Abstract
La Crosse encephalitis virus (LACV) is a mosquito-borne member of the negative-strand RNA virus family Bunyaviridae. We have previously shown that the virulence factor NSs of LACV is an efficient inhibitor of the antiviral type I interferon system. A recombinant virus unable to express NSs (rLACVdelNSs) strongly induced interferon transcription, whereas the corresponding wt virus (rLACV) suppressed it. Here, we show that interferon induction by rLACVdelNSs mainly occurs through the signaling pathway leading from the pattern recognition receptor RIG-I to the transcription factor IRF-3. NSs expressed by rLACV, however, acts downstream of IRF-3 by specifically blocking RNA polymerase II-dependent transcription. Further investigations revealed that NSs induces proteasomal degradation of the mammalian RNA polymerase II subunit RPB1. NSs thereby selectively targets RPB1 molecules of elongating RNA polymerase II complexes, the so-called IIo form. This phenotype has similarities to the cellular DNA damage response, and NSs was indeed found to transactivate the DNA damage response gene pak6. Moreover, NSs expressed by rLACV boosted serine 139 phosphorylation of histone H2A.X, one of the earliest cellular reactions to damaged DNA. However, other DNA damage response markers such as up-regulation and serine 15 phosphorylation of p53 or serine 1524 phosphorylation of BRCA1 were not triggered by LACV infection. Collectively, our data indicate that the strong suppression of interferon induction by LACV NSs is based on a shutdown of RNA polymerase II transcription and that NSs achieves this by exploiting parts of the cellular DNA damage response pathway to degrade IIo-borne RPB1 subunits.
Keywords: DNA Damage, Innate Immunity, Interferon, MRNA, Negative-strand RNA Viruses, RNA Polymerase II, Viral Protein, Interferon Antagonist, La Crosse Virus
Introduction
La Crosse virus (LACV)3 is a mosquito-borne member of the family Bunyaviridae, genus Orthobunyavirus. LACV infections are an important cause of severe encephalitis and meningitis in children and young adults in the Western United States (1–3). Around 75–100 cases per year require hospitalizations (4), and more than 10% of those patients will have long-lasting neurological deficits (1, 5). Recent observations suggest that the virus is spreading to new geographic regions (6).
Like other arboviruses, LACV cycles between vertebrate and invertebrate hosts, being able to replicate both in mammals and in insects. Depending on the host, however, the outcome of infection is different (7). In mammalian cells, infection is lytic and causes host cell shutoff and cell death. In insect cells infection is non-cytolytic and leads to long term viral persistence.
LACV is enveloped and has a tri-segmented single-stranded RNA genome of negative-sense polarity. Transcription and replication of the genome occur in the cytoplasm, and particles bud into the Golgi apparatus before being secreted. The viral genome encodes four structural proteins, the viral polymerase (L) on the large (L) segment, two glycoproteins (Gn and Gc) on the medium (M) segment, and the viral nucleocapsid protein (N) on the smallest (S) segment. In addition, LACV expresses two nonstructural proteins that are termed NSs and NSm according to their encoding genome segments (8).
We have recently shown that the NSs protein of LACV is an efficient antagonist of the antiviral type I interferon (IFN-α/β) system. A recombinant virus mutant unable to express NSs, rLACVdelNSs (9), was shown to be a strong inducer of IFN transcription both in cell culture and in vivo (10–12). The corresponding NSs-expressing wt virus (rLACV), by contrast, efficiently suppressed transcriptional up-regulation of IFNs. Moreover, ectopic expression of LACV NSs was sufficient to counteract IFN induction by transfected double-stranded RNA (dsRNA), indicating that NSs acts independent of other viral gene products.
IFNs play an important role in protection of the mammalian host from infection. Upon viral invasion of cells, transcription of IFN genes is rapidly induced, and IFNs are secreted and stimulate the expression of antiviral and immunomodulatory genes in neighboring cells (13). Initial recognition of virus infection occurs through specific cellular receptors that bind to so-called pathogen-associated molecular patterns. Two cytoplasmic RNA helicases, RIG-I and MDA5 (collectively termed RIG-like receptors), are the main intracellular receptors of viral RNA (14–16). RIG-I binds to RNA molecules containing a triphosphate group at their 5′ end, whereas MDA5 activation is more dependent on long and branched dsRNA structures (17–21). The binding of a viral RNA to RIG-I or MDA5 induces a signaling chain that eventually results in the phosphorylation and homodimerization of the transcription factor IRF-3 (22), a member of the IFN regulatory factor (IRF) family. Phosphorylated IRF-3 moves into the nucleus where it initiates IFN-β mRNA synthesis.
The best characterized bunyaviral NSs proteins are the ones of Bunyamwera virus (BUNV; genus Orthobunyavirus) and Rift Valley fever virus (RVFV; genus Phlebovirus) (reviewed in Ref. 8). These IFN antagonists are known to block IFN mRNA synthesis at the level of transcriptional initiation (RVFV NSs (23, 24)) or RNA polymerase II (RNAP II) phosphorylation (BUNV NSs (25)). For the human pathogen LACV, however, it is entirely unknown where NSs interferes with the IFN system. Connected to this is the question of which RIG-like receptors pathway would be activated by LACV. Here, we demonstrate that LACV mainly activates the RIG-I pathway and that NSs counteracts subsequent IFN gene transcription by specifically and rapidly shutting down RNAP II through a mechanism resembling a mammalian DNA damage response.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Reagents
Simian Vero cells, hamster BSR-T7/5 cells (26), and human cell lines A549, HeLa, and HEK293T were grown in Dulbecco's modified Eagle's medium supplemented with 5% fetal calf serum (FCS). Aedes albopictus C6/36 were grown at 30 °C without CO2 in Leibowitz L-15 medium (Invitrogen) supplemented with 10% FCS, antibiotics, and 4% tryptose phosphate broth (Difco). The recombinant virus strains rLACV and rLACVdelNSs were grown and maintained as described previously (9). α-Amanitin and ZVAD-fluoromethyl ketone were purchased from AppliChem and Bachem, respectively, and cycloheximide, actinomycin D, doxorubicin, and MG132 were purchased from Sigma.
shRNA Knockdowns
Gene silencing by RNAi was achieved using shRNAs expressed by retroviruses (shRNAmir constructs, Open Biosystems). The construct which efficiently targets MDA5 had the catalogue number RHS1764-9494563, whereas RIG-I was targeted with RHS1764-9499511. The control shRNA construct targets human Hsp70 interacting protein and contained the sequence RHS1764-9500871. For establishing RNAi, 293T cells grown in 6-well dishes to 80% confluency were transfected with 1 μg each of pVpack-VSV G, pVpack-GP (Stratagene), and the shRNA construct, subcultured, incubated with puromycin at day 3 post-transfection, and then incubated for another 2–3 days before usage.
Plasmid Constructs
All new plasmids were generated using standard molecular cloning techniques and confirmed by DNA sequencing. PCR for cloning purposes was carried out with AccuPrime Pfx DNA polymerase (Invitrogen). Expression plasmid pGFP-CTD expresses the C-terminal domain (CTD) of RPB1 fused to the C terminus of EGFP. The CTD sequence was PCR-amplified using pGST-CTD (27) as template (kindly provided by William S. Dynan and James Manley) and primers 5′-GAC ACA CTC GAG CTA TGT CTC CCA GCT ACT CGC CAA-3′ and 5′-GAC AGG ATC CTC AGT TCT CCT CGT CAC TGT CAT CCG GGC TGA TA-3′. These CTD forward and reverse primers contain engineered XhoI (forward primer) and a BamHI (reverse primer) restriction sites to allow in-frame cloning into the XhoI/BamHI sites of pEGFP-C1 (Clontech). Plasmids pI.18/3×FLAG_LACV NSs and pI.18/3×FLAG_ΔMx express the corresponding reading frames fused to a 3×FLAG tag sequence in the eukaryotic expression vector pI.18. To generate the pI.18/3×FLAG backbone vector, two complementary oligonucleotides encoding the 3×FLAG sequence (28) and carrying a 5′ PvuI and a 3′ BamHI recognition site were used. Sequences are 5′-GAA CGA TCG ACC ATG GAC TAC AAA GAC CAT GAC GGT GAT TAT AAA GAT CAT GAT ATC GAT TAC AAG GAT GAC GAT GAC AAG GGA TCC GGG-3′ and 5′- CCC GGA TCC CTT GTC ATC GTC ATC CTT GTA ATC GAT ATC ATG ATC TTT ATA ATC ACC GTC ATG GTC TTT GTA GTC CAT GGT CGA TCG TTC-3′. These oligonucleotides were dissolved in annealing buffer (10 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA), and the mix was first heated to 95 °C and then slowly cooled to room temperature. Subsequently, the oligonucleotides were digested with PvuI and BamHI and ligated into the plasmid pI.18 (kindly provided by Jim Robertson, National Institute for Biological Standards and Control, Hertfordshire, UK). Because the PvuI restriction site is present multiple times in pI.18, the vector was partially digested and subjected to agarose gel analysis. The band corresponding to the size of the correctly digested fragment was excised, purified, and digested with BamHI. After another round of agarose gel purification, the fragment was used for ligation with the PvuI/BamHI-digested oligonucleotides. The resulting plasmid, which carries a 3×FLAG sequence tag upstream of the multiple cloning site, was designated pI.18/3×FLAG. To construct the expression vector pI.18/3×FLAG_LACV NSs, the NSs sequence was PCR-amplified from the LACV S segment rescue plasmid (9) by using a forward primer containing a 5′ BamHI site (5′-GAG AGG ATC CTC GCA TCA ACA GGT GCA AAT GGA-3′) and a reverse primer containing an XhoI site (5′-GAC ACT CGA GCT AAA TAC CCA GAT AAT CTG TGG AT-3′). The PCR fragment was cloned into pI.18/3×FLAG via BamHI/XhoI sites present in both insert and vector. The control vector pI.18/3×FLAG_ΔMx was constructed in a similar way using appropriate primers (5′-GAG AGG ATC CGT TGT TTC CGA AGT GGA CAT CGC AAA A-3′ and 5′-GAC ACT CGA GTC ACT TGT CAT CGT CGT CCT TGT AGT CCG GGC ATC TGG TCA CGA T-3′) and a ΔMx plasmid (25) as template.
