Abstract
The premature aging and cancer-prone disease Werner syndrome is caused by loss of function of the RecQ helicase family member Werner syndrome protein (WRN). At the cellular level, loss of WRN results in replication abnormalities and chromosomal aberrations, indicating that WRN plays a role in maintenance of genome stability. Consistent with this notion, WRN possesses annealing, exonuclease, and ATPase-dependent helicase activity on DNA substrates, with particularly high affinity for and activity on replication and recombination structures. After certain DNA-damaging treatments, WRN is recruited to sites of blocked replication and co-localizes with the human single-stranded DNA-binding protein replication protein A (RPA). In this study we examined the physical and functional interaction between WRN and RPA specifically in relation to replication fork blockage. Co-immunoprecipitation experiments demonstrated that damaging treatments that block DNA replication substantially increased association between WRN and RPA in vivo, and a direct interaction between purified WRN and RPA was confirmed. Furthermore, we examined the combined action of RPA (unmodified and hyperphosphorylation mimetic) and WRN on model replication fork and gapped duplex substrates designed to bind RPA. Even with RPA bound stoichiometrically to this gap, WRN efficiently catalyzed regression of the fork substrate. Further analysis showed that RPA could be displaced from both substrates by WRN. RPA displacement by WRN was independent of its ATPase- and helicase-dependent remodeling of the fork. Taken together, our results suggest that, upon replication blockage, WRN and RPA functionally interact and cooperate to help properly resolve replication forks and maintain genome stability.
Keywords: Aging, Carcinogenesis, DNA Damage, DNA Helicase, DNA Recombination, DNA Repair, DNA Replication, RecQ Helicases, Werner Syndrome
Introduction
Accumulating evidence indicates that blocks to replication fork progression are common occurrences that must be properly dealt with to complete DNA synthesis and prevent genomic instability. Replication fork progression may be blocked by the persistence of certain types of DNA damage, by non-canonical DNA structures or by proteins tightly bound or cross-linked to parental DNA. Cells have evolved pathways that accurately resolve replication fork blockage, allowing restart and completion of DNA replication. RecQ helicases are critical for chromosome stability, hypothetically by participating in proper resolution of replication fork blockage (1, 2). RecQ family members share extensive homology in their conserved helicase domains and catalyze ATP-dependent unwinding of various DNA structures. There are five human RecQ family members, namely RECQL, BLM, WRN, RECQ4/RTS, and RECQ5. The human hereditary diseases Bloom, Rothmund-Thomson, and Werner syndromes result from the loss of function of BLM, RECQ4/RTS, and WRN, respectively (3–5). Although these diseases show genomic instability and elevated cancer predispositon, each has a distinct phenotype, suggesting that these proteins have at least some non-overlapping functions. Among these syndromes, Werner syndrome notably shows early onset and increased frequency of many age-related features (6, 7).
The basic biochemical properties of recombinant WRN protein have been characterized. In agreement with its classification as a RecQ family member, WRN is a DNA-dependent ATPase and in combination with ATP hydrolysis can unwind DNA duplexes of limited length with a 3′ to 5′ directionality. However, WRN preferentially unwinds or disrupts more complex DNA structures including forks, D-loops, G-quartets, and triplexes (8). WRN is unique among the human RecQ helicases in that it exhibits a 3′ to 5′ exonuclease activity that also acts with higher specificity toward complex DNA structures (9–11). A number of RecQ helicases, including WRN, BLM, RECQ1, RECQ4, and RECQ5 (β isoform) proteins, promote the annealing of complementary DNA strands, and coordination between the unwinding and annealing activities of WRN, BLM, and RECQ5 (β isoform) can mediate strand exchange between appropriate DNA molecules (12–17).
The biochemical properties of WRN and other RecQ helicases suggest that these proteins act on complex DNA structures such as three- or four-stranded intermediates formed during replication and recombination. In support of a replication-related role for WRN, WRN-deficient cells show replication abnormalities including slow growth, an extended S-phase, and asymmetric replication fork progression (18, 19). WRN-deficient cells are also hypersensitive to certain DNA damaging agents, particularly those known to block replication such as hydroxyurea (HU),3 topoisomerase inhibitors, and interstrand cross-linkers (20–22). In normal cells, after treatment with HU and other DNA damaging agents, WRN relocates to distinct nuclear foci where it co-localizes with replication factors, indicating that it is recruited to sites of blocked replication (23–25). Taken together, these studies indicate that WRN may play an important role in response to replication fork blockage.
According to models proposed for resolution of replication blockage, an early event in the process involves fork regression to generate a Holliday junction or “chicken foot” intermediate (26–28). Considered in conjunction with the phenotypes of RecQ-deficient cells, the unwinding and annealing activities of some RecQ enzymes suggested that they would be ideal candidates to regress replication forks. In agreement, we found that either WRN or BLM efficiently catalyzed the regression of model replication fork substrates via generation of a Holliday junction intermediate (29, 30). Moreover, the exonuclease activity of WRN contributed to enhanced regression efficiency on certain fork structures (30). In contrast to its relatively weak unwinding activity on simple duplexes, WRN-mediated fork regression occurred readily at near equimolar enzyme-DNA substrate ratios (30). Thus, WRN appears enzymatically suited to perform fork regression.
Replication protein A (RPA), is a heterotrimeric (70-, 32-, and 14-kDa subunits, hereafter referred to as RPA70, RPA32, and RPA14, respectively) single-stranded DNA (ssDNA) binding factor involved in DNA replication, recombination, and repair (31, 32). Among its many roles, RPA binds to ssDNA gaps generated by stalled replication forks, an action that not only protects this region but also plays a key role in initiating and directing downstream pathways (32, 33). Specifically, RPA promotes checkpoint signaling after replication fork stalling through its recruitment and activation of ATR·ATRIP and possibly RAD17 complexes (34–36). Furthermore, RPA binding to ssDNA overhangs at strand breaks recruits RAD51 and other recombination factors required to initiate homologous recombination processes (37, 38). In addition, RPA function is regulated by phosphorylation of the N-terminal region of the RPA32 subunit. RPA is phosphorylated during the normal cell cycle (39–41) and hyperphosphorylated in response to treatment of cells with DNA damaging agents such as camptothecin or UV (42, 43). RPA hyperphosphorylation apparently modulates multiple cellular responses to DNA damage, including possible down-regulation of DNA replication (44). Thus, hyperphosphorylation of RPA may reflect its conversion from a DNA replication factor to a DNA damage response factor.
Studies have also revealed a physical and functional interaction between RPA and WRN. RPA and WRN directly interact in vitro, and RPA enhances WRN unwinding strength (45, 46). In response to HU treatments, WRN and RPA co-localize in replication foci thought to represent sites of blocked replication (23). These studies suggest that WRN and RPA might interact at stalled replication forks, facilitating WRN function in proper resolution of replication blockage. Here, we examine the molecular and functional interaction of WRN and RPA specifically in regard to replication blockage. We have found that treatments that generate DNA damage known to block replication fork progression result in an increased association between WRN and RPA and have shown a direct interaction between purified WRN and RPA. Because WRN readily regresses model replication forks (29, 30) in perhaps an early step in resolution of fork blockage, we were interested in examining the effect of RPA on WRN-mediated enzymatic activities including fork regression. Thus, we designed replication fork and gapped duplex substrates containing ssDNA gaps sufficient to bind RPA. After confirming that RPA stably binds to these substrates, its effect on WRN action was examined. Our results demonstrate that WRN efficiently regresses our replication fork substrate even when RPA (either wild type or hyperphosphorylation mimetic RPA-32D8) is bound to the single-stranded gap. Furthermore, RPA bound to this fork substrate could be displaced by WRN, independent of its catalytic activity. Thus, these findings support the concept that RPA and WRN act in concert at stalled replication forks and suggest a reasonable model for the molecular and functional cooperation between these factors during proper resolution of replication blockage.
EXPERIMENTAL PROCEDURES
Enzymes
The WRN-E84A protein possesses normal DNA-dependent ATPase and helicase activities but contains a glutamate to alanine mutation that abolishes exonuclease activity (47). WRN-E84A was overexpressed and purified as described previously (15). Unmodified, wild type RPA (RPA-wt) and phosphomimetic RPA (RPA-32D8) were purified as described earlier (31, 48).
