Abstract
The mechanism of serine proteases prominently illustrates how charged amino acid residues and proton transfer events facilitate enzyme catalysis. Here we present an ultrahigh resolution (0.93 Å) x-ray structure of a complex formed between trypsin and a canonical inhibitor acting through a substrate-like mechanism. The electron density indicates the protonation state of all catalytic residues where the catalytic histidine is, as expected, in its neutral state prior to the acylation step by the catalytic serine. The carboxyl group of the catalytic aspartate displays an asymmetric electron density so that the Oδ2–Cγ bond appears to be a double bond, with Oδ2 involved in a hydrogen bond to His-57 and Ser-214. Only when Asp-102 is protonated on Oδ1 atom could a density functional theory simulation reproduce the observed electron density. The presence of a putative hydrogen atom is also confirmed by a residual mFobs − DFcalc density above 2.5 σ next to Oδ1. As a possible functional role for the neutral aspartate in the active site, we propose that in the substrate-bound form, the neutral aspartate residue helps to keep the pKa of the histidine sufficiently low, in the active neutral form. When the histidine receives a proton during the catalytic cycle, the aspartate becomes simultaneously negatively charged, providing additional stabilization for the protonated histidine and indirectly to the tetrahedral intermediate. This novel proposal unifies the seemingly conflicting experimental observations, which were previously seen as either supporting the charge relay mechanism or the neutral pKa histidine theory.
Keywords: Enzyme Catalysis, Enzyme Kinetics, Enzyme Mechanisms, Enzyme Structure, Enzymes, Protease, Protease Inhibitor, Proteolytic Enzymes, Serine Protease, X-ray Crystallography
Introduction
Serine proteases have been among the prime examples where the discovery of their structure led to a better understanding of enzyme catalysis. Already the very first structure of a serine protease, chymotrypsin (1), highlighted the spatial arrangement of the catalytic triad and shortly afterward a promising catalytic mechanism was proposed by Blow et al. (2). Serine proteases cleave peptide bonds in two major steps: acylation and deacylation. When a suitable substrate binds to the enzyme, a transient β-sheet forms that positions the active site next to the scissile peptide bond. The catalytic serine attacks the scissile peptide bond and a short-lived tetrahedral intermediate form, whereas the proton of the serine is transferred to the catalytic histidine. This elementary proton transfer step is supported by direct and indirect evidence (3, 4). The acylation phase is decisive for the fate of the substrate, because for good substrates the process is irreversible. On the other hand, in canonical inhibitors the acylation reaction can be reversed at low pH (5, 6). Structural details of the acylation step and its intermediates are relatively scarce; however, the putative first tetrahedral intermediate of the acylation phase has been observed by a series of intermediate trapping experiments (7).
In contrast to the well studied histidine, the role of catalytic aspartate is more elusive. It is generally assumed that this aspartate is negatively charged in the free enzyme. Blow et al. (2) initially proposed that the histidine, when it becomes protonated by the serine side chain or an attacking water molecule, subsequently transfers one of its protons to the aspartate, effectively neutralizing the active site. Criticism of this idea was focused on the protonation state of the histidine, which appeared to be directly dependent on the external pH in the free enzyme (8), but it remained in its neutral state in enzyme·inhibitor complexes (9). In the free, solvent-accessible form, the pKa of the residue appeared to be invariably ∼7–7.5, which did not motivate the transfer of a proton to a fully solvated aspartate residue (assumed pKa of ∼3). The role of the aspartate was tested by site-directed mutagenesis to neutral asparagine (10), which revealed a reduction in catalytic rate acceleration by approximately a factor of 104, further supporting the role of a negative charge near the histidine.
Nevertheless, the initial idea of a direct proton transfer from the catalytic histidine to the aspartate proved to be a long lasting one and prompted numerous experimental and theoretical investigations (for a historic review, see Ref. 11). More recently, to explain the nature of the low field proton between the charged aspartate and histidine and the unaccounted reactivity of the histidine as a general base, Frey et al. (12) suggested that this proton is participating in a low barrier hydrogen bond in certain phases of catalysis. This low barrier hydrogen bond proposal was called into question by Ash et al. (13), at least in the case of serine proteases. Neutron diffraction (14) and improvements in x-ray flux at third generation synchrotrons (15) in recent years allowed the study of hydrogens by diffraction methods directly. Although both x-ray and neutron diffraction have limitations concerning the reliable detection of hydrogens (false negative observations), when a strong signal is observed, it gives reliable and independent verification of hydrogen positions. In addition, x-ray crystallography at ultrahigh resolution also reveals the electron density distribution of covalent bonds between heavier atoms, providing indirect clues of the protonation state of catalytic (and other) residues (16). In ultrahigh resolution structures, the precision of interatomic distances is also markedly increased, and the uncertainty of the individual interatomic distances can also be estimated (17). To obtain sufficiently good quality diffraction for high resolution studies, it is necessary to grow crystals of exceptional quality. One way to improve crystal quality is to reduce the internal disorder in the unit cell. Protein complexes are particularly prone to disorder because of potential alternative binding modes of their components (18). To reduce such disorders introduced by the binding, one strategy is to improve the binding strength of the components (19).
Wild-type SGTI4 (also known as SGPI-1) (20) is a weak inhibitor of bovine trypsin with a Ki in the micromolar range (21). Directed evolution was demonstrated to be a powerful tool for identifying good substrates for serine protease (22). Using similar principles, we improved the binding constant and increased the crystallizability of the BT·SGTI complex by a phage display of SGTI (20). As a result, we obtained crystals of the complex between bovine trypsin and the evolutionary enhanced SGTI (SGPI-1-PO-2, the Schistocerca gregaria protease inhibitor 1 with T5E, N18R, T20G, P21S, T22D, and K31M mutations) diffracting to 0.93 Å resolution. Here we present the crystal structure that revealed the detailed geometry of the active site and indicated the protonation state of catalytic residues and is supported by quantum chemical interpretation.
EXPERIMENTAL PROCEDURES
Protein Complex Preparation and Crystallization
Bovine trypsin was purchased from Sigma-Aldrich and was purified by gel filtration chromatography. SGPI-1-PO-2 was synthesized, oxidized, and purified as described (21). Solutions of bovine trypsin and SGPI-1-PO-2 were mixed in a 1:2.5 molar ratio. The BT·SGPI-1-PO-2 complex was crystallized with the hanging drop vapor diffusion method. Equal amounts of protein solution (9.1 mg/ml protein complex in 0.5 mm MES, pH 6.0 buffer) and precipitant solution (30% PEG 4000, 0.3 m ammonium acetate, 0.1 m sodium acetate, pH 4.6) were mixed and equilibrated against 0.5 ml of precipitant solution. Crystals of approximately 0.5 × 0.5 × 0.2-mm size were grown in 1 week. The crystals were soaked in precipitant solutions containing 20% glycerol and flash-cooled to 100 K prior to data collection.
