Abstract
Hexaprenyl diphosphate synthase from Micrococcus luteus B-P 26 (Ml-HexPPs) is a heterooligomeric type trans-prenyltransferase catalyzing consecutive head-to-tail condensations of three molecules of isopentenyl diphosphates (C5) on a farnesyl diphosphate (FPP; C15) to form an (all-E) hexaprenyl diphosphate (HexPP; C30). Ml-HexPPs is known to function as a heterodimer of two different subunits, small and large subunits called HexA and HexB, respectively. Compared with homooligomeric trans-prenyltransferases, the molecular mechanism of heterooligomeric trans-prenyltransferases is not yet clearly understood, particularly with respect to the role of the small subunits lacking the catalytic motifs conserved in most known trans-prenyltransferases. We have determined the crystal structure of Ml-HexPPs both in the substrate-free form and in complex with 7,11-dimethyl-2,6,10-dodecatrien-1-yl diphosphate ammonium salt (3-DesMe-FPP), an analog of FPP. The structure of HexB is composed of mostly antiparallel α-helices joined by connecting loops. Two aspartate-rich motifs (designated the first and second aspartate-rich motifs) and the other characteristic motifs in HexB are located around the diphosphate part of 3-DesMe-FPP. Despite the very low amino acid sequence identity and the distinct polypeptide chain lengths between HexA and HexB, the structure of HexA is quite similar to that of HexB. The aliphatic tail of 3-DesMe-FPP is accommodated in a large hydrophobic cleft starting from HexB and penetrating to the inside of HexA. These structural features suggest that HexB catalyzes the condensation reactions and that HexA is directly involved in the product chain length control in cooperation with HexB.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Mutation, Isoprenoid, Protein Structure, X-ray Crystallography, Crystal Structure, Heterodimeric Prenyltransferase, Micrococcus luteus B-P 26
Introduction
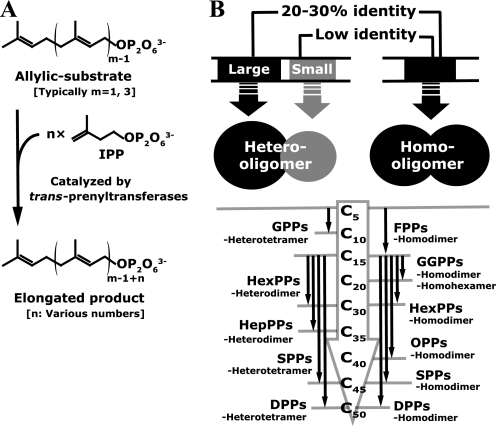
Over 50,000 structurally diverse isoprenoids, which are built from C5 isoprene units, are widely distributed in nature (1). Many kinds of isoprenoids, such as steroids, hemes, carotenoids, vitamins, quinones, and membrane lipids, are essential components of the cellular machinery of all organisms. Prenyltransferases, the so-called prenyl diphosphate synthases, catalyze consecutive head-to-tail condensations of isopentenyl diphosphates (IPP2; C5 homoallylic substrate) on an allylic substrate, such as dimethylallyl diphosphate (DMAPP; C5) or farnesyl diphosphate (FPP; C15), to form linear prenyl diphosphates with various chain lengths (Fig. 1A). The linear prenyl diphosphates are common precursors of the carbon skeletons for all isoprenoids. According to the geometry of the newly formed double bonds of the products, prenyltransferases can be divided into two major classes, trans- and cis-prenyltransferases (2–4). Furthermore, the trans-prenyltransferases can be divided into two subclasses, homo- and heterooligomeric enzymes (Fig. 1B), whereas all known cis-prenyltransferases, including farnesyl and decaprenyl diphosphate synthase from Mycobacterium tuberculosis (5) and undecaprenyl diphosphate synthases from Micrococcus luteus B-P 26 (6) and from Escherichia coli (7), are homodimeric enzymes.
FIGURE 1.
