Abstract
Mitochondrial complex II (CII) has been recently identified as a novel target for anti-cancer drugs. Mitochondrially targeted vitamin E succinate (MitoVES) is modified so that it is preferentially localized to mitochondria, greatly enhancing its pro-apoptotic and anti-cancer activity. Using genetically manipulated cells, MitoVES caused apoptosis and generation of reactive oxygen species (ROS) in CII-proficient malignant cells but not their CII-dysfunctional counterparts. MitoVES inhibited the succinate dehydrogenase (SDH) activity of CII with IC50 of 80 μm, whereas the electron transfer from CII to CIII was inhibited with IC50 of 1.5 μm. The agent had no effect either on the enzymatic activity of CI or on electron transfer from CI to CIII. Over 24 h, MitoVES caused stabilization of the oxygen-dependent destruction domain of HIF1α fused to GFP, indicating promotion of the state of pseudohypoxia. Molecular modeling predicted the succinyl group anchored into the proximal CII ubiquinone (UbQ)-binding site and successively reduced interaction energies for serially shorter phytyl chain homologs of MitoVES correlated with their lower effects on apoptosis induction, ROS generation, and SDH activity. Mutation of the UbQ-binding Ser68 within the proximal site of the CII SDHC subunit (S68A or S68L) suppressed both ROS generation and apoptosis induction by MitoVES. In vivo studies indicated that MitoVES also acts by causing pseudohypoxia in the context of tumor suppression. We propose that mitochondrial targeting of VES with an 11-carbon chain localizes the agent into an ideal position across the interface of the mitochondrial inner membrane and matrix, optimizing its biological effects as an anti-cancer drug.
Keywords: Anti-cancer Drug, Apoptosis, Cancer Therapy, Electron Transfer, Mitochondria, Mitochondrial Targeting, Vitamin E Succinate
Introduction
Mitochondria are emerging as targets for a variety of anti-cancer drugs (1–5) that belong to a group of compounds termed “mitocans” (6, 7). Of these agents, we and others have been studying the group of vitamin E (VE)2 analogs, epitomized by the “redox-silent” α-tocopheryl succinate (α-TOS) and α-tocopheryl acetyl ether (8). Both of these agents proved to be selective inducers of apoptosis in cancer cells and efficient suppressors of tumors in experimental models (9–16).
VE analogs with anti-cancer activity have been classified as mitocans (i.e. small anti-cancer agents that act by selectively destabilizing mitochondria in cancer cells) (6–8). Of the several groups of mitocans, the anti-cancer VE analogs belong to both the class of BH3 mimetics, which includes compounds interfering with the interactions of the Bcl-2 family proteins (17), as well as to the class of agents that interfere with the mitochondrial electron redox chain. The latter activity is probably the main reason for the strong apoptogenic efficacy of agents like α-TOS (18). More specifically, α-TOS interferes with the ubiquinone (UbQ)-binding site(s) of the mitochondrial complex II (CII), an event that results in generation of reactive oxygen species (ROS), in turn causing apoptosis induction (19). Moreover, CII has also been shown to be important for the anti-tumor efficacy of α-TOS (20).
Although α-TOS acts on mitochondria, it does not discriminate between the different membranous compartments within the cell. Therefore, we decided to generate a variant of the agent that would be targeted to mitochondria, anticipating that by doing so, its apoptogenic activity would be increased. This reasoning was based on the work from the group of Murphy and Smith, who prepared a series of mitochondrially targeted antioxidants by tagging them with the positively charged triphenylphosphonium group (TPP+) (21), producing very efficient redox-active compounds (22–24). Further, we assumed that the TPP+ group will be advantageous for the cancer cell specificity of the agents because cancer cell mitochondria feature greater mitochondrial inner membrane potential (ΔΨm,i) than normal cells (25–27). Our recent work3 documents that the prototypic compound of such a targeted VE analog (i.e. mitochondrially targeted vitamin E succinate (MitoVES)) indeed is some 1–2 orders of magnitude more apoptogenic than the untargeted, parental compound.
Molecular modeling and theoretical considerations suggest that tagging a hydrophobic compound with a cationic group, such as in the case of MitoVES, will dictate its position at the interface of the mitochondrial inner membrane (MIM) and the mitochondrial matrix. Therefore, we expect that it will be juxtapositioned to preferentially interact with CII more than the untargeted VE analog, such that its apoptogenic activity would be much greater. In this paper, we show that, indeed, MitoVES interacts with the proximal UbQ-binding (QP) site of CII, which endows it with greater activity for inducing cancer cell apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture
Human T lymphoma Jurkat cells were grown in RPMI medium supplemented with 10% FCS and antibiotics. Chinese hamster lung fibroblasts with a dysfunctional CII (B9 cells) as well as the parental cells (B1 cells) (28) were grown in DMEM with 10% FCS, antibiotics, 10 mg/ml glucose, and 1% non-essential amino acids. The cells were transformed to malignancy by transfection with an H-RAS vector (29), and CII in the B9 cells was reconstituted as reported elsewhere (20, 30). The human colon cancer cells HCT116ODD-GFP were cultured in DMEM with 10% FCS plus antibiotics; this subline was prepared by stable transfection of HCT116 cells using a plasmid coding for the oxygen-dependent destruction (ODD) domain of the HIF1α protein fused with GFP (31, 32).
Preparation of Submitochondrial Particles (SMPs)
Coupled bovine heart SMPs were obtained from frozen mitochondria (20–30 mg of protein/ml) incubated in the SHE medium (250 mm sucrose, 10 mm HEPES, 1 mm EGTA, pH 7.2) supplemented with 3 mm MgCl2, 3 mm ATP, and 20 mm succinate. The mitochondrial suspensions were sonicated three times in aliquots of 20 ml on ice for 15 s, with a 5-mm diameter probe tip using a Branson Sonifier 450 sonicator. The suspensions were diluted in the SHE medium and centrifuged twice at 17,370 × g for 5 min at 4 °C. The supernatant was then centrifuged at 105,000 × g for 40 min at 4 °C. The pellet was resuspended in the SHE medium plus 1% (w/v) fatty acid-free BSA and stored at −70 °C until use (33).
Synthesis of VE Analogs
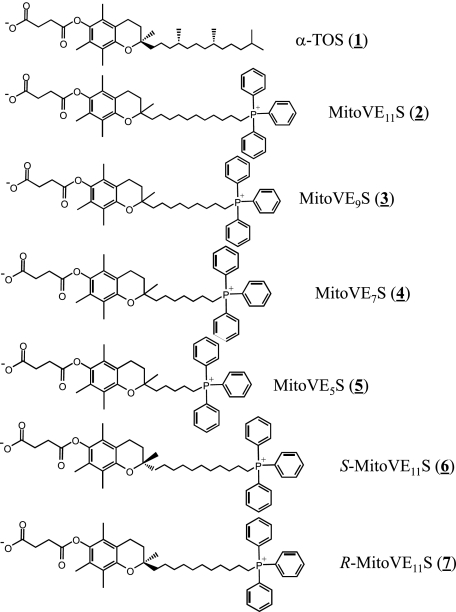
The synthesis of the prototypic, racemic MitoVE11S (2), an agent with an 11-C chain linking the TPP and the tocopheryl succinyl group, and its shorter chain homologs MitoVE9S (3), MitoVE7S (4), and MitoVE5S (5), as well as its stereoisomers S-MitoVE11S (6) and R-MitoVE11S (7), is shown in the supplemental material.
Assessment of Apoptosis, ROS Generation, and Mitochondrial Potential
Apoptosis levels were assessed using the annexin V/propidium iodide method (19). Cellular ROS levels were detected using dihydroethidium (Molecular Probes) and flow cytometry and expressed as mean fluorescence intensity or by trapping with 5,5-dimethyl-1-pyrroline N-oxide (Sigma) using EPR spectroscopy (19) and expressed in arbitrary units/mg of cellular protein. ΔΨm,i was assessed using the fluorescent probe tetramethylrhodamine methyl ester and flow cytometry according to a standard protocol.
Assessment of CI, CII, and CIII Activity
In whole cells, succinate dehydrogenase (SDH) activity was estimated using a short term (1 h) modified MTT assay with succinate as the sole source of electrons driving the respiratory system, specifically via CII, as described (18). For SMPs, CI and CII dehydrogenase activities were determined at 37 °C in 1 ml of the SHE medium that also contained 0.075–0.1 mm 2,6-dichlorophenol indophenol and 0.025 mg/ml protein. The reaction was started after a 15-min preincubation with MitoVE11S by adding succinate (0.25–2 mm) or NADH (0.1–1 mm) as CII or CI substrates, respectively. The rate of 2,6-dichlorophenol indophenol reduction was determined by measuring the absorbance change at 600 nm and using the extinction coefficient of 21.3 mm−1 cm−1. The CII dehydrogenase activity (SDH) was completely inhibited by malonate.