DNA Substrate Construction
The sequences of gel-purified oligonucleotides (Integrated DNA Technologies) are given in supplemental Table 1, with “P” and “D” designations indicating parental and daughter strands, respectively. For studying the binding of RPA-wt or RPA-32D8 to a model replication fork by electrophoretic mobility shift assays (EMSA) and their effect on WRN-mediated fork regression, the fork substrate was constructed as described below. The lagP122 and leadD52 oligomers were radiolabeled (indicated by asterisks) at their 5′ ends using [γ-32P]ATP and T4 polynucleotide kinase (New England Biolabs). In two separate annealing reactions, the labeled lagP122 and leadD52 oligomers were combined with lagD82 and leadP122 oligomers, respectively, heated to 95 °C, and subsequently slow-cooled to create lagging and leading parental-daughter partial duplexes. These partial duplexes (*lagP122/lagD82 and leadP122/*leadD52) were then incubated together for 24 h at 25 °C to generate double-labeled replication fork substrate (*lagP122/lagD82-leadP122/*leadD52). However, for DNase I footprinting studies, the fork substrate (lagP122/lagD82-*leadP122/leadD52) was labeled only on the leadP122 strand; this substrate was also used in EMSA experiments examining RPA displacement. The gapped duplex substrate was generated by annealing leadD52 and lagP38-3′ to radiolabeled leadP122. Annealing reactions containing these substrates were subject to native polyacrylamide (8%) gel electrophoresis (PAGE), bands corresponding to the fork and gapped duplex substrates were excised, and the substrates were extracted for 24 h at 4 °C into DNA elution buffer (10 mm Tris, pH 8.0, 10 mm NaCl). The 32-mer (ss32) used in the RPA displacement studies was radiolabeled and subjected to native PAGE as described above, but final purification was accomplished using a gel extraction kit (Qiagen).
EMSA
32P-Labeled fork DNA substrate (1 fmol) was incubated with or without RPA-wt (0.05–5 fmol) or RPA-32D8 (0.04–8 fmol) in WRN reaction buffer (20 μl) containing 40 mm Tris-HCl, pH 7.0, 4 mm MgCl2, 0.1 mg/ml bovine serum albumin (BSA), 5 mm dithiothreitol, 0.1% Nonidet P-40, and 1 mm ATP for 10 min at 25 °C. In assays studying the effect of WRN-E84A on RPA-wt bound to DNA substrates (Fig. 5 and supplemental Fig. 1), labeled fork DNA (∼15 fmol) or gapped duplex DNA (∼20 fmol) was preincubated with RPA-wt (10 or 20 fmol, respectively) in WRN reaction buffer for 5 min at 25 °C. Then, WRN-E84A (50 fmol) was added to the reactions, which were further incubated at 25 °C for 5 min followed by the addition of radiolabeled ss32 (∼15 or 20 fmol) as indicated and nucleotide cofactor ATPγS or ATP (1 mm) unless otherwise noted. These reactions were then incubated at 37 °C for an additional 10 min. For all EMSA experiments, ⅙ volume of 30% glycerol was added, and the samples were separated by native PAGE (3.5%-3.8%) carried out in 0.5× Tris borate EDTA at 90–100 V at 25 °C. Subsequently, the gels were dried, and DNA and DNA-protein complexes were visualized and quantitated using a Storm 860 PhosphorImager and ImageQuant software (GE Healthcare). RPA binding was quantified by comparing the amount of bound DNA with the total DNA in the reaction (% DNAbound = DNAbound/[DNAbound + DNAunbound] × 100).
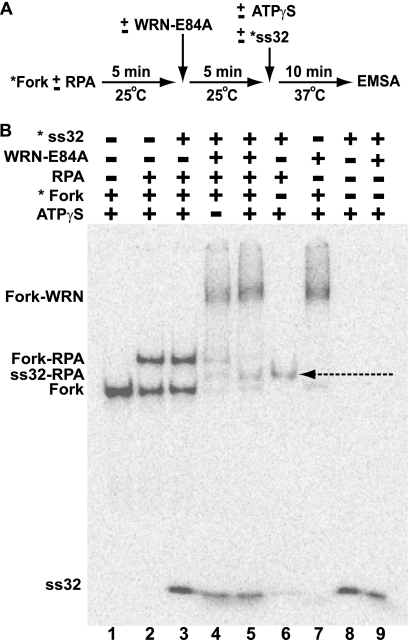
FIGURE 5.
Displacement of RPA bound to the leading arm gap of the model replication fork by WRN-E84A. A, a schematic shows the experimental strategy to examine the interaction between RPA and WRN-E84A on the replication fork substrate. B, WRN-E84A-mediated displacement of RPA from the replication fork structure is shown. Fork substrate (15 fmol) is incubated with or without RPA (10 fmol) for 5 min at 25 °C followed by further incubation with or without WRN-E84A (50 fmol) for an additional 5 min at 25 °C. This is followed by the addition of ATPγS (1 mm) and/or 15 fmol of radiolabeled 32-mer (oligo ss32) as indicated and further incubation at 37 °C for an additional 10 min. The reactions are then analyzed by EMSA as described under “Experimental Procedures.” Positions of the labeled fork DNA, labeled ss32 and fork DNA-RPA, fork DNA-WRN-E84A, and ss32-RPA (denoted by the dashed arrow) complexes are indicated at the left.
DNase I Footprinting
Fork DNA substrate (∼5 fmol), labeled on the leadP122 strand, was incubated with or without RPA-wt (0.1–5.0 fmol) or RPA-32D8 (1–32 fmol) in WRN reaction buffer for 10 min at 25 °C. DNase I (0.008 units/reaction) was then added followed by further incubation at 25 °C for 10 min. Reactions were stopped by adding an equal volume of formamide loading buffer (95% formamide, 20 mm EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol), heated to denature DNA products, and analyzed by denaturing PAGE (10%) carried out in 1× Tris borate EDTA at 45 W for 2 h. The gels were dried and subjected to phosphorimaging analysis as described above.
Fork Regression Assays
Fork substrate (1–2 fmol) was incubated with WRN-E84A (3.5–7 fmol) in WRN reaction buffer for 5 min at 4 °C and then transferred to 37 °C for the specified times. In reactions studying the effect of RPA on fork regression, the fork substrate was preincubated with either RPA-wt or RPA-32D8 for 5 min at 4 °C before WRN addition. Reactions were stopped using ⅙ volume of helicase dyes (30% glycerol, 50 mm EDTA, 0.9% SDS, 0.25% bromphenol blue, 0.25% xylene cyanol) and analyzed by native (7–9) % PAGE (acrylamide:bisacrylamide cross-linking ratio of 37.5:1) in 1× Tris borate EDTA. Gels were dried, and DNA products were visualized and quantified by phosphorimaging analysis as above. Radioactivity associated with each individual DNA species was determined for each reaction, and the percentage of each product with respect to the total DNA was calculated. Total DNA represents the amount of DNA present in each reaction after correction for the background amounts of DNA species (other than the intact fork) present in reactions without enzyme.
WRN-RPA Co-immunoprecipitation Experiments
The SV40-transformed fibroblast cell line, 1-0, used in immunoprecipitation experiments was obtained from J. Christopher States, University of Louisville (49). Methylmethanesulfonate (MMS), phenylmethylsulfonyl fluoride (PMSF), HU, and protease inhibitor mixture were purchased from Sigma, and cell culture media and reagents were purchased from Invitrogen. Cells were grown in minimal essential medium-α Glutamax medium supplemented with 10% FBS, 1% HEPES, and 1% penicillin-streptomycin at 37 °C in a humidified atmosphere containing 5% CO2. For DNA damaging treatments, cells were incubated in medium containing 1 mm MMS for 4 h or 2 mm HU for 10 h before harvesting. For co-immunoprecipitation experiments, cells were lysed by sonication in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.25% sodium deoxycholate, and 1 mm EDTA) supplemented with protease inhibitor mixture, 1 mm PMSF, and 10 units/ml of DNase I (New England Biolabs). After centrifugation at 21,000 × g for 12 min at 4 °C, supernatants were isolated, and their protein concentrations were measured. Samples (800 μg of protein each) were precleared with Protein G Plus/Protein A-agarose beads (Calbiochem) and 1 μg of normal mouse IgG (Santa Cruz) for 30 min, then incubated with mouse monoclonal anti-RPA32 antibody (Calbiochem) for 15 h at 4 °C. The samples were subsequently mixed with 30 μl of Protein G Plus/Protein A bead suspension at 4 °C for 3 h. After collection by centrifugation and removal of supernatant, the beads were then washed three times with immunoprecipitation buffer supplemented with protease mixture inhibitors, 1 mm PMSF, and 200 μg/ml ethidium bromide. After the final wash, equal portions of immunoprecipitation buffer and 2× SDS sample buffer were added to the beads, and immunoprecipitated proteins were released by heating at 95 °C for 5 min. Equal volumes of each sample were resolved by SDS-PAGE (6% or 12% for WRN or RPA, respectively). Proteins were transferred to PVDF membranes (Bio-Rad) by electroblotting. The membranes were blocked with 5% nonfat dry milk in TBST buffer (20 mm Tris, pH 7.4, 140 mm NaCl, and 0.1% Tween 20) and analyzed by Western analysis with rabbit anti-WRN (Santa Cruz) or mouse anti-RPA32 (Calbiochem) antibodies for 18 h at 4 °C followed by chemiluminescent detection using ECL Plus (GE Healthcare).
