Abstract
Protein function depends on conformational flexibility and folding stability. Loose packing of hydrophobic cores is not infrequent in proteins, as the enhanced flexibility likely contributes to their biological function. Here, using experimental and computational approaches, we show that eukaryotic pentameric ligand-gated ion channels are characterized by loose packing of their extracellular domain β-sandwich cores, and that loose packing contributes to their ability to rapidly switch from closed to open channel states in the presence of ligand. Functional analyses of GABAA receptors show that increasing the β-core packing disrupted GABA-mediated currents, with impaired GABA efficacy and slowed GABA current activation and desensitization. We propose that loose packing of the hydrophobic β-core developed as an evolutionary strategy aimed to facilitate the allosteric mechanisms of eukaryotic pentameric ligand-gated ion channels.
Keywords: Allosteric Regulation, GABA Receptors, Ion Channels, Nicotinic Acetylcholine Receptors, Protein Folding
Introduction
Proteins are conformationally dynamic and require thermodynamic stability and structural flexibility to function (1–5). Stability and flexibility are interdependent, with higher stability increasing structural rigidity, which in many cases reduces protein activity (6). Proteins are only marginally stable, with ΔGfolding values mostly between −3 and −15 kcal/mol (7). Trade-offs between thermodynamic stability and conformational flexibility are observed during protein evolution, with novel or specialized functions often acquired at the expense of stability (1, 2, 8, 9). New-function mutations, however, rarely involve active site residues (10, 11) but occur with high frequencies in hydrophobic cores (12). Core mutations, although, are highly demanding in terms of stability, because a well packed hydrophobic core is the major factor that drives protein folding and maintains a stably folded structure through entropic (“hydrophobic effect”) and enthalpic (van der Waals interactions) forces (13–16).
Protein cores are normally packed with amino acid residues with bulky hydrophobic side chains that efficiently fill the space (17, 18). Core mutations tend to loosen the packing and create “defects” (empty or water-filled cavities) that decrease thermodynamic stability up to several kcal/moles (3, 19–21). Numerous proteins (e.g. p53 (22), serpins (23), and hemagglutinin (24, 25)) have evolved localized core packing defects that enhance their biological activity at the cost of their stability.
Recently, the crystal structure of the extracellular domain of the nicotinic acetylcholine receptor (nAChR)3 α1 subunit revealed a hydrophilic water-filled cavity buried in the hydrophobic core of the protein and it was hypothesized that loose core packing is important for nAChR activity (26). nAChRs are members of the pentameric ligand-gated ion channel (pLGIC) superfamily, which includes γ-aminobutyric acid (GABAARs), serotonin (5-HT3Rs), and glycine (GlyRs) receptors. For these receptors, binding of neurotransmitter to the extracellular ligand-binding domain triggers rapid structural rearrangements (in the submillisecond to millisecond time frame) that ultimately lead to the opening of an intrinsic ion-conducting channel. Signaling in the brain and neuromuscular junction depends on their activity. pLGICs are composed of five identical or homologous subunits, each consisting of an extracellular ligand-binding domain (ECD) made of β-strands forming a β-sandwich core, a transmembrane ion pore-forming domain made of four α-helices, and a large cytoplasmic loop (27, 28). Recently, prokaryotic pLGICs were identified (29), and two were crystallized in presumed closed (Erwinia chrysanthemi ELIC) and open (Gloeobacter violaceus GLIC) channel states (30–33).
In the crystal structure of the nAChR-α1 ECD, two polar amino acids, Thr-52 and Ser-126, buried in the β-sandwich core coordinate an ordered water molecule (26) (Fig. 2). Mutation of these residues to bulky hydrophobic amino acids substantially reduced acetylcholine-activated channel current suggesting loose packing of the β-sandwich core is important for nAChR function (26). Molecular dynamics studies indicate that loose packing enhances the flexibility of nAChR-α1 ECD (34). In crystal structures of related non-channel acetylcholine-binding proteins (AChBPs), which are structural homologues of pLGICs ECDs (27), tight packing is observed with bulky valine/leucine and phenylalanine at aligned positions. Here, we examined the generality and importance of loose β-core packing for pLGIC function. Using computational approaches, we defined the residue composition of the β-cores in pLGICs and AChBPs and determined in silico how changes in packing affect their stability. Inserting less bulky or hydrophilic residues in AChBPs and GLIC destabilized their cores. Conversely, mutating nAChR-α1 Thr-52 and Ser-126 to bulkier hydrophobic residues stabilized its core. We show that loose packing of the ECD β-core is a distinctive feature of eukaryotic pLGICs. We also examined the functional consequences of altering the ECD β-core packing in related eukaryotic GABAARs. We show that increasing the β-core packing significantly decreased GABAAR activation. We argue that loose packing of eukaryotic pLGIC hydrophobic β-cores likely developed as an evolutionary strategy aimed to facilitate the allosteric transitions required for rapid switching between closed and open channel states in the presence of ligand.
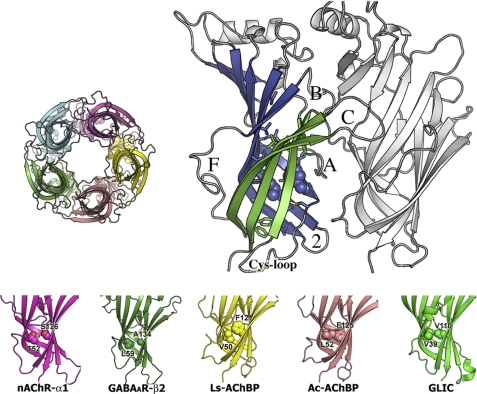
FIGURE 2.
Top, crystal structure of Ls-AChBP. Left, top view of pentameric Ls-AChBP. Right, side view of two subunits. In one subunit, outer sheet β-strands are colored green, inner sheet β-strands blue. Functionally relevant loops are labeled. Neighboring subunit at the minus-side of the ligand-binding interface is colored light gray. Hydrophobic core residues at sites 1 (Val-50) and 2 (Phe-121) are in space-filling mode. Bottom, AChBPs and pLGIC ECD structures, showing different amino acid packing at sites 1 and 2. GABAAR-β2 is a homology model based on Ac-AChBP.
MATERIALS AND METHODS
Computational Analysis
Sequences of pLGICs and AChBPs were aligned with ClustalW (35). Relative solvent accessible surface area of core-buried residues in the crystal structures of nAChR-α1 ECD (PDB code 2QC1), GLIC (3EAM), Lymnaea stagnalis Ls-AChBP (PDB code 1UV6), and Aplysia californica Ac-AChBP (PDB code 2BYN) was calculated with ASAView (36).
Thermodynamic stability changes of in silico mutants were computed based on the PDB files above using FOLDX (37). Because structural studies have extensively demonstrated that the pLGICs ECD or AChBP in monomeric form constitutes an independent folding unit (26, 27, 38), monomeric nAChR-α1 ECD and monomers of GLIC ECD and AChBPs were used. Briefly, each structure was first optimized using the repair function of FOLDX. Single amino acid mutations were then introduced with the repair position scan function. Finally, the energies (ΔGfolding) of the non-mutated and mutated structures were computed with the energy calculation function, and for each mutation the resulting ΔΔGfolding = ΔG(mutant) − ΔG(non-mutant) was obtained. The reported ΔΔGfolding values were then corrected using the normalization function ΔΔGexp = (ΔΔGFoldX + 0.078)/1.14 as described (7).
Site-directed Mutagenesis and Expression in Xenopus laevis Oocytes
Rat cDNAs encoding GABAAR subunits α1, β2, and γ2S were subcloned in pUNIV vector. Mutant GABAAR subunits were created using the QuikChange site-directed mutagenesis method (Stratagene). Capped cRNAs encoding WT and mutant GABAAR subunits were transcribed in vitro using the mMessage mMachine T7 kit (Ambion). Single X. laevis oocytes were injected with 27 nl of cRNA (2–20 ng/μl/subunit) in a 1:1:10 (α1:β2:γ2S) ratio. Injected oocytes were incubated at 16 °C in ND96 (5 mm HEPES, pH 7.2, 96 mm NaCl, 2 mm KCl, 1 mm MgCl2, 1.8 mm CaCl2) supplemented with 100 μg/ml of gentamycin and 100 μg/ml of bovine serum albumin for 2–5 days before use for electrophysiological recordings.
