papulacandin, enfumafungin, and echinocandins. The Bgs4pbr1-6 change is located 48 amino acids C-terminal from hot spot 1-1, defining a novel hot spot 1-2 of resistance to the three antifungal families.
To confirm that the Bgs4 E700V and W760S mutations were responsible for the pbr1-8 and pbr1-6 resistances, plasmids with the corresponding bgs4 substitution were made by site-directed mutagenesis and analyzed in a bgs4Δ strain. The Bgs4pbr1-8 and Bgs4pbr1-6 protein levels were similar to that of Bgs4 (data not shown). Both bgs4 mutants conferred the same resistances as those of the pbr1-8 and pbr1-6 strains. In addition, bgs4pbr1-6 generated a spherical phenotype similar to that of the sph1-1 mutant (Fig. 1A), indicating that sph1+ is also allelic to bgs4+ and that both sph1-1 and pbr1-6 phenotypes are due to the single W760S mutation. The spherical pbr1-6 (sph1-1) phenotype was unstable, originating spontaneous morphological revertants with an elongated cell shape and faster growth. Sequencing of the bgs4pbr1-6 revertants revealed the initial W760S mutation and a spontaneous A914V suppressor mutation (C2741T base change) (Fig. 1B). The suppressor function of A914V was confirmed by inserting this mutation into pbgs4pbr1-6 and observing a similar suppression (Fig. 1B). However, the antifungal resistance was not affected (Table 3), indicating that the A914V suppressor is specific to the pbr1-6 morphological and cell integrity defects.
Genetic Study of the Absence of pbr1-8 Suppression by bgs4+
The fact that the resistance of the haploid pbr1-8 strain was not suppressed but the resistance of the pbr1-8/bgs4Δ strain was suppressed by bgs4+ suggested the possibility of a second recessive resistance mutation. This second resistance should be linked to bgs4pbr1-8 because the resistance segregated 2R:2S in genetic crosses.
To test this, larger DNA fragments containing bgs4+ and adjacent ORFs were cloned. None of the fragments suppressed the pbr1-8 phenotype, but they did suppress that of cwg1-1. In ensuing experiments, total or partial 5′- or 3′-end deletions or simple deletion of the ATG start codon of bgs4pbr1-8 ORF suppressed both bgs4pbr1-8 function and antifungal resistance. These results therefore indicate that pbr1-8 resistance is only due to the bgs4pbr1-8 mutation.
Expression Level of bgs4+ Determines Its Capacity to Suppress the pbr1-8 Resistance Phenotype
To study the unexpected effect of the absence of pbr1-8 resistance suppression by bgs4+, a protocol for micro-culture assays was developed. As before, heterozygous WT/pbr1-8 showed the recessive resistance trait, and pbgs4+ was unable to suppress the haploid pbr1-8 or diploid pbr1-8/pbr1-8 resistances, except to caspofungin (Table 3). The pbr1-6 strain showed resistance to the four drugs, and in this case bgs4+ was able to suppress the four resistances (Table 3).
However, when the wild-type bgs4+ and mutant bgs4pbr1-8 plasmids were expressed in the same cell, either haploid or diploid, in the absence of endogenous bgs4 gene pbgs4+ was able to suppress the resistance due to pbgs4pbr1-8 (Table 4). Therefore, bgs4+ expressed from a plasmid is unable to suppress the endogenous pbr1-8 resistance but is able to suppress a similar resistance conferred by the bgs4pbr1-8 plasmid.
TABLE 4.
The resistances conferred by bgs4pbr1-8 plasmids to a bgs4Δ strain can be suppressed by bgs4+ plasmids
S4 indicates pJK-bgs4+ inserted into the bgs4+ promoter. R4 indicates pJK-bgs4pbr1-8 inserted into the bgs4+ promoter. SH indicates phis3-bgs4+ inserted into the his3+ promoter. RH indicates phis3-bgs4pbr1-8 inserted into the his3+ promoter. − indicates no growth; +++ indicates total growth.