The activities of both succinate-cytochrome c oxidoreductase (complex II + III) and NADH-cytochrome c oxidoreductase (complex I + III) were determined at 37 °C in 1 ml of 50 mm Hepes, pH 7.2, 1 mm cyanide, 50 μm cytochrome c (from horse heart) and SMPs at 25 μg of protein. The reaction was started after a 15-min preincubation with MitoVE11S by adding succinate (0.25–2 mm) or NADH (0.1–1 mm). The activity was determined in a dual wavelength spectrophotometer by measuring the reduction of oxidized cytochrome c over time from the absorbance difference at 550 and 540 nm and by using the extinction coefficient of 19.1 mm−1 cm−1 (34). The CII oxidoreductase activity was fully blocked by malonate or antimycin, whereas the CI oxidoreductase activity was 88% inhibited by 20 μm rotenone.
Isolation of Mitochondria and Gel Filtration Chromatography
Cells (1.2 × 108) were treated with MitoVE11S for different periods and harvested. The pellet was resuspended in 0.5 ml of ice-cold hypotonic fractionation buffer (25 mm Tris at pH 7.4, 2 mm EDTA, 5 mm MgCl2, 10 mm KCl, 125 mm sucrose, 1 mm PMSF plus a protease inhibitor mixture) and left on ice for 10 min. The swollen cells were lysed using a glass homogenizer (Kontes Glass Co.). The isotonicity of each sample was achieved by the addition of 250 μl of ice-cold hypertonic fractionation buffer with 0.5 m sucrose. Organelles and unbroken cells were centrifuged at 900 × g for 10 min, followed by centrifugation of the supernatant at 1,700 × g for 5 min. The remaining supernatant was then centrifuged at 15,000 × g for 10 min, and the mitochondrial pellet was lysed in a buffer comprising 25 mm HEPES, pH 7.5, 0.3 m NaCl, and 2% CHAPS. The mitochondrial lysates were centrifuged at 19,000 × g for 5 min and loaded onto a Superdex-200 10/300 preparation grade column (separation range 10 to ∼600 kDa; Amersham Biosciences) equilibrated with the 2% CHAPS lysis buffer (see above). Proteins were eluted at 0.3 ml/min, and fractions of 0.5 ml were collected and mixed with 3× Laemmli reducing sample buffer and boiled. The samples were then analyzed by SDS-PAGE and Western blotting for subunits of CI, CII, and CIII.
Western Blotting
Proteins in whole cell lysates or mitochondrial fractions were separated using SDS-PAGE before Western blotting was performed according to a standard protocol using the antibody to the CI 39-kDa subunit (clone 20C11), the CIII core-2 subunit (clone 16D10; both from Invitrogen), and the CII SDHC subunit (clone M01; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)).
RT-PCR and qPCR
The amount of target mRNA was assessed by RT-PCR and qPCR. Total RNA was isolated using the Aurum RNA total minikit, including the DNase treatment step (Bio-Rad), and reverse transcribed by the Revertaid First Strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions (1–5 μg of total RNA per 20 μl of reaction mixture). RT-PCR was carried out using the standard procedure. For qPCR, cDNA corresponding to 12 ng of starting total RNA was diluted with water into 4.5 μl, 0.5 μl of the combined 10 μm forward and reverse primers were added, and, finally, 5 μl of 2× SYBR Green JumpStart Taq ready mix (Sigma) was added, and the reaction was carried out on a Bio-Rad CFX96 real-time thermal cycler using three-step PCR (98 °C for 5 s, 60 °C for 15 s, and 72 °C for 25 s) for 40 cycles followed by melting curve analysis. Primers were designed on the intron/exon boundaries to prevent DNA amplification and are listed in Table 1.
TABLE 1.
Primers used for qPCR and RT-PCR
| Gene | Primers |
|
|---|---|---|
| Forward | Reverse | |
| ND1 | 5′-ATA CCC ATG GCC AAC CTC CT-3′ | 5′-GGG CCT TTG CGT AGT TGT AT-3′ |
| ND2 | 5′-GGC CCA ACC CGT CAT CTA CT3′ | 5′-GAT GCG GTT GCT TGC GTG AG-3′ |
| ND3 | 5′-CCG CGT CCC TTT CTC CAT AA-3′ | 5′-GGT AGG GGT AAA AGG AGG GC-3′ |
| ND4 | 5′-ACT ACT CAC TCT CAC TGC CC-3′ | 5′-AGT GGA GTC CGT AAA GAG GT-3′ |
| ND4L | 5′-AAC CCT CAA CAC CCA CTC CC-3′ | 5′-TAG GCC CAC CGC TGC TTC GC-3′ |
| ND5 | 5′-AAC AGA GTG GTG ATA GCG CC-3′ | 5′-CCC TAC TCC ACT CAA GCA CT-3′ |
| ND6 | 5′-CCT ACC TCC ATC GCT AAC CC-3′ | 5′-AGG GGG AAT GAT GGT TGT CT-3′ |
| CYTB | 5′-GAA ACT TCG GCT CAC TCC TT-3′ | 5′-GGC GAT TGA TGA AAA GGC GG-3′ |
| COX1 | 5′-GCC TCC GTA GAC CTA ACC AT-3′ | 5′-GTT ATG GCA GGG GGT TTT AT-3′ |
| COX2 | 5′-AGT CCT GTA TGC CCT TTT CC-3′ | 5′-GCG ATG AGG ACT AGG ATG AT-3′ |
| COX3 | 5′-CCC ACC AAT CAC ATG CCT AT-3′ | 5′-TAG GCC GGA GGT CAT TAG GA-3′ |
| ATP6 | 5′-CTG TTC GCT TCA TTC ATT GC-3′ | 5′-GAT TAG TCA TTG TTG GGT GG-3′ |
| ATP8 | 5′-TGC CCC AAC TAA ATA CTA CC-3′ | 5′-CTT TGG TGA GGG AGG TAG GT-3′ |
| P0 | 5′-TCG ACA ATG GCA GCA TCT AC-3′ | 5′-ATC CGT CTC CAC AGA CAA GG-3′ |
Site-directed Mutagenesis
The S68A and S68L substitutions were introduced by site-directed mutagenesis of human WT SDHC cDNA in the pEF-IRES-PURO expression vector using the QuikChange Lightning mutagenesis kit (Stratagene) and the following primers: S68A, 5′-TCC CAT GGC GAT GGC CAT CTG CCA CCG-3′ (forward) and 5′-CGG TGG CAG ATG GCC ATC GCC ATG GGA-3′ (reverse); S68L, 5′-CTT CCC ATG GCG ATG TTA ATC TGC CAC CGT GGC A-3′ (forward) and 5′-TGC CAC GGT GGC AGA TTA ACA TCG CCA TGG GAA G-3′ (reverse). The constructs were confirmed by sequencing and used to transfect the SDHC-deficient B9 fibroblasts using the Superfect reagent (Qiagen), followed by incubation with 2–4 μg/ml puromycin (Sigma) for 2 weeks. Clones were analyzed for the expression of human SDHC by RT-PCR, and those selected were then transformed using the pEGFP-C3-H-Ras vector as described previously (20). Total RNA was collected, and the presence of the S68A or S68L mutation was verified by cDNA sequencing.
Confocal Microscopy
HCT116ODD-GFP cells were cultured on coverslips, exposed to MitoVE11S, mounted with DAPI-containing Vectashield (Vector Laboratories), and inspected in a confocal microscope. Sections from paraffin-embedded tumors derived from control or MitoVE11S-treated mice were processed as above for the cultured cells.
Mouse Tumor Experiments
Tumors were established in immunocompromised, athymic (BALB/c nu/nu) mice by injecting HCT116ODD-GFP cells subcutaneously at 5 × 106 cells/animal. Mice were regularly checked by ultrasound imaging (USI) using the Vevo770 USI apparatus equipped with the 30-μm resolution RMV708 scan head (VisualSonics) as detailed elsewhere (18, 20, 35, 36). As soon as tumors reached ∼40 mm3, the animals were treated by intraperitoneal injection of 1–2 μmol of MitoVE11S or 15 μmol of α-TOS in corn oil containing 4% EtOH every 3–4 days. Control mice were injected with an equal volume (100 μl) of the vehicle only. Progression of tumor growth was assessed using USI, which enables three-dimensional reconstruction of tumors and precise quantification of their volume. All animal experimentation was performed according to the guidelines of the Australian and New Zealand Council for the Care and Use of Animals in Research and Teaching and was approved by the Griffith University Animal Ethics Committee.
Molecular Modeling
The binding of different MitoVE11S compounds to the mitochondrial respiratory CII protein was analyzed by means of empirical force field molecular modeling/molecular dynamics. The initial geometry of the protein was taken from the available crystal structure of the porcine heart CII (Protein Data Bank code 1ZOY). As ligands, we selected both R- and S-stereoisomeric forms of MitoVE11S as well as a series of compounds with 9, 7, and 5 carbons in the aliphatic chain.
To make the simulations feasible, the extended structure of CII was partitioned accordingly to allow closer examination of the regions of interest. The properties of the membrane-bound part of the protein and the membrane itself were analyzed by placing the hydrophobic chains (C and D) into the bilayer of phosphatidylethanolamine (POPE). For the MitoVE11S binding site study, the residues found within 10 Å from the Qp site were included. The protein/membrane study was initiated using a pre-equilibrated 6.7-nm rectangular patch of the POPE bilayer. The C and D chains of the protein were inserted into the center of the bilayer, and the overlapping POPE molecules were removed, resulting in the complex containing 76 POPE molecules.