Characterization and classification of trans-prenyltransferases. A, reactions catalyzed by trans-prenyltransferases. Prenyltransferases condense IPP molecules consecutively (n times) on an allylic substrate. The elongated product is widely used for various isoprenoid biosyntheses. The HexPPs here investigated catalyzes the condensation of IPP three times (n = 3) on FPP (m = 3). B, two subclasses of trans-prenyltransferases. Several enzymes belonging to each subclass are listed together with the starting substrate and the final product. The large components of the heterooligomeric enzymes show 20–30% identities to the homooligomeric enzymes. The small components shown in gray have little similarity with the homooligomeric enzymes. The HexA and HexB subunits correspond with the small and the large components, respectively. The detailed list of well characterized enzymes is shown in supplemental Fig. 1. SPPs, solanesyl diphosphate synthase.
Homooligomeric trans-prenyltransferases are widely known and well characterized (Fig. 1B). Most of them exist as a homodimer under physiological conditions. So far, the crystal structures of many of the homooligomeric trans-prenyltransferases have been determined as a homodimer with a 2-fold axis at the interface (8–20). All of them typically possess a bundle of anti-parallel α-helices in a monomer. Each subunit has a hydrophobic cleft at each center surrounded by the helices. Other homooligomeric trans-prenyltransferases are tetrameric, hexameric, octameric, or much larger oligomeric enzymes (21–25). The crystal structure of geranylgeranyl diphosphate (GGPP; C20) synthase from Homo sapiens (Hs-GGPPs) (PDB code 2Q80), a homohexameric enzyme, reveals that the hexamer is an assembly of the three homodimer units (22). These structural and biological works suggest that the functional unit of the homooligomeric enzymes is a homodimer. Each subunit of the homodimer in the homooligomeric enzymes has two highly conserved aspartate-rich DDXX2–4D and DDXXD motifs as well as other characteristic GKXXR, RRG, G(Q/E), KT, and (F/Y)Q motifs (2, 3). Many structural and mutagenesis studies have shown that all of these motifs are located around the entrance of the hydrophobic cleft and are involved in the recognition of the diphosphate part of the two substrates or product and in the condensation reactions (10, 13–15, 17, 18, 22, 26–32). The crystal structure of FPP synthase from E. coli (Ec-FPPs) with its two ligands (DMAPP analog and IPP) elucidates the detailed binding geometry of the substrates (10). On the other hand, the interior part of the cleft accommodates the hydrophobic tail of the reaction product. The size of the cleft is strongly involved in the regulation of the final product chain length (9, 11, 12, 26, 33–39). The crystal structure of GGPP synthase from Saccharomyces cerevisiae (Sc-GGPPs) with its final product GGPP revealed that the hydrophobic tail of the product just fits the cleft in both of the two homodimeric subunits (18). Many mutagenesis investigations at the interior of the hydrophobic cleft have reported that the deeper cleft synthesizes the longer final product (12). The cleft shape and size are also the key factor to develop the inhibitors of these enzymes, such as bisphosphonates, to medicate osteoporosis, Paget disease, and hypercalcemia (13, 14, 16–20, 40–42). It should be noted that almost all mutational investigations to extend the product chain length are designed to make the cleft deeper along with the antiparallel α-helices because the hydrophobic chain is commonly believed to extend along the helices (9, 11, 12, 33, 37, 43).
In contrast, the function of heterooligomeric enzymes is not well understood. Only limited numbers of the enzymes in this class are known as heterodimeric or heterotetrameric enzymes (Fig. 1B) (44–57). The heterooligomeric enzymes are composed of two different components, large and small subunits, and are shown to be unable to catalyze the chain elongation reaction without either of the two components (49, 51, 52, 54–56, 58). The amino acid sequence similarity between the large components and homooligomeric enzymes is less than 30% (Fig. 1B), but the catalytically important motifs in homooligomeric enzymes are still conserved in the large components of these heterooligomeric enzymes (2, 44, 51, 52). Recently, the crystal structure of geranyl diphosphate (GPP; C10) synthase from Mentha piperita (Mp-GPPs), a heterotetrameric enzyme, has been determined (59). The structure showed that the large subunits participate in the substrate recognition and the condensation reactions using the catalytic motifs as seen in homooligomeric trans-prenyltransferases. On the other hand, the small components do not possess the catalytic motifs and show little sequence similarity (up to 10%) with the homooligomeric enzymes and with the large components (27, 46–48, 50). The overall architectures of the small subunits of heterotetrameric Mp-GPPs (SSU) are quite similar to those of the large subunits and of known homooligomeric trans-prenyltransferases. So far, the role of the small component is still unclear due to the lack of links between the structure and mutational analyses.