X-ray Data Collection and Analysis
X-ray data were collected at Beamline ID14–1 at the European Synchrotron Radiation Facility. The high resolution data set (resolution range, 2.5–0.93 Å) consisted of 180 images with 1° oscillation and 1-s exposure at full transmission. This corresponds to an absorbed dose of ∼1.5 × 106 Gy. The low resolution data set (resolution range, 43.8–1.95 Å) was collected at separate positions on the crystal using 5.8% attenuation 180 images with 1° oscillation and 1-s exposure. The total absorbed dose in the low resolution pass was estimated to be 8.8 × 104 Gy. The absorbed dose was estimated with the program RADDOSE (23).
Crystallographic intensities were integrated and scaled to a resolution of 0.93 Å using XDS and XSCALE (24). Completeness of the data was 91.3% after merging the low and high resolution data sets. 2037 unique reflections were randomly selected as the Rfree set.
The structure was solved by molecular replacement using the program PHASER (25) of the Collaborative Computing Project 4 (26). As a search model, the structure of bovine trypsin was used, which was derived from Protein Data Bank (27) entry 1K1J (28). Molecular replacement revealed that the asymmetric unit contains one trypsin-inhibitor 1:1 complex. From here automated model building was carried out with Arp/wArp (29) using amino acid sequences of both SGPI-1-PO-2 and bovine trypsin. Arp/wArp docked 239 amino acid residues into the sequence corresponding to 99% sequence coverage. After the addition of 459 water molecules, automated model building was completed with r = 21.5% and Rfree = 23.4%. The model was then systematically improved using iterative cycles of manual rebuilding with the program Coot (30) and restrained least squares refinement with SHELXL (31). For the refinement in SHELX, the diffraction intensities were used as targets. Atomic B factors were refined anisotropically, and this step reduced the R factor and Rfree by 5.5 and 4.4%, respectively.
All except for hydroxyl oxygen and His Nϵ2 and Nδ1 atoms riding hydrogens were added to the structure. Finally, at the methyl, amino, and hydroxyl hydrogens, which were supported by clear mFobs − DFcalc density at higher than 2.5 σ, a riding model with torsional freedom was introduced (with the command AFIX 137, 138, 147, and 148). The scissile peptide bond of the inhibitor and residues 57, 102, 189, 193–195, and 214 of the enzyme were not geometrically restrained in the final rounds of the refinement. The final model contains residues 16–245 (chymotrypsin numbering system) (32) of bovine trypsin and residues I2-I34 of SGPI-1-PO-2. The stereochemistry of the structure was assessed with WHATCHECK and PROCHECK (33). The distribution of anisotropic B factors was monitored with the program PARVATI (34). The data collection and refinement statistics are shown in Table 1.
TABLE 1.
Crystallographic data and refinement statistics
| Resolution (Å)a | 43.8-0.93 (0.98-0.93) |
| Space group, unit cell parameters | P21, a = 36.9 Å, b = 63.6 Å, c = 43.9 Å, β = 93.8° |
| Total absorbed dose (Gy) | 1.6 × 106 |
| Number of unique reflectionsa | 123,475 (10,579) |
| Completeness (%)a | 91.3 (54.0) |
| Multiplicitya | 3.4 (1.8) |
| <I/σ>a | 17.9 (2.2) |
| Rsym (%)a,b | 3.8 (34.2) |
| R/Rfree (%)c | 11.6/14.0 |
| Root mean square deviation from ideal bond length (Å) and angles (°) | 0.018/2.23 |
| No. of modeled non-hydrogen atom positions (protein/waters/hetero) | 1958/350/5 |
| Ramachandran plot (% by PROCHECK) | |
| Most favored | 92.0 |
| Additionally allowed | 7.5 |
| Generously allowed | 0.4 |
| Disallowed | 0.1 |
| Average B factors (Å2) | |
| All atoms | 10.5 ± 8.6 |
| Protein | 8.3 ± 5.6 |
| Waters | 23.0 ± 11.3 |
| Anisotropy | |
| All atoms | 0.37 ± 0.13 |
| Protein | 0.38 ± 0.13 |
| Waters | 0.29 ± 0.14 |
a The values in parentheses indicate statistics for the highest resolution shell.
b Rsym = Σ||Io − <I>|/Σ Io × 100%, where Io is the observed intensity of a reflection and <I> is the average intensity obtained from multiple observations of symmetry related reflections.
c R factor = Σ ||Fobs| − |Fcalc||/Σ |Fobs| × 100%.
DFT and Continuum Electrostatic Calculations
The DFT calculations were performed using the program PC-GAMESS/Firefly (35). The B3LYP functional was employed with the 6-31G** basis set for the model optimization and calculating the optimized theoretical electron density. pKa values of Asp-102 and His-57 in the BT·SGPI-1-PO-2 complex were calculated with the program suite MEAD (36). Other titratable side chains were assigned with the program PROPKA (37), assuming a pH of 7. The atoms of the protein were charged by the program PDB 2PQR (38) using the AMBER force field. The calcium ion was also included in the model with a point charge of +2, but the water molecules were not modeled explicitly. The dielectric constant inside the protein was defined as ϵ = 10, and the solvent was modeled with an ϵ of 80. Electron density and structural models were visualized with Pymol (39), Molden (40), povscript+ (41), and Povray.
RESULTS AND DISCUSSION
Optimization of protein contact surfaces often leads to improved binding and less dynamic complexes. To this aim, a phage display assay selected a randomized set of SGTI mutants that showed improved binding to bovine trypsin (20). SGTI in its wild-type form is a poor binder to BT (21). A unique combination of mutations (T5E, N18R, T20G, P21S, T22D, and K31M) improved the inhibitory constant to the picomolar range (20), which made it possible to crystallize the complex formed between the optimized mutant inhibitor (SGPI-1-PO-2) and BT and solve its x-ray crystal structure.