For the MitoVE11S complexes, the protein residues within 10 Å from the Qp site were first selected. In those situations where only one to two residues were missing between consecutive residues, these residues were also added into the selection. Terminal residues of the selection were capped with acetamide and N-methyl groups.
The molecular dynamics study was performed employing the AMBER force field. The parm99 force field (37) was used for the standard protein residues, whereas for the MitoVES molecules, the general AMBER force field (GAFF) (38) parameters were used. The MitoVE11S point charges were determined by a restrained fit to the electrostatic potential according to recommended procedures (39).
The modeled complexes were placed in a periodic rectangular box 1 nm larger than the complex along all three axes. For the study of the interaction of the hydrophobic chains (C and D) of the protein with the membrane, a 1-nm extension of the box size was used only in the “out of plane” coordinate, resulting in periodic lipid bilayer slabs. The box was filled with TIP3P water molecules. Chlorine ions were added to neutralize the system, placed at the positions with the lowest electrostatic potential. A molecular dynamics simulation was then conducted employing the GROMACS suite of programs (40). The equilibration procedure used consists of heating the water molecules separately to 300 K during 20 ps with the system held at 10 K, followed by a 20-ps heating of the whole system to 300 K while applying position restraints on the heavy atoms of the solute. After heating, the position restraints only on the carbonyl carbons of protein were used (allowing for side chain rearrangement while keeping the backbone fixed), and the 2-ns simulation at constant temperature of 300 K and constant pressure of 1 atm was performed. A time step of 2 fs with van der Waals and electrostatic cut-offs of 1 nm were used throughout the simulations.
For calculating interaction energies, the energy groups were introduced, the periodic conditions were removed while increasing the cut-offs to 3 nm, and the energies were calculated using the rerun switch of the mdrun program operating on the previously obtained trajectories. The group energies were collected for the (equilibrated) second half of the trajectories and analyzed with the g_analyze program.
For assessment of changes in the structure of CII upon replacing Ser68 with Leu, the original geometry of the protein was taken from the available crystal structure (Protein Data Bank code 1ZOY). Modeling of the three-dimensional structure was performed by Asmara version 8.12.26 (41). The cell boundaries were defined as 80 × 80 × 120 Å, which were filled with a water density of 1.0 g/ml, and the AMBER99 force field was then applied. When modeling of the three-dimensional structure was completed, structural alignment to establish equivalences between the original structure 1ZOY and the structure with the mutated residue, based on their shape and three-dimensional conformation, was carried out. It is common for structural alignment methods to use only the backbone atoms included in the peptide bond. For simplicity and efficiency, only the Cα positions were considered because the peptide bond has a minimally variant planar conformation. The root mean square deviation is the measure of the average distance between the backbones of superimposed proteins. In the study of globular protein conformations, one customarily measures the similarity of the three-dimensional structure by the root mean square deviation of the Cα atomic coordinates after optimal rigid body superposition (42).
Statistics
All data shown are mean values of three independent experiments (unless stated otherwise) ± S.D. Statistical significance was assessed using Student's t test, and differences were considered significant at p < 0.05.
RESULTS
Induction of Apoptosis by MitoVE11S Is Dependent on Mitochondrial Potential and Complex II
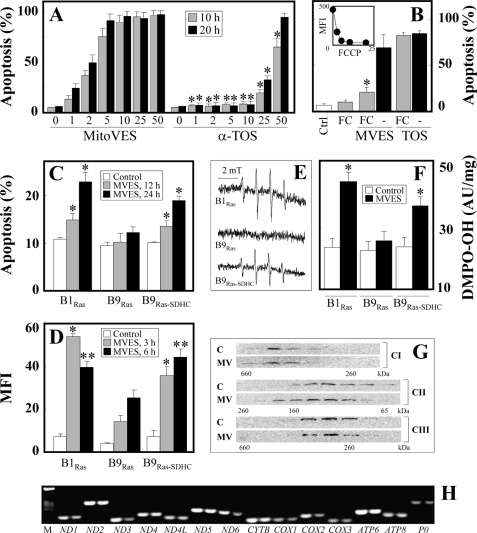
We recently found that the mitochondrially targeted vitamin E analog, MitoVE11S, is some 20–50-fold more efficient than the untargeted α-TOS,3 as documented in Fig. 2A for Jurkat cells. This is in good agreement with the IC50 value of ∼0.5 μm for MitoVES for apoptosis induction in Jurkat cells, which was ∼20 μm for α-TOS. Mitochondrial accumulation of MitoVE11S can be ascribed to the TPP+ tag and the high ΔΨm,i. To prove this, we carried out experiments in which Jurkat cells were exposed to MitoVE11S or α-TOS in the absence or presence of the mitochondrial uncoupler carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone (FCCP) and assessed for apoptosis. Fig. 2B documents that FCCP considerably lowered apoptosis induced by MitoVE11S but not by α-TOS, documenting that ΔΨm,i is important for high biological activity of the mitochondrially targeted vitamin E analog, which is not the case for α-TOS lacking the TPP+ tag.
FIGURE 2.
MitoVE11S is superior to α-TOS, requires high ΔΨm,i, and acts by targeting the mitochondrial respiratory CII. A, Jurkat cells were exposed to MitoVE11S or α-TOS at the concentrations shown (μm) for 10 or 20 h and assessed for apoptosis by flow cytometry. B, Jurkat cells were exposed to 4 μm MitoVE11S (MVES) for 4 h or 100 μm α-TOS (TOS) for 6 h in the presence or absence of 5 μm FCCP (FC) and assessed for apoptosis. The inset shows that exposure of Jurkat cells to increasing doses (μm) of FCCP for 6 h results in dissipation of ΔΨm,i, as assessed using the fluorescent probe tetramethylrhodamine methyl ester and flow cytometry (mean fluorescence intensity (MFI)). B1Ras, B9Ras, and B9Ras-SDHC cells were exposed to 5 μm MitoVE11S for the times shown (C and D) or for 2 h (E and F) and assessed for apoptosis induction (C) and ROS accumulation by flow cytometry (D) and EPR spectroscopy (E, EPR spectra; F, double integration evaluation of the signal; AU, arbitrary units). G, Jurkat cells were exposed to 5 μm MitoVE11S (MV) for 6 h, their mitochondria were lysed, and the lysates were fractionated using size exclusion chromatography. Individual fractions were probed by Western blotting for the presence of CI–CIII using specific antibodies. H, Jurkat cells were exposed to 5 μm MitoVE11S for 6 h, and mRNA levels of the mtDNA genes coding subunits of mitochondrial complexes were assessed using RT-PCR (left lanes, control; right lanes, MitoVE11S; M, markers). The data shown are mean ± S.D. (error bars) (n = 3); the images are representative of three independent experiments. * (A), statistically significant differences between corresponding treatments with MitoVE11S and α-TOS; * (B), statistically significant differences between treatments in the absence and presence of FCCP; * and ** (C–F), significant differences between B1Ras or B9Ras-SDHC cells and B9Ras cells treated with MitoVE11S for 3 and 6 h, respectively (p < 0.05).
Due to the superior activity of MitoVE11S, we investigated its molecular target. Given the fact that α-TOS, which lacks specific mitochondrial targeting, was shown to cause apoptosis by interacting with CII lying at the interphase of the MIM and the mitochondrial matrix (18), it was reasoned that MitoVE11S (compound 2 in Fig. 1, where it is referred to as MitoVE11S due to the 11-C chain spanning the TPP+ and the tocopheryl succinyl headgroup) would be more ideally located to induce apoptosis due to its accumulation in close proximity to the target. Therefore, the capacity of MitoVE11S to induce apoptosis in parental, CII-dysfunctional and CII-reconstituted B1Ras, B9Ras, and B9Ras-SDHC cells, respectively, was tested (20). Fig. 2C documents that the parental as well as CII-reconstituted cells were susceptible to MitoVE11S, whereas the CII-dysfunctional cells were resistant. Notably, the capacity of the three cell lines to generate radicals in response to MitoVE11S exposure correlated with their susceptibility to apoptosis, as shown by EPR spectroscopy using a radical trap and flow cytometry using a fluorescent probe (Fig. 2, C–E).
FIGURE 1.
Structures of the compounds used in this study.
We next tested whether the generation of radicals was due to an effect of MitoVE11S on the integrity of the mitochondrial electron transport complexes. This was particularly important given that CI-CIII are considered to be major sources of mitochondrially derived ROS (43). To ascertain this point, Jurkat cells, which are very sensitive to MitoVE11S,3 were used. The cells were treated with 5 μm MitoVE11S for 6 h before mitochondria were isolated, and their extracts were separated by size exclusion chromatography, followed by Western blotting of the individual fractions for subunits of CI, CII, and CIII. Fig. 2G documents virtually no effect of the agent on the integrity of any of the three complexes. We did, however, observe a slight shift toward higher molecular weight for CI and CII. Further, it was examined whether MitoVE11S affects the expression of any of the 13 proteins coded by mtDNA as subunits of mitochondrial electron redox chain complexes. A 6-h exposure of Jurkat cells to 5 μm MitoVE11S followed by RT-PCR revealed a very small, if any, effect on expression of the mtDNA-coded genes at the level of mRNA, with a mild effect on ND3, ND6, COX1, and COX2 (Fig. 2G). Analysis by qPCR confirmed this mild effect (data not shown). This indicates that generation of ROS in cancer cells exposed to MitoVE11S is not due to an effect of the agent on the levels or integrity of the mitochondrial complexes.