Far Western Assays
Purified RPA (60 and 120 ng or 0.5 and 1 pmol, respectively) or recombinant WRN (30, 60, and 90 ng or 180, 360, and 540 fmol, respectively) and corresponding concentrations of BSA were applied directly onto nitrocellulose membranes. After allowing the applied samples to dry for 15 min at 4 °C, membranes were blocked for 1 h at 4 °C with 5% nonfat dry milk in TBST. The membranes were then incubated in 5 ml of TBST, 5% milk solution (including 25 mm NaCl or 100 mm NaCl as indicated) containing purified WRN (400 ng = 2.4 pmol) or RPA (360 ng = 3 pmol) for 3 h at 4 °C. After washing 3 times for 10 min each with TBST, membranes were subjected to immunodetection by 1) incubation for 1 h with anti-WRN or anti-RPA32 antibody, 2) incubation for 1 h with appropriate HRP-linked secondary antibodies, 3) chemiluminescent development using ECL Plus (GE Healthcare), and 4) visualization by autoradiography. Films were scanned to assess the level of protein binding, with comparison to RPA standards spotted separately on the same membranes.
RESULTS
DNA Damage Enhances Intracellular Association of WRN with RPA
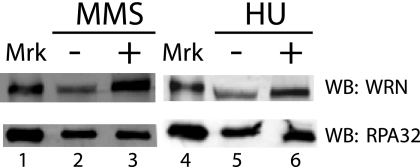
A significant amount of evidence indicates that WRN functions in a pathway that responds to replication blockage to maintain chromosomal stability. A previous study (23) showing WRN and RPA colocalization in nuclear foci in response to treatment with HU suggests that these proteins might interact at blocked replication forks. RPA is widely believed to bind and protect ssDNA formed as a result of blockage of replicative DNA synthesis (32, 33). We reasoned that it should be possible to detect interactions between these two proteins within cells, particularly in response to DNA damaging treatments. To investigate this possibility, co-immunoprecipitation experiments were conducted with human fibroblasts treated either for 10 h with or without HU, which blocks DNA replication by depleting deoxynucleotide pools, or for 4 h with or without MMS, an alkylating agent that generates lesions that block DNA synthesis (50) and causes modification of WRN and its relocalization to nuclear foci (25). The buffers for this protocol included DNase I during lysis and immunoprecipitation and ethidium bromide during washing of the immunoprecipitate to ensure that protein interactions were not mediated through DNA bridging. Using a monoclonal antibody to the RPA32 subunit, it is clear that RPA is present in equal amounts in the immunoprecipitated fraction in HU- and MMS-treated and untreated cells (Fig. 1, lower panel). Although some association of WRN with RPA is detectable even in untreated cells, the amount of WRN precipitated with RPA is significantly increased in cells treated with MMS or HU (Fig. 1, upper panel). Quantitation of data from multiple independent experiments indicates that MMS and HU treatments result in 4.4- and 2.0-fold increases, respectively, in the levels of WRN that are pulled down with RPA. Thus, these data indicate that DNA damaging treatments that block replication increase the association of WRN with RPA and suggest that these proteins not only co-localize at blocked replication forks but also interact within a complex at these sites.
FIGURE 1.
DNA damage enhances the co-immunoprecipitation of WRN with RPA. Cells incubated with or without MMS (1 mm) for 4 h or with HU (2 mm) for 10 h were prepared for immunoprecipitation with anti-RPA32 subunit monoclonal antibody as described under “Experimental Procedures.” Aliquots (40 μl) of the resuspended immunoprecipitated fractions from untreated (lanes 2 and 5), MMS-treated (lane 3), and HU-treated (lane 6) lysates were analyzed by SDS-PAGE (6%) and Western blotting (WB) using anti-WRN antibody and chemiluminescent detection (upper panel). In parallel, aliquots (2.5 μl) of these same immunoprecipitated fractions were analyzed by SDS-PAGE (12%) and Western blotting using anti-RPA32 antibody (lower panel). Purified WRN and RPA were loaded as protein markers (Mkr, lanes 1 and 4).
Direct Interaction between WRN and RPA
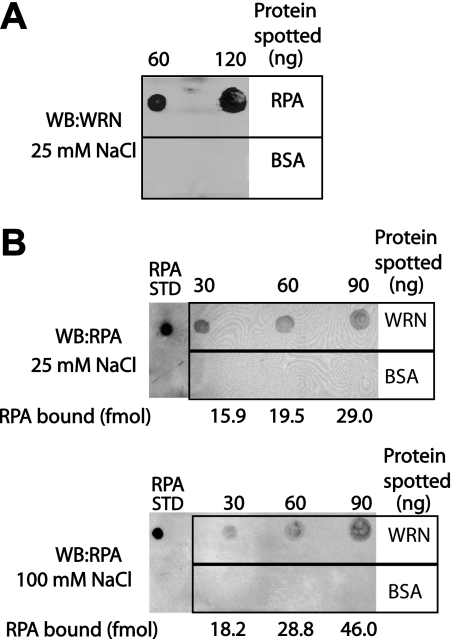
Although the experiments above indicate an association between WRN and RPA in vivo, we wanted to determine whether this association might occur via a direct interaction between these two proteins. To this end, we employed a Far Western dot-blotting technique to probe for binding between purified RPA and recombinant WRN. In these experiments, several concentrations of one protein were immobilized on nitrocellulose membranes; as a control for nonspecific binding, the same amounts of BSA were separately spotted onto the membrane. These membranes were then incubated in buffer containing the other protein, and immunodetection was used to assess binding of this protein. When RPA was spotted onto the membrane, binding of WRN to RPA (in a manner dependent on the RPA concentration) was detected (Fig. 2A). We also obtained a similar result when WRN was spotted onto the membrane, i.e. that RPA bound specifically to WRN in generally a concentration-dependent manner (Fig. 2B). In these assays, little or no nonspecific binding of WRN or RPA to BSA was observed. Furthermore, we tested the stability of this WRN-RPA interaction by challenging with increasing concentrations of NaCl. Our results show that the amounts of RPA bound to WRN at NaCl concentrations of 25 and 100 mm are comparable (Fig. 2B). These results demonstrate a direct, salt tolerant interaction between WRN and RPA.
FIGURE 2.
RPA and WRN directly interact with each other. For Far Western analysis (WB), purified RPA (A), WRN-E84A (B), and BSA (both, as a control for nonspecific binding) were immobilized on nitrocellulose membranes at the indicated concentrations. After blocking, membranes were then incubated in buffer containing 25 or 100 mm NaCl as indicated and either WRN-E84A (400 ng = 2.4 pmol) (A) or RPA (360 ng = 3 pmol) (B). As described under “Experimental Procedures,” immunodetection and autoradiography were used to assess binding of the protein in solution to the immobilized protein on the membrane. The amounts of RPA bound to immobilized WRN (indicated below corresponding spots in B) were determined by comparison to an RPA standard (50 fmol) spotted on each membrane.