The T7-driven expression constructs for ΔMx and LACV NSs are described elsewhere (29). The firefly luciferase reporter construct for monitoring IFN-β promoter activation (p125Luc) was kindly provided by Takashi Fujita, Kyoto University, Japan (30). The control plasmid pRL-SV40 (Promega) contains the Renilla luciferase gene under control of the constitutively active SV40 promoter.
IRF-3 Dimerization Assay
Cells were lysed in buffer containing 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, protease inhibitors, and phosphatase inhibitors, vortexed, incubated on ice for 10 min, and then centrifuged at 4 °C for 5 min at 10,000 × g. Proteins were then separated by electrophoresis in a 10% nondenaturing polyacrylamide gel with 1% deoxycholate in the cathode buffer (31). The proteins were transferred to a PVDF membrane (Millipore) followed by incubation in saturation buffer (PBS containing 5% nonfat dry milk and 0.05% Tween). Membranes were incubated overnight with the polyclonal anti-IRF-3 antibody FL-425 (Santa Cruz Biotechnology) diluted 1:1000 in saturation buffer and then washed 3 times with 0.05% PBS, Tween followed by incubation with a horseradish peroxidase-conjugated secondary antibody. After three additional washing steps, detection was performed using the SuperSignal West Femto chemiluminescence kit (Pierce).
Transient Transfections and Reporter Gene Assays
Subconfluent 293T cells (reporter assays) or HeLa cells (immunofluorescence analyses) grown in six-well dishes were transfected with plasmids at the indicated concentrations. Nanofectin (PAA Laboratories GmbH, Coelbe, Germany) was used as the transfection reagent. At 18 h post-transfection, cells were harvested and either lysed in 100 μl of passive lysis buffer for reporter gene assays (Promega) or fixed and permeabilized for immunofluorescence as outlined below.
RNA Extraction and Real-time RT-PCR
Total cellular RNA was harvested at the indicated times post-infection using the Nucleospin kit (Macherey-Nagel) according to the manufacturer's recommendation. An amount of 600 ng to 1 μg of RNA was used to synthesize cDNA with the Quantitect reverse transcription kit (Qiagen). mRNA levels of human γ-actin, pak6, and p53 were detected by QuantiTect primers QT00996415, QT00089117, and QT00060235 (Qiagen) using the QuantiTect SYBR Green RT-PCR kit (Qiagen) and a LightCycler 1.5 (Roche Applied Science). Custom primers were used to detect γ-actin intron 3 (5′-GCT GTT CCA GGC TCT GTT CC-3′ and 5′-ATG CTC ACA CGC CAC AAC ATG C) (32), LACV N RNA (5′-GGG TAT ATG GAC TTC TGT G-3′ and 5′-GCC TTC CTC TCT GGC TTA-3′), and the 5.8 S rRNA precursor (5′-GCG TGT GGC GTG CGC CCC GC-3′ and 5′-CGC CGC CGG GTC TGC GCT T-3′). All values obtained were normalized against the γ-actin mRNA signal using the ddCT method (33). Up-regulation of inducible genes is depicted in relation to non-stimulated or non-infected (mock) cells. The absence of contaminating genomic DNA was verified by control reactions omitting reverse transcriptase during cDNA synthesis.
Bromouridine Labeling of Nascent RNA Transcripts
HeLa cells seeded in 6-well plates were infected with rLACV, rLACVdelNSs, or left uninfected (mock) for 8 h. For bromouridine (BrU) incorporation into nascent RNAs, the protocol as described in Wansink et al. (34) was used. Cells were washed once with PBS and once with glycerol buffer (20 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 0.5 mm PMSF, 0.5 mm EGTA, 25% glycerol) and subsequently incubated with permeabilization buffer (glycerol buffer containing 0.1% Triton X-100) for 3 min at room temperature. Labeling was performed by incubating the permeabilized cells with transcription buffer (100 mm KCl, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 1 mm PMSF, 0.5 mm EGTA, 25% glycerol, 2 mm ATP, 0.5 mm CTP, 0.5 mm GTP, 0.2 mm Br-UTP, 25 units/ml RNasin) for 30 min at 37 °C. To quench RNA polymerase II transcription, 1 μg/ml α-amanitin was added to the transcription buffer. The wells were rinsed twice with PBS, and immunofluorescence analysis detecting incorporated BrU was performed as described below.
Immunofluorescence Analysis
Cells were grown on coverslips to 30–50% confluency and incubated for the indicated times. Then cells were fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100 dissolved in PBS, and washed three times with PBS containing 1% FCS. Primary antibodies were diluted in PBS containing 1% FCS. The following antibodies were used: mouse monoclonal antibody H5 recognizing RPB1 CTD phosphorylated on serine 2 (Covance, 1:200), rabbit polyclonal recognizing the LACV N protein (10) (1:1000), and mouse monoclonal BMC9318 detecting BrU (Roche Applied Science, 1:100). After incubation at room temperature for 1 h, the coverslips were washed three times in PBS then treated with the secondary antibody goat anti-mouse Cy3 conjugated, goat anti-rabbit Cy2 conjugated, or goat anti-rabbit Cy5 conjugated at a dilution of 1:200, and cells were again washed three times in PBS and mounted using Fluorsave solution (Calbiochem). Chromatin was stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize nuclei during secondary antibody incubation in a 1:200 dilution. Stained cell samples were examined using a Leica confocal laser scanning microscope.
Western Blot Analysis
Cells were lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 7.6, 150 mm NaCl, 1% Nonidet P-40) containing protease inhibitors (Complete protease inhibitor, Roche Applied Science) and phosphatase inhibitors (phosphatase inhibitor mixture II, Calbiochem). A total of 10 μg of protein was separated on a 5% polyacrylamide gel by electrophoresis and blotted onto an Immobilon-P membrane (Millipore) following the manufacturer's instructions (Bio-Rad). The membrane was incubated with dilutions of mouse antibodies H5 recognizing CTD-p-Ser-2 (Covance, 1:500), H14 H5 recognizing CTD-p-Ser-5 (Covance, 1:500), N-20 recognizing the N terminus of RPB1 (Santa Cruz Biotechnology, 1:500), mouse monoclonal DO-1 recognizing p53 (Calbiochem, 1:500), rabbit polyclonal recognizing phospho-Ser-15 p53 (Cell Signaling, 1:1000), mouse monoclonal JBW301 recognizing phospho-Ser-139 H2A.X (Millipore, 1:1000), rabbit anti-LACV N serum (1:1000), or mouse monoclonal anti-tubulin (Sigma, 1:1000). Protein bands were visualized using the ECL plus method (Amersham Biosciences).