MitoVE11S Inhibits the Oxidoreductase Activity of Complex II
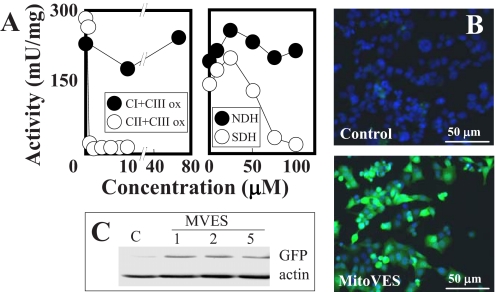
Next, the effects of MitoVE11S on the enzymatic and oxidoreductase activities of CI and CII were examined. For these studies, heart tissue-derived mitochondrial preparations as SMPs were used because of their ready availability and because they would be directly accessible to drug testing on the function of the respiratory chain. MitoVE11S was examined for its capacity to disrupt the electron transfer between CI and CIII, and between CII and CIII. Fig. 3A reveals that the oxidoreductase activity of CII (transfer of electrons from CII to CIII) was very sensitive to low levels of the compound (IC50 values of 1.5 and 10 μm at high and low levels of succinate, respectively) (Table 2), whereas no effect was evident on the electron transfer activity from CI to CIII (Fig. 3A). This suggests that MitoVE11S preferentially binds to the fully active CII. At low succinate, the addition of detergent to the reaction mixture only minimally affected the MitoVE11S IC50 value, whereas at high succinate, the detergent's effects became more pronounced, revealing the importance of the natural and fully active conformation of the CII tetramer to allow for maximal effects of MitoVE11S (Table 2). The SDH enzymatic activity of CII, on the other hand, was only inhibited by high MitoVE11S concentrations (Fig. 3A) (IC50 values for conversion of succinate to fumarate by CII were 80 μm at high and 76 μm at low succinate; Table 2), whereas the NADH dehydrogenase (NDH) activity of CI was not affected (Fig. 3A).
FIGURE 3.
MitoVE11S efficiently inhibits the oxidoreductase activity of CII. A, SMPs were assessed for the transfer of electrons from CI to CIII and from CII to CIII (left) and the activity of CI (NDH) and CII (SDH) (right) in the presence of MitoVE11S. HCT116ODD-GFP cells were assessed for stabilization of GFP by Western blotting after exposure to MitoVE11S at the levels shown (μm) for 24 h by Western blotting (B) or by fluorescence microscopy following 24-h exposure to 5 μm MitoVE11S (C). The data in A are average values from two independent experiments; the images are representative of three independent experiments.
TABLE 2.
IC50 values of MitoVE11S for CII activities
| Compound | Activity | Tween | IC50a |
|
|---|---|---|---|---|
| 0.25 mm succinate | 2 mm succinate | |||
| μm | ||||
| MitoVE11S | SDHb | 76 | 80 | |
| MitoVE11S | e− transferc | − | 10 | 1.5 |
| MitoVE11S | e− transferc | + | 20 | 18.5 |
| MitoVE5S | e− transfer | − | 35 | 24 |
a The IC50 values are given in μm and are average values of two independent experiments with the individual values differing by not more than 10%.
b The SDH activity refers to conversion of succinate to fumarate.
c The electron transfer activity refers to transfer of e− from CII to CIII.
Inhibition of SDH can cause the state of pseudohypoxia, leading to activation of prolyl hydroxylase that modifies two proline residues flanking the ODD domain of HIF1α, resulting in the accumulation of this transcription factor (31). To find out whether this process was invoked by MitoVE11S, HCT116 cells stably transfected with ODD-GFP were used. Prolonged exposure of the cells under normoxia to MitoVE11S at 1–5 μm resulted in the expression of the ODD-GFP fusion protein (Fig. 3, B and C). This indicates that even at relatively low drug levels, sufficient SDH inhibition can occur over the long term to enable succinate levels to accumulate that activate the pseudohypoxic state and further documents the role for CII as a target for MitoVE11S. In support of this premise, we have recently found that exposure of cancer cells to vitamin E analogs results in succinate accumulation.4
MitoVE11S Targets the Proximal UbQ-binding Site of Complex II
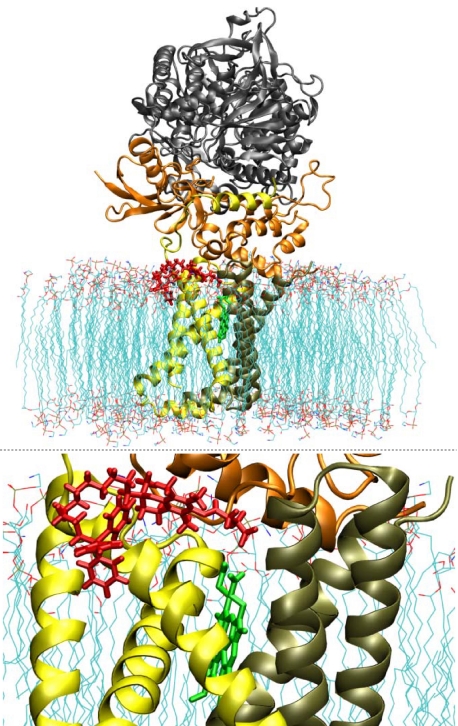
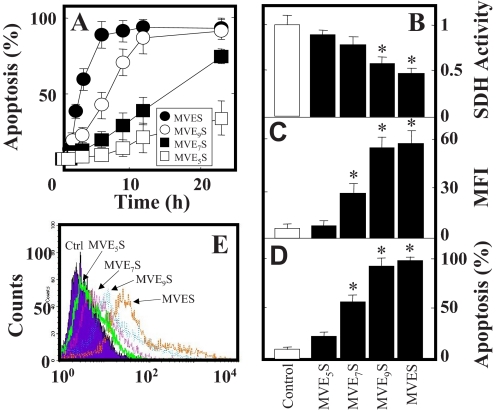
In order to characterize the interaction of MitoVE11S with CII in more detail, computer modeling was used, based on the published crystal structure of CII (44), which we inserted within a POPE bilayer simulating the environment of the MIM. Our model predicts that the TPP+ group of MitoVE11S is located at the matrix interface of the MIM, whereas the active succinyl group of the drug is buried within the membrane, interacting with the Qp site of CII (Fig. 4). This requires the aliphatic linker separating the chromanol succinate and TPP+ moieties to be of a certain length. Indeed, when serially shorter linkers were used in the simulation, the model-derived binding energies of these MitoVES homologs were correspondingly successively reduced (Table 3). Experimental results confirmed these predictions. Reducing the length of the linker lowered the biological activity of the MitoVES homologs, and the shortest variant, MitoVE5S (compound 5), lost most of its activity, as shown by low levels of apoptosis in Jurkat cells (Fig. 5A) as well as in HCT116ODD-GFP cells examined for SDH activity inhibition, ROS accumulation, apoptosis induction, and ODD-GFP stabilization (Fig. 5, B–E). Similar results were obtained for the reduction of CII oxidoreductase activity in isolated SMPs, thereby excluding the possibility of a reduction in cellular uptake of the shorter MitoVES homologs (Table 2).
FIGURE 4.
Molecular modeling predicts binding of MitoVE11S at the QP site. The CII model was refined by the addition of POPE molecules simulating the MIM. The position of the heme group is indicated in green, and the predicted position of MitoVE11S is shown in red. The bottom panel shows the detail of the interaction of MitoVE11S with CII.
TABLE 3.
Relative interaction energies for MitoVES homologues and MitoQ binding the Qp site of CII
| Compound | Relative interaction energy |
|---|---|
| % | |
| R-MitoVE11Sa | 100b |
| S-MitoVE11S | 79.5 |
| R-MitoVE9S | 79.3 |
| R-MitoVE7S | 70.5 |
| R-MitoVE5S | 58.6 |
| MitoQ | 51.2 |
a Except for S-MitoVE11S, all other homologues used for calculations of their relative interaction energies with Qp of CII were in the R conformation.
b The calculated interaction energy of R-MitoVE11S was set as 100%.
FIGURE 5.
Shortening the aliphatic chain of MitoVE11S successively reduces its cancer cell cytotoxic activity. A, Jurkat cells were exposed to homologs of MitoVES at 5 μm and evaluated for the level of apoptosis. HCT116ODD-GFP cells were exposed to MitoVES homologs at 5 μm for 3 h and assessed for SDH activity (B) and ROS accumulation (C) or exposed for 12 h and assessed for the level of apoptosis (D) and expression of GFP (E). The data in A–D are mean ± S.D. (error bars) (n = 3), and data in E are representative of three independent experiments. *, significant difference (p < 0.05) between control cells and cells treated with different MitoVES homologs.