RPA Binding to a Model Replication Fork
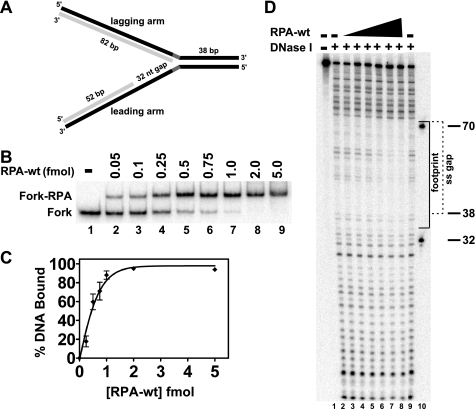
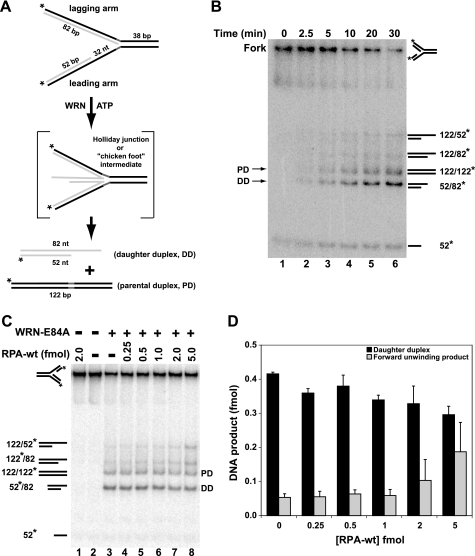
The results above suggest that WRN and RPA directly interact at blocked replication forks. RPA is a heterotrimer with subunits of 70, 32, and 14 kDa (RPA70, RPA32, and RPA14, respectively) that binds ssDNA gaps generated by stalled replication forks; these RPA-DNA complexes play a role in signaling the DNA damage response (32, 33). In vitro experiments have examined the interaction of RPA with a variety of ssDNA substrates, and several distinct RPA-ssDNA binding modes have been described (31–32, 51), with RPA binding stably to a single-stranded region of 30 nt (52, 53). WRN preferentially acts on forked DNA structures (54), and our previous studies have shown that WRN efficiently regresses model replication fork structures (29, 30), perhaps as an early step in overcoming replication blockage. Because RPA not only interacts with WRN but also binds at single-stranded gaps resulting from blocked replication, we thought that RPA might influence WRN activity at replication forks. Thus, we constructed a model replication fork with a single-stranded gap on the leading arm that would permit efficient fork regression by WRN, with the gap being of sufficient size to stably bind RPA. This particular fork structure reflects the situation when DNA synthesis is blocked by a lesion in the leading parental strand. Our model fork substrate (see “Experimental Procedures” for construction details and supplemental Table 1 for nucleotide sequences) contained a 38-bp parental duplex region and leading and lagging parental-daughter arms of equal length, but the leading parental-daughter arm contained a 32-nt single-stranded gap at the fork junction to allow RPA binding (Fig. 3A). The lagging and leading arms of this substrate are entirely homologous to facilitate pairing between daughter strands and repairing of parental strands during fork regression except for 5 nt of noncomplementary sequence incorporated on each arm at the fork junction to prevent spontaneous branch migration. EMSA experiments demonstrated that RPA-wt formed very discrete, stable complexes with this replication fork substrate (Fig. 3B). The amount of RPA-DNA complex formed was dependent upon RPA-wt concentration, with essentially complete binding being observed at approximately equimolar (and higher) RPA-wt to fork substrate ratios (Fig. 3, B and C). These experiments indicate that one molecule of RPA-wt (heterotrimer) binds stably to one molecule of the model replication fork substrate.
FIGURE 3.
RPA-wt binding to a replication fork substrate containing a single-stranded gap on the leading arm. A, the model replication fork substrate generated by two-step annealing of leadP122, lagP122, leadD52, and lagD82 oligonucleotides as indicated that contains a 32-nt single-stranded gap on the leading arm is shown, with parental and daughter strands depicted in black and light gray, respectively (for construction details and oligo sequences, see “Experimental Procedures” and supplemental Table 1, respectively). The parental-daughter arms were completely homologous except for a 5-nt non-complementary region (dark gray) at the fork junction to prevent spontaneous branch migration. The relevant lengths of the duplex and single-stranded regions are indicated. B, RPA-wt (0–5.0 fmol) was incubated with the fork substrate (1 fmol) for 10 min at 25 °C, and binding of RPA to the substrate was analyzed by EMSA as described under “Experimental Procedures.” The positions of the RPA-fork complexes and the fork substrate are indicated at the left. C, graphic representation of RPA binding to the fork substrate, derived from experiments performed as in B, calculated as the percentage of Fork-RPA complex compared with the total DNA for each reaction is shown. Data points are the mean of three independent experiments, except for values at 0.5 and 1 fmol of RPA-wt (four independent experiments). D, shown is RPA-wt (0.1–5 fmol) binding to the fork substrate (5 fmol) analyzed by DNase I footprinting as described under “Experimental Procedures.” The sizes of the markers (lane 10) and the boundaries of the leading arm gap (dashed bracket) and of the area of protection by RPA-wt (solid bracket) are denoted at the right.
Although EMSA analysis identified stable complexes between RPA-wt and the fork DNA substrate and it would be expected that RPA would bind to the single-stranded gap, we used DNase I footprinting to determine the precise site of RPA binding. DNase I digestion of the fork DNA substrate (labeled on the leading parental strand, leadP122) without RPA-wt is shown in Fig. 3D (lanes 2 and 9); the digestion pattern is weak in the region of the 32-nt gap (Fig. 3D, denoted by a dashed bracket) because of the known weaker activity of DNase I on ssDNA. However, increasing concentrations of RPA further inhibited DNase I incision in the 32-nt gap region near the fork junction (Fig. 3D, lanes 3–8). In agreement with EMSA, at an equimolar RPA to fork DNA ratio (∼5 fmol each), a distinct footprint was observed on the fork substrate (Fig. 3D, lane 7). On this substrate, RPA protects a region of ∼36 nt (Fig. 3D, denoted by a solid bracket), encompassing the entire single-stranded gap and extending slightly into the parental duplex region.
Effect of RPA on Fork Regression by WRN
We have shown that WRN acts on model replication fork structures to generate (via a Holliday junction intermediate) parental and daughter duplexes, consistent with the notion that it may regress replication forks in vivo (29, 30). Our earlier results indicated that the 3′ to 5′ exonuclease activity of WRN increases regression efficiency on substrates with a gap size of ≤11 nt by targeted digestion of the leading daughter strand (30). For the replication fork structure used in the current study, the length of the lagging and leading arms was significantly longer than in our earlier substrates (122 versus 70 nt), and the ssDNA gap on the leading arm was much larger (32 nt). As this gap size was >11 nt, we expected that WRN would efficiently regress this fork substrate independent of its exonuclease activity; furthermore, use of exonuclease-deficient WRN-E84A also simplifies analysis of DNA products. Therefore, we examined the action of WRN-E84A on our model fork substrate. Regression of this fork substrate (labeled on both the lagging parental and leading daughter strands) by WRN-E84A should generate parental (*lagP122/leadP122) and daughter (lagD82/*leadD52) duplexes (Fig. 4A), with the latter being diagnostic for fork regression. Alternatively, WRN-mediated forward unwinding of the fork would produce parental-daughter partial duplexes (*lagP122/lagD82 and leadP122/*leadD52). To obtain adequate separation of these DNA products, we used native polyacrylamide (7–9%) gels with an acrylamide to bisacrylamide ratio of 37.5:1. Under these conditions, the daughter duplex (denoted DD or 52*/82) migrated faster than the parental duplex (PD or 122/122*), with even slower migration for the forward unwinding products (*122/82 and 122/*52). In a kinetic analysis of WRN-mediated activity on this substrate, WRN-E84A regressed the replication fork to yield significant amounts of the parental and daughter duplex products within 5 min, and these DNA species accumulated further over time (Fig. 4B). In contrast, only minor amounts of other DNA species, including forward unwinding products, were generated over time. These results demonstrate that WRN-E84A readily and preferentially catalyzes regression of this model replication fork with the 32-nt single-stranded gap on the leading arm.
FIGURE 4.