Overall Description of the Structure
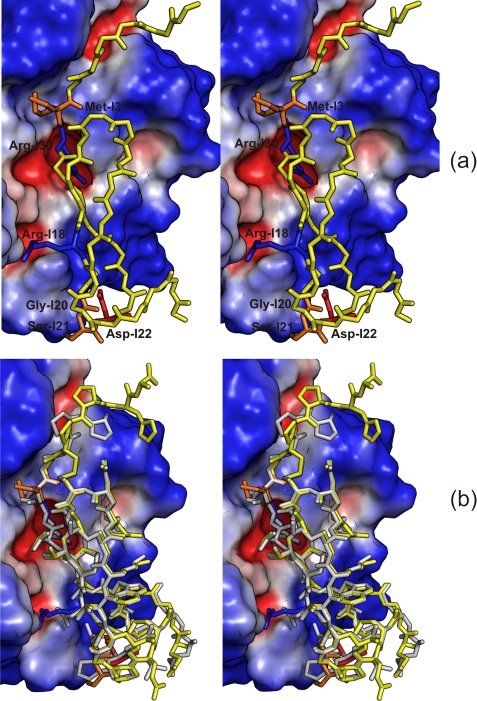
Data sets to 0.93 Å resolution were collected from crystals of BT·SGPI-1-PO-2 complex using a total absorbed x-ray dose of ∼7.6% of the Henderson limit (42). The final model has R and Rfree factors of 11.7 and 13.9%, respectively, and the data and refinement statistics are summarized in Table 1. The binding of SGPI-1-PO-2 to bovine trypsin is remarkably similar to the binding mode observed in the CFT3·SGTI complex (43, 44) (Fig. 1). Main differences are on the level of side chains, and the main chain is almost identical to SGTI with an root mean square deviation of 0.72 Å for all 32 Cα atoms and only 0.41 Å, when considering the Cα atoms of residues I4-I32 core of the inhibitor, which is flanked by rigid disulfide bridges. The mutations enhance the electrostatic complementary between the strongly cationic bovine trypsin and the insect inhibitor (Fig. 1).
FIGURE 1.
Stereo diagram showing SGPI-1-PO-2 bound to the surface of bovine cationic trypsin colored by the electrostatic potential (red negative, blue positive). a, the Glu-I5, Arg-I18, Gly-I20, Ser-I21, Asp-I22, Arg-I30, and Met-I31 residues of the inhibitor are highlighted. Basic residues (Arg-I18 and Arg-I30) and acidic residue Asp-I22 are marked as blue and red, respectively, whereas neutral residues Gly-I20, Ser-I21, and Met-I31 are colored orange. For other amino acid residues of the inhibitor, only the backbone is displayed (yellow). It is evident that the mutations N18R, T22D, and K31M improve the complementarity to the surface charges of bovine cationic trypsin. b, SGPI-1-PO-2 (yellow) and SGTI (gray) in the enzyme-bound form (43) are superimposed and reveal a similar fold. The mutation sites are highlighted with the same colors as in a.
As a result of the high resolution, electron density of hydrogen atoms appeared in the mFobs − DFcalc difference Fourier maps. In the final mFobs − DFcalc maps calculated with the riding hydrogen atoms omitted, 824 difference Fourier peaks above 2.5 σ appeared within 0.5 Å of the expected hydrogen positions (of a potential 1845). The visibility of hydrogen classes vary; for example, only 36 are visible of all 93 aromatic hydrogens, whereas of 245 amid hydrogens, 171 are visible, and of the remaining 1510 hydrogen atoms, 617 are visible. In general, hydrogen atoms with rotational freedom (methyl and hydroxyl) are less frequently observed above 2.5 σ in the difference Fourier maps, unless those hydrogen atoms are locked by steric constraints or in the case of hydroxyl hydrogens stabilized by hydrogen bonds.
Electron Density of Asp-102
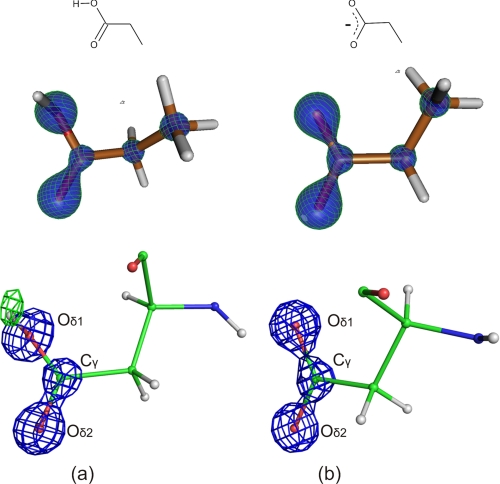
In addition to the electrons immediately surrounding the hydrogen atom that appear in the mFobs − DFcalc maps, the molecular orbitals also rearrange in the carboxyl group, which facilitates the identification of protonation state of aspartate and glutamate residues. If the proton is bound to the carboxyl group, the delocalized symmetric electron density of the deprotonated carboxylate ion is perturbed such that the π electrons localize at higher probability at one of the carbonyl bonds, which can be recognized as a continuous density between the carbonyl carbon and oxygen opposite to the hydroxyl group (15). In the BT·SGPI-1-PO-2 complex, aspartate residues (Asp-102 and Asp-189) are well ordered and occupy a single position, and in the case of Asp-102 mFobs − DFcalc, difference Fourier peaks of more than 2.5 σ are visible where hydroxyl hydrogens are poised to appear. Bond length difference between the two O-C pairs is 0.07 ± 0.02 Å when isotropic displacement parameters are used. This is accompanied by an asymmetric distribution of π orbitals of the carboxylate group (Fig. 2a; see also supplemental Movie S1). The hourglass-shaped continuous electron density of the carbonyl group (in the 4.2–5.2 σ range of 2mFobs − DFcalc map) is in stark contrast to the isolated density peak of a putative hydroxyl group. On the other hand, the carboxyl group of Asp-189 (Fig. 2b) displays symmetric electron distribution, which argues for its deprotonated form in the salt bridge to the P1 arginine residue.
FIGURE 2.
Comparison of the theoretical protonated and deprotonated carboxyl groups to experimental electron density in the crystal structure at 0.93-Å resolution. a, electron density of Asp-102 compared with theoretical protonated propionic acid. In the DFT calculation, the H-O-C-C torsional angle was constrained to 109° to match the putative hydrogen position observed in the mFobs − DFcalc electron density. b, electron density of Asp-189 compared with theoretical deprotonated propionic acid. Theoretical electron densities of propionic acid are contoured at 2.35 e−/Å3, whereas the experimental 2mFobs − DFcalc electron density map (blue) is contoured at 3.01 e−/Å3 (4.5 σ), and the positive mFobs − DFcalc density map (green) is contoured at 0.22 e−/Å3 (2.5 σ).