Two-electrode Voltage Clamp
Oocytes were perfused continuously with ND96 at a flow rate of 5 ml/min, while being held under two-electrode voltage clamp at −80 mV in a bath volume of 200 μl. Borosilicate glass electrodes (Warner Instruments) used for recordings were filled with 3 m KCl and had resistances of 0.4 to 1.0 mΩ. Electrophysiological data were collected using Oocyte Clamp OC-725C (Warner Instruments) interfaced to a computer with an ITC-16 A/D device (Instrutech) and were recorded using the Whole Cell Program, version 4.0.9 (kindly provided by J. Dempster, University of Strathclyde, Glasgow, UK). Stock solutions of GABA (Sigma), pentobarbital (PB) (Sigma), and piperidine-4-sulfonic acid (P4S) (Sigma) were prepared fresh daily in ND96.
Concentration-Response Analysis
GABA, PB, and P4S concentration responses were scaled to a low, non-desensitizing concentration of agonist (EC2 to EC10) applied just before the test concentration to correct for any slow drift in current responsiveness over the course of the experiment. Currents elicited by each test concentration were normalized to the corresponding low concentration current before curve fitting. At high micromolar concentrations and above, PB blocks GABAAR. Relief of channel block upon drug wash yields a characteristic tail current. For PB concentration-response curves, currents measured at high micromolar concentrations and above included tail current measurements. For currents recorded on the same oocyte with saturating concentrations of GABA and PB or P4S, extensive washes (6–10 min) were performed before each application of the different drugs. GraphPad Prism 4 software was used for data analysis and fitting. Concentration-response data were fit to the equation: I = Imax/(1 + (EC50/[A])n), where I is the peak response to a given concentration of agonist, Imax is the maximum amplitude of current, [A] is the agonist concentration, and n is the Hill coefficient.
Statistical Analysis
Log(EC50) values for GABA and PB concentration responses, max current ratios, and GABA-elicited current activation and desensitization rates were analyzed using one-way analysis of variance, followed by a post hoc Dunnett's test to determine the level of significance between WT and mutant receptors. Natural logarithm (ln) transformed values of WT and mutant EC50 values were used for computing interaction free energies, such that ΔGmutant = RT × [ln(EC50mutant/EC50WT)], and the free energy changes due to interaction ΔΔGINT = RT × [ln(EC50WT) + ln(EC50mut1,mut2) − ln(EC50mut1) − ln(EC50mut2)], with propagated errors reported in S.E.
RESULTS
Packing Defects of Eukaryotic pLGICs ECD β-Cores
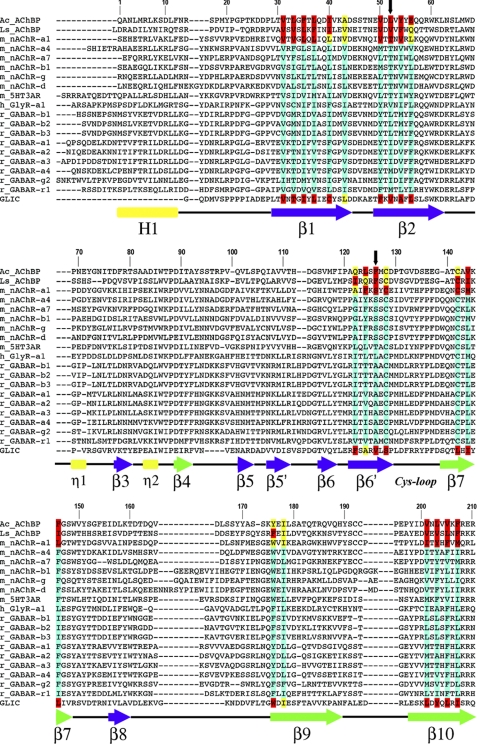
Using the available structures of the nAChR α1 subunit (26, 28), AChBPs (27, 39), and prokaryotic GLIC (32, 33), we analyzed and compared the amino acid composition of their ECD β-core buried residues (accessible surface area ≤5%) to identify possible differences in β-core packing. As expected, their β-cores are very similar and are largely formed by bulky hydrophobic amino acids (valine, leucine, isoleucine, methionine, phenylalanine, tyrosine, tryptophan) (Fig. 1, supplemental Figs. S1–S2, and supplemental Table S1). Notable exceptions are the two conserved polar residues corresponding to Thr-52 and Ser-126 in the nAChR α1 subunit (henceforth called sites 1 and 2, respectively, Fig. 1), which create a hydrophilic water filled cavity (26) and loosen the β-core packing (Fig. 2). AChBPs and GLIC have hydrophobic residues (valine, leucine, and phenylalanine) at these positions (Fig. 1).
FIGURE 1.
Sequence alignment of AChBPs and representative pLGIC ECDs. The core residues, in crystal structures, with accessible surface area (ASA) ≤5 and ≤25% are highlighted in red and yellow, respectively. Based on sequence alignments, putative core residues in other pLGICs are highlighted in turquoise. Black downward arrows mark the positions of core residues aligned with nAChR-α1 Thr-52 (site 1) and Ser-126 (site 2). Amino acid numbering refers to the mouse nAChR-α1 sequence. Secondary structure elements are indicated below the sequences and are labeled in Greek letters based on the crystal structure of nAChR-α1 ECD (26). β-Strands are blue (inner sheet) or green (outer sheet) arrows, helices are rectangles (the N-terminal α-helix is labeled in uppercase, the 310-helices in lowercase), and connecting loops are black lines.
We used FOLDX (37) to analyze and compare in silico the folding stability of the ECD β-core of nAChR-α1, GLIC, and AChBP. In particular, we aimed to determine how the loose β-core packing in nAChR affects its thermodynamic stability compared with AChBPs and GLIC. FOLDX estimates the energetic contribution of amino acids to the stability of a protein based on a computer algorithm that introduces a mutation at a given site and calculates the ΔΔGfolding of mutant relative to wild-type. Namely, each core-buried residue in nAChR-α1 (PDB code 2QC1), GLIC (PDB 3EAM), and AChBP (L. stagnalis Ls-AChBP, PDB 1UV6, and A. californica Ac-AChBP, PDB 2BYN) was individually mutated to alanine and the effects on ΔΔGfolding calculated (supplemental Table S2). Because the mutations to alanine (the residue with the smallest hydrophobic side chain of all natural amino acids) involve removal of hydrophobic side chains, no steric clash is expected and major protein reorganization is unlikely to occur (19, 40). Therefore, the calculated ΔΔGfolding energy differences can be considered approximately the contribution of the wild-type residue to the folding stability of the protein. Hence, large positive changes in ΔΔGfolding would indicate that introduction of the alanine mutations were destabilizing at specific sites and the wild-type residues are highly stabilizing to the protein fold.
Mutating the conserved bulky, hydrophobic β-core residues of each of the proteins into alanines was highly destabilizing, with ΔΔGfolding values > +2 kcal/mol and in most cases exceeding +3 kcal/mol (supplemental Table S2). This observation is consistent with the idea that introducing non-bulky residues into the β-core loosens packing and decreases stability (19, 20, 40–42). Moreover, the data indicate that residues contributing to β-core packing stability are remarkably similar in nAChR, AChBPs, and GLIC. The major differences between nAChR-α1, AChBPs, and GLIC in the effects of the alanine mutations were observed at sites 1 and 2 (supplemental Table S2). In nAChR-α1, introducing an alanine at site 1 (T52A) had little effect (+0.68 kcal/mol) and the alanine at site 2 (S126A) was even stabilizing (−0.95 kcal/mol), whereas the alanines at sites 1 and 2 in AChBPs and GLIC were highly destabilizing (ΔΔGfolding from +2 kcal/mol to +4.5 kcal/mol). This indicates that loose packing at sites 1 and 2 in nAChR-α1 results in an intrinsically less stable fold compared with the high fold stability conferred to GLIC and AChBP by bulky hydrophobic residues at the corresponding sites.