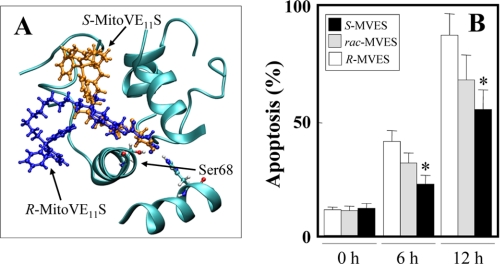
The model also predicted that the R enantiomer of MitoVE11S (compound 7 in Fig. 1) would bind CII ∼20% more efficiently than the S enantiomer (compound 6) (Fig. 6A and Table 3). In support of this, the R enantiomer induced apoptosis more efficiently in Jurkat cells than the S enantiomer, with the results for the racemic mixture of MitoVE11S between the two (Fig. 6B). This difference in the apoptotic efficacy of the stereoisomers was apparent at relatively low levels of MitoVE11S (e.g. the concentration of 2.5 μm shown in Fig. 6B), and it was lost at 5 μm MitoVE11S (data not shown).
FIGURE 6.
Apoptogenic efficacy of MitoVE11S depends on its chirality. A, molecular modeling documents the predicted interaction of R- (blue) and S-MitoV11ES (brown) with the UbQ-interacting Ser68 in the QP site of SDHC. B, Jurkat cells were exposed to rac-, R-, or S-MitoVE11S at 2.5 μm for the times shown and assessed for apoptosis induction. The data are mean ± S.D. (error bars) (n = 3). *, significant difference (p < 0.05) between cells treated with S- and R-MitoVE11S.
Mutation of the CII UbQ-binding Ser68 in the QP Site Causes Resistance to MitoVE11S
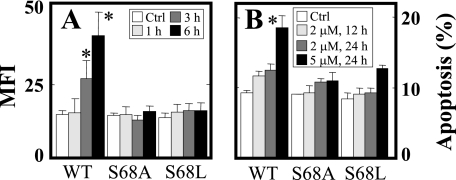
In order to verify the significance of the Qp site for the biological activity of MitoVE11S, we reconstituted the SDHC-deficient B9 cells with SDHC variants mutated at Ser68, a residue important for UbQ binding (44) (Ser27 in E. coli (45)). Substitution of Ser68 by either Ala or Leu, which according to the model should cause only minor shifts within the binding site (the root mean square deviation being 44 Å for S68A and 52 Å for S68L) abrogated the capacity of MitoVE11S to induce ROS production and apoptosis (Fig. 7, A and B), confirming the importance of the Qp site and, more specifically, the UbQ-binding S68 of SDHC for the activity of MitoVE11S.
FIGURE 7.
The UbQ-binding Ser68 in the QP site of CII is important for the effect of MitoVE11S. B9Ras-SDHC (WT), B9Ras-SDHCS68A (S68A), or B9Ras-SDHCS68L cells (S68L) were exposed to MitoVE11S at 2 μm for the times shown and assessed for ROS accumulation (A) and apoptosis levels (B). The data are mean ± S.D. (error bars) (n = 3). *, significant difference (p < 0.05) between MitoVE11S-treated B9Ras cells stably transfected with WT SDHC or with S68A or C68L mutant SDHC.
MitoVE11S Suppresses Tumor Progression and Causes Pseudohypoxia in Tumor Cells
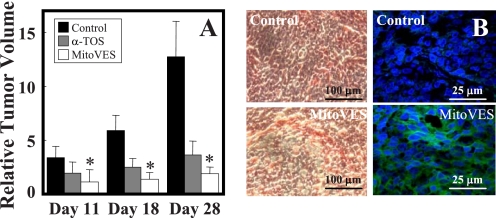
To assess the effect of MitoVE11S on tumors and obtain insight into the molecular mechanism of its anti-cancer effects, nude mice with xenografts derived from the HCT116ODD-GFP cells were used. Once palpable tumors appeared (volume of ∼40 mm3), the animals were treated by intraperitoneal administration of either MitoVE11S at 1–2 μmol or α-TOS at 15 μmol. Fig. 8A shows that MitoVE11S, applied at 10-fold lower concentration than the untargeted α-TOS, suppressed the growth of colorectal carcinomas and was, therefore, much more efficient. At the end of the experiment, the mice were sacrificed, and the isolated tumors were used for preparation of paraffin-embedded sections, which were inspected by light microscopy for their morphology and by confocal microscopy for the appearance of the ODD-GFP green fluorescence (Fig. 8B). Unlike tumors from the control mice, tumors from the MitoVE11S-treated animals showed abundant green fluorescence in the tumor cell cytoplasm, documenting the stable expression of the ODD-GFP transgene. This result suggests that the molecular mechanism by which MitoVE11S induces apoptosis in cultured tumor cells and in experimental tumors to suppress their growth involves targeting of CII, resulting in generation of ROS, and culminates in the state of cell pseudohypoxia.
FIGURE 8.
MitoVE11S suppresses tumor progression and causes the state of pseudohypoxia. A, BALB/c nu/nu mice with xenografts derived from HCT116ODD-GFP cells were treated by intraperitoneal injection of 1–2 μmol of MitoVE11S or 15 μmol of α-TOS per mouse every 3–4 days, and tumors were visualized and their volume was quantified using USI on days 11, 18, and 28. B, tumors were paraffin-embedded, sectioned, and stained with H&E and photographed in a light microscope (left images) or mounted in DAPI-containing Vectashield and imaged using a confocal microscope (right images). The data are mean ± S.D. (error bars) (n = 6–7). *, significant difference between corresponding control and MitoVE11S-treated animals (p < 0.05).
DISCUSSION
In this paper, we present data on the molecular mechanism of generation of ROS in cancer cells exposed to the novel VE analog, MitoVE11S. We show that (i) MitoVE11S requires a functional CII for ROS generation and apoptosis induction; (ii) MitoVE11S very efficiently suppresses electron transfer from CII to CIII while only mildly inhibiting the SDH headgroup enzymatic activity of CII; (iii) the length of the hydrophobic chain of MitoVE11S is critical to allow the biologically active succinate moiety of the agent to reach the QP of CII; (iv) the UbQ-interacting Ser68 of the QP is important for the biological activity of MitoVE11S; and (v) the molecular mechanism by which MitoVE11S triggers apoptosis in cultured cells in vitro and suppresses tumor progression in vivo can be closely correlated to involve the CII QP binding and downstream effects. These major findings document that MitoVE11S, as an efficient anti-cancer agent, has the propensity to interact with CII, which plays a role in the pro-apoptotic activity of several anti-cancer drugs (46) and has been proposed as a novel target for mitocans from the group of VE analogs (18, 20).
Although CII has only now been identified as a target for anti-cancer drugs, its subunits SDHB, SDHC, and SDHD have been reported as tumor suppressor genes (47–49), whose mutations give rise to relatively rare neoplastic diseases, including familial paragangliomas and pheochromocytomas (50, 51). The molecular mechanism of tumorigenicity arising from mutations in SDHB, SDHC, or SDHD is not completely clear at present, although it has been suggested that mutations resulting in impaired expression of the protein(s), imperfect assembly, or lack of binding of UbQ may give rise to slightly increased generation of ROS, probably promoting the malignant transformation (30, 52, 54–57). In light of these findings, it is interesting that CII can also serve as a target for anti-cancer drugs, an intriguing paradigm corroborated by the notion that CII mutates mostly in relatively benign neoplastic diseases, whereas, for example, only one of 1 million breast cancer patients is positive for a mutation in a CII subunit (51).
Our recent finding of CII as a target for anti-cancer drugs comes from experiments with α-TOS, which showed that a mutation in the SDHC gene, whereby the protein is not expressed, renders the cells resistant to the VE analog. Because CII appears to be a highly intriguing, invariant target (mutating very rarely in major carcinomas), especially considering the findings that cancers are extremely promiscuous and feature different sets of mutations even within the same type of the disease (58), our aim was to maximize the efficacy of CII-targeting agents. To achieve this, the strategy of Murphy and Smith (21), used previously to deliver antioxidants to the mitochondria of cultured cells and tissues in vivo was adapted here. For their studies, a lipophilic cationic group was used to tag and modify antioxidants (21, 59), thereby endowing them with profound biological activity (60, 61) while not jeopardizing the normal mitochondrial physiology (62). Further, incorporation of lipophilic cationic compounds inside tumor mitochondria is favored by the inherent nature of their much greater electrochemical gradient (by 20–60 mV versus normal cell mitochondria). This promotes increased lipophilic cation accumulation at the matrix face of the MIM, imparting higher selectivity for such modified drugs to target cancer cells (63). The premise that ΔΨm,i is greater in cancer cells when compared with their non-malignant counterparts (25–27) would also indicate that TPP+-tagged compounds would be selective for cancer cells. Indeed, we found that the IC50 for killing of cancer cells by MitoVES is ∼0.5–3 μm for cancer cells and ∼20–60 μm for non-malignant cells,3 further supporting the intriguing nature of such compounds.