Hexaprenyl diphosphate (HexPP; C30) synthase from M. luteus B-P 26 (Ml-HexPPs) is known to be a heterodimeric trans-prenyltransferase catalyzing three consecutive condensations of IPP on FPP to produce HexPP. Several mutational studies have been performed on this enzyme and a related heterodimeric enzyme, heptaprenyl diphosphate (HepPP; C35) synthase from Bacillus subtilis and Bacillus stearothermophilus (60–62). The small component of Ml-HexPPs (HexA) is composed of only 143 amino acids, which is approximately half the size of its large component (HexB; 325 amino acids), half the size of the small and the large components of heterotetrameric Mp-GPPs (SSU, 266 aa; LSU, 295 aa), or half the size of the typical homooligomeric trans-prenyltransferases (∼300 aa). The small and simple architecture of HexA together with the information of mutational analyses make Ml-HexPPs an attractive target to elucidate the detailed molecular mechanisms of heterooligomeric trans-prenyltransferases, especially the role of the small components in the overall function. Thus, we have determined the crystal structure of Ml-HexPPs in both the substrate-free and ligand-bound forms. The structure implies that HexA is directly involved in the product chain length regulation together with the other component HexB.
MATERIALS AND METHODS
Cloning, Expression, and Purification
The EK/LIC cloning kit (Novagen) was utilized to construct the expression systems. The gene fragment encoding 143 aa of HexA (AB003188) was amplified with polymerase chain reaction (PCR) using hexs-a_pUC119 (50) as the template with the forward primer 5′-GACGACGACAAGATGCGTTATTTACATAAAATTGAACTAGAA-3′ and the reverse primer 5′-GAGGAGAAGCCCGGTTACTCATAAGACACCACTTTCTCATTG-3′. The amplified gene was inserted into an ampicillin-resistant vector pET32Ek/LIC (Novagen). The created plasmid named hexs-a_pET32 expresses HexA with thioredoxin, His, and S tags in front of the N terminus. The enterokinase protease failed to cut the S tag from the expressed HexA at the specific digestion site. Thus, we made a new construct (hexs-a_pET32ΔS) without the S tag and enterokinase-specific site in HexA with an inverse PCR method using the forward primer 5′-ATGCGTTATTTACATAAAATTGAACTAG-3′ and the reverse primer 5′-GACCACGGTGCGCCAAGACCATAC-3′. The resultant vector, named hexs-a_pET32ΔS, expresses HexA with thioredoxin and His tags in front of its N terminus. The tags on the new construct can be removed only with thrombin. The HexB (325 aa; AB003188) gene fragment was amplified from the template plasmid hexs-b_pUC119 (50), with the forward primer 5′-GACGACGACAAGATGATTGCTTTGAGTTATAAAGCGTTTTTA-3′ and the reverse primer 5′-GAGGAGAAGCCCGGTTAATAAACACGTTTTAACATTTTTTCGTG-3′. The amplified gene was inserted into the LIC site in a kanamycin-resistant vector, pET30Ek/LIC (Novagen). The resultant vector, named hexs-b_pET30, expresses HexB with His and S tags in front of its N-terminal. The tags on the construct can be removed with thrombin and recombinant enterokinase.