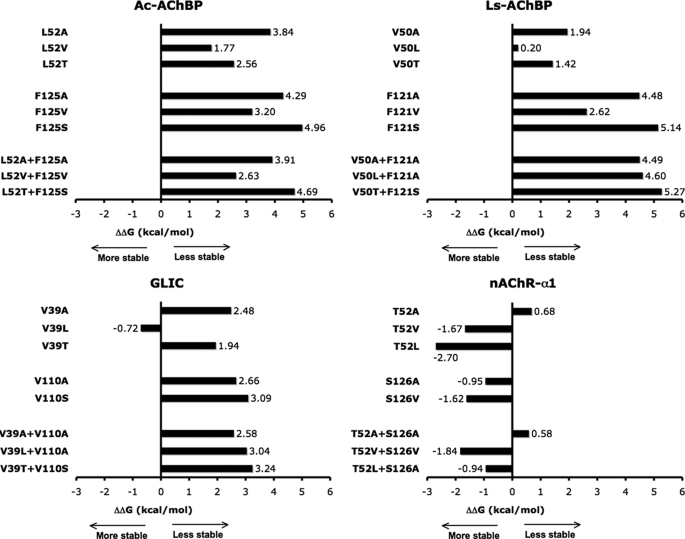
We then used FOLDX to explore how changing the packing at sites 1 and 2 would affect folding stability. In each structure, we mutated sites 1 and 2 to aligned residues found in other pLGICs and AChBPs, and computed the ΔΔGfolding (Fig. 3). Inserting less bulky or hydrophilic residues in the AChBP or GLIC destabilized the cores (most ΔΔGfolding ≥ 3 kcal/mol). Conversely, mutating nAChR-α1 Thr-52 and Ser-126 to bulkier hydrophobic residues stabilized the core, with ΔΔGfolding values from −0.95 to −2.70 kcal/mol. Overall, the FOLDX analysis suggests that the contribution of core-forming amino acids to the folding stability of pLGIC ECDs and AChBP is very similar, except for sites 1 and 2 where marked differences in the thermodynamic stability were estimated between nAChR-α1, GLIC, and AChBP. Namely, tight core packing makes AChBPs very stable, whereas the loose packing in the nAChR ECD locally decreases its stability likely resulting in increased flexibility of the core (34).
FIGURE 3.
Plots of ΔΔGfolding values. In available crystal structures of AChBPs (top) and pLGIC ECDs (bottom), β-core residues at sites 1 and 2 were mutated and ΔΔGfolding values were computed with FOLDX. In AChBPs or GLIC, substituting less bulky or hydrophilic residues at sites 1 and 2 resulted in significant thermodynamic destabilization, with values of ΔΔG ≥ 3 kcal/mol particularly at site 2. In contrast, mutations in nAChR-α1 that replaced the hydrophilic Thr-52 and Ser-126 with bulkier hydrophobic residues made its core more stable. Overall, the data suggest that the tight core packing makes AChBPs stable, whereas loose packing in the nAChR ECD decreases its folding stability.
Using structure based sequence alignment we scrutinized other eukaryotic pLGIC subunits for potential ECD β-core loose packing residues (Fig. 1). All nAChR subunits and 5-HT3R subunits contain a threonine at site 1 and a serine at site 2 indicating that loose packing at these positions may be a general feature for these receptors. Interestingly, the GABAAR subunits contain a hydrophobic leucine or isoleucine at site 1 (as observed in tightly packed cores) but a strictly conserved alanine at site 2 (Fig. 1). Alanines (100 Å3 smaller than a phenylalanine) buried in hydrophobic cores tend to create loose packing through cavities that decrease protein stability (19, 40). Thus, for GABAARs, a potential loose packing defect is also present but a conserved alanine at site 2 may be responsible (Fig. 2, bottom). GlyR subunits appear to be the only eukaryotic pLGICs without potential packing defects at sites 1 or 2, as bulky hydrophobic residues (valine and leucine) are found at these positions. Whether GlyR subunits contain other unique packing defects will need to await direct structural analysis. In prokaryotic GLIC, packing at sites 1 and 2 is similar to AChBPs as both residues are hydrophobic valines. However, in the GLIC ECD β-core, an alanine at position 108 (two residues away from site 2) may contribute to a loose packing defect. Finally, it should be noted that in limited subsets of eukaryotic pLGICs a handful of other residues at different positions from sites 1 or 2 could contribute to loose packing. These residues include threonine in strand β6′ of GABAAR β-subunits, and alanine (GABAAR-α2, GlyR-α1) or threonine (GABAAR-α1,3) in strand β10. However, differently from sites 1 and 2 these potentially core-loosening residues are not absolutely conserved in pLGIC families.
Altering the Packing at Site 1 and Site 2 Affects GABAAR Function
To further test the generality of the idea that loose packing of the β-core contributes to pLGIC function, we examined how altering GABAAR β-core packing at sites 1 and 2 affects its function. We mutated the alanine at site 2 in the GABAAR α1, β2, and γ2 subunits to bulkier hydrophobic residues (valine, as in GLIC, or phenylalanine, as in AChBP). In addition, we replaced site 1 leucine/isoleucine with smaller residues (valine or alanine). Double mutants with valine introduced at sites 1 and 2 (GLIC-like) were also engineered. Wild-type and mutant subunits were expressed in X. laevis oocytes to form α1β2γ2 GABAARs and functionally characterized using two-electrode voltage clamp.
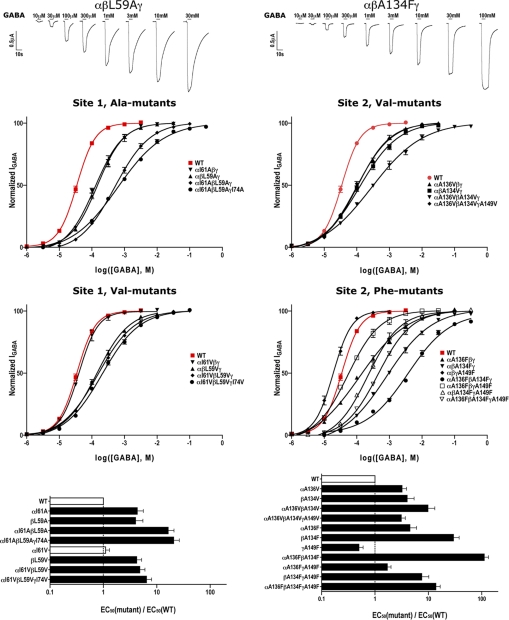
All of the mutant subunits assembled into receptors that responded to GABA. In general, the mutations shifted the GABA dose-response curves to the right. Representative current traces (top panel) and GABA dose-response curves (middle panels) are shown in Fig. 4. Replacing site 2 alanine in the α1 and β2 subunits with bulky residues (valine and phenylalanine) increased GABA EC50 values 3–30-fold (Fig. 4, bottom, and Table 1). In the β2 subunit, as the size of the substituting residue increased, the effect on GABA EC50 increased. The largest increase in GABA EC50 values occurred when α1 and β2 site 2 phenylalanine mutant subunits were co-expressed (111-fold). Interestingly, co-expressing the site 2 mutant γ2 subunit with mutant α1 and β2 subunits reduced the magnitude of the GABA EC50 changes observed. At site 1, replacing the bulky leucine/isoleucine with smaller residues (alanine and valine) also significantly increased GABA EC50 values, with the largest increase (21-fold) occurring when the alanine mutation was present in each of the subunits, αI61AβL59AγI74A. Overall, the data suggest that the combination of a bulky residue at site 1 (leucine/isoleucine) and a small alanine at site 2 is required for efficient GABA-induced channel activation. At site 2, replacing alanine with bulky side chains (like in AChBP) likely fills a cavity, which alters β-core packing, reduces flexibility, and impairs GABAAR activation. At site 1, reducing side chain volume also results in impaired GABA-mediated channel activation, likely due to destabilized β-core packing and/or local changes in the arrangement of nearby functionally important parts of the protein.
FIGURE 4.
Altering the β-core packing at sites 1 (left panels) and 2 (right panels) affects GABA activation of GABAARs. Representative GABA activated current traces (top), GABA dose-response curves (middle), and plots of EC50mutant/EC50WT ratios (bottom) are shown. Wild-type (WT) α1β2γ2 GABAAR dose-response curves are shown in red (EC50 = 34.59 ± 4.02 μm, n = 5). Black bars in bottom panels indicate values significantly different from WT (p < 0.01). EC50 values and Hill coefficients are reported in Table 1. At site 1, replacing the bulky hydrophobic (leucine/isoleucine) in GABAAR subunits with smaller residues (alanine and valine) right-shifted the GABA dose-response curves. At site 2, replacing the conserved alanine, which likely creates a cavity in the hydrophobic β-core, with larger residues (valine and phenylalanine) also significantly right-shifted the GABA dose-response curves.