| Strain | Papulacandin (20 μg/ml) | Enfumafungin (20 μg/ml) | Echinocandin (20 μg/ml) | Caspofungin (20 μg/ml) |
|---|---|---|---|---|
| bgs4Δ S4 | − | − | − | ± |
| bgs4Δ SH | − | − | − | ± |
| bgs4Δ R4 | +++ | +++ | +++ | ++ |
| bgs4Δ RH | +++ | +++ | +++ | ++ |
| bgs4Δ S4 SH | − | − | − | ± |
| bgs4Δ R4 RH | +++ | +++ | +++ | +++ |
| bgs4Δ S4 RH | − | − | − | − |
| bgs4Δ R4 SH | ± | ± | − | − |
| bgs4Δ S4/bgs4Δ SH | − | − | − | − |
| bgs4Δ R4/bgs4Δ RH | +++ | +++ | +++ | +++ |
| bgs4Δ S4/bgs4Δ RH | − | − | − | − |
| bgs4Δ R4/bgs4Δ SH | − | − | − | − |
The absence of pbr1-8 suppression by pbgs4+ was opposite to the suppression observed with p81X-bgs4+ (Fig. 6C). To study this discrepancy, p81X-bgs4+ was analyzed in the pbr1-8 and pbr1-8/pbr1-8 strains that had previously showed an absence of suppression by pbgs4+ (Table 3). Surprisingly, induced p81X-bgs4+ was able to suppress the resistance of both strains (Table 5). This suggests that the ability of bgs4+ to suppress the pbr1-8 resistance depends on the expression level of the corresponding bgs4+ plasmid. This was confirmed by expressing pbgs4+ in hemi-dosage pbr1-8/bgs4Δ diploid cells. Whereas pbgs4+ did not suppress pbr1-8/pbr1-8 resistance (Table 3), it did suppress pbr1-8/bgs4Δ resistance (Table 5). This shows that the ectopic gene expression may be less efficient than the chromosomal gene expression, sufficient to suppress some (cwg1-1, cwg1-2, orb11-56, and pbr1-6) but not other mutations (pbr1-8).
DISCUSSION
S. pombe is an attractive model to study the effect of antifungals specific to β(1,3)glucan synthesis. Its special rod shape makes the fission yeast an excellent organism to explore morphological defects. Any minor cell wall defect will result in detectable morphological changes. In addition, S. pombe has no detectable chitin, and therefore β(1,3)glucan acquires more relevance as a structural cell wall polysaccharide. Most fungi contain a single essential Fks protein or two redundant Fks subunits, which are only essential when both are affected. In this sense, S. pombe is also special because it contains four Fks homologs, Bgs1 to Bgs4, and three of them, Bgs1, Bgs3, and Bgs4, are essential for vegetative cells. Therefore, study of the effect of GS inhibitors on S. pombe cells, GS activity, and Bgs subunits could provide important information about the mechanism of action of each family of specific GS inhibitors.
Analysis of the resistance of fission yeast to GS inhibitors could also provide interesting information. Resistance to GS inhibitors in other fungi is associated with mutations in their single Fks protein. In the case of S. pombe, a mutation in only one of the three essential Bgs proteins would not alter the antifungal sensitivity of the two other Bgs subunits, and the cell would remain sensitive to the antifungals. Therefore, which mechanism confers antifungal resistance to S. pombe cells? Initially, one possibility was that the resistance was due to a general regulator of the three Bgs proteins. However, cloning of the gene involved in the resistance revealed that it was exclusively associated with Bgs4, implying that Bgs1 and Bgs3 are not affected by the antifungals. This opens interesting new questions about the intrinsic resistance of Bgs1 and Bgs3, even though their sequences harbor all the residues that are determinant for the Bgs4/Fks sensitivity to GS antifungals. The identification of Bgs1 and Bgs3 as new antifungal targets opens new possibilities in the search for new GS inhibitors with a broader mechanism of action.