For signal quantification, proteins were transferred to nitrocellulose membranes (GE Healthcare) and incubated with 1:10 diluted hybridoma supernatant of Pol 3/3 mouse mAb for RPB1 (35) or mouse mAb against α-tubulin (Sigma, 1:10,000) at 4 °C overnight. The membranes were stained with affinity-purified, IR-labeled secondary antibodies against mouse (IR Dye 800 CW anti-mouse IgG (H+L); Rockland Inc.). Signals were imaged and quantified with a Odyssey (Li-Cor) system.
RESULTS
NSs of LACV Inhibits RIG-I-dependent IFN Induction Downstream of IRF-3
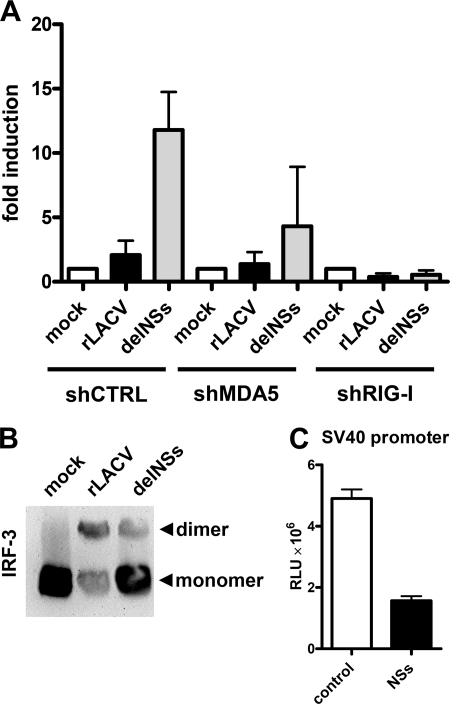
As part of our studies on the LACV-IFN system interactions, we wanted to know which intracellular pathogen recognition pathway would be activated by LACV. Therefore, IFN induction was measured in dependence of MDA5 and RIG-I, the main intracellular pathogen recognition receptors driving the IFN response against RNA viruses. MDA5 or RIG-I expression was knocked down using a previously established shRNA protocol (36), and IFN induction was determined with a reporter assay based on an IFN-β promoter luciferase construct. Fig. 1A shows that the NSs-expressing wt virus, rLACV, did not up-regulate the IFN-β promoter as expected. The NSs-deleted mutant virus rLACVdelNSs, by contrast, strongly activated the IFN-β promoter. IFN induction by rLACVdelNSs was severely affected when RIG-I was down-regulated, whereas MDA5 knockdown appears to have a weaker effect. In a similar vein, the RNA of concentrated LACV particles was found to contain the RIG-I-activating 5′ triphosphate group and to induce IFN transcription in a RIG-I-dependent manner (data not shown). These data demonstrate that in infected cells the viral RNA of LACV strongly triggers IFN induction via RIG-I and that NSs counteracts this signaling pathway.
FIGURE 1.
Activation of the RIG-I pathway by LACV and blockade by NSs downstream of IRF-3. A, RIG-I-dependent IFN induction and suppression by NSs is shown. Human 293T cells were treated with retroviral shRNA constructs directed against either MDA5 or RIG-I or against the heat shock 70 interacting protein as control (shCTRL) as described (36). Knockdown cells were transfected for 5 h with 500 ng of p125luc reporter construct encoding the firefly luciferase under control of the IFN-β promoter and 100 ng of pRL-SV40 reporter construct encoding the Renilla luciferase under control of the constitutively active SV40 promoter. Then cells were infected for 18 h with rLACV or rLACVdelNSs or were left uninfected (mock). Specific IFN-β promoter activity was determined by normalizing firefly to Renilla luciferase activities and setting the mock-infected control as 1. Mean values and S.D. from three independent experiments are shown. B, IRF-3 homodimerization is shown. Extracts from cells infected with rLACV or rLACVdelNSs for 16 h were analyzed by nondenaturing gel electrophoresis followed by an immunoblot to detect IRF-3. Positions of IRF-3 monomers and dimers are indicated on the right. C, influence of NSs on the constitutively active SV40 promoter is shown. 293T cells grown in 12 well plates were transfected with 100 ng of pRL-SV40 and 1 μg of either pI.18/3×FLAG_LACV NSs (NSs) or the plasmid pI.18/3×FLAG_ΔMx (control). Renilla luciferase activity was measured after overnight incubation. Mean values and S.D. from three independent experiments are shown. RLU, Renilla luciferase units.
We assessed whether the block in IFN induction imposed by NSs occurs on the level of the IFN-specific signaling chain, which begins with RIG-I and ends with the transcription factor IRF-3. In uninfected cells, IRF-3 is present in the cytoplasm as a monomer, whereas activated IRF-3 dimerizes and moves to the nucleus. To investigate the influence of NSs on IRF-3 activation, we infected cells with recombinant wt and mutant viruses and monitored the dimerization state of IRF-3 by native gel electrophoresis followed by Western blot analysis. Fig. 1B shows that both rLACV and LACVdelNSs triggered dimerization of IRF-3 in a similar manner, suggesting that the NSs-imposed block on IFN induction does not occur at the level of IRF-3 but further downstream. In support of this assumption, LACV NSs expressed from a cDNA plasmid strongly inhibited a co-transfected reporter gene that is under control of the constitutively active SV40 promoter (Fig. 1C). Our data, thus, indicate that LACV activates RIG-I by the 5′-triphosphate group of its genomic RNA and that NSs counteracts IFN induction downstream of IRF-3, most likely at the level of cellular RNA transcription.
NSs Is a Global Inhibitor of RNA Polymerase II
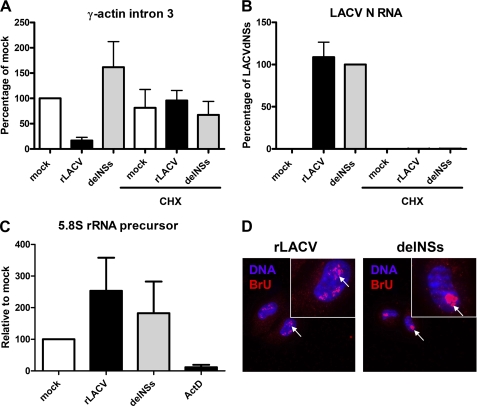
We further explored the possibility that NSs may affect cellular RNA synthesis in general. Simply determining overall mRNA levels in infected cells would not give reliable information about the rate of transcription, as many RNA molecules have long half-lives. For introns and pre-mRNA species, however, it is assumed that their half-life is within the range of minutes (37). Therefore, we used intron abundance as a readout of de novo mRNA synthesis by RNAP II, measured by real-time RT-PCR. A similar strategy has been described for analysis on the human insulin gene transcription (38). For our analyses we chose an intron of the γ-actin transcription unit because this gene is strongly and constitutively transcribed (32). Cells were infected with either rLACV or the delNSs mutant or were left uninfected. After 4 h total RNA was prepared, and real-time RT-PCR analyses were performed using primers specific for intron 3 of the γ-actin gene. As shown in Fig. 2A, the relative abundance of γ-actin intron 3 was reduced by ∼80% in rLACV-infected cells as compared with the mock control. Mutant virus LACVdelNSs, by contrast, was unable to reduce the amount of introns. Importantly, preincubation of cells with the translation inhibitor cycloheximide completely abrogated the effect of rLACV on intron abundance, indicating that NSs needs to be synthesized upon infection. Moreover, cycloheximide treatment was shown to inhibit transcription of the viral N gene (Fig. 2B), as expected, as Bunyavirus transcription is strictly coupled to translation (39). Similar results were obtained with primers specific for intron 4 and for the intron 5-exon 6 border of the γ-actin gene (data not shown). Thus, NSs expressed by rLACV efficiently and rapidly suppresses mRNA synthesis by RNAP II.
FIGURE 2.
NSs specifically suppresses RNAP II. A–C, real-time RT-PCR analysis is shown. 293T cells were seeded in 12-well plates to 80% confluency. At 1 h before infection, cells were either pretreated with 50 μg/ml cycloheximide (CHX) or left untreated. Then cells were either infected with rLACV or rLACVdelNSs (m.o.i. = 5) or mock-infected or treated with 1 μg/ml actinomycin D (ActD). After 4 h of incubation, total cell RNA was extracted, and real-time RT-PCR was performed using primers specific for γ-actin intron 3 (A), LACV N RNA (B), or the 5.8 S rRNA precursor (C). All values obtained were normalized against γ-actin mRNA levels. Up-regulation of inducible genes is depicted in relation to mock-infected cells. Mean values and S.D. from three independent experiments are shown. D, an in situ run-on assay is shown. HeLa cells were grown on coverslips and infected with rLACV or rLACVdelNSs (m.o.i. = 1) for 8 h. After permeabilization with buffer containing Triton X-100, cells were incubated with buffer containing Br-UTP to label newly synthesized RNA and α-amanitin to quench RNAP II. After 30 min of labeling, cells were fixed and analyzed by immunofluorescence using an antibody against BrU (red signal) and counterstained with DAPI (blue signal). Insets show a magnification of individual nuclei, and arrows indicate the nucleoli containing de novo synthesized RNAP I transcripts.