In addition to the protonation state, electron density is also modified by the environment of the side chains. This may lead to a shift of electron density in the absence of full proton transfer. As an attempt to qualitatively rationalize the electron density observed at Asp-102, the experimental coordinates of the simplified active site were optimized using DFT in the program PC-GAMESS/Firefly (see supplemental materials). Within the limits of the applied theory, only protonation at the Oδ1 equivalent atom of the carboxyl group introduced the experimentally observed electron density distortion.
Most crystal structures solved at subatomic resolution so far revealed symmetric (deprotonated) carboxyl group in the catalytic triad of free serine proteases (16, 45–47). There is a notable exception, however, when the electron distribution of the carboxylate group appears to be asymmetric, suggesting a protonated side chain even if the location of hydroxyl hydrogen could not be identified; in the 0.78 Å resolution structure of subtilisin, the Asp 32 Cγ-Oδ2 bond at 4 σ is continuous, whereas the density for Oδ1 is isolated (Fig. 3 in Ref. 48). Subtilisin was crystallized at pH 5.9, 1.4 units higher than the crystal reported here. Therefore, it is not inconceivable that serine proteases have a neutral aspartate in their active site close to the neutral pH.
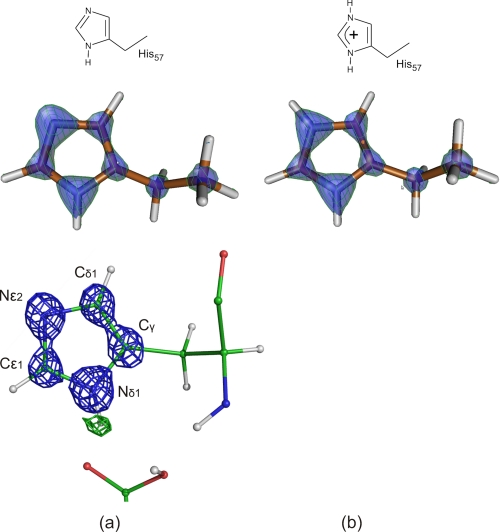
FIGURE 3.
Alternative protonation states of the imidazole ring as calculated by density functional theory and experimentally observed at residue His-57 in the electron density maps at 0.93-Å resolution. The DFT electron density of 4-ethyl-imidazole is contoured at 2.01 e−/Å3, whereas the 2mFobs − DFcalc (blue) and mFobs − DFcalc (green) electron density maps at 2.34 e−/Å3 (3.5 σ) and 0.26 e−/Å3 (3.0 σ), respectively. The experimentally observed electron density is more similar to an unprotonated imidazole ring.
His-57 in the Enzyme Inhibitor Complex
Most discussion in the literature focuses on the nature of the hydrogen bond between Asp-102 and His-57. In the BT·SGPI-1-PO-2 complex, a strong mFobs − DFcalc peak appears at 0.94-Å distance from the Nδ1 atom of His-57 side chain and 1.95 Å from Oδ2 atom of Asp-102. The peak does not lie on the line connecting Asp-102 Oδ2 and His-57 Nδ1 atom; rather it is displaced toward Oδ1 atom of Asp-102 (2.55 Å). The Nδ1-H bond distance is in good accordance with other aromatic bond distances unlike in the 0.78-Å subtilisin structure (48), and the distance from the hydrogen bond acceptor is also longer. The Oδ2–H-Nδ1 angle is 137.8°, much sharper than 180° because of the displacement of the hydrogen atom, which may participate in a slightly bifurcated hydrogen bond involving Oδ1 as well. Other aromatic hydrogens on His-57 did not appear in the difference Fourier maps, but the electron density of the side chain reveals the location of the double bonds (Fig. 3). The distribution of resonance forms in the neutral and the protonated states of the histidine side chain are different. Both states are aromatic; however, the distribution of the π electrons are skewed toward the Cϵ1–Nϵ2 bond in the neutral form, whereas it is more symmetrically distributed between the Cϵ1–Nϵ2 and Cϵ1–Nδ1 bonds when both nitrogens are protonated. Fig. 3b shows the simulated electron density of protonated and unprotonated histidine, and Fig. 3c shows the experimentally determined electron density. At a 3.5-σ level, the 2mFobs − DFcalc electron density map appears to be asymmetric, which indicates that His-57 is predominantly unprotonated. The putative Cϵ–H–O bond between the carbonyl oxygen of Ser-214 and the side chain of His-57 was also suggested to influence the tautomeric forms of the histidine (49). The donor acceptor bond length is 3.25 ± 0.02 Å, which indicates that the hydrogen bond character is only weakly present in the BT·SGPI-1-PO-2 complex.
The histidine has a pKa of 7 in the free active site of trypsin (9). This value reportedly varies according to the experimental conditions as judged by the direct observation of histidine electron density at 100 K and residual trypsin activity at pH 5 at room temperature (16). The histidine may be preferentially in the deprotonated form in the enzyme·substrate complex because of steric or electrostatic reasons. In this case, the protonation state of the histidine is unaffected by the bulk solvent in the Michaelis complex, and it becomes the only suitable proton acceptor for the serine hydroxyl proton when the substrate approaches the serine and a tetrahedral intermediate forms.
Geometry and Electron Density of the Scissile Peptide Bond
The scissile peptide bond appears to be perfectly planar where the deviation of Cα, N, and carbonyl O and C atoms from planarity is only 0.004 Å. The serine hydroxyl group is positioned perpendicularly to the plane of the scissile bond. The hydroxyl group and the carbonyl carbon of the scissile peptide are much closer than their combined van der Waal's distance (2.64 ± 0.01 Å). This unusually short interatomic distance may be interpreted as a partial, stretched covalent bond or a repulsive van der Waal's interaction. On the other hand a covalent bond would imply: 1) the presence of bonding electrons and corresponding electron density between the bound atoms or 2) distortion of the scissile peptide bond from planarity as the carbonyl carbon assumes an sp3 character. Neither of these possibilities can be confirmed from the crystal structure. Therefore the interaction appears to be a repulsive one, before the traversal of the activation barrier leading to the formation of a covalent bond. This repulsive interaction is countered by the hydrogen bonding and electrostatic interactions between the enzyme and the inhibitor, yet other interactions within the inhibitor hold back the scissile peptide bond (44). This deadlock is occasionally broken by the dynamics of the enzyme·inhibitor complex.