There are several antifungal families that specifically inhibit β(1,3)glucan synthesis and GS activity (2, 12, 44, 45), but little is known about their mechanisms of action. In this work, we compared the effect of three GS inhibitor families on S. pombe wild-type and papulacandin-resistant cells and GS activities, and we found interesting differences both in vivo and in vitro. In wild-type cells, papulacandin and enfumafungin cause generalized cell lysis, mainly during cell separation7 and subsequent total cell growth arrest, whereas resistant cells are not affected. Echinocandins are different; the lysis of wild-type cells is incomplete, and the surviving cells become rounded and maintain a residual cell growth. The resistant cells are partially affected in morphology and cell growth, and their resistance is variable, depending on the type of mutant and echinocandin, ranging from high (pbr1-8 with pneumocandin), to low (caspofungin), to nonresistant (pbr1-6 with aculeacin). This effect could be due to a distinct permeability to the different drugs, but in that case the cell should present similar or higher resistance than its in vitro GS resistance, and in our case we observed the opposite. It should be noted that whereas pbr1-8 and pbr1-6 were highly resistant to papulacandin and enfumafungin, they exhibited opposite behaviors with respect to aculeacin; pbr1-8 was highly resistant and pbr1-6 was sensitive. The finding of this variability in the resistance to echinocandins was possible because the mutants were selected for their resistance to papulacandin (25). Most of the mutants described have been isolated as resistant to echinocandins (12, 21, 22, 37–43). We have inserted some of these mutations into bgs4+, and none of them altered the in vivo sensitivity of the cells to papulacandin and enfumafungin but produced variable degrees of resistance to echinocandins,7 showing that each antifungal family acts through different sites of the GS enzyme and that some resistances may be specific to a given antifungal family. We have also analyzed the GS inhibitor Aerothricin3/FR901469 (generous gift from Dr. Osamu Kondoh, Chugai Pharmaceutical) (46) on wild-type and resistant cells, and we found no differences. GS antifungals such as Aerothricin3, acting on different sites of the Bgs4/Fks proteins, could be good candidates for a combined therapy to minimize the appearance of resistant strains.
The MIC values of papulacandin and aculeacin described for S. cerevisiae and Candida are similar to the values for S. pombe (11, 25, 47, 48). In other cases, the MIC value of a specific inhibitor fluctuate, depending on the method of analysis, the strain, or the organism. The MIC values of caspofungin described for C. albicans (0.0125–0.8 μg/ml) and other Candida species (0.01–8.0 μg/ml) are highly variable, although overall they denote a lower MIC compared with that obtained for S. pombe (11, 23, 49). This could be due to a lower susceptibility of Bgs4 or to a compensatory mechanism of the intrinsic resistant Bgs1 and Bgs3 subunits.
We compared the in vitro effect of each antifungal family on wild-type GS activity. Papulacandin exerted an in vitro effect that was 103–104-fold stronger than that of the other drugs, except caspofungin. This effect has not been described in previous studies (25), probably due to the method used to prepare the GS enzyme. However, this in vitro difference was not correlated with a similar increase in in vivo susceptibility. This could be due to other antifungal interactions with the cell or to the in vivo accessibility of the enzyme. A similar case was observed with caspofungin, with an IC50 102–103-fold lower than that of the other drugs, except papulacandin, with no correlation with its in vivo inhibitory capacity. By contrast, some in vitro mutant GS resistances to pneumocandin, aculeacin, or caspofungin are much higher than the resistances detected in the cells. Furthermore, some bgs4 mutants are not resistant in vivo but do show in vitro GS resistance to echinocandins.7 These differences show that in vitro GS resistance may be insufficient to confer resistance to cells and suggest the presence of other targets modulating the GS activity in vivo.
Interestingly, we observed that some of the antifungals tested were able to increase the mutant GS activity above the initial activity. This activating effect was observed in pbr1-8 but not in pbr1-6 GS activity, denoting specific and antagonistic drug effects that depend on the interacting GS conformation. pbr1-8 is activated by enfumafungin and echinocandins, but other bgs4 mutants are also activated by papulacandins or only one antifungal.7 No GS activation by GS inhibitors has been reported, except for papulacandin (50). In that case, the drug activation affected the wild-type GS and was dependent on low substrate concentrations. It is possible that some mutations, such as that of pbr1-8, in the presence of an antifungal could mimic the proposed preferential binding of substrate to the active form of the enzyme.