TABLE 1.
Summary of GABA dose responses from wild-type (WT) and mutant GABAARs
Values are mean ± S.E. from n experiments. GABA EC50 values, Hill coefficients (nH, maximal GABA-activated current range (Imax), and mutant/WT (mut/wt) EC50 ratios are indicated. Mutants indicated with ‘VV’ have a valine mutations at both sites 1 and 2.
| Receptor | EC50 | nH | Imax range | n | mut/WT |
|---|---|---|---|---|---|
| μm | μA | ||||
| WT | 34.59 ± 4.02 | 1.53 ± 0.08 | 7.35–15.71 | 5 | |
| αI61Aβγ | 151.65 ± 28.16a | 1.09 ± 0.10 | 9.57–15.28 | 4 | 4.3 ± 1.3 |
| αβL59Aγ | 140.65 ± 37.72a | 1.08 ± 0.12 | 1.68–2.80 | 4 | 4.0 ± 1.5 |
| αI61AβL59Aγ | 564.80 ± 98.78a | 0.86 ± 0.06 | 0.16–1.70 | 5 | 16.3 ± 4.7 |
| αI61AβL59AγI74A | 717.15 ± 115.50a | 0.64 ± 0.04 | 0.10–0.18 | 4 | 20.7 ± 5.7 |
| αI61Vβγ | 40.43 ± 3.37 | 1.50 ± 0.32 | 10.67–20.32 | 3 | 1.1 ± 0.2 |
| αβL59Vγ | 146.07 ± 18.85a | 0.91 ± 0.04 | 3.27–12.00 | 4 | 4.2 ± 1.0 |
| αI61VβL59Vγ | 166.05 ± 24.32a | 0.82 ± 0.03 | 4.96–17.63 | 4 | 4.8 ± 1.2 |
| αI61VβL59VγI74V | 222.82 ± 31.72a | 0.81 ± 0.05 | 4.10–13.82 | 4 | 6.4 ± 1.6 |
| αA136Vβγ | 110.74 ± 14.13a | 0.90 ± 0.07 | 2.19–6.38 | 4 | 3.2 ± 0.7 |
| αβA134Vγ | 138.67 ± 32.76a | 0.80 ± 0.07 | 3.13–10.78 | 4 | 4.0 ± 1.4 |
| αA136VβA134Vγ | 341.30 ± 79.25a | 0.63 ± 0.09 | 1.81–14.98 | 4 | 9.8 ± 3.4 |
| αA136VβA134VγA149V | 108.63 ± 8.25a | 0.88 ± 0.02 | 1.14–6.70 | 3 | 3.1 ± 0.6 |
| αA136Fβγ | 157.67 ± 33.79a | 0.69 ± 0.04 | 2.38–7.14 | 4 | 4.5 ± 1.5 |
| αβA134Fγ | 1,023.74 ± 167.70a | 0.72 ± 0.06 | 1.11–3.36 | 5 | 29.5 ± 8.2 |
| αβγA149F | 19.23 ± 3.33a | 1.57 ± 0.02 | 13.75–14.71 | 3 | 0.5 ± 0.1 |
| αA136FβA134Fγ | 3,842.50 ± 351.84a | 0.65 ± 0.07 | 0.15–0.90 | 4 | 111.0 ± 23.0 |
| αA136FβγA149F | 59.41 ± 6.17a | 0.96 ± 0.13 | 4.29–4.64 | 3 | 1.7 ± 0.3 |
| αβA134FγA149F | 262.02 ± 65.77a | 0.96 ± 0.18 | 0.89–4.41 | 4 | 7.5 ± 2.7 |
| αA136FβA134FγA149F | 480.40 ± 47.09a | 0.88 ± 0.09 | 0.82–5.65 | 5 | 13.8 ± 2.9 |
| α'VV'βγ | 723.83 ± 15.00a | 0.58 ± 0.01 | 1.39–6.04 | 3 | 20.9 ± 2.8 |
| αβ'VV'γ | 2,360.00 ± 344.41a | 0.54 ± 0.03 | 0.91–3.78 | 3 | 68.2 ± 17.8 |
| α'VV'β'VV'γ | 2,475.25 ± 511.70a | 0.56 ± 0.03 | 1.95–10.08 | 4 | 71.5 ± 23.1 |
| α'VV'β'VV'γ'VV' | 1,832.50 ± 351.65a | 0.47 ± 0.13 | 0.14–0.45 | 4 | 52.9 ± 16.3 |
a p < 0.01, values statistically different from WT.
We also swapped the residues at sites 1 and 2 in the GABAAR β2 subunit. The swap αβ(L59A,A134F)γ mutant did not rescue function, but rather showed cumulative effects of the individual mutations (low GABA-activated current amplitudes with IGABAmax range 0.48–0.07 μA, GABA EC50 >100 mm). Our data are consistent with previous studies on a model system, the bacteriophage f1 gene V protein (16, 18), which showed that the contribution of amino acids to the folding stability of a protein strongly depends on their position within the core, and that the effect of swapping buried residues is usually cumulative. Our results suggest that the effect of the putative cavity created by alanine strongly depends on its position within the core, being functionally favorable only at site 2. We postulate that only when alanine is at site 2 is there an appropriate balance between loss of thermodynamic stability and gain of molecular function (through structural flexibility).
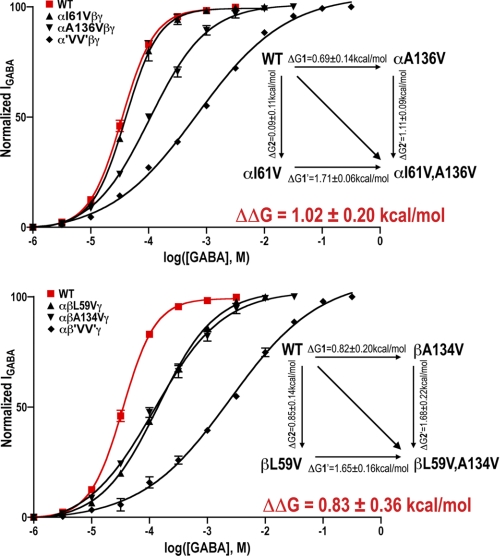
In the α and β subunits, introducing valines at both site 1 and site 2 (α'VV'βγ, αβ'VV'γ) increased GABA EC50 values 21- and 68-fold, respectively (Table 1, Fig. 5). The effects of the double Val mutations on GABA EC50 values were substantially larger than the product of the individual valine mutations effects suggesting that the residues interact. If the mutation at one site had no impact at the second site (sites independent), the effect of the double mutations would increase GABA EC50 values 3.5-fold (α subunit 'VV' mutants) and 16.8-fold (β subunit 'VV' mutants). We used mutant cycle analysis (43) to estimate the coupling and interaction energy between residues at sites 1 and 2 (Fig. 5, insets). Coupling parameters (Ω = (EC50(WT) × EC50(mut.1,2))/(EC50(mut.1) × EC50(mut.2)) significantly different from 1 indicate the residues are energetically coupled (44). Mutant cycle analysis yielded Ω of 5.6 ± 1.1 (α'VV'βγ) and 4.0 ± 1.7 (αβ'VV'γ) corresponding to interaction energies of +1.02 ± 0.20 kcal/mol (α subunit) and +0.83 ± 0.36 kcal/mol (β subunit), suggesting that the β-core residues at sites 1 and 2 are energetically coupled.
FIGURE 5.
Mutant cycle analysis suggests that GABAAR residues at sites 1 and 2 are energetically coupled. GABA dose-response curves for single and double valine mutations in the α1 (top) and β2 (bottom) subunits. WT dose-response curves are shown in red. The double valine mutation (GLIC-like packing) increased GABA EC50 21-fold in α1 subunit, and 68-fold in β2 subunit, which are larger than the product of the individual valine mutation effects. EC50 values and Hill coefficients are reported in Table 1. The insets depict mutant cycles, changes in free energies due to mutations (ΔG), and the overall interaction energies (ΔΔG).