Effect of RPA-wt on fork regression by WRN-E84A. A, WRN-E84A-mediated conversion of the fork substrate via Holliday junction intermediate to parental and daughter duplexes is depicted. B, a fork regression reaction (40 μl) containing the replication fork substrate (2 fmol) was incubated with WRN-E84A (7 fmol) at 37 °C, and aliquots were removed at the indicated times and analyzed as described under “Experimental Procedures.” The migration of specific DNA species is indicated on the right and for parental (PD) and daughter duplexes (DD) also on the left. C, regression reactions (20 μl) containing fork substrate (1 fmol) with or without RPA (0.25–5.0 fmol) were incubated for 5 min at 4 °C followed by the addition of WRN-E84A (3.5 fmol), except where noted, and further incubation at 37 °C for 15 min. Positions of relevant DNA species are indicated as above. D, for WRN-mediated fork regression reactions analyzed in C, the amount of daughter duplex and the product of forward unwinding was quantitated with respect to the total DNA as described under “Experimental Procedures” and plotted as a bar graph showing the amounts of daughter duplex (black) and forward unwinding product (gray) at various concentrations of RPA (0–5 fmol).
We then examined the effect of RPA on regression of our replication fork substrate by WRN-E84A. For these experiments, RPA was preincubated with the fork substrate for 5 min at 4 °C before the addition of WRN-E84A and further incubation at 37 °C for 15 min. A representative experiment is shown in Fig. 4C, whereas Fig. 4D shows data from multiple regression assays comparing the formation of daughter duplex (which specifically reflects fork regression) and a forward unwinding product. RPA alone cannot regress or unwind the replication fork substrate, whereas, as above, WRN-E84A alone primarily yields parental and daughter duplexes (Fig. 4C, lanes 1 and 3). Because WRN is known to bind ssDNA (55), it might be expected that RPA prebound to the ssDNA gap of the fork substrate would obstruct WRN-E84A binding and inhibit its regression activity. However, in such reactions containing both RPA-wt and WRN-E84A, the dominant products are the daughter and parental duplexes (Fig. 4C, lanes 3–7). Importantly, at concentrations of RPA sufficient to completely bind all fork substrate molecules, WRN-E84A still preferentially catalyzes fork regression with similar efficiency as compared with reactions without RPA (compare Fig. 4, C and D, to Fig. 3, B and C). This result also suggests that WRN may displace bound RPA from the ssDNA gap to carry out fork regression. At substantially higher RPA to fork DNA (5:1) ratios, some increase in forward unwinding of the fork was observed, as evidenced by increased production of parental-daughter partial duplexes *122/82 and 122/*52 (Fig. 4, C, lane 8, and D). Based on our experience, this effect is due to the inherent strand melting property associated with excess RPA (56) that likely enhances the weak forward unwinding activity of WRN on these types of replication fork substrates. Because of these issues associated with excess RPA in the reaction, we believe that the molecular events (in this case, WRN-catalyzed fork regression) taking place at the minimal concentrations of RPA sufficient to completely bind the substrate are the most relevant (Fig. 4, C and D).
Displacement of RPA Bound to the Fork and Gapped Duplex DNA by WRN
Our results above demonstrate that WRN-E84A readily regresses the model replication fork even when RPA is bound in a very stable complex to the single-stranded gap on this substrate. Because regression generates DNA products that eliminate this gap, it stands to reason that RPA must become displaced by the completion of the regression reaction. We were very interested to determine whether the displacement of RPA by WRN-E84A occurred before or during the regression process and if the ATPase/helicase function of WRN-E84A was required for displacement. To address these questions, we modified our experimental protocol and used EMSA to track changes in Fork-RPA complexes (Fig. 5A). Because regression requires the ATPase/helicase activities of WRN-E84A (29), we deliberately excluded ATP from our reactions to determine whether RPA displacement occurred before regression, instead sometimes substituting the non-hydrolyzable analog ATPγS. Importantly, a labeled 32-mer (*ss32, with identical sequence to the gap) was included (at equimolar amounts with respect to the fork substrate) in some reactions to specifically bind displaced RPA, forming a complex with unique mobility that could also be visualized on a gel. In these experiments it was also crucial that the RPA concentration was limiting with respect to the fork substrate, so that there could be little or no free RPA at the time WRN-E84A was added. This protocol allowed us to visually monitor possible displacement of bound RPA from the fork substrate by its transfer to the labeled 32-mer.
For these reactions (Fig. 5A), RPA-wt was preincubated with the fork as before to first generate stable Fork-RPA complexes, then WRN-E84A was added where indicated with the temperature kept at 25 °C. Subsequently, ATPγS and/or *ss32 oligomer was added, the temperature was shifted to 37 °C for 10 min, and then the reactions were immediately subject to EMSA to assess the resulting DNA-protein complexes. As expected with fork DNA and RPA-wt alone, specific and stable complexes (Fork-RPA) were formed (Fig. 5B, lane 2); about half the DNA was bound in these complexes, indicating that RPA was indeed limiting, i.e. little or no free RPA remained in solution. Importantly, the addition of *ss32 alone has no effect on these Fork-RPA complexes; specifically, the amount of Fork-RPA complex remains the same without detectable formation of RPA-ss32 complexes (Fig. 5B, lane 3). This result indicates that, once bound, RPA-wt does not spontaneously dissociate from the fork, and *ss32 alone cannot displace RPA-wt. Moreover, the lack of RPA-ss32 complexes in this reaction also confirms that there is no free RPA-wt at the time other components are added. When the fork DNA was incubated with only WRN-E84A, another distinct, much slower-migrating DNA-protein complex (Fork-WRN) was formed (Fig. 5B, lane 7). The addition of WRN-E84A to reactions preincubated with RPA-wt resulted in a DNA-protein complex that co-migrated with the Fork-WRN complex, whereas the amount of the Fork-RPA complex dropped precipitously (Fig. 5B, lanes 4 and 5). This confirmed that in the reactions containing RPA-wt, WRN-E84A still formed a stable complex with the fork DNA and suggested that WRN displaced RPA from the fork. Indeed, when labeled *ss32 was added to reactions containing the fork, RPA-wt, and WRN-E84A, a new band (indicated by a dashed arrow) appears that exactly corresponds to the RPA-ss32 complex (Fig. 5B, lane 6), demonstrating that RPA was displaced from the fork by WRN-E84A and subsequently became bound to *ss32. Although displacement of RPA by WRN-E84A was achieved without ATP (Fig. 5B, lane 4), it was markedly enhanced with ATPγS (Fig. 5B, lane 5), suggesting that ATP binding by WRN-E84A stimulated its ability to displace RPA from the gap of the fork substrate. These results clearly demonstrate that although both RPA and WRN-E84A can bind independently to our model replication fork, WRN-E84A displaces bound RPA from the single-stranded gap of this fork substrate without the need for ATP hydrolysis or helicase activity. This displacement of RPA from the fork paves the way for WRN to, concomitant with ATP hydrolysis, efficiently catalyze regression.
We used a similar EMSA strategy to test the interplay between WRN-E84A and RPA on another relevant DNA substrate (leadD52 + lagP38-3′/*leadP122) containing two duplex regions (of 52 and 38 bp) surrounding a ssDNA gap of 32 nt (supplemental Fig. 1A). Importantly, these types of gapped DNA structures would result from the blockage of lagging strand synthesis and, thus, may be physiologically relevant targets for RPA binding. As for the fork substrate, RPA forms a stable complex with the gapped substrate (Gap-RPA), and the addition of *ss32 had no effect on this Gap-RPA complex (supplemental Fig. 1B, lanes 2 and 3). When this Gap-RPA complex was challenged with WRN-E84A in the presence of *ss32, the amount of Gap-RPA was dramatically reduced and two new species appeared (supplemental Fig. 1B, lane 4); one of these species co-migrated with a Gap-WRN complex (lane 6), whereas the other co-migrated with a *ss32-RPA complex (lane 5). This result indicates that WRN-E84A displaces RPA from the gapped substrate, resulting in its transfer to oligomeric *ss32. Thus, WRN displacement of RPA from ssDNA occurs in at least two DNA structural contexts representative of RPA-DNA complexes formed subsequent to replication blockage.