The single transformed E. coli cells (Rosetta2(DE3)pLysS) with either hexs-a_pET32ΔS or hexs-b_pET30 express HexA or HexB as an inclusion body, respectively. In contrast, the co-transformed cells with both of the two vectors express a decent amount of the soluble HexA and HexB as described below. The co-transformants were plated on Luria-Bertani (LB) agar plates containing 34 μg/ml chloramphenicol, 50 μg/ml carbenicillin, and 20 μg/ml kanamycin at 37 °C. The resultant colonies were separately picked up and precultured at 37 °C with overnight shaking at 180 rpm. volume of preculture was inoculated into LB medium containing the same antibiotics with the plate. The cells were cultured at 37 °C for about 2.5 h with shaking at 180 rpm until the absorbance at 600 nm reached 0.6. Isopropyl β-d-thiogalactopyranoside was added to a final concentration of 1 mm, and the induction of the recombinant protein was continued for an additional 3 h at 25 °C.
The harvested cell pellet was suspended in suspending buffer (50 mm sodium phosphate (pH 7.0), 300 mm NaCl, 1 mm DTT) containing 10 μl/ml protease/inhibitor cocktail (NACALAI TESQUE). The suspension was sonicated on ice, and the cell debris was removed by centrifugation for 60 min. at 18,000 rpm (30,000 × g). After filtration by a 1.2-μm pore filter, the supernatant was applied to TALON His tag affinity resin (Clontech) equilibrated with equilibration buffer (50 mm sodium phosphate (pH 7.0), 300 mm NaCl). The resin was washed with equilibration buffer, and the stepwise imidazole concentrations (50, 100, and 150 mm) were used for elution. The buffer of the fractions containing Ml-HexPPs (100 and 150 mm imidazole in equilibration buffer) was exchanged with equilibration buffer. The tags were removed simultaneously with a treatment with thrombin (GE Healthcare; 10 units for 1 mg of protein) and recombinant enterokinase (Novagen; 2 units for 1 mg of protein) in cleavage solution (50 mm sodium phosphate (pH 7.0), 300 mm NaCl, 5 mm MgCl2, 100 μm FPP) for 40 h at 4 °C. The digestion was stopped by adding 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride to a final concentration of 1 mm. The reaction mixture was applied to another TALON column equilibrated with equilibration buffer. The flow-through was collected, and its buffer was exchanged to buffer A (20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 1 mm DTT, 10% (v/v) glycerol). The sample was applied to an anion exchange column (MiniQ 4.6/50 PE, 0.8 ml; GE Healthcare) equilibrated with buffer A and eluted at a flow rate of 0.3 ml/min. with a linear NaCl gradient (50–1000 mm) with buffer B (20 mm Tris-HCl (pH 7.5), 1000 mm NaCl, 1 mm DTT, 10% (v/v) glycerol). The Ml-HexPPs fraction was concentrated to a volume of 500 μl in buffer C (20 mm Tris-HCl (pH 7.5), 300 mm NaCl, 1 mm DTT, 10% (v/v) glycerol) and applied to a size exclusion column (Superdex 200 10/300 GL, 24 ml; GE Healthcare) equilibrated with buffer C at a flow rate of 0.4 ml/min. All purification steps were carried out at 4 °C. Approximately 8 mg of the purified Ml-HexPPs was obtained from 3 g of the wet cells (co-transformed with hexs-a_pET32ΔS and hexs-b_pET30) that had been harvested from 1.5 liters of the culture. The purity of the final Ml-HexPPs sample for crystallization was more than 95% (SDS-PAGE).
Prenyltransferase Activity Assay
The prenyltransferase activity assay was performed as reported previously with small modifications (63). The assay mixture (10 mm Tris-HCl (pH 7.5), 2.5 μm [14C]IPP (2.18 GBq/mmol), 2.5 μm FPP, 5 mm MgCl2, and 30 μg of recombinant Ml-HexPPs) was incubated in a final volume of 200 μl for 1 h at 37 °C, and the reaction was stopped by chilling in an ice bath. The product was extracted by shaking with 600 μl of 1-butanol saturated with water. The butanol layer was washed with 200 μl of water saturated with NaCl and treated with potato acid phosphatase. The hydrolysates were extracted with pentane and analyzed by reversed phase TLC using a precoated plate, LKC-18F (Whatman), developed with acetone/H2O (9:1) (64). Authentic standard alcohols were visualized with iodine vapor, and the distribution of radioactivity was detected using a Fuji BAS 1000 Mac bioimaging analyzer (Fujifilm, Tokyo, Japan).