Implication of a Protonated Catalytic Aspartate Residue
Following the pioneering x-ray diffraction studies of serine proteases the charge relay proposal (2) was put forward to explain the role of the buried aspartate residue in the active site. This proposal by Blow et al. (2) speculated that the proton extracted from the catalytic serine or water molecule is transferred to the catalytic histidine, which in turn donates its proton to the negatively charged aspartate. In other words, the negative charge of the aspartate is ultimately transferred to the oxyanion of the tetrahedral intermediate through the catalytic triad. NMR experiments and other evidence argued against the proposal assuming that the buried aspartate is negatively charged at pH higher than 3. It is important to mention that small molecular models (13) are the main source of experimental information on the pKa of the carboxyl group, even though they lack the specific environment of the serine protease active site. The technically very challenging direct titration of Asp-102 showed a pKa of 6.8 (50). Early proton inventory experiments that followed the catalytic reaction on extended substrates implied two protons being transferred at each catalytic cycle (51). The BT·SGPI-1-PO-2 complex was crystallized at pH 4.6. The pH of the crystallization buffer is negatively correlated with the temperature by changing 0.015 ± 0.001 pH units/K in the 282–296 K range (see supplemental materials). Prototropic proton mobility has been observed even at cryogenic temperatures (52). Extrapolating the temperature dependence to 100 K, the buffer alone raises the pH of the crystal by ∼2.8 ± 0.1 units up to ∼7.5 ± 0.2. Protonated Asp-102 may mean significantly higher proton affinity in the enzyme·inhibitor complex compared with that observed in the small molecule model (12), at least at low temperature, which can partly be explained by the buried nature of the residue. Simultaneously His-57 is observed in its deprotonated form, suggesting that the active site may be catalytically competent.
Thurlkill et al. (53) estimated that the pKa of a buried carboxylate groups can be as high as 8.8. The surrounding hydrogen bonds and the microenvironment of Asp-102 appear to have an influence on its protonation state as theoretical calculations already suggested (54). Asp-102 most likely does not have a well defined proton affinity throughout the entire catalytic cycle. The assumption of invariable pKa of serine protease catalytic residues is at odds with mechanisms in other proteins, for example bacteriorhodopsin (55) and bacterial photosynthetic reaction center (56) where one or more residues change proton affinity during the course of catalytic cycle. The driving force of pKa change is often the introduction of a new charge close to the residue in question or a conformational change that exposes the side chain to a new environment for example by changing its solvent accessibility. In serine proteases, His-57 becomes positively charged in conjunction with the nucleophilic attacks, and the solvent accessibility of Asp-102 changes between the native and substrate-bound form of the enzyme. As indirect evidence, the catalytic aspartate appears to be very sensitive to its microenvironment because other low pH ultrahigh resolution crystal structures (as low as pH 5) (45–47, 57) do not show signs of protonation.
Suggested Interplay between the Catalytic Residues
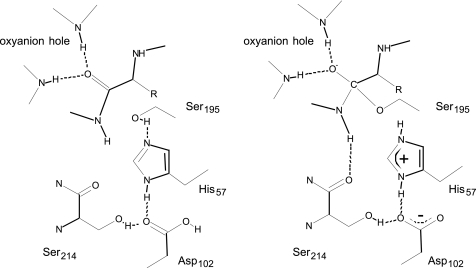
Although the presence of a protonated Asp-102 appears to support the charge relay mechanism (2), there is an important distinction: the hydrogen of the Asp-His hydrogen bond is located closer to the histidine and does not appear to be transferred to the aspartate. Moreover, the changes in the protonation state of the histidine and aspartate do not need to be coupled by direct proton transfer via the connecting hydrogen bond as illustrated in Fig. 4. Instead the proton may be transferred from aspartate to the buried group of water molecules of the active site (Wat-201 and Wat-202), and independently, the histidine becomes protonated either from the solvent in the free enzyme as observed by NMR titration experiments (8, 9) or from Ser-195 in the Michaelis complex. However, this mechanism does not exclude changes in the strength of the Asp-His hydrogen bond, depending on the protonation state of Asp-102 and His-57 (12, 58).
FIGURE 4.
Schematic representation of the proposed catalytic mechanism. Left, Michaelis complex with both His-57 and Asp-102 in the neutral state. Right, the tetrahedral intermediate state when both His-57 and Asp-102 becomes charged.
The transfer of an ion from bulk water (ϵ = 80) to a nonpolar medium with a dielectric constant of 4–10 (interior of proteins) costs significant energy, as follows from Born equation (59). However, residues can remain charged even in the nonpolar environment if they are involved in intramolecular ion pairs. Such ion pairs are frequently observed between stronger acids (glutamates and aspartates) and bases (lysines and arginines) even in the hydrophobic core of the proteins. However, histidine is a relatively weak base; therefore an aspartate-histidine pair can exist in a mutually neutral form over a wider range of pH while buried in the substrate·enzyme complex. His-57 is particularly resistant to protonation when titrated in the enzyme inhibitor complex (9). Therefore Asp-102 in our view acts as part of an inducible solvation layer around protonated and buried His-57. In this mechanism, protonation of the histidine triggers deprotonation of Asp-102 and the formation of the ion pair. To explore this scenario, we performed a continuum electrostatic calculation using MEAD (36) to determine how the introduction of a positive charge on His-57 changes the pKa of Asp-102 in the BT·SGPI-1-PO-2 complex. The calculation shows that the protonation of His-57 induces a negative shift of 3.8 pKa units for Asp-102 (assuming an ϵ = 10 inside the protein). It means that even in a buried position Asp-102 is more likely to deprotonate when His-57 is positively charged. The indirect indication that the stabilization of His-57 is of primarily electrostatic origin comes from mutagenesis studies showing that the exchange of the side chains of residues Asp-102 and Ser-214 still provide acceptable catalytic efficiency (60).
During the catalytic cycle, His-57 becomes protonated, and in that case the pKa of Asp-102 drops. Even if Asp-102 is in the protonated form and the drop in pKa is sufficiently large, a proton acceptor with higher pKa is still required. In the crystal structure, the most likely acceptor of its proton is a buried water molecule that is coordinated by Ser-214, another well conserved part of the active site. In the protein structure, the side chain of Ser-214 is in hydrogen bonding distance to Asp-102 and also through its carbonyl oxygen backbone with the substrate in the acyl enzyme and presumably in the tetrahedral intermediate state; therefore it may be able to assist the synchronization of the Asp-102 protonation state with the position of the substrate on the reaction coordinate. Despite the strong evolutionary conservation of Ser-214, the S214A mutation actually increases the catalytic efficiency of the enzyme on small synthetic substrates at the expense of slightly increased Km (61). Again, because proteases have evolved to act on proteins or large peptides, using small synthetic substrates as model systems might lead to misleading results.