Increasing Packing at Site 2 Reduces GABA Efficacy
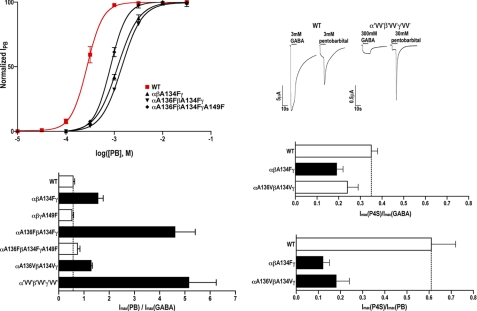
The shifts in GABA EC50 values observed following mutation of β-core sites 1 and 2 could be due to changes in GABA binding, GABA efficacy (channel gating), or both (45). The maximal GABA current amplitudes (IGABAmax) for several of the mutant GABAARs, particularly the site 2 Phe mutants and double Val mutants, were smaller when compared with WT receptors (Table 1) suggesting changes in GABA efficacy and/or GABAAR expression. To test if the mutations affected GABA efficacy, we measured and compared currents induced by a saturating GABA concentration to those induced by a saturating concentration of PB in the same oocyte. PB is an allosteric modulator that binds to a different site than GABA (46) and at high concentrations can directly gate the channel with a similar conductance as GABA (47). For WT receptors, the ratio of currents elicited by saturating concentrations of PB and GABA (IPBmax/IGABAmax) was 0.57 ± 0.06 (Fig. 6, Table 2). Because at saturating concentrations of ligand, all the receptors are fully bound, a change in the IPBmax/IGABAmax ratio would provide evidence that the mutation altered channel open probability (gating). If the mutations only affected protein folding and expression levels, we should observe comparable effects on IPBmax and IGABAmax. In oocytes expressing αβA134Fγ, αA136FβA134Fγ, αA136VβA134Vγ, and α'VV'β'VV'γ'VV' receptors, the currents elicited by saturating concentration of GABA were smaller than currents induced by PB indicating a reduction in GABA efficacy (Fig. 6, bottom left).
FIGURE 6.
Introducing a bulky residue at site 2 (stabilizing packing) reduces GABA efficacy suggesting loose packing at site 2 (alanine) promotes GABA full agonist actions. Top left, PB dose-response curves for WT (red) and mutant GABAARs with phenylalanine introduced at site 2 (WT PB EC50 = 275.35 ± 51.58 μm, n = 4). EC50 values and Hill coefficients are reported in Table 2. Top right, representative GABA and PB currents from oocytes expressing WT and α'VV'β'VV'γ'VV' GABAARs. Bottom, currents elicited by sequential applications of saturating concentrations of GABA and PB (left) or P4S (right) measured from the same oocyte expressing either WT or mutant GABAARs. Maximum current ratios are plotted and represent mean ± S.E. from >3 independent experiments. Black bars represent ratios significantly different from WT (p < 0.05).
TABLE 2.
PB EC50 values from wild-type (WT) and mutant GABAARs
Values are mean ± S.E. from n experiments. Ranges in IPBmax and IGABAmax currents elicited from oocytes expressing WT or mutant GABAARs are indicated. IPBmax/IGABAmax ratios from the same oocyte are mean ± S.E. from n experiments. Mutants indicated with ‘VV’ have a valine mutation at both sites 1 and 2.
| Receptor | EC50 | nH | n | IPBmax | IGABAmax | IPBmax/IGABAmax | n |
|---|---|---|---|---|---|---|---|
| μm | μA | ||||||
| WT | 275.35 ± 51.58 | 2.77 ± 0.41 | 4 | 4.14–9.32 | 6.93–15.58 | 0.57 ± 0.06 | 7 |
| αβA134Fγ | 1,171.33 ± 69.00a | 2.00 ± 0.32 | 3 | 0.96–6.39 | 0.67–4.16 | 1.57 ± 0.19b | 4 |
| αβγA149F | 608.93 ± 22.21a | 3.70 ± 0.41 | 3 | 4.84–6.49 | 8.39–11.61 | 0.52 ± 0.07 | 6 |
| αA136FβA134Fγ | 1,424.33 ± 53.57a | 2.06 ± 0.16 | 3 | 0.52–2.29 | 0.11–0.60 | 4.62 ± 0.79a | 5 |
| αA136FβA134FγA149F | 840.00 ± 71.29a | 2.50 ± 0.05 | 3 | 1.73–3.38 | 1.96–5.00 | 0.74 ± 0.10 | 5 |
| αA136VβA134Vγ | 243.70 ± 11.47 | 2.65 ± 0.11 | 3 | 6.06–15.03 | 4.73–11.85 | 1.30 ± 0.03b | 4 |
| α'VV'β'VV'γ'VV' | 1,744.66 ± 120.66a | 2.23 ± 0.32 | 3 | 0.81–1.64 | 0.20–0.27 | 5.17 ± 1.07a | 4 |
a p < 0.01, values statistically different from WT.
b p < 0.05, values statistically different from WT.
The site 2 phenylalanine mutations and the double Val mutations also significantly increased PB EC50 values compared with WT (2–6-fold, Table 2, Fig. 6, top) suggesting that increasing side chain volume at site 2 not only affects GABA EC50 values but also adversely alters the ability of PB to activate the receptor. The differences in the magnitude of the mutations effects on GABA EC50 and PB EC50 values (e.g. for αA136FβA134Fγ receptors, Δ in GABA EC50 = 111-fold, PB EC50 5-fold, Tables 1 and 3) likely reflect the fact that GABA and PB bind to different sites in the receptor and trigger different activation pathways and movements (48). Some of the mutations with larger effects on GABA EC50 may also be due to changes in GABA binding.
TABLE 3.
Maximal GABA-activated current rise times (10%–90%) and percent current desensitization at 20 s for WT and mutant GABAARs
Values are mean ± S.E. for n experiments.
| Receptor | Rise time, 10 → 90% | Desensitization (Imax – I20 sec)/Imax | n |
|---|---|---|---|
| s | |||
| WT | 0.28 ± 0.10 | 45.8 ± 3.9% | 3 |
| αI61Aβγ | 0.85 ± 0.32 | 24.2 ± 7.5%a | 4 |
| αβL59Aγ | 0.62 ± 0.40 | 54.7 ± 14.6% | 4 |
| αI61AβL59Aγ | 1.01 ± 0.48b | 34.4 ± 2.8% | 3 |
| αI61AβL59AγI74A | 1.08 ± 0.31b | 7.2 ± 2.0%a | 3 |
| αI61Vβγ | 0.48 ± 0.20 | 41.4 ± 3.4% | 3 |
| αβL59Vγ | 0.43 ± 0.24 | 38.4 ± 10.2% | 4 |
| αI61VβL59Vγ | 0.91 ± 0.35b | 26.1 ± 4.0%a | 5 |
| αI61VβL59VγI74V | 0.76 ± 0.28 | 18.9 ± 4.7%a | 5 |
| αA136Vβγ | 0.91 ± 0.34 | 25.0 ± 3.2%b | 4 |
| αβA134Vγ | 0.49 ± 0.26 | 29.2 ± 4.6%b | 5 |
| αA136VβA134Vγ | 1.13 ± 0.13b | 21.1 ± 3.7%a | 3 |
| αA136VβA134VγA149V | 0.39 ± 0.11 | 51.1 ± 2.7% | 3 |
| αA136Fβγ | 0.60 ± 0.13 | 27.2 ± 4.7%a | 4 |
| αβA134Fγ | 1.23 ± 0.47a | 22.8 ± 2.1%a | 4 |
| αβγA149F | 0.27 ± 0.02 | 43.9 ± 5.5% | 2 |
| αA136FβA134Fγ | 1.01 ± 0.26b | 5.6 ± 1.6%a | 3 |
| αA136FβγA149F | 0.25 ± 0.08 | 52.0 ± 5.9% | 4 |
| αβA134FγA149F | 0.13 ± 0.02 | 51.0 ± 15.7% | 3 |
| αA136FβA134FγA149F | 0.24 ± 0.10 | 53.6 ± 4.8% | 6 |
| α'VV'βγ | 0.79 ± 0.40 | 19.4 ± 3.4%a | 3 |
| αβ'VV'γ | 0.61 ± 0.16 | 32.4 ± 2.7%b | 3 |
| α'VV'β'VV'γ | 3.05 ± 1.40a | 15.9 ± 1.4%a | 6 |
| α'VV'β'VV'γ'VV' | 1.09 ± 0.21b | 18.8 ± 4.6%a | 3 |
a p < 0.01, values statistically different from WT.
b p < 0.05, values statistically different from WT.