We devised a similar real time RT-PCR assay to see whether LACV infection has an effect on RNAP I. As for RNAP II transcripts, rRNA precursors are rapidly processed after their transcription, and their abundance, thus, reflects transcriptional processivity by RNAP I (40). The cells infected for 4 h were assayed for the presence of a typical RNAP I transcript, the precursor of the 5.8 S rRNA. As shown in Fig. 2C, 5.8 S rRNA precursor abundance was not down-regulated upon LACV infection. As control, we showed that the pan DNA-dependent RNA polymerase inhibitor actinomycin D was able to substantially reduce the relative amount of 5.8 S rRNA precursors. To bolster these findings, we performed an in situ RNAP I run-on assay. Cells were infected for 8 h with the viruses, permeabilized, and incubated with a buffer containing ATP, CTP, GTP, bromo-UTP, and α-amanitin. The NTPs and Br-UTP are incorporated into nascent transcripts, whereas α-amanitin serves to quench transcription by RNAP II. After 30 min of incubation, cells were fixed and immunostained with an antibody recognizing BrU and with DAPI as a nuclear counterstain. Fig. 2D shows that BrU-labeled transcripts were detected in cells infected with both viruses, although they are more dispersed upon rLACV infection. The addition of actinomycin D to the NTP mix completely abrogated Br-UTP incorporation in uninfected cells, as expected (data not shown). Because RNAP I is responsible for 80% of the nuclear transcription (41), the presence of de novo RNA transcripts in cells treated with α-amanitin and infected with rLACV, thus, again indicates that RNA polymerase I activity is not compromised by NSs. Taken together, our results demonstrate that infection with wt rLACV (but not with the delNSs mutant) strongly and swiftly inhibits RNAP II transcription. This indicates that the IFN antagonist NSs of LACV is a specific inhibitor of RNAP II.
NSs Affects the Hyperphosphorylated Form of Mammalian RNAP II
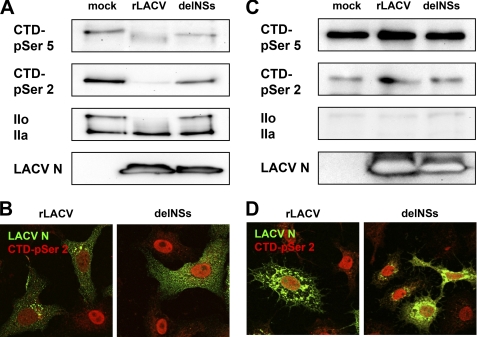
The large subunit of RNAP II, RPB1, contains a long, flexible CTD that consists of 52 heptad repeats with the consensus sequence 1YSPTSPS7 (42). Phosphorylation and dephosphorylation of the serine 2 and serine 5 residues in these heptad repeats drive the RNAP II transcription cycle, although a contribution of serine 7 is also established (43). RNAP II recruited to the promoter site is first phosphorylated on CTD-serine 5 of RPB1, whereas elongating RNAP II bears phosphate groups on both CTD serine 2 and serine 5. We investigated whether wt rLACV infection has an influence on the phosphorylation status of RPB1. To this end, Vero cells were infected for 12 h with viruses, and Western blot analysis was performed using phosphospecific antibodies for the CTD heptad residue serine 5 (CTD-p-Ser-5) or serine 2 (CTD-p-Ser-2). Moreover, we analyzed the presence of the hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of RNAP II. This is possible through using an antibody directed against the N terminus of RPB1, as IIo and IIa strongly differ in their molecular weight. Strikingly, the signals for CTD-p-Ser-2 signal as well for RNAP IIo completely disappeared after wt rLACV infection, whereas the rLACVdelNSs mutant had no effect (Fig. 3A). Also, the CTD-p-Ser-5 signal was affected to some extent, again exclusively by the NSs-expressing rLACV. The same results were obtained by immunofluorescence analysis. When infected cells were stained for CTD-p-Ser-2, a clear signal was only detected in cells infected with the delNSs mutant, whereas rLACV infection yielded only poor staining (Fig. 3B).
FIGURE 3.
RPB1 subunit of RNAP II in LACV-infected mammalian and insect cells. A, Vero cells were either mock-infected or infected with rLACV or rLACVdelNSs at 5 plaque-forming units per cell. At 12 h post-infection, cells were lysed and analyzed by Western blot using either phosphospecific antibodies for the CTD heptad residue serine 5 (CTD-p-Ser-5) or serine 2 (CTD-p-Ser-2), an antibody recognizing the N terminus of RPB1 to visualize hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms or an antiserum against the LACV N protein. B, Vero cells were infected with viruses at an m.o.i. of 1 and analyzed by immunofluorescence for the presence of RPB1 CTD-p-Ser-2 (red) or LACV N (green) using specific antibodies. C and D, shown are A. albopictus C6/36 cells infected for 24 h at an m.o.i. of 10 and analyzed by Western blot (C) and immunofluorescence (D) as described for A and B, respectively. Note that the N signal is much stronger in insect cells due to the prolonged infection time.
In contrast to the lytic infection of mammalian cells, infection of mosquito cells by LACV is persistent. When we performed the same analyses of RNAP II in A. albopictus mosquito cells, no changes in the state of RPB1 were observed independent of the infecting virus (Figs. 3, C and D).
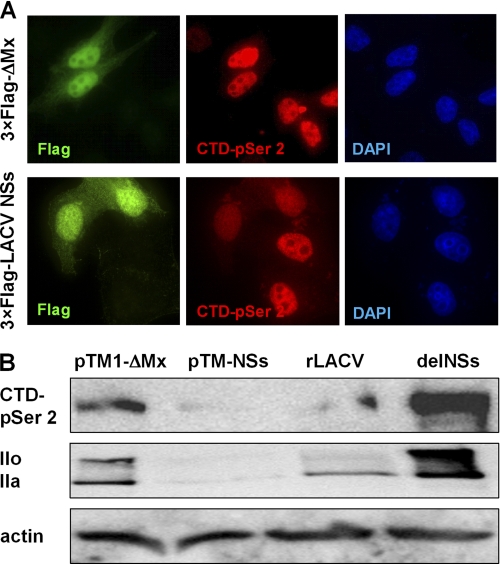
To investigate whether NSs is sufficient to obtain the changes in RNAP II, we monitored the state of RNAP II in cells transfected with an NSs-expressing cDNA construct. Of note, robust detection of LACV NSs expressed from an RNAP II-driven promoter required usage of a high level plasmid vector containing a 5′ splice site (pI.18) and fusion of the NSs reading frame to an N-terminal 3×FLAG epitope tag (28). HeLa cells were transfected either with the 3×FLAG NSs pI.18 plasmid or with a control expressing the biologically inactive 3×FLAG-tagged ΔMx protein. After 18 h of incubation, cells were fixed and processed for double immunofluorescence analysis using antisera specific for the FLAG tag and for CTD-p-Ser-2. Fig. 4A shows that in cells expressing 3×FLAG-tagged NSs, the CTD-p-Ser-2 signal is drastically reduced, whereas the ΔMx control had no such effect. To reinforce these observations on the total cell level, we had to bypass the autoinhibitory effect of NSs on RNAP II by switching to a T7 expression system. The Western blot shown in Fig. 4B, upper lane, confirms that overexpression of NSs abrogated the CTD-p-Ser-2 signal. Strikingly, T7-driven overexpression of NSs also leads to a disappearance not only of IIo, as in the case of rLACV infection, but also of IIa (Fig. 4B, middle panel). Taken together, the data indicate that NSs preferentially targets the phosphorylated form of RNAP II in a manner specific for mammalian cells.
FIGURE 4.
Influence of ectopically expressed LACV NSs on RPB1. A, HeLa cells were seeded on coverslips in 6-well plates and transfected with 1 μg of cDNA expression constructs pI.18/3×FLAG_ΔMx or pI.18/3×FLAG_LACV NSs. After 18 h of incubation, cells were fixed and immunostained for the FLAG tag (green) and the RPB1 CTD-p-Ser-2 (red), and nuclear DNA was counterstained with DAPI (blue). B, BSR-T7/5 cells were transfected with 1 μg of expression constructs pTM-ΔMx or pTM-LACV NSs or infected with LAC viruses (m.o.i. = 5) and analyzed by Western blot after 18 h incubation as indicated for Fig. 3A.