Oligomeric State Analysis
The molecular weight of samples in solution was analyzed using a size exclusion column (Superdex 200 10/300 GL) equilibrated with buffer C at a flow rate of 0.4 ml/min. The retention volume of Ml-HexPPs was compared with those of marker proteins. The samples and markers applied to the column were purified Ml-HexPPs (3.5 mg/ml, 500 μl), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), and ribonuclease A (14 kDa). All marker proteins were purchased from GE Healthcare.
Crystallization and Data Collection
Initial crystallization screening was performed at 4 °C with the sitting drop vapor diffusion method using typical commercial crystal screen kits (Hampton Research) with over 400 conditions. The crystals of Ml-HexPPs were obtained in a mixture of 1 μl of the sample solution (10 mg/ml recombinant Ml-HexPPs, 20 mm Tris-HCl (pH 7.5), 50 mm NaCl, 1 mm DTT, 10% (v/v) glycerol) and 1 μl of the reservoir solution (0.08 m Tris-HCl (pH 8.5), 0.16 m magnesium chloride hexahydrate, 24% (w/v) PEG 4000, 20% (v/v) glycerol), equilibrated with 100 μl of the reservoir solution by the sitting drop vapor diffusion method at 4 °C. The crystals were mounted on a cryoloop (Hampton Research) and flash cryocooled in a nitrogen stream at 95 K without using any specific cryoprotectant solutions. Data sets of x-ray diffraction were collected at 95 K using synchrotron radiation at the BL5A (ADSC Quantum 315 CCD detector) beamline at the Photon Factory, KEK, Japan, under the following conditions: an oscillation range, an exposure time per frame, and a crystal-to-detector distance of 0.2°, 5.0 s, and 280.1 mm, respectively. The derivative crystals were prepared by soaking in a reservoir solution containing 1 mm thimerosal (a mercury compound) for 14 h. The data sets were collected at the BL17A (ADSC Quantum 270 CCD detector) at the Photon Factory, under the following conditions: an oscillation range, an exposure time per frame, and a crystal-to-detector distance of 0.5°, 5.0 s, and 331.3 mm, respectively. The crystals in complex with 7,11-dimethyl-2,6,10-dodecatrien-1-yl diphosphate ammonium salt (3-DesMe-FPP) (65) were prepared by soaking in reservoir solution with 1 mm analog for 12 h. The compound 3-DesMe-FPP was a kind gift from Prof. Y. Maki (Yamagata University) (65). The data sets were collected at NW12A (ADSC Quantum 270 CCD detector) at the Photon Factory Advanced Ring under the following conditions: an oscillation range, an exposure time per frame, and a crystal-to-detector distance of 0.25°, 1.0 s, and 235.1 mm, respectively. All diffraction images were integrated and scaled using the HKL2000 program suite (66).
Phasing, Model Building, and Refinement
Phases were determined at the resolution range of 50–3.0 Å by the SIRAS (single isomorphous replacement with anomalous scattering) method using the program SOLVE (67) with the native and thimerosal derivative data sets. The phases were improved, and an initial model was constructed using the program RESOLVE (68) at the same resolution range. The initial 2Fo − Fc electron density map after the RESOLVE treatment clearly showed electron density for most amino acid residues. The final model structure consists of two heterodimers in a crystallographic asymmetric unit. Each of the heterodimers is constructed from subunits S1 (HexA) and L1 (HexB) as well as S2 (HexA′) and L2 (HexB′). Residues 1–4 in subunit L1 as well as the C terminus of 135–143 in subunits S1 and S2 were disordered. The model was manually modified at the 50–3.0 Å resolution range using the program COOT (69). The refinement was finalized at the resolution of 50–2.4 Å for the native data set using the program REFMAC5 (70) in CCP4i (Collaborative Computational Project 4 interface) (71). The 2Fo − Fc electron density map was significantly improved after the refinement. The Rfree value, which was calculated with randomly selected 5% reflections, was used for monitoring the quality of the model. The final figure of merit, Rwork, and Rfree values were 0.79, 24.0, and 27.6%, respectively. The data set of the 3-DesMe-FPP complex was directly phased with the final substrate-free Ml-HexPPs model and refined at 50–2.6 Å resolution. The Ramachandran analyses for the structures were performed using the program MOLPROBITY (72, 73). The statistics for data collection, phasing, refinement, and the final model are summarized in Table 1.