If the mutations adversely altered the gating equilibrium, then one would also expect the efficacy and maximum response to a partial agonist would decrease. Changes in efficacy are much more readily detected using partial agonists than full agonists (45). Thus, we looked at the effect of β-core mutations at site 2 on the relative efficacy of the partial agonist P4S macroscopic currents (49). We measured and compared currents induced by a saturating P4S concentration (10 mm for WT, 300 mm for mutants) to those induced by a saturating concentration of GABA (3 mm for WT, 300 mm for mutants) in the same oocyte for WT, αβA134Fγ, and αA136VβA134Vγ receptors (Fig. 6, supplemental Table S4). For WT GABAAR, P4S elicited a maximum response only a third of that elicited by GABA, IP4Smax/IGABAmax = 0.35 ± 0.03. For αβA134Fγ and αA136VβA134Vγ receptors, the IP4Smax/IGABAmax ratio decreased to 0.19 ± 0.03 and 0.24 ± 0.05, respectively (Fig. 6, bottom right). The efficacy of P4S relative to PB was also measured and was significantly decreased for both mutants (0.12 ± 0.03 for αβA134Fγ, and 0.18 ± 0.06 for αA136VβA134Vγ) compared with WT (0.61 ± 0.11) (Fig. 6, bottom right). Taken together, the data indicate that increasing the packing at site 2 reduces agonist efficacy.
Packing at Sites 1 and 2 Influences GABAAR Current Activation and Macroscopic Desensitization
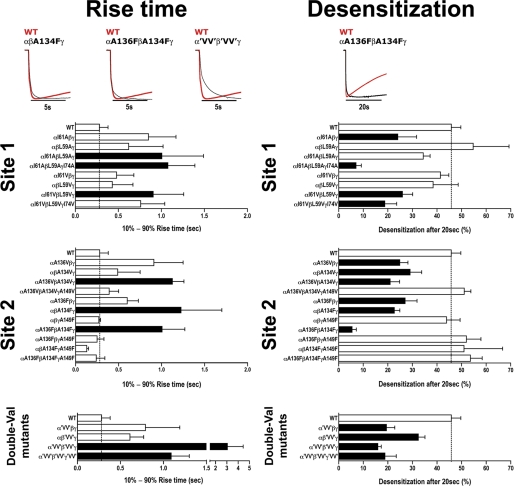
We also measured the current 10–90% apparent rise times at saturating GABA concentrations for mutant and WT receptors (Fig. 7 and Table 3). Although measuring current onset is limited by the slow solution-exchange times when recording from oocytes (≈300 ms), a slowed GABA current apparent rise time compared with WT would suggest a change in channel gating. GABA rise times for αI61AβL59Aγ, αI61AβL59AγI74A, αI61VβL59Vγ, αA136VβA134Vγ, αβA134Fγ, αA136FβA134Fγ, α'VV'β'VV'γ and α'VV'β'VV'γ'VV' receptors were significantly slower than WT receptors (0.28 ± 0.1 s for WT, Fig. 7, Table 3) with the double Val mutations having the largest effect (11-fold) suggesting that disrupting the packing at β-core sites 1 and 2 reduced channel opening. The mutations also decreased the extent of macroscopic current desensitization measured after a 20-s application of saturating GABA (Fig. 7, Table 3).
FIGURE 7.
Altering packing at sites 1 and 2 slows GABA current rise time and desensitization. Representative saturating GABA current traces from oocytes expressing WT and mutant GABAARs are peak normalized to highlight the slowing in GABA 10–90% rise times (left) and current desensitization (right) for mutant receptors. Values are reported in Table 3. Bars represent mean ± S.E. of 10–90% rise times (s) and percent current desensitization after 20 s (((Imax − I20 sec)/Imax) × 100) from WT and mutant receptors (n > 3 individual experiments). Black bars indicate values significantly different from WT (p < 0.05).
DISCUSSION
Although packing defects (cavities) in hydrophobic cores account for as little as 1% of the volume of all globular proteins, they are often functionally relevant (50). Here, we provide evidence that a localized loose packing defect in the ECD β-cores of eukaryotic pLGICs (namely nAChRs, 5-HT3Rs, and GABAARs) is a key structural element for receptor function, which likely facilitates ligand-induced activation.
pLGICs are allosteric proteins that exist in at least three distinct, interconvertible states: resting (unliganded, closed channel), activated (liganded, open channel), and desensitized (liganded, closed channel). These receptors respond to neurotransmitter binding with allosteric transitions that switch the protein from a non-conducting closed conformation to an ion-conducting open conformation rapidly and with high probability. Our understanding of the protein movements underlying these conformational transitions is limited. Receptor activation is initiated by agonist binding in the ECD, and propagates ∼60 Å to eventually gate the membrane-embedded ion-conducting pore. One model of activation suggests that neurotransmitter binding induces movements of the inner and outer β-sheets of the ECD relative to each other (51, 52). This, in turn, changes the orientation and interactions of the flexible loops at the ECD-transmembrane ion pore-forming domain interface resulting in a repositioning of the M2 channel-lining helices, such that the channel pore opens. A second model suggests that a concerted quaternary twist of the ECD drives channel activation (32, 53). Experiments using rate-equilibrium free energy relationships suggest that transduction of binding to gating occurs as a conformational wave from the transmitter binding site to the channel (54). Regardless of the model, any conformational change required for channel activation would be facilitated by enhanced flexibility of the ECD, as this would decrease the energy barrier necessary for the conformational switch to take place (3–5).
Flexibility of hydrophobic cores is usually the result of buried polar or non-bulky residues that loosen the core packing at the expense of thermodynamic stability. Here, we identify a localized loose packing defect in the ECD β-cores of eukaryotic pLGICs that decreases the stability of the hydrophobic core. Moreover, we demonstrate that loose packing facilitates ligand-induced receptor activation supporting the idea that local β-core flexibility helps drive pLGIC activation.
Structural and computational studies show that pLGIC ECDs and AChBPs share the same fold, with the main chain traces of their β-sandwich cores nearly superimposable (26, 31–33). Our computational analyses demonstrate that pLGICs ECDs and AChBP β-cores are tightly packed with bulky hydrophobic amino acids at most positions (Figs. 1 and 2, supplemental Figs. S1 and S2), which confer thermodynamic stability (supplemental Tables S1 and S2). The β-core packing of the nAChR, 5-HT3R, and GABAAR, however, each contains a localized packing defect not observed in AChBPs or GLIC (Figs. 1 and 2). In nAChRs and 5-HT3Rs, two conserved polar residues corresponding to Thr-52 and Ser-126 in the nAChR α1 subunit (sites 1 and 2, respectively) are present that loosen the β-core packing (Figs. 1 and 2). In GABAARs, a strictly conserved alanine at site 2 creates a potential packing defect. These residues are unusual in hydrophobic cores as they decrease thermodynamic stability (4, 18–20).
Using FOLDX, we determined that the ECD β-core of nAChR-α1 is less stable at sites 1 and 2 compared with AChBP and GLIC (supplemental Tables S1 and S2). Consistent with our results, in the crystal structure of nAChR-α1 ECD, the side chains of Thr-52 and Ser-126 are buried in the β-core and form a water-filled cavity (26). This packing defect is energetically costly because placing two polar side chains and a water molecule into a hydrophobic environment has high entropic penalty (∼20 kcal/mol), which is only partly compensated by the formation of water-protein hydrogen bonds (55, 56). In AChBP and GLIC, the cores are conventionally packed at both sites with bulky hydrophobic residues (valine, leucine, and phenylalanine). As expected, introducing residues that potentially create packing defects (threonine, serine, and alanine) at these sites was highly destabilizing, with ΔΔGfolding increasing 3–5 kcal/mol (Fig. 3).
Mutation of Thr-52 and Ser-126 in the nAChR α1 subunit to bulky hydrophobic amino acids substantially reduces acetylcholine-activated channel current (26). When we used FOLDX and mutated nAChR-α1 Thr-52 and Ser-126 to bulkier hydrophobic residues to pack the core, it stabilized the core (Fig. 3). Previous work has shown that mutating Ser-126 to alanine or valine had little effect on gating equilibrium and is consistent with our results, as the smaller side chain of alanine and valine would likely still leave a cavity (57). Together, the data indicate that loose packing of the β-sandwich core is important for nAChR function. Because 5-HT3R subunits have the same loose packing defect as the nAChR-α1 (threonine at site 1, and threonine or serine at site 2), we reason that it also plays a critical role in 5-HT3R channel activation.