No Apparent Influence of NSs on the CTD Phosphorylation System
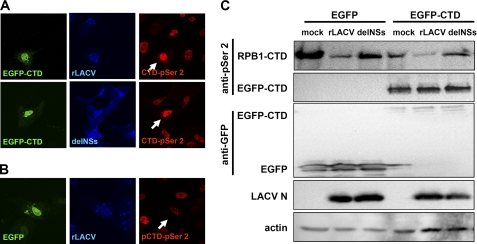
An obvious explanation for the observed influence of NSs on CTD-phosphorylated RNAP II would be the inhibition of the relevant kinases or the activation of a CTD phosphatase. As a tool to investigate this, we fused the human CTD sequence to EGFP and monitored its phosphorylation state in infected cells. CTD sequences expressed in isolation are known to be phosphorylated in a similar manner as the RNAP II-borne CTD (27). For immunofluorescence, we used the CTD-p-Ser-2-specific antibody as a sensitive readout. In cells transfected with EGFP-CTD, wt rLACV did not decrease the CTD-p-Ser-2 signal (Fig. 5A, upper panel), and the signal remained as strong as after rLACVdelNSs infection (Fig. 5A, lower panel) or mock infection (data not shown). A control experiment demonstrated that EGFP without a fused CTD had no influence on the reduction of the CTD-p-Ser-2 signal by rLACV (Fig. 5B). Most likely, the CTD-p-Ser-2 signal observed in EGFP-CTD/rLACV cells (see Fig. 5A) is derived from the recombinant CTD fused to EGFP and not from the cellular CTD. To investigate this in a more direct manner, infected cells that had been transfected with the EGFP constructs were analyzed by Western blot. Fig. 5C shows that in cells infected with rLACV, the CTD-p-Ser-2 signal of EGFP-CTD is unaffected by rLACV infection, whereas the CTD-p-Ser-2 signal of RNAP II is reduced to the same extent as in EGFP-transfected cells. Thus, an exogenous CTD is inert to the action of NSs and does not serve as a decoy target. In line with this, overexpression of EGFP-CTD did not turn rLACV into an IFN inducer like the delNSs virus (supplemental Fig. S1). Moreover, overexpression of the CTD kinases CDK7, CDK8, or CDK9 along with their respective cyclins could not rescue the inhibitory effect of NSs on IFN induction (data not shown). Together, these results are not compatible with the idea that LACV NSs reduces the RNAP IIo form by influencing the CTD phosphorylation/dephosphorylation system.
FIGURE 5.
Phosphorylation of exogenous CTD sequences in LACV-infected cells. A and B, Vero cells were transfected for 18 h with a cDNA construct expressing the RNAP II CTD fused to EGFP (A) or an EGFP-only plasmid as control (B). Then cells were infected for 24 h with rLACV or rLACVdelNSs (m.o.i. = 0.5), fixed, and permeabilized and immunostained for the presence of RPB1 CTD-p-Ser-2 (red) and LACV N (blue) using specific antibodies. GFP was detected via its autofluorescence activity. Arrows indicate individual cells that are both transfected and infected. C, 293T cells were transfected and infected as in A and analyzed by Western blot using antibodies specific for CTD-p-Ser-2, EGFP, LACV N, and cellular actin. CTD-p-Ser-2 signals of RPB2 and EGFP could be distinguished by their different molecular weights. Note the higher expression levels of GFP compared with GFP-CTD. Most likely, CTD overexpression detracts basal CTD phosphorylation factors away from the RNAP II machinery, as RNAP II CTD phosphorylation is generally reduced in GFP-CTD-transfected cells (compare the first through third lanes with the fourth through sixth lanes in the upper panel).
NSs Triggers Degradation of RNAP IIo
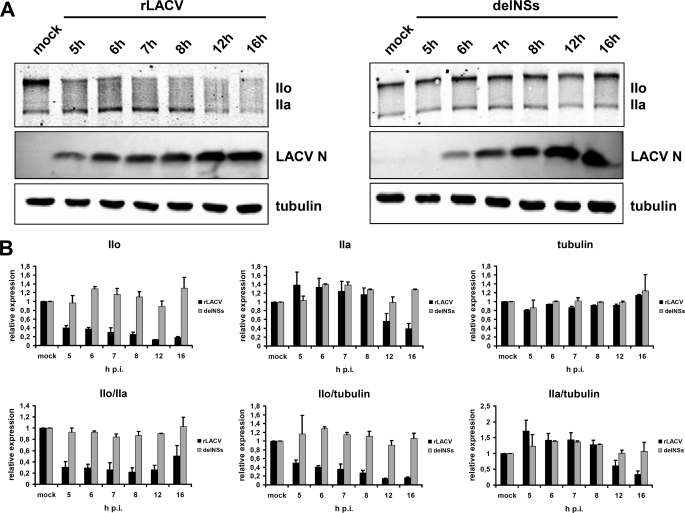
Close examination of the IIo/IIa Western blots in rLACV-infected cells (see Figs. 3A and 4B) indicate that the hypophosphorylated IIa signal is not increasing at the expense of the hyperphosphorylated IIo form. This would again argue against dephosphorylation as the explanation for the disappearance of IIo. To examine whether proteolytic degradation may be involved, we performed a time-course analysis of the RNAP IIo and IIa signals. Vero cells were infected with the viruses, and samples were taken every hour from 5 h post-infection (p.i.) to 8 h p.i. and additionally at 12 h and 16 h p.i. Cell extracts were examined by Western blot analysis for the presence of IIo and IIa and for tubulin and the viral N protein as controls (Fig. 6A). To measure the relative amounts of respective proteins, Western blot signals of the cellular proteins were quantified. As is illustrated in Fig. 6B, upper panel, levels of IIo rapidly decreased upon rLACV infection, whereas the delNSs mutant had no such effect. IIa levels, by contrast, remained similar between the two virus infections for at least 8 h but were also specifically reduced by rLACV from 12 h p.i. on. Tubulin levels were not affected by either virus. We also set the different protein signals in relation to each other (Fig. 6B, lower panel). This analysis showed again that in rLACV-infected cells the IIo/IIa ratio and the IIo/tubulin ratio were decreased from 5 h p.i. on and the IIa/tubulin ratio from 12 h p.i. on. In rLACVdelNSs-infected cells, all signal ratios were comparable with mock-infected cells. We noted, however, a modest increase in IIa levels in rLACV-infected cells at 5 h p.i., which is caught up in delNSs-infected cells at 6 h p.i. Because multiplication of the delNSs virus is slightly delayed compared with rLACV (see LACV N blots in Fig. 6A), we assume that the slight increase in IIa levels represent a general reaction to virus infection. Taken together, the protein quantification experiments demonstrate that IIa levels remain largely constant during the first few hours of wt virus infection, whereas IIo levels go down. From 12 h p.i. on, IIo has largely disappeared, and IIa levels are also affected. These data suggest that NSs expressed by rLACV triggers the specific disappearance of IIo, the transcriptionally active form of RNAP II. Because IIo is recruited from a pool of IIa polymerases, the constant depletion of IIo eventually results in the disappearance of IIa as well.
FIGURE 6.
Time course of RNP IIo and IIa under LACV infection. A, Western blot analysis is shown. Vero cells were either mock-infected or infected with rLACV or rLACVdelNSs (m.o.i. = 5). Samples were taken from 5 to 8 h p.i. every hour and in addition at 12 h and at 16 h p.i. Western blot analysis was performed using an antibody recognizing the N terminus of RPB1 to visualize hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms and antibodies recognizing LACV N or tubulin as infection and loading controls, respectively. B, Western blot signals obtained in A were quantified using an Odyssey imaging system, and relative protein levels (upper panel) and ratios (lower panel) of individual proteins in infected and mock cells were determined. Shown are the mean values and S.D. from two independent Western blots and quantifications.