Among the inhibitors studied, caspofungin was seen to be the best candidate for antifungal therapy. The caspofungin MIC of resistant strains was much lower than that of the other drugs, and it produced a reduction in cell growth and an altered morphology of resistant cells. Caspofungin also showed special properties as regards the in vitro GS activity, with an IC50 102–103-fold lower than that of other drugs, except papulacandin, and with two previously unreported inhibitory effects of high (<0.7 μg/ml) and low (>70 μg/ml) affinity. This suggests the presence of two GS interaction sites with caspofungin. Both pbr1-8 and pbr1-6 GS were resistant to the high affinity inhibition but not to the low affinity inhibition, suggesting that the resistance hot spots 1-1 and 1-2 would be located in the region involved in this high affinity interaction with caspofungin. One candidate for the low affinity interaction site could be a short hot spot 2 located in the predicted second transmembrane region (12, 41). Alternatively, the low affinity inhibition could be due to interactions between caspofungin and Bgs1 or Bgs3. Both hypotheses are currently under investigation.
The interesting effect of caspofungin in that it produces two separate inhibitory effects is different from the paradoxical growth effect, or Eagle effect, described for Candida (51, 52), which consists of an in vivo attenuation of growth inhibition at drug concentrations above the inhibitory concentration. The result is growth inhibition followed by a resumption of growth at higher antifungal concentrations and a new inhibitory effect when the drug concentration increases. This phenomenon has been associated with compensatory mechanisms from the cell integrity and calcineurin pathways. In our case, the effect was observed in the GS activity in vitro, and therefore it cannot be explained in terms of cell compensatory mechanisms. In addition, the GS activity did not increase but remained constant over a broad concentration range of antifungals until a new inhibition appeared.
All the mutants selected as resistant to echinocandins have been described as dominant or semi-dominant (12, 20, 33, 34, 36, 37, 53). Only mutants isolated as resistant to papulacandin have shown a recessive trait (25). S. cerevisiae pbr1-1 resistance is suppressed by the PBR1/FKS1 gene. However, although S. pombe pbr1-8 resistance was clearly recessive and caused by a bgs4+ mutation, bgs4+ expressed from plasmids was unable to suppress it. Our studies show that chromosomal and plasmid expression may differ, at least in their resulting phenotype; a recessive chromosomal trait can appear as dominant when the complementing gene is expressed ectopically. However, the complementing gene remains dominant over the recessive allele when both are expressed ectopically or when the chromosomal recessive gene dosage decreases, as is the case of hemi-dosage diploid cells. This explains the ability of bgs4+ plasmids to suppress, in pbr1-8 cells, the low resistance to caspofungin but not higher resistances to the other inhibitors. These results show that in some cases typical gene cloning by suppression of the recessive mutant phenotype may not be possible, and hence other techniques must be used.
All the echinocandin resistance mutations have been localized in a 9-amino acid resistance hot spot 1. In this study, we found interesting new mutations that conferred resistance to the three antifungal families and were external to the hot spot 1. One of these mutations extends the hot spot to 13 amino acids in the resistance hot spot 1-1. The other mutation, although in the same region, is distant and defines a new resistance hot spot 1-2. The use of other GS-specific drugs, such as papulacandin, and the use of other species, such as S. pombe, have been shown to be helpful to define new amino acids and regions in GS that are important for interaction with antifungals. Studying these and other new resistances will help to develop more efficient antifungals, such as caspofungin, which is able to inhibit resistant mutants at concentrations closer to those effective for wild-type cells.
This work was supported in part by Grants BIO2006-13566 from Ministerio de Educación y Ciencia, BIO2009-10597 from Ministerio de Ciencia e Innovación, and GR231 from Junta de Castilla y León, Spain.
I. M. Martins, M. Ramos, J. Muñoz, M. B. Moreno, J. A. Clemente-Ramos, J. C. G. Cortés, A. Durán, and J. C. Ribas, unpublished results.