Effect of Phosphomimetic RPA (RPA-32D8) on WRN-mediated Fork Regression
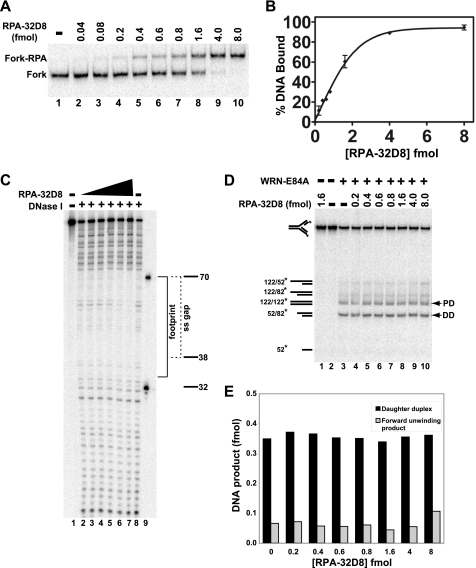
RPA becomes phosphorylated on its RPA32 subunit in response to cell cycle progression or DNA damage. The N-terminal domain of RPA32 contains multiple serine and threonine residues that can be modified to give rise to differentially phosphorylated forms of RPA (57–59). Treatment with DNA damaging agents or replication inhibitors results in (hyper)phosphorylation of multiple serines and threonines in RPA32 that appears to modulate DNA replication after DNA damage (60–62). These findings suggest that a hyperphosphorylated form of RPA might be present at single-stranded regions generated by replication fork blockage. This notion has been further supported by studies using a synthetic mutant form of RPA (RPA-32D8) in which both of the cyclin-cdk2 sites (Ser-23 and Ser-29) and six other N-terminal phosphorylation sites (Ser-8, Ser-11, Ser-12, Ser-13, Thr-21, and Ser-33) in the RPA32 subunit were replaced by aspartate to reflect ionic changes associated with phosphorylation. This substitution of multiple serine and threonine residues with aspartate has been shown to have similar effects on protein structure and function as hyperphosphorylation and RPA-32D8 supports mismatch repair in vitro (37, 60, 63). Thus, the hyperphosphorylated form of RPA is mimicked by RPA-32D8, which we used here to examine its effect on fork regression by WRN-E84A. As before, we used EMSA to study the binding of RPA-32D8 to our replication fork substrate. As in the case of RPA-wt, RPA-32D8 also formed stable and specific complexes with this fork substrate, although approximately a 4-fold excess of RPA-32D8 was needed to achieve complete binding (Fig. 6, A and B). When DNase I footprinting was used, no qualitative differences between the binding patterns of RPA-wt and RPA-32D8 to the fork substrate were observed, but again, a significantly higher ratio of RPA-32D8 (∼6-fold) over the fork DNA was needed to significantly inhibit DNase I cleavage in the single-stranded region (compare Figs. 3D and 6C). Under the conditions used in these assays, RPA binding to the DNA substrate is stoichiometric. Thus, complex formation would only depend upon the amounts of active RPA complex in each reaction. Notably, RPA-wt and RPA-32D8 were shown to have comparable affinities for ssDNA substrates (64). Thus, the higher amounts of RPA-32D8 needed here to saturate the substrate is most likely due to some inactive protein in these preparations, as noted previously (65). Regardless, these results indicate that both RPA-wt and RPA-32D8 bind quite stably to the 32-nt gap of our model replication fork.
FIGURE 6.
Effect of phosphomimetic RPA-32D8 on WRN-E84A-mediated fork regression. A, RPA-32D8 (0.04–8.0 fmol) was incubated with the fork substrate (1 fmol) for 10 min at 25 °C, and DNA binding of RPA-32D8 was analyzed by EMSA as described. The positions of the RPA-32D8-fork complexes and the fork substrate are indicated at the left. B, for experiments as presented in A, the amount of fork substrate bound was determined by comparing the amount of bound DNA to the total DNA for each reaction and plotted versus RPA-32D8 concentration. Each data point is the average of two independent experiments. C, binding of RPA-32D8 (1.0–32 fmol) to the fork substrate (5 fmol) is analyzed by DNase I footprinting as described under “Experimental Procedures.” Position of the markers, the boundary of the leading arm gap (dashed bracket), and the area of protection by RPA-32D8 (solid bracket) is denoted on the right. D, in fork regression assays, shown is the fork substrate (1 fmol) with or without RPA-32D8 (0.2–8.0 fmol) for 5 min at 4 °C followed by the addition of WRN-E84A (3.5 fmol), except where indicated, and further incubation at 37 °C for 15 min. The position of individual DNA species is noted on the left and for parental (PD) and daughter duplexes (DD) by arrowheads also on the right. E, for WRN-mediated fork regression reactions as depicted in D, amounts of daughter duplexes and products of forward unwinding were quantitated as described, and the data are plotted as a bar graph showing the amounts of these products with respect to RPA-32D8 concentration.
Next, we examined the effect of RPA-32D8 on WRN-mediated fork regression. The presence of RPA-32D8 alone had no effect on the fork substrate (Fig. 6D, lane 1), whereas WRN-E84A alone produced primarily the expected parental and daughter duplex products characteristic for fork regression (Fig. 6D, lane 3). At RPA-32D8 concentrations at which partial and complete binding to the fork substrate was observed, WRN-E84A-dependent production of parental and daughter duplexes was essentially identical to that of WRN-E84A alone (Fig. 6, D and E). At the highest RPA-32D8 concentrations, a very modest increase in forward unwinding was observed, as seen by the increased production of parental-daughter partial duplexes (Fig. 6, D and E). These results suggest that prebound RPA-wt and RPA-32D8 have very similar effects on WRN-mediated regression of fork substrate. Although both RPA-wt and RPA-32D8 can bind and protect single-stranded gaps on the leading arm of our model replication fork, their presence does not substantially alter the enzymatic preference of WRN to perform fork regression or interfere with the strength of the regression reaction.
DISCUSSION
The progression of replication forks is often stalled or blocked in vivo, and cells have evolved mechanisms to deal with these situations so that DNA replication can be resumed and completed accurately. RecQ family members including WRN have been postulated to participate in proper resolution of replication blockage. In support of such a role for WRN, treatment of cells with HU or certain DNA damaging agents results in WRN relocalization to nuclear foci, where it colocalizes with replication factors including RPA (23–25). Notably, improper resolution of blocked replication forks leads to genomic instability, a well established feature of cells lacking WRN function. At the molecular level, blockage of replication by DNA lesions or other obstacles results in formation and persistence of regions of ssDNA. Specifically, blockage of lagging strand synthesis results in persistence of ssDNA gaps between Okazaki fragments; when leading strand synthesis is blocked, synthesis of lagging strand may temporarily continue, leading to the formation of ssDNA gaps on the leading arm adjacent to the fork junction. RPA is widely thought to bind and protect regions of ssDNA created by stalled replication forks and help to activate the DNA damage response pathway (35, 36). It is also well established that RPA undergoes hyperphosphorylation on its RPA32 subunit in response to DNA damage (37), theoretically shifting its role from DNA replication to repair. These studies suggest possible molecular interactions between WRN and RPA relevant to resolution of replication fork blockage, a scenario specifically examined in this study.
If WRN and RPA act cooperatively in response to blocked replication, it should be possible to detect interactions between these two factors, particularly in response to treatment of cells with DNA damaging agents. Our co-immunoprecipitation experiments clearly demonstrate an association of WRN with RPA that is substantially increased after MMS or HU treatment. To our knowledge, this is the first time that co-immunoprecipitation of RPA and WRN from cell lysates has been demonstrated as well as its enhancement by DNA damaging treatments. These experiments indicate that, in response to MMS or HU treatment, WRN and RPA associate with each other within a protein complex but do not necessarily prove a direct interaction between the two proteins. Therefore, we tested whether purified WRN and RPA could bind to each other by Far Western analysis. These experiments demonstrate a direct, specific interaction between WRN and RPA that is stable even at physiologically relevant salt concentrations. These experiments confirm and extend previous studies showing a direct interaction between purified WRN and RPA that maps to the N-terminal region of WRN (aa239–499) and the RPA70 subunit (amino acids 100–300) (45, 66, 67). Taken together, this evidence strongly suggests that co-localization and co-immunoprecipitation of WRN and RPA observed after HU and other DNA damaging treatments is mediated by a direct interaction between these two proteins.