NSs Action Is Proteasome-dependent and Partially Resembles a DNA Damage Response
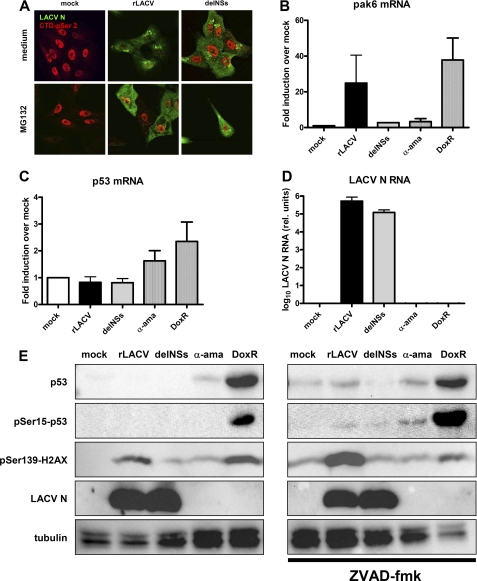
It is known that DNA damage, which obstructs RNAP II progression, results in proteolysis of the large subunit of RNAP II, RPB1. This degradation is commonly mediated by the ubiquitin-proteasome pathway and confined to the IIo form of RPB1, which carries out transcriptional elongation (44–50). As our data suggest specific degradation of IIo by NSs, we wanted to investigate an involvement of the proteasome using the specific inhibitor MG132. Unfortunately, global analysis of RNAP II states by Western blot turned out to be unreliable, as proteasomal inhibitors substantially delay LACV replication. Using immunofluorescence analysis, however, we were able to detect viral gene expression in individual MG132-treated cells (Fig. 7A, lower panel). Strikingly, in these rLACV-infected, MG132-treated cells the CTD-p-Ser-2 signal was as strong as in the mock- or delNSs-infected cells. Because normally in rLACV-infected cells the signals for LACV N and for CTD-p-Ser-2 are mutually exclusive (Fig. 7A, upper panel), we conclude that NSs triggers the disappearance of RNAP IIo via the proteasomal degradation pathway.
FIGURE 7.
Similarities between LACV NSs action and the DDR. A, proteasome inhibition and the NSs effect on RNAP II is shown. Vero cells grown on coverslips were either infected with rLACV or rLACVdelNSs at an m.o.i. of five or left uninfected (mock). During the infection period of 16 h, cells were either left untreated (upper panel) or kept under 0.5 μm MG132 (lower panel). Then cells were fixed and analyzed by indirect immunofluorescence with antibodies directed against CTD-p-Ser-2 (red) or LACV N (green). B–D, real-time RT-PCR analysis is shown. HeLa cells were either mock-infected, infected with rLACV or rLACVdelNSs (m.o.i. = 5), or treated with 10 μg/ml α-amanitin (α-ama) or 0.5 μg/ml doxorubicin (DoxR). After 8 h of incubation, total cell RNA was extracted, and real-time RT-PCR was performed using primers specific for pak6 mRNA (B), p53 mRNA (C), or LACV N RNA (D). All values obtained were normalized against γ-actin mRNA levels. Results shown are triplicates represented as the mean ± S.D. E, Western blot analysis is shown. Vero cells were mock-infected, infected with rLACV, or rLACVdelNSs or treated with α-amanitin or DoxR as indicated above. After 8 h of incubation, cells were lysed, and protein extracts were tested for the presence of p53 protein, p53 Ser-15 phosphorylation, histone H2A.X Ser-139 phosphorylation, LACV N expression, and tubulin levels using appropriate antisera (left panel). In a parallel experiment, infected and treated cells were kept under 100 μm ZVAD-fluoromethyl ketone (fmk) during the whole incubation period (right panel).
We directly addressed possible similarities between NSs action and the DNA damage response (DDR) on the transcriptional and signaling levels. It is known that some genes of the DDR can be transcribed by the non-phosphorylated IIa form of RNAP II (70). We evaluated the transcriptional state of pak6, a sensitive DDR gene (51), using real-time RT-PCR. Fig. 7B shows that pak6 transcripts are clearly up-regulated by rLACV infection but not by the delNSs mutant. Importantly, pak6 mRNA levels were unaffected by the specific RNAP II inhibitor α-amanitin but up-regulated by the DNA-damaging agent doxorubicin. p53 mRNA levels, by contrast, remained unchanged by infection with any virus but were slightly up-regulated by doxorubicin (Fig. 7C). Real-time RT-PCR analysis for the LACV N mRNA confirmed infection of cells (Fig. 7D). Thus, apparently, the DDR gene pak6 escapes the strong inhibitory effect of NSs on RNAP II transcription.
We also monitored several classical DDR markers on the protein and signaling level, namely p53 protein, p53 Ser-15 phosphorylation, and histone H2A.X Ser-139 phosphorylation (52). Western blot analysis demonstrates that p53 protein levels and p53 Ser-15 phosphorylation are not detectably activated by an 8-h infection with any LACV variant, whereas the DNA-damaging agent doxorubicin up-regulated these DDR markers (Fig, 7E, left panel). In a similar vein, BRCA1 phosphorylation on Ser-1524, another important DDR marker, is not activated by wt rLACV (supplemental Fig. S2). Strikingly, however, wt LACV triggered Ser-139 phosphorylation of histone H2A.X at a similar level as doxorubicin, whereas the delNSs mutant had no such effect. LACV NSs is known to activate caspase-dependent apoptosis (53). To rule out NSs-mediated apoptosis as the cause of H2A.X Ser-139 phosphorylation, we performed a parallel infection experiment with added pan-caspase inhibitor ZVAD- fluoromethyl ketone. Fig. 7E, right panel, demonstrates that also under caspase-inhibited conditions, wt LACV, but not the delNSs mutant, strongly activates phosphorylation of histone H2A.X. Together with the fact that NSs triggers degradation of IIo in a proteasome-dependent manner, we conclude that the virulence factor NSs of LACV exploits parts of the DDR machinery to impose the blocking and degradation of RNAP II.
DISCUSSION
LACV is capable of causing severe encephalitis in humans and experimental animals (1, 2, 54). We have previously demonstrated that virus capacity to rapidly spread in the host is largely dependent on the NSs protein and that NSs is only important if the host has a functional IFN system (10). Moreover, NSs-expressing wt LACV was shown to be capable of suppressing the induction of IFN-β in both virus-infected cells and animals. Nonetheless, as we have shown here, wt LACV strongly activates the IFN-β transcription factor IRF-3. This is, however, counterbalanced by the fast and efficient block in RNAP II-dependent transcription imposed by NSs. Thus, NSs is different from most other viral IFN antagonists (15, 55) by acting downstream of the canonical RIG-I/IRF-3 signaling pathway.
Bunyaviruses like LACV use host cell mRNAs as primers for viral transcription (56). However, blocking cellular RNA synthesis by actinomycin D has no effect on the multiplication of LACV (57). This indicates that the cytoplasmic pool of host mRNAs is sufficient for viral transcription and that LACV does not require any nuclear function for transcription or replication of its genome. Thus, LACV can afford to express an IFN antagonist, which completely shuts down RNAP II. Similar observations apply to the bunyaviruses BUNV and RVFV. Both these viruses encode NSs proteins, which, although being different in sequence, size, and mode of expression, act on the level of general host cell transcription. BUNV (which is closely related to LACV) expresses an NSs, which interferes with the phosphorylation on CTD-Ser-2 (25) and interacts with MED8, a component of the Mediator transcription complex (58). The Mediator complex is involved in RNAP II regulation (59) and contacts the CTD-Ser-2 kinase P-TEFb (60). BUNV NSs may, thus, utilize MED8 to down-regulate CTD-Ser-2 phosphorylation and, hence, RNAP II transcription. The NSs of RVFV (which is more remotely related to LACV) inhibits IFN transcription by two different means. First, RVFV specifically inhibits the IFN-β promoter by recruiting the repressor protein SAP30 (24). Moreover, it interacts with and degrades the p44 subunit of the general transcription factor TFIIH (23). Different to the RNAP II-specific NSs of LACV, the actions of RVFV NSs lead to a decrease in RNA synthesis by both RNAP I and RNAP II (23). For BUNV it has not been investigated so far whether its NSs also affects RNAP I. The bunyaviruses LACV, BUNV, and RVFV, thus, encode NSs proteins that either degrade transcriptionally active RNAP II (LACV), interfere with RNAP II CTD-Ser-2 phosphorylation (BUNV), or target TFIIH (RVFV). It should be mentioned that bunyaviruses replicate in the cytoplasm and, due to the segmented nature of their genome, contain three RIG-I ligands per intruding particle (Ref. 36 and data not shown). The high virion content of these immunogenic molecules may have driven the evolution of IFN antagonists, which quickly and radically shut down host cell transcription in a general manner.
LACV infection of mammalian cells is lytic, whereas infection of insect cells is non-cytolytic and persistent. Our data indicate that NSs only affects RNAP II of mammalian cells but not of mosquito cells. The host specificity of NSs action, thus, parallels the presence (mammals) or absence (insects) of an antiviral IFN system and supports our claim that NSs has evolved as an IFN antagonist, although it inhibits host transcription in a general manner.