We therefore synthesized analogs of anti-cancer agents, with the mitochondrially targeted vitamin E succinate, MitoVE11S, as the prototypic compound by tagging VES with the TPP+ group (cf. Fig. 1) because such a modification is expected to cause preferential compartmentalization of the compounds in mitochondria, enhancing their bioactivity (64). We have shown that, indeed, MitoVE11S causes a greater level of apoptosis in cancer cells, some 1–2 log greater than does α-TOS, and that it does partition to mitochondria while retaining the cancer cell selectivity of the untargeted α-TOS.3 Due to the structure of MitoVE11S and the data shown for the mitochondrially targeted UbQ (MitoQ) (21, 23, 24), we reasoned that MitoVE11S would be positioned so that its TPP+ group is at the matrix face of the MIM and the tocopheryl succinyl group buried in the MIM, potentially in the vicinity or inside the QP site of CII. To test whether MitoVE11S induces ROS generation and apoptosis via CII, we used the parental, CII-dysfunctional and CII-reconstituted, RAS-transformed, Chinese hamster lung fibroblasts (20) and found that the agent was relatively inefficient in inducing the two processes in the CII-dysfunctional cells compared with cells with normal SDH activity.
The role of CII as a target for MitoVE11S is further documented by the stabilization of the ODD-GFP protein expressed in HCT116ODD-GFP cells when exposed to the agent. This is because inhibiting SDH, even with low levels of the drug over extended periods, probably results in an increase in the succinate/fumarate ratio, diffusion of succinate to the cytosol, and inhibition of prolyl hydroxylases, invoking the state of pseudohypoxia (31, 32). This, in turn, stabilizes the ODD domain of the HIF1α, which in the ODD-GFP construct regulates the stability of GFP. Because we did not observe GFP fluorescence in HCT116ODD-GFP cells exposed to MitoVE11S in less than 6 h (data not shown), whereas significant ROS accumulation occurred within relatively short periods, the pseudohypoxic state is probably secondary to the importance of ROS in promoting the onset of apoptosis in cancer cells exposed to MitoVE11S. To corroborate this premise, using live confocal microscopy and cells transfected with the pHyPer-dMito plasmid coding for the redox sensor OxyR (65), we observed generation of ROS in cancer cells as early as in 5 min following the addition of MitoVE11S.3 This further supports the primary role of the CII UbQ-binding site in the molecular action of the agent and is consistent with the notion that inhibition of CII can result in HIF1α stabilization (31, 32, 66, 67).
MitoVE11S-induced ROS did not significantly affect the expression of individual mtDNA-encoded subunits examined or the stability of the CI–CIII assemblies, although some shift toward the higher molecular weight was observed for CII and CIII (cf. Fig. 2E). It is unlikely that this has a direct effect, such as on the generation of ROS, because this is observed within minutes after the addition of MitoVE11S (see above). It is likely that the observed increase in the molecular weight of CI and CII is a consequence of the early ROS generation and the ensuing apoptosis. Most importantly, generation of ROS is not a result of destabilization of the mitochondrial complexes. However, MitoVE11S did specifically affect CII, inhibiting the conversion of succinate to fumarate and transfer of electrons from CII to CIII, which is normally accomplished by the endogenous UbQ of CII. The IC50 values for the two activities indicate a much stronger inhibition of electron transfer than that of the SDH enzymatic activity (1.5 versus 80 μm, respectively). Because MitoVE11S very efficiently blocks electron transfer from CII to CIII, electrons will be redirected to produce superoxide anion radicals, triggering the apoptotic pathway (68). At the same time, the relatively mild inhibition of the SDH activity of CII allows for succinate conversion to fumarate, resulting in generation of electrons to form superoxide levels high enough for apoptosis induction.
Because TPP+ acts as a charged anchor excluded from the lipid bilayer, it cannot be incorporated into the MIM (23, 24, 69). Shortening the aliphatic chain of MitoVE11S is proposed to restrict the access of the tocopheryl succinate headgroup penetrating down into the bilayer, causing loss of CII binding and apoptotic activity. Consistent with this premise, the short chain MitoVE11S homologs were much less efficient in ROS generation and apoptosis induction. In addition, the shorter chain compounds were less effective in causing GFP stabilization in the HCT116ODD-GFP cells and in inhibiting the electron transfer from CII to CIII. Hence, the data are consistent with the molecular mechanism of MitoVE11S with its biologically active moiety most likely binding into the Qp site, thereby affecting the function of UbQ. This is consistent with a report on MitoQ, a TPP-tagged analog of UbQ with a 10-C hydrophobic chain, whose quinone headgroup interacts with CII but not CI or CIII, such that it becomes the acceptor of electrons coming from the conversion of succinate to fumarate. The authors also showed that shortening the hydrophobic chain resulted in a lower level of its reduction by CII (23, 24).
The above findings were verified by molecular modeling, which was based on the published crystal structure of mammalian CII (44). Phospholipids were introduced to mimic the MIM in which the SDHC and SDHD subunits of CII are buried, enabling calculations of the interaction energies. MitoVES homologs with TPP+ at the surface of the mitochondrial face of the MIM and succinate lying at the Qp site showed affinities paralleling their relative activities in killing cancer cells as well as in causing ROS accumulation. The results of the modeling were also consistent with the higher apoptogenic activity obtained for R-MitoVE11S over the S-isomer. The data for the calculated affinity of MitoVE11S binding in the Qp site were supported by the results of an experiment in which we used SDHC-deficient cells transfected with the WT SDHC gene (18, 20) or with an SDHC gene in which the UbQ-binding Ser68 (44) was replaced with Ala or Leu. Cells expressing mutant SDHC showed greater resistance to ROS generation and apoptosis induction in response to MitoVE11S, supporting the importance of the UbQ-binding of Ser68 in the QP site for the biological activity of the agent.
Finally, we assessed the effect of MitoVE11S on a preclinical model of cancer, based on nude mice with HCT116ODD-GFP cell-derived xenografts. Although MitoVE11S was >10-fold more efficient than the non-targeted α-TOS, it caused stabilization of GFP in the tumors, as revealed by their sectioning followed by confocal microscopy. This result suggests that the molecular basis for induction of apoptosis by MitoVE11S in cultured cells, involving the state of pseudohypoxia, is also operational in suppression of tumor growth in preclinical, experimental carcinomas.
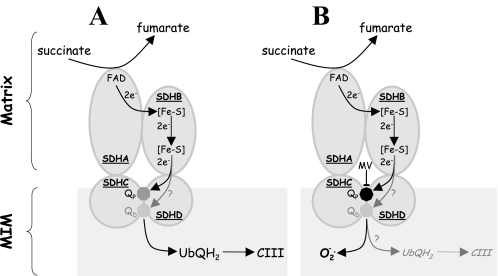
We propose that the molecular mechanism by which MitoVE11S affects cancer cells is based on its strong interaction with the binding of UbQ to the CII QP site. Consequently, the agent very efficiently blocks transfer of electrons from CII to CIII while only mildly suppressing the SDH activity of CII, which is important for generation of electrons from conversion of succinate to fumarate at the CII SDHA. Because the electrons are blocked from transfer to CIII, they then recombine with molecular oxygen to give rise to the apoptosis-inducing ROS. Further, it has been shown that the mammalian CII comprises two UbQ sites, the proximal (QP) and the distal (QD) sites (44). We suggested earlier that α-TOS is likely to interact with both QP and QD because it is not restricted in dissolving in the MIM (18). MitoVE11S, on the other hand, can only reach the QP site. The molecular mechanism for its effect on CII is indicated in Fig. 9, which depicts that MitoVE11S interferes with the QP site, blocking the movement of electrons from CII to CIII, which yields superoxide that acts as a signal triggering apoptosis. It is possible that in the presence of MitoVE11S, some electrons may proceed to CIII via the QD site. However, because this site is less well defined than QP and because its biological relevance is not defined, the potential flow of electrons via the QD is questionable at this stage.
FIGURE 9.
Model for the molecular mechanism of action of MitoVE11S on CII. A, in unstimulated cells, electrons generated from the conversion of succinate to fumarate at SDHA move via a series of Fe-S clusters down SDHB to the QP and, possibly, QD made up by amino acid residues of SDHC and SDHD. UbQ undergoes a two-electron reduction, when its affinity for the Q site(s) of CII is low, and it directs the two electrons to CIII, where it reoxidizes and shuttles back to CII. B, in the presence of MitoVE11S, the SDH activity of CII is only mildly reduced (IC50 ∼80 μm). Therefore, electrons are formed by conversion of succinate to fumarate, moving to the QP (and possibly QD) within the MIM. However, MitoVE11S, which is positioned so that its TPP+ group lies at the matrix face of the MIM and the tocopheryl succinyl group, interacts with the QP, displacing access to UbQ and very efficiently suppresses transfer of electrons from QP of CII to CIII (IC50 ∼1.5 μm). Consequently, this situation is highly unstable and gives rise to generation of superoxide as a by-product.