We show that loose β-core packing is also important for GABAAR function. In GABAAR subunits, a hydrophobic leucine or isoleucine is at site 1 (as observed in tightly packed cores) but a strictly conserved alanine is at site 2, which may create loose packing. Leucine and isoleucine are canonical side chains for a hydrophobic core. In our FOLDX analysis, mutating any β-core leucine or isoleucine to alanine in pLGIC or AChBP structures caused destabilization, with ΔGfolding increments of 3–5 kcal/mol (supplemental Table S2). Thus, we expected that mutating leucine/isoleucine at site 1 in GABAAR to a smaller residue would decrease the stability of the β-core even further (which is constitutively less stable than AChBP because of the site 2 alanine) and exert an adverse effect on receptor function. As anticipated, replacing site 1 leucine/isoleucine with smaller residues (valine or alanine) significantly increased GABA EC50 values 4–20-fold (Fig. 4, left, and Table 1). The effects on GABA EC50 were inversely proportional to the size of the side chain substitution, and increased with co-expression of multiple mutant subunits.
Replacing site 2 alanine with bulky residues (valine or phenylalanine) to stabilize the core resulted in significantly larger disruptions in GABAAR activation with increased GABA EC50 values (3–111-fold, Fig. 4, right, and Table 1), and decreased GABA efficacy, GABA current apparent rise times, and macroscopic current desensitization (Fig. 7, Tables 2 and 3). Depending on the subunit, the mutations had different effects. The valine and phenylalanine mutations in the α1 and β2 subunits had the largest effects, which increased with co-expression. On the other hand, increasing the packing of the γ2 subunit β-core at site 2 had little effects on GABA EC50, GABA current apparent rise time, and macroscopic current desensitization (Table 1, Fig. 7) suggesting that GABAAR activation has distinct subunit requirements for ECD flexibility. The data are consistent with the fact that agonist binding occurs at the interfaces between GABAAR α and β subunit ECDs and that movements in the ECD triggered by agonist binding appear asymmetrical (51, 58). Interestingly, when the γ mutant subunits were co-expressed with the α and β mutant subunits the magnitude of the effects on GABAAR activation were reduced (Table 1) suggesting that decreasing the flexibility of the γ2 subunit can act as a positive modulator of GABAAR activation.
The Ala → Phe mutation, which replicates in GABAAR the packing at sites 1 and 2 of Ac-AChBP (leucine and phenylalanine), had the largest effects on GABAAR function (Tables 1 and 3). Compared with WT GABAAR, the αA136FβA134Fγ mutant displayed a 111-fold increase in GABA EC50, a ∼4-fold slower current rise time, and a ∼8-fold slower current desensitization rate (Fig. 7). It is unlikely that these effects are due to improper ECD folding. First, the IgG-fold common to pLGICs ECD and AChBP can accommodate at sites 1 and 2 a variety of residue pairs (from threonine-serine to leucine-phenylalanine) with little adjustments of protein backbone, because the distance (Cα-Cα and Cβ-Cβ) between sites 1 and 2 residues changes very little from structure to structure, either pLGIC or AChBP (supplemental Table S3). The GABA maximal elicited currents from oocytes expressing αA136FβA134Fγ were smaller compared with oocytes expressing WT receptors (Table 1), which could be explained by decreased expression of surface receptors. However, when we compared the currents elicited by a saturating concentration of GABA to those induced by a saturating concentration of PB in the same oocyte, the mutations had much larger effects of IGABAmax demonstrating that the mutations reduced GABA efficacy (Fig. 6, bottom left, and Table 2). The Ala → Phe mutation also decreased the efficacy of partial agonist P4S in the αβA134Fγ mutant (Fig. 6, bottom right, and supplemental Table S4), further supporting the idea that tight packing at site 2 adversely affects receptor activation.
Similar results were seen for the double Val mutants, which mimic GLIC β-core packing with valines at sites 1 and 2. Along with increasing GABA EC50, the β-core packing reconfiguration slowed current activation and desensitization rates (Fig. 7). The α'VV'β'VV'γ and α'VV'β'VV'γ'VV' mutant receptors displayed current profiles reminiscent of the slow activating, slow desensitizing GLIC (30), suggesting β-core packing influences current response properties. Again, for 'VV' mutant receptors, IPBmax were decreased to a much less extent than IGABAmax confirming that amino acid composition at sites 1 and 2 affects GABA efficacy.
It is important to note that increasing the rigidity of the β-core packing in GABAARs or nAChRs does not abolish function but rather hinders the ability of agonist to efficiently activate the receptor. In a similar fashion, whereas the common scaffold of AChBP apparently possesses the basic ability to trigger ion pore gating when fused to the 5-HT3AR channel transmembrane domain and its loops 2, 7, and 9 are replaced with corresponding 5-HT3AR loops, it is not as efficient as an eukaryotic pLGIC ECD in coupling binding to channel activation (59, 60). Channel gating of the prokaryotic pLGIC homologue, GLIC, is also different compared with eukaryotic heteromeric pLGICs, with slow channel activation kinetics and little apparent desensitization (30). Taken together, the data suggest that loose packing was evolutionarily developed to increase ECD flexibility, which facilitates agonist-induced channel activation in nAChR, 5-HT3R, and GABAAR. Support for this idea comes from a recent study on glutamate receptors (GluRs). Like pLGICs, eukaryotic GluRs have much faster activation and desensitization rates than prokaryotic GluRs. This is likely due to loose packing of the hydrophobic core of Lobe 2, which confers to the ligand-binding domain of eukaryotic GluRs greater flexibility that speeds up the receptor response to agonist (61). Furthermore, a recent study on the GlyR proposes channel activation is accompanied by reorganization of the hydrophobic core (62).
Enhanced ECD flexibility due to loose packing of the β-core is also supported by the structural information available. In the crystal structures of AChBP in complex with agonists, no significant ligand-dependent conformational changes are observed other than movement of loop C to cap the ligand-binding pocket (63, 64). Similarly, comparing the recent crystal structures of GLIC at pH 7 (corresponding to closed state (38)) and pH 4.6 (presumably open state (32)) limited reorganization of the two core-forming β-sheets is observed. Larger conformational changes are observed in eukaryotic receptors. In the low-resolution electron microscopy analysis of the Torpedo nAChR, agonist-induced rotations of the β-core inner and outer β-sheets about an axis normal to the membrane plane were detected in the α-subunits (51). Molecular dynamics simulations also find larger flexibility of the nAChR-α1 ECD at sites 1 and 2 compared with the prokaryotic receptor ELIC, which has valine and phenylalanine at the two sites like Ls-AChBP (34).
This structural information correlates well with our computational analysis. The packing of GLIC at sites 1 and 2 (valine and valine, for a total volume of 160Å3 of buried side chains) is more rigid than that of nAChR-α1 (threonine and serine, 85 Å3), but less rigid than that of AChBP (valine/leucine and phenylalanine, 210–235 Å3) (Fig. 2, bottom). In our FOLDX simulations, mutations that reduce side chain volume at both sites in GLIC are less energetically costly compared with AChBP (about 3 versus 5 kcal/mol, see Fig. 3), suggesting a slightly higher propensity to structural flexibility of the ECD β-core, in agreement with the crystallographic evidence above.
Overall, the efficiency of allosteric activation seems to depend on the flexiblity of the β-core. Taken together, our results support the idea that during the evolution of pLGIC loose packing of the ECD β-sandwich core was acquired to increase flexibility to facilitate the allosteric activation of nAChRs, GABAARs, and 5-HT3Rs. This provides novel insight into the activation mechanism of pLGICs and other related allosteric proteins.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S4.
This paper is dedicated to the memory of Dr. Zuo-Zhong Wang (1963–2008).
- nAChR
- nicotinic acetylcholine receptor
- pLGIC
- pentameric ligand-gated ion channel
- ECD
- extracellular domain
- AChBP
- acetylcholine-binding protein
- PB
- pentobarbital
- P4S
- piperidine-4-sulfonic acid
- GluR
- glutamate receptors
- PDB
- Protein Data Bank.