Two other viruses, herpes simplex virus and influenza A virus, were previously shown to trigger degradation of RNAP II (61–64). However, for both these viruses, levels of total RNAP II or of IIa are affected rather than preferentially IIo as in the case of LACV. Herpes simplex virus achieves RNAP II degradation through the proteasomal system (61, 62), whereas for influenza A virus the proteolytic viral protein PA is responsible (63, 64). It should be noted that, in contrast to LACV, these viruses are directly dependent on RNAP II activity. RNAP II degradation by herpesvirus (a DNA virus) is discussed as being a side effect of massive RNAP II transcription of viral genes (61) or as a means to prefer viral transcription over cellular transcription (62). For influenza A virus, RNAP II degradation is possibly a mechanism to shift the balance from viral transcription (which is RNAP II-dependent) to genome replication late in infection (65).
Some aspects of LACV NSs action have striking similarity to the response of cells to damaged DNA, the DDR. Productively elongating RNAP II normally requires an RPB1 subunit with a phosphorylated CTD-Ser-2. DNA lesions cause a transcriptional arrest of RNAP II, which in turn triggers the degradation of CTD-Ser-2-phosphorylated RPB1 via the ubiquitin-proteasome system (48, 66–69). Removal of the RPB1 subunit from the transcribing RNAP IIo complex then results in a disassembly of the whole complex (47). DDR genes, however, can bypass the requirement for CTD-Ser-2 phosphorylation and, thus, escape the transcriptional arrest imposed by IIo degradation (51, 70). We found that LACV NSs triggered up-regulation of the DDR gene pak6 despite its usually deleterious effect on RNAP II activity. Moreover, NSs triggered Ser-139 phosphorylation of histone H2A.X, one of the first events during the DDR. On the other hand, neither p53 nor BRCA1, two other important DDR markers, are influenced by NSs. Apparently, LACV NSs exploits parts of the DDR machinery to attack CTD-Ser-2 phosphorylated RNAP II.
It is well known that many DNA viruses and retroviruses counteract or utilize the DDR machinery to promote infection (71). Moreover, the RNA viruses simian virus 5 and human parainfluenza virus 2 were shown to utilize the damage-specific DNA-binding protein 1 to degrade the IFN signaling molecule STAT1 in the cytoplasm (72–74). However, we are not aware of any example of an RNA virus that has directly subjugated immediate early steps of the DDR. LACV may, thus, pursue a unique strategy for shutting down the antiviral host response by exploiting a rapidly reacting cellular pathway, which normally ensures genome integrity. How exactly NSs interacts with the DDR will be subject to our future investigations.
The cellular interaction partners of LACV NSs remain to be identified. LACV NSs does not interact with MED 8 or p44 (data not shown), the host cell factors mediating RNAP II inhibition by the NSs proteins of BUNV and RVFV, respectively (23, 58). Moreover, unlike what was shown for herpes simplex virus (75), a dominant-negative form of HSC70 cannot rescue RNAP IIo degradation by LACV infection (data not shown). This indicates that RNAP II inhibition or degradation by LACV NSs is different from the mechanisms of other viruses. Because the early steps of DDR signaling are not fully understood yet, identifying cellular interaction partners of NSs may also help to shed more light onto the initiation and execution of the DDR.
It is becoming increasingly clear that regulation of RNAP II activity occurs not only on the level of RNAP II recruitment and transcription initiation but also on the level of mRNA elongation (76, 77). In addition, more and more genes are identified on which RNAP II is preloaded and phosphorylated on CTD-Ser-5, but is paused until being activated by the CTD-Ser-2 kinase P-TEFb (76, 78–80). Those genes often belong to classes reacting to external stimuli or developmental signals. Recently, innate immunity genes were also shown to be regulated on the level of transcriptional elongation (81). Thus, interfering with elongating RNAP IIo by LACV NSs may not only be an efficient way to achieve a general host mRNA shut-off but also to ensure a certain selectivity for genes involved in the antiviral response.
Supplementary Material
Acknowledgments
We thank Valentina Wagner for excellent technical assistance and Otto Haller for constant support and advice. Moreover, we are indebted to Takashi Fujita, William S. Dynan, James Manley, and Jim Robertson for providing essential reagents.
This work was supported by Grants We2616/2-3 and SFB 593 (to F. W.) and SFB-Transregio5 (to D. E.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- LACV
- La Crosse virus
- BrU
- bromouridine
- BUNV
- Bunyamwera virus
- DDR
- DNA damage response
- IRF
- IFN regulatory factor
- m.o.i.
- multiplicity of infection
- NSs
- nonstructural protein encoded by the S segment
- p.i.
- post-infection
- rLACV
- recombinant La Crosse virus NSs deletion mutant
- RVFV
- Rift Valley fever virus
- RNAP II
- RNA polymerase II
- N
- nucleocapsid protein
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. McJunkin J. E., de los Reyes E. C., Irazuzta J. E., Caceres M. J., Khan R. R., Minnich L. L., Fu K. D., Lovett G. D., Tsai T., Thompson A. (2001) N. Engl. J. Med. 344, 801–807 [DOI] [PubMed] [Google Scholar]
- 2. Rust R. S., Thompson W. H., Matthews C. G., Beaty B. J., Chun R. W. (1999) J. Child Neurol. 14, 1–14 [DOI] [PubMed] [Google Scholar]
- 3. Whitley R. J., Gnann J. W. (2002) Lancet 359, 507–513 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention (2005) Confirmed and Probable California Serogroup Viral (Mainly La Crosse) Encephalitis Cases, Human, United States, 1964–2005, by State, www.cdc.gov
- 5. McJunkin J. E., Khan R. R., Tsai T. F. (1998) Infect. Dis. Clin. North. Am. 12, 83–93 [DOI] [PubMed] [Google Scholar]
- 6. Lambert A. J., Blair C. D., D'Anton M., Ewing W., Harborth M., Seiferth R., Xiang J., Lanciotti R. S. (2010) Emerg. Infect. Dis. 16, 856–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borucki M. K., Kempf B. J., Blitvich B. J., Blair C. D., Beaty B. J. (2002) Microbes Infect. 4, 341–350 [DOI] [PubMed] [Google Scholar]
- 8. Elliott R. M., Weber F. (2009) Viruses 1, 1003–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blakqori G., Weber F. (2005) J. Virol. 79, 10420–10428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blakqori G., Delhaye S., Habjan M., Blair C. D., Sánchez-Vargas I., Olson K. E., Attarzadeh-Yazdi G., Fragkoudis R., Kohl A., Kalinke U., Weiss S., Michiels T., Staeheli P., Weber F. (2007) J. Virol. 81, 4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delhaye S., Paul S., Blakqori G., Minet M., Weber F., Staeheli P., Michiels T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7835–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lienenklaus S., Cornitescu M., Zietara N., £yszkiewicz M., Gekara N., Jabłónska J., Edenhofer F., Rajewsky K., Bruder D., Hafner M., Staeheli P., Weiss S. (2009) J. Immunol. 183, 3229–3236 [DOI] [PubMed] [Google Scholar]
- 13. Samuel C. E. (2001) Clin. Microbiol. Rev. 14, 778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pichlmair A., Reis e Sousa C. (2007) Immunity 27, 370–383 [DOI] [PubMed] [Google Scholar]
- 15. Randall R. E., Goodbourn S. (2008) J. Gen. Virol. 89, 1–47 [DOI] [PubMed] [Google Scholar]
- 16. Yoneyama M., Fujita T. (2009) Immunol. Rev. 227, 54–65 [DOI] [PubMed] [Google Scholar]
- 17. Kato H., Takeuchi O., Mikamo-Satoh E., Hirai R., Kawai T., Matsushita K., Hiiragi A., Dermody T. S., Fujita T., Akira S. (2008) J. Exp. Med. 205, 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pichlmair A., Schulz O., Tan C. P., Näslund T. I., Liljeström P., Weber F., Reis e Sousa C. (2006) Science 314, 997–1001 [DOI] [PubMed] [Google Scholar]
- 19. Pichlmair A., Schulz O., Tan C. P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. (2009) J. Virol. 83, 10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schlee M., Roth A., Hornung V., Hagmann C. A., Wimmenauer V., Barchet W., Coch C., Janke M., Mihailovic A., Wardle G., Juranek S., Kato H., Kawai T., Poeck H., Fitzgerald K. A., Takeuchi O., Akira S., Tuschl T., Latz E., Ludwig J., Hartmann G. (2009) Immunity 31, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt A., Schwerd T., Hamm W., Hellmuth J. C., Cui S., Wenzel M., Hoffmann F. S., Michallet M. C., Besch R., Hopfner K. P., Endres S., Rothenfusser S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiscott J. (2007) Cytokine Growth Factor Rev. 18, 483–490 [DOI] [PubMed] [Google Scholar]