Present MitoVE11S results provide very strong evidence for mitochondrial CII as a bona fide target for potential cancer treatment and considerably extend our previous data obtained with α-TOS (18, 20). We conclude that mitochondrial targeting, achieved by tagging hydrophobic compounds with cationic groups, epitomized by VES modified with the TPP+ group, endows such agents with superior efficacy in apoptosis induction, translating into efficient anti-cancer activity, while retaining selectivity for malignant cells. Given the bleak outlook for cancer management (70, 71), finding novel, highly efficient, and selective drugs is of the utmost importance. Our proposal to target drugs that relay their activity by interfering with the mitochondrial function (mitocans) (1–5, 53) to these organelles to increase their concentration at their molecular target is a highly intriguing paradigm that has great potential for clinical benefit.
Supplementary Material
Acknowledgments
We thank I. Scheffler for B1 and B9 cells, E. Gottlieb for HCT116ODD-GFP cells, D. Spitz and F. Domann for pCR3.1-SDHC, and J. Hancock for pEGFP-C3-H-Ras.
This work was supported by grants from the Australian Research Council, the Cancer Council Queensland, the National Breast Cancer Foundation, Grant Agency of the Czech Republic Grants 204/08/0811 and P301/10/1937, Grant Agency of the Academy of Sciences of the Czech Republic Grants IAA500520702, KJB500970904, and KAN200520703) (to J. N.), the Griffith Health Institute (to L.-F. D.), the Otago University Research Grants Committee (to R. A. J. S.), Consejo Nacional de Ciencia y Tecnologia-México Grants 80534 and 20675 (to R. M.-S. and S. R.-E.), and National Institutes of Health, NCRR, Grant P20RR016456 (to E. M. and B. A. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental material describing the synthesis of VE analogs.
L.-F. Dong, V. J. A. Jameson, D. Tilly, L. Prochazka, J. Rohlena, K. Valis, J. Truksa, R. Zobalova, E. Mahdavian, K. Kluckova, M. Stantic, J. Stursa, X.-F. Wang, R. Freeman, P. K. Witting, E. Norberg, J. Goodwin, B. A. Salvatore, J. Novotna, J. Turanek, M. Ledvina, P. Hozak, B. Zhivotovsky, M. J. Coster, S. J. Ralph, R. A. J. Smith, and J. Neuzil, submitted for publication.
J. Rohlena and J. Neuzil, unpublished data.
- VE
- vitamin E
- CI
- CII, and CIII, complex I, II, and III, respectively
- FCCP
- carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone
- MIM
- mitochondrial inner membrane
- MitoQ
- mitochondrially targeted coenzyme Q
- MitoVES
- mitochondrially targeted vitamin E succinate
- MitoVEnS
- MitoVES homologue with n carbons linking the tocopheryl headgroup and the triphenyphosphonium group
- ODD
- oxygen-dependent destruction
- POPE
- phosphatidylethanolamine
- ROS
- reactive oxygen species
- SDH
- succinate dehydrogenase
- SMP
- submitochondrial particle
- α-TOS
- α-tocopheryl succinate
- TPP+
- triphenylphosphonium
- UbQ
- ubiquinone
- USI
- ultrasound imaging
- VES
- vitamin E succinate
- ΔΨm,i
- mitochondrial inner transmembrane potential
- QP
- proximal UbQ site
- QD
- distal UbQ site
- qPCR
- quantitative PCR
- mtDNA
- mitochondrial DNA.
REFERENCES
- 1. Fantin V. R., Leder P. (2006) Oncogene 25, 4787–4797 [DOI] [PubMed] [Google Scholar]
- 2. Moreno-Sánchez R., Rodríguez-Enríquez S., Marín-Hernández A., Saavedra E. (2007) FEBS J. 274, 1393–1418 [DOI] [PubMed] [Google Scholar]
- 3. Gogvadze V., Orrenius S., Zhivotovsky B. (2008) Trends Cell Biol. 18, 165–173 [DOI] [PubMed] [Google Scholar]
- 4. Trachootham D., Alexandre J., Huang P. (2009) Nat. Rev. Drug Discov. 8, 579–591 [DOI] [PubMed] [Google Scholar]
- 5. Fulda S., Galluzzi L., Kroemer G. (2010) Nat. Rev. Drug Discov. 9, 447–464 [DOI] [PubMed] [Google Scholar]
- 6. Neuzil J., Wang X. F., Dong L. F., Low P., Ralph S. J. (2006) FEBS Lett. 580, 5125–5129 [DOI] [PubMed] [Google Scholar]
- 7. Neuzil J., Dong L. F., Ramanathapuram L., Hahn T., Chladova M., Wang X. F., Zobalova R., Prochazka L., Gold M., Freeman R., Turanek J., Akporiaye E. T., Dyason J. C., Ralph S. J. (2007) Mol. Aspects Med. 28, 607–645 [DOI] [PubMed] [Google Scholar]
- 8. Neuzil J., Tomasetti M., Zhao Y., Dong L. F., Birringer M., Wang X. F., Low P., Wu K., Salvatore B. A., Ralph S. J. (2007) Mol. Pharmacol. 71, 1185–1199 [DOI] [PubMed] [Google Scholar]
- 9. Neuzil J., Weber T., Gellert N., Weber C. (2001) Br. J. Cancer 84, 87–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neuzil J., Weber T., Schröder A., Lu M., Ostermann G., Gellert N., Mayne G. C., Olejnicka B., Nègre-Salvayre A., Stícha M., Coffey R. J., Weber C. (2001) FASEB J. 15, 403–415 [DOI] [PubMed] [Google Scholar]
- 11. Weber T., Lu M., Andera L., Lahm H., Gellert N., Fariss M. W., Korinek V., Sattler W., Ucker D. S., Terman A., Schröder A., Erl W., Brunk U. T., Coffey R. J., Weber C., Neuzil J. (2002) Clin. Cancer Res. 8, 863–869 [PubMed] [Google Scholar]
- 12. Malafa M. P., Fokum F. D., Mowlavi A., Abusief M., King M. (2002) Surgery 131, 85–91 [DOI] [PubMed] [Google Scholar]
- 13. Yu W., Sanders B. G., Kline K. (2003) Cancer Res. 63, 2483–2491 [PubMed] [Google Scholar]
- 14. Stapelberg M., Gellert N., Swettenham E., Tomasetti M., Witting P. K., Procopio A., Neuzil J. (2005) J. Biol. Chem. 280, 25369–25376 [DOI] [PubMed] [Google Scholar]
- 15. Lawson K. A., Anderson K., Menchaca M., Atkinson J., Sun L., Knight V., Gilbert B. E., Conti C., Sanders B. G., Kline K. (2003) Mol. Cancer Ther. 2, 437–444 [PubMed] [Google Scholar]
- 16. Hahn T., Szabo L., Gold M., Ramanathapuram L., Hurley L. H., Akporiaye E. T. (2006) Cancer Res. 66, 9374–9378 [DOI] [PubMed] [Google Scholar]
- 17. Shiau C. W., Huang J. W., Wang D. S., Weng J. R., Yang C. C., Lin C. H., Li C., Chen C. S. (2006) J. Biol. Chem. 281, 11819–11825 [DOI] [PubMed] [Google Scholar]
- 18. Dong L. F., Low P., Dyason J. C., Wang X. F., Prochazka L., Witting P. K., Freeman R., Swettenham E., Valis K., Liu J., Zobalova R., Turanek J., Spitz D. R., Domann F. E., Scheffler I. E., Ralph S. J., Neuzil J. (2008) Oncogene 27, 4324–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weber T., Dalen H., Andera L., Nègre-Salvayre A., Augé N., Sticha M., Lloret A., Terman A., Witting P. K., Higuchi M., Plasilova M., Zivny J., Gellert N., Weber C., Neuzil J. (2003) Biochemistry 42, 4277–4291 [DOI] [PubMed] [Google Scholar]
- 20. Dong L. F., Freeman R., Liu J., Zobalova R., Marin-Hernandez A., Stantic M., Rohlena J., Valis K., Rodriguez-Enriquez S., Butcher B., Goodwin J., Brunk U. T., Witting P. K., Moreno-Sanchez R., Scheffler I. E., Ralph S. J., Neuzil J. (2009) Clin. Cancer Res. 15, 1593–1600 [DOI] [PubMed] [Google Scholar]
- 21. Murphy M. P., Smith R. A. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 629–656 [DOI] [PubMed] [Google Scholar]
- 22. Kelso G. F., Porteous C. M., Coulter C. V., Hughes G., Porteous W. K., Ledgerwood E. C., Smith R. A., Murphy M. P. (2001) J. Biol. Chem. 276, 4588–45896 [DOI] [PubMed] [Google Scholar]
- 23. James A. M., Cochemé H. M., Smith R. A., Murphy M. P. (2005) J. Biol. Chem. 280, 21295–21312 [DOI] [PubMed] [Google Scholar]
- 24. James A. M., Sharpley M. S., Manas A. R., Frerman F. E., Hirst J., Smith R. A., Murphy M. P. (2007) J. Biol. Chem. 282, 14708–14718 [DOI] [PubMed] [Google Scholar]