TABLE 1.
Statistics for data collection and structure refinement
| Native | Thimerosala | 3-DesMe-FPP | |
|---|---|---|---|
| Data collection statistics | |||
| X-ray source | PF-BL5A | PF-BL17A | PFAR-NW12A |
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0000 |
| Space group | I213 | I213 | I213 |
| Cell parameter a (Å) | 189.6 | 189.6 | 189.8 |
| Resolution range (Å)b | 50–2.40 (2.49–2.40) | 50–2.75 (2.85–2.75) | 50–2.60 (2.69–2.60) |
| No. of reflections, total/unique | 503,756/44,225 | 326,388/29,490 | 553,102/34,831 |
| Completeness (%) (I/σ(I) > 0)b | 100 (100) | 100 (100) | 100 (100) |
| Redundancyb | 11.4 (11.5) | 11.1 (10.5) | 15.9 (13.3) |
| I/σ(I)b | 29.3 (5.4) | 20.3 (7.0) | 27.5 (9.1) |
| Rsym (%)b,c | 8.3 (49.4) | 9.5 (36.8) | 9.9 (34.2) |
| Rr.i.m. (%)b,d | 8.6 (48.6) | 9.8 (39.8) | 7.1 (33.5) |
| Rp.i.m. (%)b,e | 2.6 (14.3) | 3.0 (12.2) | 1.6 (8.5) |
| Mosaicity range | 0.38–0.47 | 0.38–0.51 | 0.94–1.48 |
| Phasing statistics | |||
| Riso (%)f | 27.9 | ||
| Figure of merit after SOLVE/RESOLVE | 0.45/0.58 | ||
| Refinement statistics | |||
| Resolution range (Å) | 50–2.4 | 50–2.6 | |
| Rwork (%)g/Rfree (%)h | 24.0/27.6 | 23.8/28.2 | |
| Figure of merit after refinement | 0.79 | 0.78 | |
| Model statistics | |||
| No. of atoms | |||
| Protein | 7436 | 7436 | |
| Ligand | 46 | ||
| Metal ion | 4 | 6 | |
| Solvent | 120 | 69 | |
| Average B factor (Å2) | 40.8 | 46.3 | |
| r.m.s. deviation | |||
| Bond lengths (Å) | 0.003 | 0.003 | |
| Bond angles (degrees) | 0.616 | 0.676 | |
| Ramachandran plot | |||
| Favored (%) | 97.5 | 98.0 | |
| Allowed (%) | 2.5 | 2.0 | |
a C9H9HgNaO2S.
b Values in parentheses indicate statistics for the highest resolution shells.
c Rsym = Σ|Ii − 〈I〉|/Σ〈I〉, where Ii is the observed intensity and 〈I〉 is the average intensity over symmetry-equivalent measurements.
d Rr.i.m. = Σ(N/(N − 1))½|Ii − 〈I〉|/Σ〈I〉, where Ii is the observed intensity, 〈I〉 is the average intensity over symmetry-equivalent measurements, and N is the redundancy; calculated by SCALA (81, 82) in CCP4i.
e Rp.i.m. = Σ(1/(N − 1))½|Ii − 〈I〉|/Σ〈I〉, where Ii is the observed intensity, 〈I〉 is the average intensity over symmetry-equivalent measurements, and N is the redundancy; calculated by SCALA (81, 82) in CCP4i.
f Riso = Σ‖FPH| − |FP‖/Σ|FP|, where FPH and FP are the derivative and native structure factors, respectively.
g Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
h Rfree is the same as Rwork except for a 5% subset of all reflections that were never used in crystallographic refinement.