- 23. Le May N., Dubaele S., Proietti, De Santis L., Billecocq A., Bouloy M., Egly J. M. (2004) Cell 116, 541–550 [DOI] [PubMed] [Google Scholar]
- 24. Le May N., Mansuroglu Z., Léger P., Josse T., Blot G., Billecocq A., Flick R., Jacob Y., Bonnefoy E., Bouloy M. (2008) PLoS Pathog. 4, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas D., Blakqori G., Wagner V., Banholzer M., Kessler N., Elliott R. M., Haller O., Weber F. (2004) J. Biol. Chem. 279, 31471–31477 [DOI] [PubMed] [Google Scholar]
- 26. Buchholz U. J., Finke S., Conzelmann K. K. (1999) J. Virol. 73, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peterson S. R., Dvir A., Anderson C. W., Dynan W. S. (1992) Genes Dev. 6, 426–438 [DOI] [PubMed] [Google Scholar]
- 28. Hernan R., Heuermann K., Brizzard B. (2000) Biotechniques 28, 789–793 [DOI] [PubMed] [Google Scholar]
- 29. Blakqori G., Kochs G., Haller O., Weber F. (2003) J. Gen. Virol. 84, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 30. Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. (1998) EMBO J. 17, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwamura T., Yoneyama M., Yamaguchi K., Suhara W., Mori W., Shiota K., Okabe Y., Namiki H., Fujita T. (2001) Genes Cells 6, 375–388 [DOI] [PubMed] [Google Scholar]
- 32. Cheng C., Sharp P. A. (2003) Mol. Cell. Biol. 23, 1961–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 34. Wansink D. G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. (1993) J. Cell Biol. 122, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kontermann R. E., Liu Z., Schulze R. A., Sommer K. A., Queitsch I., Dübel S., Kipriyanov S. M., Breitling F., Bautz E. K. (1995) Biol. Chem. Hoppe-Seyler 376, 473–481 [DOI] [PubMed] [Google Scholar]
- 36. Habjan M., Andersson I., Klingström J., Schümann M., Martin A., Zimmermann P., Wagner V., Pichlmair A., Schneider U., Mühlberger E., Mirazimi A., Weber F. (2008) PLoS ONE 3, e2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Clement J. Q., Qian L., Kaplinsky N., Wilkinson M. F. (1999) RNA 5, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans-Molina C., Garmey J. C., Ketchum R., Brayman K. L., Deng S., Mirmira R. G. (2007) Diabetes 56, 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kolakofsky D., Bellocq C., Raju R. (1987) Cold Spring Harbor Symp. Quant. Biol. 52, 373–379 [DOI] [PubMed] [Google Scholar]
- 40. Schneider D. A., Michel A., Sikes M. L., Vu L., Dodd J. A., Salgia S., Osheim Y. N., Beyer A. L., Nomura M. (2007) Mol. Cell 26, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sollner-Webb B., Tower J. (1986) Ann. Rev. Biochem. 55, 801–830 [DOI] [PubMed] [Google Scholar]
- 42. Chapman R. D., Heidemann M., Hintermair C., Eick D. (2008) Trends Genet. 24, 289–296 [DOI] [PubMed] [Google Scholar]
- 43. Chapman R. D., Heidemann M., Albert T. K., Mailhammer R., Flatley A., Meisterernst M., Kremmer E., Eick D. (2007) Science 318, 1780–1782 [DOI] [PubMed] [Google Scholar]
- 44. Anindya R., Aygün O., Svejstrup J. Q. (2007) Mol. Cell 28, 386–397 [DOI] [PubMed] [Google Scholar]
- 45. Bregman D. B., Halaban R., van Gool A. J., Henning K. A., Friedberg E. C., Warren S. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11586–11590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee K. B., Wang D., Lippard S. J., Sharp P. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 4239–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Malik S., Bagla S., Chaurasia P., Duan Z., Bhaumik S. R. (2008) J. Biol. Chem. 283, 6897–6905 [DOI] [PubMed] [Google Scholar]
- 48. Ratner J. N., Balasubramanian B., Corden J., Warren S. L., Bregman D. B. (1998) J. Biol. Chem. 273, 5184–5189 [DOI] [PubMed] [Google Scholar]
- 49. Somesh B. P., Sigurdsson S., Saeki H., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2007) Cell 129, 57–68 [DOI] [PubMed] [Google Scholar]
- 50. Svejstrup J. Q. (2007) Trends Biochem. Sci. 32, 165–171 [DOI] [PubMed] [Google Scholar]
- 51. Zhang M., Siedow M., Saia G., Chakravarti A. (2010) Prostate 70, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huen M. S., Sy S. M., Chen J. (2010) Nat. Rev. Mol. Cell Biol. 11, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Colón-Ramos D. A., Irusta P. M., Gan E. C., Olson M. R., Song J., Morimoto R. I., Elliott R. M., Lombard M., Hollingsworth R., Hardwick J. M., Smith G. K., Kornbluth S. (2003) Mol. Biol. Cell 14, 4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bennett R. S., Cress C. M., Ward J. M., Firestone C. Y., Murphy B. R., Whitehead S. S. (2008) Virol. J. 5, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Weber F., Haller O. (2007) Biochimie 89, 836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Patterson J. L., Holloway B., Kolakofsky D. (1984) J. Virol. 52, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raju R., Kolakofsky D. (1988) J. Virol. 62, 27–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Léonard V. H., Kohl A., Hart T. J., Elliott R. M. (2006) J. Virol. 80, 9667–9675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Casamassimi A., Napoli C. (2007) Biochimie 89, 1439–1446 [DOI] [PubMed] [Google Scholar]
- 60. Yang Z., Yik J. H., Chen R., He N., Jang M. K., Ozato K., Zhou Q. (2005) Mol. Cell 19, 535–545 [DOI] [PubMed] [Google Scholar]
- 61. Dai-Ju J. Q., Li L., Johnson L. A., Sandri-Goldin R. M. (2006) J. Virol. 80, 3567–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fraser K. A., Rice S. A. (2007) J. Virol. 81, 5091–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rodriguez A., Pérez-González A., Nieto A. (2007) J. Virol. 81, 5315–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodriguez A., Pérez-González A., Hossain M. J., Chen L. M., Rolling T., Pérez-Breña P., Donis R., Kochs G., Nieto A. (2009) J. Virol. 83, 11166–11174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vreede F. T., Chan A. Y., Sharps J., Fodor E. (2010) Virology 396, 125–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Beaudenon S. L., Huacani M. R., Wang G., McDonnell D. P., Huibregtse J. M. (1999) Mol. Cell. Biol. 19, 6972–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Harreman M., Taschner M., Sigurdsson S., Anindya R., Reid J., Somesh B., Kong S. E., Banks C. A., Conaway R. C., Conaway J. W., Svejstrup J. Q. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20705–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Heine G. F., Horwitz A. A., Parvin J. D. (2008) J. Biol. Chem. 283, 9555–9561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Somesh B. P., Reid J., Liu W. F., Søgaard T. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2005) Cell 121, 913–923 [DOI] [PubMed] [Google Scholar]
- 70. Gomes N. P., Bjerke G., Llorente B., Szostek S. A., Emerson B. M., Espinosa J. M. (2006) Genes Dev. 20, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lilley C. E., Schwartz R. A., Weitzman M. D. (2007) Trends Microbiol. 15, 119–126 [DOI] [PubMed] [Google Scholar]
- 72. Andrejeva J., Poole E., Young D. F., Goodbourn S., Randall R. E. (2002) J. Virol. 76, 11379–11386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Precious B., Childs K., Fitzpatrick-Swallow V., Goodbourn S., Randall R. E. (2005) J. Virol. 79, 13434–13441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ulane C. M., Horvath C. M. (2002) Virology 304, 160–166 [DOI] [PubMed] [Google Scholar]
- 75. Li L., Johnson L. A., Dai-Ju J. Q., Sandri-Goldin R. M. (2008) PLoS One 3, e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brookes E., Pombo A. (2009) EMBO Rep. 10, 1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fuda N. J., Ardehali M. B., Lis J. T. (2009) Nature 461, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. (2007) Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007) Nature Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Saunders A., Core L. J., Lis J. T. (2006) Nat. Rev. Mol. Cell Biol. 7, 557–567 [DOI] [PubMed] [Google Scholar]
- 81. Hargreaves D. C., Horng T., Medzhitov R. (2009) Cell 138, 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.