- 25. Davis S., Weiss M. J., Wong J. R., Lampidis T. J., Chen L. B. (1985) J. Biol. Chem. 260, 13844–13850 [PubMed] [Google Scholar]
- 26. Modica-Napolitano J. S., Aprille J. R. (2001) Adv. Drug Deliv. Rev. 49, 63–70 [DOI] [PubMed] [Google Scholar]
- 27. Fantin V. R., Berardi M. J., Scorrano L., Korsmeyer S. J., Leder P. (2002) Cancer Cell 2, 29–42 [DOI] [PubMed] [Google Scholar]
- 28. Oostveen F. G., Au H. C., Meijer P. J., Scheffler I. E. (1995) J. Biol. Chem. 270, 26104–26108 [DOI] [PubMed] [Google Scholar]
- 29. Prior I. A., Harding A., Yan J., Sluimer J., Parton R. G., Hancock J. F. (2001) Nat. Cell Biol. 3, 368–375 [DOI] [PubMed] [Google Scholar]
- 30. Slane B. G., Aykin-Burns N., Smith B. J., Kalen A. L., Goswami P. C., Domann F. E., Spitz D. R. (2006) Cancer Res. 66, 7615–7620 [DOI] [PubMed] [Google Scholar]
- 31. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., Gottlieb E. (2005) Cancer Cell 7, 77–85 [DOI] [PubMed] [Google Scholar]
- 32. MacKenzie E. D., Selak M. A., Tennant D. A., Payne L. J., Crosby S., Frederiksen C. M., Watson D. G., Gottlieb E. (2007) Mol. Cell. Biol. 27, 3282–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jasso-Chávez R., Torres-Márquez M. E., Moreno-Sánchez R. (2001) Arch. Biochem. Biophys. 390, 295–303 [DOI] [PubMed] [Google Scholar]
- 34. Covián R., Moreno-Sánchez R. (2001) Eur. J. Biochem. 268, 5783–5790 [DOI] [PubMed] [Google Scholar]
- 35. Dong L. F., Swettenham E., Eliasson J., Wang X. F., Gold M., Medunic Y., Stantic M., Low P., Prochazka L., Witting P. K., Turanek J., Akporiaye E. T., Ralph S. J., Neuzil J. (2007) Cancer Res. 67, 11906–11913 [DOI] [PubMed] [Google Scholar]
- 36. Wang X. F., Birringer M., Dong L. F., Veprek P., Low P., Swettenham E., Stantic M., Yuan L. H., Zobalova R., Wu K., Ledvina M., Ralph S. J., Neuzil J. (2007) Cancer Res. 67, 3337–3344 [DOI] [PubMed] [Google Scholar]
- 37. Wang J. M., Cieplak P., Kollman P. A. (2000) J. Comput. Chem. 21, 1049–1074 [Google Scholar]
- 38. Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. (2004) J. Comput. Chem. 25, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 39. Bayly C. I., Cieplak P., Cornell W. D., Kollman P. A. (1993) J. Phys. Chem. 97, 10269–10280 [Google Scholar]
- 40. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 41. Krieger E., Koraimann G., Vriend G. (2002) Proteins 47, 393–402 [DOI] [PubMed] [Google Scholar]
- 42. Maiorov V. N., Crippen G. M. (1994) J. Mol. Biol. 235, 625–634 [DOI] [PubMed] [Google Scholar]
- 43. Adam-Vizi V., Chinopoulos C. (2006) Trends Pharmacol. Sci. 27, 639–645 [DOI] [PubMed] [Google Scholar]
- 44. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. (2005) Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 45. Yang X., Yu L., He D., Yu C. A. (1998) J. Biol. Chem. 273, 31916–31923 [DOI] [PubMed] [Google Scholar]
- 46. Albayrak T., Scherhammer V., Schoenfeld N., Braziulis E., Mund T., Bauer M. K., Scheffler I. E., Grimm S. (2003) Mol. Biol. Cell 14, 3082–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eng C., Kiuru M., Fernandez M. J., Aaltonen L. A. (2003) Nat. Rev. Cancer 3, 193–202 [DOI] [PubMed] [Google Scholar]
- 48. Gottlieb E., Tomlinson I. P. (2005) Nat. Rev. Cancer 5, 857–866 [DOI] [PubMed] [Google Scholar]
- 49. King A., Selak M. A., Gottlieb E. (2006) Oncogene 25, 4675–4682 [DOI] [PubMed] [Google Scholar]
- 50. Schiavi F., Boedeker C. C., Bausch B., Peçzkowska M., Gomez C. F., Strassburg T., Pawlu C., Buchta M., Salzmann M., Hoffmann M. M., Berlis A., Brink I., Cybulla M., Muresan M., Walter M. A., Forrer F., Välimäki M., Kawecki A., Szutkowski Z., Schipper J., Walz M. K., Pigny P., Bauters C., Willet-Brozick J. E., Baysal B. E., Januszewicz A., Eng C., Opocher G., Neumann H. P. (2005) JAMA 294, 2057–2063 [DOI] [PubMed] [Google Scholar]
- 51. Peczkowska M., Cascon A., Prejbisz A., Kubaszek A., Cwikła B. J., Furmanek M., Erlic Z., Eng C., Januszewicz A., Neumann H. P. (2008) Nat. Clin. Pract. Endocrinol. Metab. 4, 111–115 [DOI] [PubMed] [Google Scholar]
- 52. Ishii T., Yasuda K., Akatsuka A., Hino O., Hartman P. S., Ishii N. (2005) Cancer Res. 65, 203–209 [PubMed] [Google Scholar]
- 53. Galluzzi L., Morselli E., Kepp O., Vitale I., Rigoni A., Vacchelli E., Michaud M., Zischka H., Castedo M., Kroemer G. (2010) Mol. Aspects Med. 31, 1–20 [DOI] [PubMed] [Google Scholar]
- 54. Szeto S. S., Reinke S. N., Sykes B. D., Lemire B. D. (2007) J. Biol. Chem. 282, 27518–27526 [DOI] [PubMed] [Google Scholar]
- 55. Cervera A. M., Apostolova N., Crespo F. L., Mata M., McCreath K. J. (2008) Cancer Res. 68, 4058–4067 [DOI] [PubMed] [Google Scholar]
- 56. Guzy R. D., Sharma B., Bell E., Chandel N. S., Schumacker P. T. (2008) Mol. Cell. Biol. 28, 718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lemarie A., Grimm S. (2009) Mitochondrion 9, 254–260 [DOI] [PubMed] [Google Scholar]
- 58. Hayden E. C. (2008) Nature 455, 158 [Google Scholar]
- 59. Smith R. A., Porteous C. M., Gane A. M., Murphy M. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5407–5412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Adlam V. J., Harrison J. C., Porteous C. M., James A. M., Smith R. A., Murphy M. P., Sammut I. A. (2005) FASEB J. 19, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 61. Neuzil J., Widén C., Gellert N., Swettenham E., Zobalova R., Dong L. F., Wang X. F., Lidebjer C., Dalen H., Headrick J. P., Witting P. K. (2007) Redox Rep. 12, 148–162 [DOI] [PubMed] [Google Scholar]
- 62. Rodriguez-Cuenca S., Cochemé H. M., Logan A., Abakumova I., Prime T. A., Rose C., Vidal-Puig A., Smith A. C., Rubinsztein D. C., Fearnley I. M., Jones B. A., Pope S., Heales S. J., Lam B. Y., Neogi S. G., McFarlane I., James A. M., Smith R. A., Murphy M. P. (2010) Free Radic. Biol. Med. 48, 161–172 [DOI] [PubMed] [Google Scholar]
- 63. Rodríguez-Enríquez S., Marín-Hernández A., Gallardo-Pérez J. C., Carreño-Fuentes L., Moreno-Sánchez R. (2009) Mol. Nutr. Food Res. 53, 29–48 [DOI] [PubMed] [Google Scholar]
- 64. Biassutto L., Dong L. F., Neuzil J., Zoratti M. (2010) Mitochondrion, in press [DOI] [PubMed] [Google Scholar]
- 65. Belousov V. V., Fradkov A. F., Lukyanov K. A., Staroverov D. B., Shakhbazov K. S., Terskikh A. V., Lukyanov S. (2006) Nat. Methods 3, 281–286 [DOI] [PubMed] [Google Scholar]
- 66. Piantadosi C. A., Suliman H. B. (2008) J. Biol. Chem. 283, 10967–10977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hawkins B. J., Levin M. D., Doonan P. J., Petrenko N. B., Davis C. W., Patel V. V., Madesh M. (2010) J. Biol. Chem. 285, 26494–26505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Prochazka L., Dong L. F., Valis K., Freeman R., Ralph S. J., Turanek J., Neuzil J. (2010) Apoptosis 15, 782–794 [DOI] [PubMed] [Google Scholar]
- 69. Biasutto L., Sassi N., Mattarei A., Marotta E., Cattelan P., Toninello A., Garbisa S., Zoratti M., Paradisi C. (2010) Biochim. Biophys. Acta 1797, 189–196 [DOI] [PubMed] [Google Scholar]
- 70. Twombly R. (2005) J. Natl. Cancer Inst. 97, 330–331 [DOI] [PubMed] [Google Scholar]
- 71. Jemal A., Siegel R., Xu J., Ward E. (2010) CA Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.