In light of these findings, it is relevant to consider the molecular consequences of the interaction between WRN and RPA at blocked replication fork structures. According to some proposed models for processing blocked replication forks (26–28, 68, 69), fork regression may be an early step in dealing with repair and resolution of damaged forks. Fork regression involves unwinding of parental-daughter duplexes followed by pairing of the nascent daughter strands and re-pairing of the parental strands to form a four-stranded Holliday junction or chicken foot intermediate. One scenario suggests that the Holliday junction is then further processed to generate a double-strand break that may initiate recombination pathways to re-establish a functional replication fork. Alternatively, once regression has occurred, either repair mechanisms can access and remove the blocking lesion followed by reestablishment of the replication fork by reverse branch migration or a strand switching mechanism is employed in which the blocking lesion is bypassed in an error-free manner after reverse branch migration. Previous studies from our laboratory have shown that WRN readily regresses specific model replication forks in vitro (29, 30). Because RPA (unmodified or hyperphosphorylated) is widely believed to bind ssDNA gaps in the vicinity of forks blocked by DNA damage, we constructed a model replication fork substrate containing a ssDNA gap on the leading arm sufficient to bind RPA. We then examined binding of RPA (wild type or RPA-32D8 hyperphosphorylation mimetic) to this substrate and its effects on the WRN-E84A-mediated regression reaction. EMSA and DNase I footprinting analysis confirmed that both RPA-wt and phosphomimetic RPA-32D8 bound stably to this substrate and precisely at the 32-nt gap on the leading arm. Because WRN is a DNA-binding protein with some specificity for ssDNA (55, 70), the binding of RPA to this ssDNA region might have been expected to inhibit or alter the activity of WRN on this fork substrate. On the contrary, our studies indicated that WRN-E84A was still able to efficiently regress the replication fork substrate even with either form of RPA stoichiometrically bound at the leading arm gap. This result suggests two (not mutually exclusive) alternatives; 1) binding of RPA at the leading arm gap does not significantly influence the affinity or binding mode of WRN for this substrate, or 2) RPA bound to the substrate might actually help in WRN recruitment and its subsequent action on this substrate. Importantly, the latter explanation is consistent with the observed direct interaction between RPA and WRN and parallels the established function of RPA in protecting ssDNA and subsequently directing downstream events (recruitment of ATR·ATRIP or recombination factors). Similarly, our results suggest that RPA may help recruit WRN to stalled replication forks in vivo.
Because WRN-mediated regression of our model fork substrate generates DNA products that eliminate the ssDNA gap to which RPA binds, these results also indicated that the action of WRN-E84A leads to displacement of the bound RPA either before or during the regression reaction. Upon further examination, we found that WRN-E84A displaced bound RPA from the single-stranded region of our fork substrate by observing the transfer of RPA from the fork to a 32-mer only upon the addition of WRN-E84A. Intriguingly, this WRN-mediated displacement of RPA occurred in the absence of ATP and was enhanced in the presence of the non-hydrolyzable analog ATPγS. Although the ATP-bound conformation of WRN-E84A was more efficient in displacing RPA, its ATPase and helicase activities were not required. This result indicated that RPA was dislodged from the single-stranded gap by WRN-E84A independent of ATPase-and helicase-mediated remodeling of the DNA substrate. Furthermore, these findings strongly suggest that the direct interactions observed between these proteins were involved in the displacement of RPA by WRN and implied a scenario in which RPA displacement likely precedes the WRN-E84A-mediated regression reaction. We also found that WRN displaced RPA from a gapped duplex substrate that is reflective of the structure of ssDNA gaps that persist after blockage of lagging strand synthesis and are also targets for RPA binding. These results indicate that this molecular cooperation between RPA and WRN can be observed with other DNA structures relevant to replication blockage. Taken together, our experiments strongly suggest that the displacement of RPA by WRN is mediated (at least in part) by the direct protein-protein interaction between these proteins. These findings indicate that RPA and WRN may cooperate during fork regression and/or perhaps other DNA remodeling events subsequent to replication fork blockage. Consistent with our findings, a very recent study suggests that the WRN and RPA orthologs in Caenorhabditis elegans act cooperatively at replication forks stalled by HU (71). Because both RPA and WRN are modified in response to DNA damage or replication blockage and modification of WRN correlates with its relocalization to replication foci that contain RPA, it also seems likely that modified forms of each protein may mediate or enhance this interaction or coordination. Although we have determined here that a phosphomimetic form of RPA bound to the ssDNA gap of our fork substrate also readily permits WRN-mediated regression, further investigation is needed to clarify the molecular roles of WRN and RPA modifications in these events.
Although RPA-WRN interactions have been reported previously, the results in this study provide (for the first time) a logical scenario as to how these proteins might function cooperatively at the molecular level in vivo that is also highly consistent with putative physiological roles for both factors. Based on our findings, we propose the following model related to resolution of stalled replication forks resulting from DNA damage or other circumstances. Subsequent to replication fork blockage, RPA is initially recruited to single-stranded gaps proximal to the forks. In addition to protecting these regions from degradation, the presence of RPA-ssDNA complexes may then mediate the recruitment and activation of other DNA damage response proteins including the ATR·ATRIP complex and WRN. Recruitment of ATR initiates the replication checkpoint pathway, whereas specific recruitment of WRN allows displacement of RPA from these gaps and facilitates the DNA remodeling reactions (including fork regression) necessary for downstream steps and proper resolution of the blocked replication fork. Within the framework of this model, ATR may also have a role in regulation of WRN function at blocked replication forks, as suggested by recent studies (72). By this reasoning, proper resolution of replication blockage is compromised in cells lacking WRN, the replication fork collapses resulting in double-strand breakage, and a more error-prone recombination pathway is likely utilized. In agreement with this notion, WRN-deficient cells show increased spontaneous levels of RAD51 foci (21, 73) and chromosomal aberrations (74). These concepts are highly consistent with the DNA damage sensitivities, replication abnormalities, and genomic instability associated with WRN-deficient cells and Werner syndrome.
Supplementary Material
Acknowledgments
We thank Nelson Chan for valuable reagents and Deanna Edwards for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA113371 and by AG027258 (NIA) (to D. K. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
- HU
- hydroxyurea
- ATPγS
- adenosine 5′-O-(thio)triphosphate
- MMS
- methylmethanesulfonate
- RPA
- human replication protein A
- WRN
- Werner syndrome protein
- nt
- ss32, 32-mer
- EMSA
- electrophoretic mobility shift assay.