For this mechanism to work, the pH has to be near or below the resting state pKa of Asp-102 in the Michaelis complex. If the pH is high enough, nucleophilic catalysis is favored already, partly because the histidine is predominantly in the neutral form irrespective of the negatively charged Asp-102. On the other hand, enzymes are often forced to operate outside their catalytic optimum. For example, they can usually perform better at higher temperature than the temperature inside the host organism according to the Arrhenius equation. The optimal pH might as well be far from the actual pH at which the enzyme operates. Concerning intestinal serine proteases, the pH of their environment seldom increases above 7.5 units in the entire intestinal system in a healthy individual (62, 63), and the enzymes acting in the intestine had to adapt to this environment. At the mildly acidic or neutral pH level (pH 5.5–7) typically present in the duodenum, where the strongly acidic content of the stomach mixes with the pancreatic juice, the presence of an aspartate in the active site provides a compelling example of adaptation. This residue in its neutral form supports the presence of an unprotonated histidine and therefore primes it for being a good proton acceptor in the enzyme·substrate complex. When the histidine does become protonated in the tetrahedral intermediate state, the aspartate can still provide stabilization by becoming negatively charged.
Interactions between Enzyme and Inhibitor and the Result of Directed Evolution
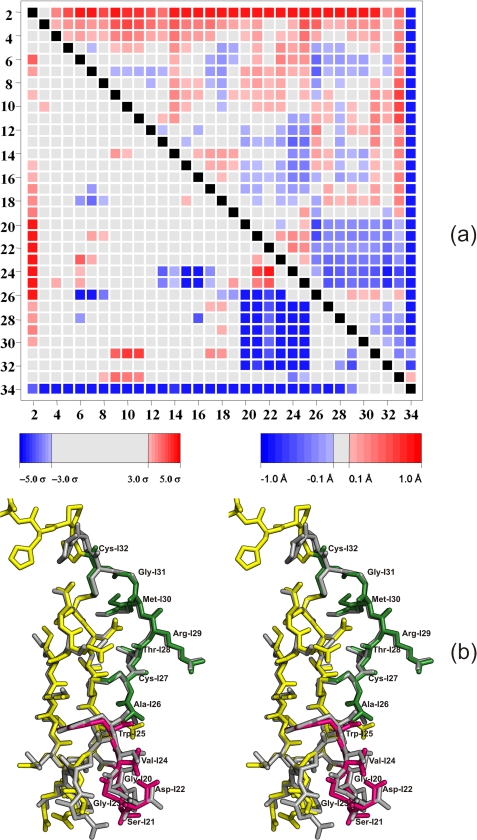
Compared with SGTI, its variant SGPI-1-PO-2 derived by directed evolution exhibits highly improved inhibitory efficiency on BT with a Ki value in the 10−12 m range (20). The comparison of binding strength suggested that the mutations contribute a number of favorable interactions between SGPI-1-PO-2 and BT. The structure of the mutant inhibitor complexed with BT helped to identify such potential interactions and revealed important clues of the inhibitor action. The mutant and wild-type inhibitor scaffold in different trypsin complexes superposes with an root mean square deviation of 0.41 Å when considering the Cα atoms of residues I4-I32 core. No significant van der Waal's clashes are observed in the interface region of the two enzyme inhibitor complex. Using an unbiased ESCET comparison (64), the most pronounced, significant differences in the structures apart from the C- and N-terminal residues are located between residues I20-I25 and I26–32 (Fig. 5a). It is important to note that four of the six mutations are located in these two regions, which constitute approximately one-third of the inhibitor. It appears that these two regions, which are part of the antiparallel β-sheet interacting with the enzyme, slide toward one another in the BT-bound SGPI-1-PO-2. The conformational change that allows this shift is focused in the I25-I27 region where the backbone changes more than 25° in the ψ angle in contrast to the core residues (I4-I30) of the structure where the deviation is 7.7 ± 7.8° on average. The most significant backbone movement is observed at Trp-I25, which forms a new hydrogen bond with Ser-217. In BT, Tyr at the same position does not have a suitable hydrogen bond donor for the wild-type inhibitor.
FIGURE 5.
a, ESCET structural comparison of SGTI and SGPI-1-PO-2. Blue in the matrix marks Cα atoms that are closer to one another in the SGPI-1-PO-2 structure than in SGTI (44), whereas red represents Cα atom pairs that are further away from one another. Apart from the C and N termini, two extended regions (residues ranges I26-I32 and I20-I25) appear to be closer to one another in SGPI-1-PO-2. b, these regions are illustrated in a stereo diagram of the superpositioned SGTI and SGPI-1-PO-2 inhibitors. The coloring scheme follows Fig. 1 with SGPI-1-PO-2 (yellow) and SGTI (gray), but in addition regions I20-I25 and I26-I32 in SGPI-1-PO-2 are highlighted as magenta and green, respectively.
Intermolecular hydrogen bond donors of the inhibitor are equivalent in the CFT3·SGTI and BT·SGPI-1-PO-2 complexes and mainly restricted to the substrate-enzyme β-sheet and the P1 side chain. The length of these hydrogen bonds is identical within experimental error. On the other hand, there is a marked difference between the hydrogen bond acceptors in the two inhibitors. In addition to the four conserved hydrogen bond acceptors, the BT·SGPI-1-PO-2 complex has a strong hydrogen bond and ionic interaction between Asp-I22 and Lys-224. There is no equivalent interaction between Thr-I22 and Tyr-224 in the CFT3·SGTI complex because of the large distance and the lack of electrostatic complementarily. The hydrogen bond between the backbone oxygen of Trp-I25 (Fig. 5b) and the hydroxyl group of Ser-217 also contribute to the stability of the complex. In principle, this hydrogen bond may form between BT and wild-type SGTI, even though the binding strength of the complex is substantially weaker.