- GS
- β(1,3)glucan synthase
- MIC
- minimal inhibitory concentration
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Shao P. L., Huang L. M., Hsueh P. R. (2007) Int. J. Antimicrob. Agents 30, 487–495 [DOI] [PubMed] [Google Scholar]
- 2. Vicente M. F., Basilio A., Cabello A., Peláez F. (2003) Clin. Microbiol. Infect. 9, 15–32 [DOI] [PubMed] [Google Scholar]
- 3. Chapman S. W., Sullivan D. C., Cleary J. D. (2008) Trans. Am. Clin. Climatol. Assoc. 119, 197–215; discussion 215–216 [PMC free article] [PubMed] [Google Scholar]
- 4. Montravers P., Jabbour K. (2006) Int. J. Antimicrob. Agents 27, 1–6 [DOI] [PubMed] [Google Scholar]
- 5. Bal A. M. (2010) Int. J. Antimicrob. Agents 35, 13–18 [DOI] [PubMed] [Google Scholar]
- 6. Denning D. W. (2003) Lancet 362, 1142–1151 [DOI] [PubMed] [Google Scholar]
- 7. Eschenauer G., Depestel D. D., Carver P. L. (2007) Ther. Clin. Risk Manag. 3, 71–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lesage G., Bussey H. (2006) Microbiol. Mol. Biol. Rev. 70, 317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klis F. M., Boorsma A., De Groot P. W. (2006) Yeast 23, 185–202 [DOI] [PubMed] [Google Scholar]
- 10. Durán A., Pérez P. (2004) in Molecular Biology of Schizosaccharomyces pombe (Egel R. ed) pp. 269–279, Springer Verlag, Berlin [Google Scholar]
- 11. Onishi J., Meinz M., Thompson J., Curotto J., Dreikorn S., Rosenbach M., Douglas C., Abruzzo G., Flattery A., Kong L., Cabello A., Vicente F., Pelaez F., Diez M. T., Martin I., Bills G., Giacobbe R., Dombrowski A., Schwartz R., Morris S., Harris G., Tsipouras A., Wilson K., Kurtz M. B. (2000) Antimicrob. Agents Chemother. 44, 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perlin D. S. (2007) Drug Resist. Updat. 10, 121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Humbel B. M., Konomi M., Takagi T., Kamasawa N., Ishijima S. A., Osumi M. (2001) Yeast 18, 433–444 [DOI] [PubMed] [Google Scholar]
- 14. Cortés J. C., Konomi M., Martins I. M., Muñoz J., Moreno M. B., Osumi M., Durán A., Ribas J. C. (2007) Mol. Microbiol. 65, 201–217 [DOI] [PubMed] [Google Scholar]
- 15. Cortés J. C., Carnero E., Ishiguro J., Sánchez Y., Durán A., Ribas J. C. (2005) J. Cell Sci. 118, 157–174 [DOI] [PubMed] [Google Scholar]
- 16. Martín V., García B., Carnero E., Durán A., Sánchez Y. (2003) Eukaryot. Cell 2, 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martín V., Ribas J. C., Carnero E., Durán A., Sánchez Y. (2000) Mol. Microbiol. 38, 308–321 [DOI] [PubMed] [Google Scholar]
- 18. Verma D. P., Hong Z. (2001) Plant Mol. Biol. 47, 693–701 [DOI] [PubMed] [Google Scholar]
- 19. Walker L. A., Gow N. A., Munro C. A. (2010) Fungal Genet. Biol. 47, 117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rocha E. M., Garcia-Effron G., Park S., Perlin D. S. (2007) Antimicrob. Agents Chemother. 51, 4174–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Katiyar S. K., Edlind T. D. (2009) Antimicrob. Agents Chemother. 53, 1772–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katiyar S., Pfaller M., Edlind T. (2006) Antimicrob. Agents Chemother. 50, 2892–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia-Effron G., Katiyar S. K., Park S., Edlind T. D., Perlin D. S. (2008) Antimicrob. Agents Chemother. 52, 2305–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ribas J. C., Roncero C., Rico H., Durán A. (1991) FEMS Microbiol. Lett. 63, 263–267 [DOI] [PubMed] [Google Scholar]
- 25. Castro C., Ribas J. C., Valdivieso M. H., Varona R., del Rey F., Duran A. (1995) J. Bacteriol. 177, 5732–5739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Egel R. (1984) Curr. Genet. 8, 199–203 [DOI] [PubMed] [Google Scholar]
- 27. Alfa C., Fantes P., Hyams J., McLeod M., Warbrick E. (eds) (1993) Experiments with Fission Yeast: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Moreno M. B., Durán A., Ribas J. C. (2000) Yeast 16, 861–872 [DOI] [PubMed] [Google Scholar]