RESULTS AND DISCUSSION
Overall Structure of Ml-HexPPs and Structural Comparison
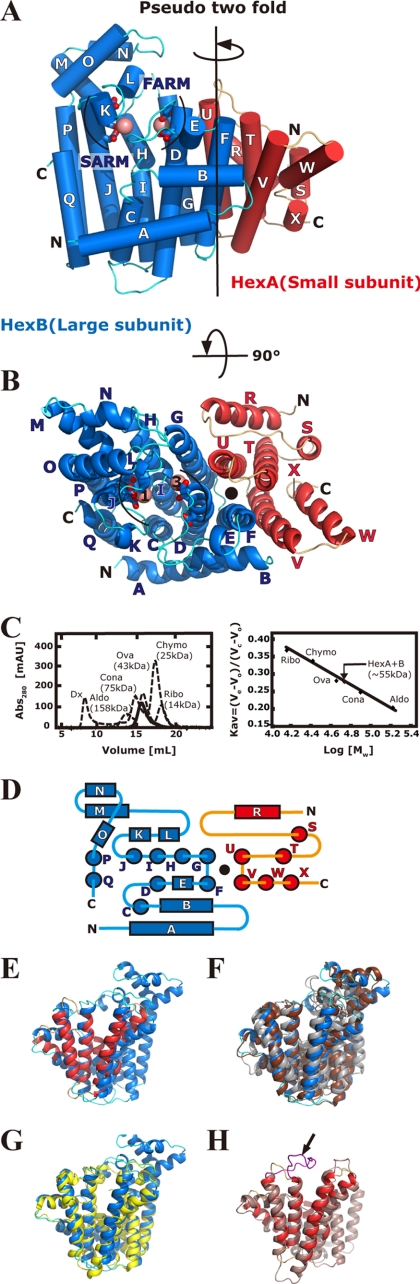
The crystal structure of Ml-HexPPs was determined at 2.4 Å resolution (Fig. 2, A and B, and Table 1). Ml-HexPPs is composed of the small component HexA (143 aa, 17 kDa) and the large component HexB (325 aa, 37 kDa). Two heterodimers (HexA-HexB and HexA′-HexB′) are found in an asymmetric unit of the crystal (supplemental Fig. 2, A and B). The two heterodimers are related with a non-crystallographic 2-fold axis. The intraheterodimer interactions (between subunits S1 (HexA) and L1 (HexB) as well as S2 (HexA′) and L2 (HexB′)) are apparently stronger than the interheterodimer interactions (Table 2). The large intraheterodimer interaction area and ΔiG value (solvation free energy upon formation of the interface) are comparable with those of the intrahomodimer and intraheterodimer interfaces found in various enzymes (Table 2). The strong interaction between HexA and HexB (∼1800 Å2; Table 2) probably contributes to the stabilization of the heterodimers, as seen in the dimeric unit of trans-prenyltransferases. The size exclusion column chromatography profile of Ml-HexPPs estimated its molecular mass in solution as ∼55 kDa (Fig. 2C), which was very close to the sum of the calculated molecular masses of HexA and HexB (54 kDa). The topology of the Ml-HexPPs heterotetrameric assembly in the crystallographic asymmetric unit is different from that of a heterotetrameric enzyme, Mp-GPPs (supplemental Fig. 2, B and C). These aspects combined with the prenyltransferase activity assay (supplemental Fig. 3) support that Ml-HexPPs functions as a heterodimer in solution, as previously suggested (50, 53, 54, 56). It should be noted that Ml-HexPPs and Mp-GPPs show the equivalent interaction area and ΔiG value for their interheterodimer interfaces (Table 2). Thus, it is also possible to consider that Ml-HexPPs forms a heterotetramer like Mp-GPPs (59). Nevertheless, because the two present heterodimers are well superposed on each other (r.m.s. deviation value 0.66 Å for all 455 Cα atom pairs in HexA (subunits S1 and S2) and HexB (subunits L1 and L2)), we use one of the heterodimers (subunits S1 and L1) for the following discussion.