The mutations and the binding to BT instead of CFT3 also change the intramolecular hydrogen bond pattern of SGPI-1-PO-2 as compared with wild-type SGTI. In the BT·SGPI-1-PO-2 complex SGPI-1-PO-2 has in total 24 intramolecular hydrogen bonds, whereas in the CFT3·SGTI complex there are only 21. In addition the strength of the hydrogen bonds also changes substantially. The largest shortening (0.45 Å) is observed between the side chain of Glu-I3 and amid nitrogen Trp-I25. This hydrogen bond appears to contribute to the change in the backbone conformation in amino acid residues I25-I27. Other notable hydrogen bond shortening occurs between the amid nitrogen of residue I20 and the carbonyl group of Val-I23 (0.17 Å). This is one of the consequences of the three mutations (T20G, P21S, and T22D) in the turn between β-strands. As a result, the β-turn becomes sharper, and Val-I23 tugs the β-strand toward itself, again contributing to the convergence of regions I20-I25 and I26-I32.
Electrostatic complementarity is also an important contributing factor to the increased affinity of SGPI-1-PO-2 binding to BT (compared with wild type) (20). Four of the mutations potentially change the charge distribution of the inhibitor. T5E, T22D, and K31M mutations are favorable for binding to the cationic bovine trypsin, because they increase the negative net charge, whereas N18R is potentially destabilizing. Of the mutations, Asp-I22, Met-I31, and Arg-I18 are located in the interface region (Fig. 1a). Asp-I22, as discussed before, forms a strong salt bridge with Lys-224 and is probably the strongest contributor to the complex stability. Met-I31 replaces the wild-type Lys residue, which faces a strongly positive electrostatic surface (Fig. 1). The neutral residue at this position prevents a potentially destabilizing electrostatic repulsion. Arg-I18 is surrounded by neutral hydrophobic residues, and its Coulombic interaction is relatively weak. On the other hand its side chain forms a hydrogen bond with the aromatic π electrons of Trp-215, which probably offsets the potentially destabilizing Coulombic interaction with the cationic bovine trypsin.
Conclusion
Serine proteases continue to be an inspiration for mechanistic studies of enzyme catalysis. Using directed evolution, we enhanced the inhibitory power and binding strength of SGTI, a naturally occurring trypsin inhibitor. We solved the ultrahigh resolution crystal structure of its complex with bovine trypsin, which led to the unique observation of potentially protonated Asp-102 residue in a protease·inhibitor complex. Canonical inhibitors in complex with serine proteases still provide the best model for the enzyme·substrate (Michaelis) complex, even if they are hydrolyzed tantalizingly slowly. Although the mechanistic implication may require further investigations, we found that the temporary, synchronized deprotonation of Asp-102 may contribute to the stabilization of protonated His-57 and (indirectly) the tetrahedral intermediate by becoming negatively charged. In line with the original proposal by Blow et al. (2), we suggest that protonation states of the active site residues are coupled. In contrast to the charge relay mechanism, the mechanism presented here implies that a − + − charge distribution is responsible for the ultimate stabilization of the tetrahedral intermediate. The electrostatic environment is shown to account for major part of catalytic rate acceleration (65); therefore the comprehensive description of the protonation state of an enzyme is crucial at the pH, where the enzyme operates. We hope that this refined model will initiate further experimental and theoretical studies and fruitful discussions in the field of serine proteases.
Supplementary Material
Acknowledgments
We are grateful to Olivia Slater and the staff at Beamline ID14-1 at the European Synchrotron Radiation Facility for expert assistance. We also thank Katalin A. Kékesi for the mass spectrometry analysis of the chemically synthesized SGPI-1-PO-2 inhibitor.
This work was supported by the Swedish Science Research Council, the Harald and Greta Jeanssons Foundation, and OTKA Grants 68408 and 81950.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Movie S1, and Figs. S1–S3.
The atomic coordinates for the crystal structure of this protein are available in the Molecular Modeling Database under MMDB accession number 86047.
- SGTI
- S. gregaria protease (trypsin) inhibitor 1
- BT
- bovine trypsin
- CFT3
- crayfish trypsin isoform 3
- DFT
- density functional theory
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1. Matthews B. W., Sigler P. B., Henderson R., Blow D. M. (1967) Nature 214, 652–656 [DOI] [PubMed] [Google Scholar]
- 2. Blow D. M., Birktoft J. J., Hartley B. S. (1969) Nature 221, 337–340 [DOI] [PubMed] [Google Scholar]
- 3. Markley J. L., Ibañez I. B. (1978) Biochemistry 17, 4627–4640 [DOI] [PubMed] [Google Scholar]
- 4. Bachovchin W. W., Roberts J. D. (1978) J. Am. Chem. Soc. 100, 8041–8047 [Google Scholar]
- 5. Zakharova E., Horvath M. P., Goldenberg D. P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11034–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Finkenstadt W. R., Laskowski M., Jr. (1967) J. Biol. Chem. 242, 771–773 [PubMed] [Google Scholar]
- 7. Liu B., Schofield C. J., Wilmouth R. C. (2006) J. Biol. Chem. 281, 24024–24035 [DOI] [PubMed] [Google Scholar]
- 8. Robillard G., Shulman R. G. (1972) J. Mol. Biol. 71, 507–511 [DOI] [PubMed] [Google Scholar]
- 9. Markley J. L., Porubcan M. A. (1976) J. Mol. Biol. 102, 487–509 [DOI] [PubMed] [Google Scholar]
- 10. Sprang S., Standing T., Fletterick R. J., Stroud R. M., Finer-Moore J., Xuong N. H., Hamlin R., Rutter W. J., Craik C. S. (1987) Science 237, 905–909 [DOI] [PubMed] [Google Scholar]
- 11. Kraut J. (1977) Annu. Rev. Biochem. 46, 331–358 [DOI] [PubMed] [Google Scholar]
- 12. Frey P. A., Whitt S. A., Tobin J. B. (1994) Science 264, 1927–1930 [DOI] [PubMed] [Google Scholar]