REFERENCES
- 1. Atkinson J., McGlynn P. (2009) Nucleic. Acids Res. 37, 3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sidorova J. M. (2008) DNA Repair 7, 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. (1995) Cell 83, 655–666 [DOI] [PubMed] [Google Scholar]
- 4. Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G. M., Mulligan J., Schellenberg G. D. (1996) Science 272, 258–262 [DOI] [PubMed] [Google Scholar]
- 5. Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999) Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- 6. Goto M. (1997) Mech. Ageing Dev. 98, 239–254 [DOI] [PubMed] [Google Scholar]
- 7. Martin G. M., Oshima J. (2000) Nature 408, 263–266 [DOI] [PubMed] [Google Scholar]
- 8. Opresko P. L., Cheng W. H., Bohr V. A. (2004) J. Biol. Chem. 279, 18099–18102 [DOI] [PubMed] [Google Scholar]
- 9. Huang S., Beresten S., Li B., Oshima J., Ellis N. A., Campisi J. (2000) Nucleic. Acids Res. 28, 2396–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Machwe A., Xiao L., Theodore S., Orren D. K. (2002) J. Biol. Chem. 277, 4492–4504 [DOI] [PubMed] [Google Scholar]
- 11. Orren D. K., Theodore S., Machwe A. (2002) Biochemistry 41, 13483–13488 [DOI] [PubMed] [Google Scholar]
- 12. Cheok C. F., Wu L., Garcia P. L., Janscak P., Hickson I. D. (2005) Nucleic Acids Res. 33, 3932–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. (2004) EMBO J. 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P. (2006) Nucleic. Acids Res. 34, 5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Machwe A., Xiao L., Groden J., Matson S. W., Orren D. K. (2005) J. Biol. Chem. 280, 23397–23407 [DOI] [PubMed] [Google Scholar]
- 16. Macris M. A., Krejci L., Bussen W., Shimamoto A., Sung P. (2006) DNA Repair 5, 172–180 [DOI] [PubMed] [Google Scholar]
- 17. Sharma S., Sommers J. A., Choudhary S., Faulkner J. K., Cui S., Andreoli L., Muzzolini L., Vindigni A., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 28072–28084 [DOI] [PubMed] [Google Scholar]
- 18. Poot M., Hoehn H., Rünger T. M., Martin G. M. (1992) Exp. Cell Res. 202, 267–273 [DOI] [PubMed] [Google Scholar]
- 19. Rodríguez-López A. M., Jackson D. A., Iborra F., Cox L. S. (2002) Aging Cell 1, 30–39 [DOI] [PubMed] [Google Scholar]
- 20. Lebel M., Leder P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13097–13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pichierri P., Franchitto A., Mosesso P., Palitti F. (2001) Mol. Biol. Cell 12, 2412–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Poot M., Yom J. S., Whang S. H., Kato J. T., Gollahon K. A., Rabinovitch P. S. (2001) FASEB J. 15, 1224–1226 [DOI] [PubMed] [Google Scholar]
- 23. Constantinou A., Tarsounas M., Karow J. K., Brosh R. M., Bohr V. A., Hickson I. D., West S. C. (2000) EMBO Rep. 1, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pichierri P., Rosselli F., Franchitto A. (2003) Oncogene 22, 1491–1500 [DOI] [PubMed] [Google Scholar]
- 25. Karmakar P., Bohr V. A. (2005) Mech. Ageing Dev. 126, 1146–1158 [DOI] [PubMed] [Google Scholar]
- 26. Cox M. M. (2002) Mutat. Res. 510, 107–120 [DOI] [PubMed] [Google Scholar]
- 27. Haber J. E. (1999) Trends Biochem. Sci. 24, 271–275 [DOI] [PubMed] [Google Scholar]
- 28. Kowalczykowski S. C. (2000) Trends Biochem. Sci. 25, 156–165 [DOI] [PubMed] [Google Scholar]
- 29. Machwe A., Xiao L., Groden J., Orren D. K. (2006) Biochemistry 45, 13939–13946 [DOI] [PubMed] [Google Scholar]
- 30. Machwe A., Xiao L., Lloyd R. G., Bolt E., Orren D. K. (2007) Nucleic. Acids Res. 35, 5729–5747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iftode C., Daniely Y., Borowiec J. A. (1999) Crit. Rev. Biochem. Mol. Biol. 34, 141–180 [DOI] [PubMed] [Google Scholar]
- 32. Wold M. S. (1997) Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 33. Raderschall E., Golub E. I., Haaf T. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1921–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dart D. A., Adams K. E., Akerman I., Lakin N. D. (2004) J. Biol. Chem. 279, 16433–16440 [DOI] [PubMed] [Google Scholar]
- 35. Zou L., Elledge S. J. (2003) Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 36. Zou L., Liu D., Elledge S. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Binz S. K., Sheehan A. M., Wold M. S. (2004) DNA Repair 3, 1015–1024 [DOI] [PubMed] [Google Scholar]
- 38. Jackson S. P. (2002) Carcinogenesis 23, 687–696 [DOI] [PubMed] [Google Scholar]
- 39. Din S., Brill S. J., Fairman M. P., Stillman B. (1990) Genes. Dev. 4, 968–977 [DOI] [PubMed] [Google Scholar]
- 40. Dutta A., Stillman B. (1992) EMBO J. 11, 2189–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitsis P. G. (1995) Dev. Biol. 170, 445–456 [DOI] [PubMed] [Google Scholar]
- 42. Oakley G. G., Loberg L. I., Yao J., Risinger M. A., Yunker R. L., Zernik-Kobak M., Khanna K. K., Lavin M. F., Carty M. P., Dixon K. (2001) Mol. Biol. Cell 12, 1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shao R. G., Cao C. X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. (1999) EMBO J. 18, 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Anantha R. W., Borowiec J. A. (2009) Cell Cycle 8, 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brosh R. M., Jr., Orren D. K., Nehlin J. O., Ravn P. H., Kenny M. K., Machwe A., Bohr V. A. (1999) J. Biol. Chem. 274, 18341–18350 [DOI] [PubMed] [Google Scholar]
- 46. Shen J. C., Gray M. D., Oshima J., Loeb L. A. (1998) Nucleic. Acids Res. 26, 2879–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang S., Li B., Gray M. D., Oshima J., Mian I. S., Campisi J. (1998) Nat. Genet. 20, 114–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 49. States J. C., Quan T., Hines R. N., Novak R. F., Runge-Morris M. (1993) Carcinogenesis 14, 1643–1649 [DOI] [PubMed] [Google Scholar]
- 50. Wyatt M. D., Pittman D. L. (2006) Chem. Res. Toxicol. 19, 1580–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nasheuer H. P., Smith R., Bauerschmidt C., Grosse F., Weisshart K. (2002) Prog. Nucleic Acid Res. Mol. Biol. 72, 41–94 [DOI] [PubMed] [Google Scholar]
- 52. Bastin-Shanower S. A., Brill S. J. (2001) J. Biol. Chem. 276, 36446–36453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bochkareva E., Belegu V., Korolev S., Bochkarev A. (2001) EMBO J. 20, 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brosh R. M., Jr., Waheed J., Sommers J. A. (2002) J. Biol. Chem. 277, 23236–23245 [DOI] [PubMed] [Google Scholar]
- 55. Machwe A., Xiao L., Orren D. K. (2006) BMC Mol. Biol. 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lao Y., Lee C. G., Wold M. S. (1999) Biochemistry 38, 3974–3984 [DOI] [PubMed] [Google Scholar]
- 57. Liu Y., Kvaratskhelia M., Hess S., Qu Y., Zou Y. (2005) J. Biol. Chem. 280, 32775–32783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oakley G. G., Patrick S. M., Yao J., Carty M. P., Turchi J. J., Dixon K. (2003) Biochemistry 42, 3255–3264 [DOI] [PubMed] [Google Scholar]
- 59. Zernik-Kobak M., Vasunia K., Connelly M., Anderson C. W., Dixon K. (1997) J. Biol. Chem. 272, 23896–23904 [DOI] [PubMed] [Google Scholar]
- 60. Vassin V. M., Wold M. S., Borowiec J. A. (2004) Mol. Cell. Biol. 24, 1930–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carty M. P., Zernik-Kobak M., McGrath S., Dixon K. (1994) EMBO J. 13, 2114–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu V. F., Weaver D. T. (1993) Mol. Cell. Biol. 13, 7222–7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Guo S., Zhang Y., Yuan F., Gao Y., Gu L., Wong I., Li G. M. (2006) J. Biol. Chem. 281, 21607–21616 [DOI] [PubMed] [Google Scholar]
- 64. Binz S. K., Lao Y., Lowry D. F., Wold M. S. (2003) J. Biol. Chem. 278, 35584–35591 [DOI] [PubMed] [Google Scholar]
- 65. Kim C., Paulus B. F., Wold M. S. (1994) Biochemistry 33, 14197–14206 [DOI] [PubMed] [Google Scholar]
- 66. Doherty K. M., Sommers J. A., Gray M. D., Lee J. W., von Kobbe C., Thoma N. H., Kureekattil R. P., Kenny M. K., Brosh R. M., Jr. (2005) J. Biol. Chem. 280, 29494–29505 [DOI] [PubMed] [Google Scholar]
- 67. Shen J. C., Lao Y., Kamath-Loeb A., Wold M. S., Loeb L. A. (2003) Mech. Ageing Dev. 124, 921–930 [DOI] [PubMed] [Google Scholar]
- 68. McGlynn P., Lloyd R. G. (2002) Nat. Rev. Mol. Cell Biol. 3, 859–870 [DOI] [PubMed] [Google Scholar]
- 69. Higgins N. P., Kato K., Strauss B. (1976) J. Mol. Biol. 101, 417–425 [DOI] [PubMed] [Google Scholar]
- 70. Orren D. K., Brosh R. M., Jr., Nehlin J. O., Machwe A., Gray M. D., Bohr V. A. (1999) Nucleic Acids Res. 27, 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee S. J., Gartner A., Hyun M., Ahn B., Koo H. S. (2010) PLoS. Genet. 6, e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ammazzalorso F., Pirzio L. M., Bignami M., Franchitto A., Pichierri P. (2010) EMBO J. 29, 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Franchitto A., Pirzio L. M., Prosperi E., Sapora O., Bignami M., Pichierri P. (2008) J. Cell Biol. 183, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fukuchi K., Martin G. M., Monnat R. J., Jr. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5893–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.