- 30. Cortés J. C., Ishiguro J., Durán A., Ribas J. C. (2002) J. Cell Sci. 115, 4081–4096 [DOI] [PubMed] [Google Scholar]
- 31. Ishiguro J., Saitou A., Durán A., Ribas J. C. (1997) J. Bacteriol. 179, 7653–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ribas J. C., Diaz M., Duran A., Perez P. (1991) J. Bacteriol. 173, 3456–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Douglas C. M., D'Ippolito J. A., Shei G. J., Meinz M., Onishi J., Marrinan J. A., Li W., Abruzzo G. K., Flattery A., Bartizal K., Mitchell A., Kurtz M. B. (1997) Antimicrob. Agents Chemother. 41, 2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Douglas C. M., Marrinan J. A., Li W., Kurtz M. B. (1994) J. Bacteriol. 176, 5686–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Douglas C. M., Foor F., Marrinan J. A., Morin N., Nielsen J. B., Dahl A. M., Mazur P., Baginsky W., Li W., el-Sherbeini M., Clemas J. A., Mandala S. M., Frommer B. R., Kurtz M. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12907–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. El-Sherbeini M., Clemas J. A. (1995) Antimicrob. Agents Chemother. 39, 200–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Balashov S. V., Park S., Perlin D. S. (2006) Antimicrob. Agents Chemother. 50, 2058–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Desnos-Ollivier M., Bretagne S., Raoux D., Hoinard D., Dromer F., Dannaoui E. (2008) Antimicrob. Agents Chemother. 52, 3092–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia-Effron G., Lee S., Park S., Cleary J. D., Perlin D. S. (2009) Antimicrob. Agents Chemother. 53, 3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garcia-Effron G., Park S., Perlin D. S. (2009) Antimicrob. Agents Chemother. 53, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park S., Kelly R., Kahn J. N., Robles J., Hsu M. J., Register E., Li W., Vyas V., Fan H., Abruzzo G., Flattery A., Gill C., Chrebet G., Parent S. A., Kurtz M., Teppler H., Douglas C. M., Perlin D. S. (2005) Antimicrob. Agents Chemother. 49, 3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cleary J. D., Garcia-Effron G., Chapman S. W., Perlin D. S. (2008) Antimicrob. Agents Chemother. 52, 2263–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thompson J. R., Douglas C. M., Li W., Jue C. K., Pramanik B., Yuan X., Rude T. H., Toffaletti D. L., Perfect J. R., Kurtz M. (1999) J. Bacteriol. 181, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Odds F. C., Brown A. J., Gow N. A. (2003) Trends Microbiol. 11, 272–279 [DOI] [PubMed] [Google Scholar]
- 45. Barrett D. (2002) Biochim. Biophys. Acta 1587, 224–233 [DOI] [PubMed] [Google Scholar]
- 46. Kondoh O., Takasuka T., Arisawa M., Aoki Y., Watanabe T. (2002) J. Biol. Chem. 277, 41744–41749 [DOI] [PubMed] [Google Scholar]
- 47. Font de Mora J., Gil R., Sentandreu R., Herrero E. (1991) Antimicrob. Agents Chemother. 35, 2596–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bartizal K., Abruzzo G., Trainor C., Krupa D., Nollstadt K., Schmatz D., Schwartz R., Hammond M., Balkovec J., Vanmiddlesworth F. (1992) Antimicrob. Agents Chemother. 36, 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stone E. A., Fung H. B., Kirschenbaum H. L. (2002) Clin. Ther. 24, 351–377 [DOI] [PubMed] [Google Scholar]
- 50. Kang M. S., Szaniszlo P. J., Notario V., Cabib E. (1986) Carbohydr. Res. 149, 13–21 [DOI] [PubMed] [Google Scholar]
- 51. Fleischhacker M., Radecke C., Schulz B., Ruhnke M. (2008) Eur. J. Clin. Microbiol. Infect. Dis. 27, 127–131 [DOI] [PubMed] [Google Scholar]
- 52. Wiederhold N. P. (2007) Curr. Opin. Infect. Dis. 20, 574–578 [DOI] [PubMed] [Google Scholar]
- 53. Kurtz M. B., Abruzzo G., Flattery A., Bartizal K., Marrinan J. A., Li W., Milligan J., Nollstadt K., Douglas C. M. (1996) Infect. Immun. 64, 3244–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]