- 13. Ash E. L., Sudmeier J. L., De Fabo E. C., Bachovchin W. W. (1997) Science 278, 1128–1132 [DOI] [PubMed] [Google Scholar]
- 14. Kossiakoff A. A., Spencer S. A. (1981) Biochemistry 20, 6462–6474 [DOI] [PubMed] [Google Scholar]
- 15. Ahmed H. U., Blakeley M. P., Cianci M., Cruickshank D. W., Hubbard J. A., Helliwell J. R. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 906–922 [DOI] [PubMed] [Google Scholar]
- 16. Schmidt A., Jelsch C., Ostergaard P., Rypniewski W., Lamzin V. S. (2003) J. Biol. Chem. 278, 43357–43362 [DOI] [PubMed] [Google Scholar]
- 17. Vrielink A., Sampson N. (2003) Curr. Opin. Struct. Biol. 13, 709–715 [DOI] [PubMed] [Google Scholar]
- 18. Burgess S. G., Messiha H. L., Katona G., Rigby S. E., Leys D., Scrutton N. S. (2008) Biochemistry 47, 5168–5181 [DOI] [PubMed] [Google Scholar]
- 19. Williams D. H., Stephens E., O'Brien D. P., Zhou M. (2004) Angew Chem. Int. Ed. Engl. 43, 6596–6616 [DOI] [PubMed] [Google Scholar]
- 20. Szenthe B., Patthy A., Gáspári Z., Kékesi A. K., Gráf L., Pál G. (2007) J. Mol. Biol. 370, 63–79 [DOI] [PubMed] [Google Scholar]
- 21. Malik Z., Amir S., Pál G., Buzás Z., Várallyay E., Antal J., Szilágyi Z., Vékey K., Asbóth B., Patthy A., Gráf L. (1999) Biochim. Biophys. Acta 1434, 143–150 [DOI] [PubMed] [Google Scholar]
- 22. Matthews D. J., Wells J. A. (1993) Science 260, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 23. Paithankar K. S., Owen R. L., Garman E. F. (2009) J. Synchrotron Radiat. 16, 152–162 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 26. Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dullweber F., Stubbs M. T., Musil D., Stürzebecher J., Klebe G. (2001) J. Mol. Biol. 313, 593–614 [DOI] [PubMed] [Google Scholar]
- 29. Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Sheldrick G. M., Schneider T. R. (1997) Methods Enzymol. 277, 319–343 [PubMed] [Google Scholar]
- 32. Shotton D. M., Hartley B. S. (1970) Nature 225, 802–806 [DOI] [PubMed] [Google Scholar]
- 33. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 34. Merritt E. A. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 35. Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S. J., Windus T. L., Dupuis M., Montgomery J. A. (1993) J. Comput. Chem. 14, 1347–1363 [Google Scholar]
- 36. You T. J., Bashford D. (1995) Biophys. J. 69, 1721–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li H., Robertson A. D., Jensen J. H. (2005) Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 38. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 40. Schaftenaar G., Noordik J. H. (2000) J. Comput. Aided Mol. Des. 14, 123–134 [DOI] [PubMed] [Google Scholar]
- 41. Fenn T. D., Ringe D., Petsko G. A. (2003) J. Appl. Crystallogr. 36, 944–947 [Google Scholar]
- 42. Henderson R. (1990) Proc. R. Soc. Lond. B241, 6–8 [Google Scholar]
- 43. Fodor K., Harmat V., Hetényi C., Kardos J., Antal J., Perczel A., Patthy A., Katona G., Gráf L. (2005) J. Mol. Biol. 350, 156–169 [DOI] [PubMed] [Google Scholar]
- 44. Fodor K., Harmat V., Neutze R., Szilágyi L., Gráf L., Katona G. (2006) Biochemistry 45, 2114–2121 [DOI] [PubMed] [Google Scholar]
- 45. Fuhrmann C. N., Kelch B. A., Ota N., Agard D. A. (2004) J. Mol. Biol. 338, 999–1013 [DOI] [PubMed] [Google Scholar]
- 46. Katona G., Wilmouth R. C., Wright P. A., Berglund G. I., Hajdu J., Neutze R., Schofield C. J. (2002) J. Biol. Chem. 277, 21962–21970 [DOI] [PubMed] [Google Scholar]
- 47. Wurtele M., Hahn M., Hilpert K., Hohne W. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 520–523 [DOI] [PubMed] [Google Scholar]
- 48. Kuhn P., Knapp M., Soltis S. M., Ganshaw G., Thoene M., Bott R. (1998) Biochemistry 37, 13446–13452 [DOI] [PubMed] [Google Scholar]
- 49. Derewenda Z. S., Derewenda U., Kobos P. M. (1994) J. Mol. Biol. 241, 83–93 [DOI] [PubMed] [Google Scholar]
- 50. Koeppe R. E., 2nd, Stroud R. M. (1976) Biochemistry 15, 3450–3458 [DOI] [PubMed] [Google Scholar]
- 51. Elrod J. P., Hogg J. L., Quinn D. M., Venkatasubban K. S., Schowen R. L. (1980) J. Am. Chem. Soc. 102, 3917–3922 [Google Scholar]
- 52. Uritski A., Presiado I., Erez Y., Gepshtein R., Huppert D. (2009) J. Phys. Chem. C 113, 10285–10296 [Google Scholar]
- 53. Thurlkill R. L., Grimsley G. R., Scholtz J. M., Pace C. N. (2006) J. Mol. Biol. 362, 594–604 [DOI] [PubMed] [Google Scholar]
- 54. Warshel A., Naray-Szabo G., Sussman F., Hwang J. K. (1989) Biochemistry 28, 3629–3637 [DOI] [PubMed] [Google Scholar]
- 55. Neutze R., Pebay-Peyroula E., Edman K., Royant A., Navarro J., Landau E. M. (2002) Biochim. Biophys. Acta 1565, 144–167 [DOI] [PubMed] [Google Scholar]
- 56. Wöhri A. B., Katona G., Johansson L. C., Fritz E., Malmerberg E., Andersson M., Vincent J., Eklund M., Cammarata M., Wulff M., Davidsson J., Groenhof G., Neutze R. (2010) Science 328, 630–633 [DOI] [PubMed] [Google Scholar]
- 57. Radisky E. S., Lee J. M., Lu C. J., Koshland D. E., Jr. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6835–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Frey P. A. (2004) J. Phys. Org. Chem. 17, 511–520 [Google Scholar]
- 59. Born M. (1920) Z. Phys. 1, 45–48 [Google Scholar]
- 60. Corey D. R., Mcgrath M. E., Vasquez J. R., Fletterick R. J., Craik C. S. (1992) J. Am. Chem. Soc. 114, 4905–4907 [Google Scholar]
- 61. McGrath M. E., Vásquez J. R., Craik C. S., Yang A. S., Honig B., Fletterick R. J. (1992) Biochemistry 31, 3059–3064 [DOI] [PubMed] [Google Scholar]
- 62. Maqbool S., Parkman H. P., Friedenberg F. K. (2009) Dig. Dis. Sci. 54, 2167–2174 [DOI] [PubMed] [Google Scholar]
- 63. Borgstrom B., Dahlqvist A., Lundh G., Sjovall J. (1957) J. Clin. Invest. 36, 1521–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schneider T. R. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 714–721 [DOI] [PubMed] [Google Scholar]
- 65. Warshel A. (1998) J. Biol. Chem. 273, 27035–27038 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.