REFERENCES
- 1. DePristo M. A., Weinreich D. M., Hartl D. L. (2005) Nat. Rev. Genet. 6, 678–687 [DOI] [PubMed] [Google Scholar]
- 2. Pál C., Papp B., Lercher M. J. (2006) Nat. Rev. Genet. 7, 337–348 [DOI] [PubMed] [Google Scholar]
- 3. Pakula A. A., Sauer R. T. (1989) Annu. Rev. Genet. 23, 289–310 [DOI] [PubMed] [Google Scholar]
- 4. Fields P. A. (2001) Comp. Biochem. Physiol. A Mol. Integr. Physiol. 129, 417–431 [DOI] [PubMed] [Google Scholar]
- 5. Daniel R. M., Dunn R. V., Finney J. L., Smith J. C. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 69–92 [DOI] [PubMed] [Google Scholar]
- 6. Somero G. N. (1995) Annu. Rev. Physiol. 57, 43–68 [DOI] [PubMed] [Google Scholar]
- 7. Tokuriki N., Stricher F., Schymkowitz J., Serrano L., Tawfik D. S. (2007) J. Mol. Biol. 369, 1318–1332 [DOI] [PubMed] [Google Scholar]
- 8. Tokuriki N., Tawfik D. S. (2009) Curr. Opin. Struct. Biol. 19, 596–604 [DOI] [PubMed] [Google Scholar]
- 9. Tokuriki N., Tawfik D. S. (2009) Science 324, 203–207 [DOI] [PubMed] [Google Scholar]
- 10. Aharoni A., Gaidukov L., Khersonsky O., McQ Gould S., Roodveldt C., Tawfik D. S. (2005) Nat. Genet. 37, 73–76 [DOI] [PubMed] [Google Scholar]
- 11. Morley K. L., Kazlauskas R. J. (2005) Trends Biotechnol. 23, 231–237 [DOI] [PubMed] [Google Scholar]
- 12. Tokuriki N., Stricher F., Serrano L., Tawfik D. S. (2008) PLoS Comput. Biol. 4, e1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chothia C., Gough J. (2009) Biochem. J. 419, 15–28 [DOI] [PubMed] [Google Scholar]
- 14. Dill K. A. (1990) Biochemistry 29, 7133–7155 [DOI] [PubMed] [Google Scholar]
- 15. Kauzmann W. (1959) Adv. Protein Chem. 14, 1–63 [DOI] [PubMed] [Google Scholar]
- 16. Sandberg W. S., Terwilliger T. C. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1706–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chothia C. (1984) Annu. Rev. Biochem. 53, 537–572 [DOI] [PubMed] [Google Scholar]
- 18. Sandberg W. S., Terwilliger T. C. (1989) Science 245, 54–57 [DOI] [PubMed] [Google Scholar]
- 19. Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. (1992) Science 255, 178–183 [DOI] [PubMed] [Google Scholar]
- 20. Kellis J. T., Jr., Nyberg K., Sali D., Fersht A. R. (1988) Nature 333, 784–786 [DOI] [PubMed] [Google Scholar]
- 21. Sonavane S., Chakrabarti P. (2008) PLoS Comput. Biol. 4, e1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cañadillas J. M., Tidow H., Freund S. M., Rutherford T. J., Ang H. C., Fersht A. R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2109–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee C., Park S. H., Lee M. Y., Yu M. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7727–7731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. (1994) Nature 371, 37–43 [DOI] [PubMed] [Google Scholar]
- 25. Chen J., Lee K. H., Steinhauer D. A., Stevens D. J., Skehel J. J., Wiley D. C. (1998) Cell 95, 409–417 [DOI] [PubMed] [Google Scholar]
- 26. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 27. Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 28. Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 29. Tasneem A., Iyer L. M., Jakobsson E., Aravind L. (2005) Genome Biol. 6, R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bocquet N., Prado de Carvalho L., Cartaud J., Neyton J., Le Poupon C., Taly A., Grutter T., Changeux J. P., Corringer P. J. (2007) Nature 445, 116–119 [DOI] [PubMed] [Google Scholar]
- 31. Hilf R. J., Dutzler R. (2008) Nature 452, 375–379 [DOI] [PubMed] [Google Scholar]
- 32. Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 33. Hilf R. J., Dutzler R. (2009) Nature 457, 115–118 [DOI] [PubMed] [Google Scholar]
- 34. Cheng X., Ivanov I., Wang H., Sine S. M., McCammon J. A. (2009) Biophys. J. 96, 4502–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahmad S., Gromiha M., Fawareh H., Sarai A. (2004) BMC Bioinformatics 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guerois R., Nielsen J. E., Serrano L. (2002) J. Mol. Biol. 320, 369–387 [DOI] [PubMed] [Google Scholar]
- 38. Nury H., Bocquet N., Le Poupon C., Raynal B., Haouz A., Corringer P. J., Delarue M. (2010) J. Mol. Biol. 395, 1114–1127 [DOI] [PubMed] [Google Scholar]
- 39. Hansen S. B., Sulzenbacher G., Huxford T., Marchot P., Taylor P., Bourne Y. (2005) EMBO J. 24, 3635–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eriksson A. E., Baase W. A., Matthews B. W. (1993) J. Mol. Biol. 229, 747–769 [DOI] [PubMed] [Google Scholar]
- 41. Shortle D., Stites W. E., Meeker A. K. (1990) Biochemistry 29, 8033–8041 [DOI] [PubMed] [Google Scholar]
- 42. Milla M. E., Brown B. M., Sauer R. T. (1994) Nat. Struct. Biol. 1, 518–523 [DOI] [PubMed] [Google Scholar]
- 43. Horovitz A. (1996) Fold Des. 1, R121–126 [DOI] [PubMed] [Google Scholar]
- 44. Gleitsman K. R., Shanata J. A., Frazier S. J., Lester H. A., Dougherty D. A. (2009) Biophys. J. 96, 3168–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Colquhoun D. (1998) Br. J. Pharmacol. 125, 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amin J., Weiss D. S. (1993) Nature 366, 565–569 [DOI] [PubMed] [Google Scholar]
- 47. Jackson M. B., Lecar H., Mathers D. A., Barker J. L. (1982) J. Neurosci. 2, 889–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mercado J., Czajkowski C. (2008) J. Biol. Chem. 283, 15250–15257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mortensen M., Kristiansen U., Ebert B., Frølund B., Krogsgaard-Larsen P., Smart T. G. (2004) J. Physiol. 557, 389–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Williams M. A., Goodfellow J. M., Thornton J. M. (1994) Protein Sci. 3, 1224–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Unwin N., Miyazawa A., Li J., Fujiyoshi Y. (2002) J. Mol. Biol. 319, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 52. McLaughlin J. T., Fu J., Rosenberg R. L. (2007) Mol. Pharmacol. 71, 1312–1318 [DOI] [PubMed] [Google Scholar]
- 53. Taly A., Delarue M., Grutter T., Nilges M., Le Novère N., Corringer P. J., Changeux J. P. (2005) Biophys. J. 88, 3954–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Auerbach A. (2010) J. Physiol. 588, 573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fischer S., Verma C. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9613–9615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olano L. R., Rick S. W. (2004) J. Am. Chem. Soc. 126, 7991–8000 [DOI] [PubMed] [Google Scholar]
- 57. Chakrapani S., Bailey T. D., Auerbach A. (2004) J. Gen. Physiol. 123, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Law R. J., Henchman R. H., McCammon J. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 6813–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bouzat C., Gumilar F., Spitzmaul G., Wang H. L., Rayes D., Hansen S. B., Taylor P., Sine S. M. (2004) Nature 430, 896–900 [DOI] [PubMed] [Google Scholar]
- 60. Grutter T., Prado de Carvalho L., Virginie D., Taly A., Fischer M., Changeux J. P. (2005) C. R. Biol. 328, 223–234 [DOI] [PubMed] [Google Scholar]
- 61. Maltsev A. S., Oswald R. E. (2010) J. Biol. Chem. 285, 10154–10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miller P. S., Topf M., Smart T. G. (2008) Nat. Struct. Mol. Biol. 15, 1084–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Celie P. H., van Rossum-Fikkert S. E., van Dijk W. J., Brejc K., Smit A. B., Sixma T. K. (2004) Neuron 41, 907–914 [DOI] [PubMed] [Google Scholar]
- 64. Hibbs R. E., Sulzenbacher G., Shi J., Talley T. T., Conrod S., Kem W. R., Taylor P., Marchot P., Bourne Y. (2009) EMBO J. 28, 3040–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.