FIGURE 2.
Overall structure of Ml-HexPPs and its analyses. A, front view of the heterodimer structure (cylindrical helices). HexB, the large subunit of Ml-HexPPs, is shown by blue helices (A–Q) and cyan loops. The first and the second aspartate-rich motifs, FARM (DDXX2–4D) and SARM (DDXXD), in HexB are shown by stick models. The other subunit, HexA, is shown by red helices (R–X) and orange loops. Magnesium ions are shown by pink spheres. The black line represents the pseudo-2-fold axis at the center of the heterodimer. B, top view of the heterodimer structure. The color coding is the same as in A. The closed black circle indicates the pseudo-2-fold axis. The pink spheres labeled with 1 and 3 correspond to Mg1 and Mg3 under “Results and Discussion,” respectively. C, molecular weight analysis of Ml-HexPPs in solution by gel filtration chromatography. The molecular mass is estimated as 55 kDa based on the retention volume of Ml-HexPPs with the following protein molecular markers: blue dextran 2000 (Dx; 2000 kDa), aldolase (Aldo; 158 kDa), conalbumin (Cona; 75 kDa), ovalbumin (Ova; 43 kDa), chymotrypsinogen A (Chymo; 25 kDa), and ribonuclease A (Ribo; 14 kDa). The estimated molecular mass is ∼55 kDa, which is equivalent to the calculated molecular mass of the heterodimer (∼54 kDa). The Kav value is defined using the equation Kav = (Ve − Vo)/(Vc − Vo), where Ve, Vo, and Vc are the elution, column void, and geometric column volumes, respectively. D, a topology diagram of the secondary structures of the Ml-HexPPs. Helices of HexA and HexB are colored in red and blue, respectively. The closed black circle indicates the pseudo-2-fold axis as in B. E, HexA superposed on HexB. The color code is the same as in A, B, and D. F, HexB, Ph-GGPPs (PDB code 1WY0), and Ss-HexPPs (PDB code 2AZJ) colored in blue, dark brown, and gray, respectively. Ph-GGPPs (chain A) and Ss-HexPPs (chain A) are superposed on HexB. G, HexB (blue) superposed on the large subunit LSU (chain D) of Mp-GPPs (yellow) (PDB code 3KRP) (59). H, HexA (red) superposed on the small subunit SSU (chain C) of Mp-GPPs (light brown) (PDB code 3KRP) (59). The purple loop indicated by the black arrow is described under “Results and Discussion.” Figs. 2–5 and supplemental Fig. 2 were prepared using the program PyMOL (available on the World Wide Web).
TABLE 2.
Analyses of subunit-interface interactions
| Enzyme | Oligomeric type | Subunits | Interface type | PDB code | Interface areaa,b | ΔiGa,c |
|---|---|---|---|---|---|---|
| Å2 | kcal/mol | |||||
| Ml-HexPPsd | Heterodimer | S1-L1 | Intraheterodimer | 3AQB | 1871.7 | −24.7 |
| S2-L2 | 1830.9 | −27.1 | ||||
| L1-L2 | Interheterodimer | 354.2 | −2.0 | |||
| S1-L2 | 194.0 | −2.4 | ||||
| S2-L1 | 199.0 | −3.5 | ||||
| Mp-GPPsd | Heterotetramer | S1-L1 | Intraheterodimer | 3KRP | 1928.3e | −36.3 |
| S2-L2 | 1963.4e | −36.0 | ||||
| L1-L2 | Interheterodimer | 70.2 | −1.1 | |||
| S1-L2 | 376.1e | 0.2 | ||||
| S2-L1 | 341.7e | −1.2 | ||||
| Gg-FPPsf | Homodimer | A-A | Intrahomodimer | 1FPS (dimerized A) | 1489.8 | −16.6 |
| Sc-GGPPsf | Homodimer | A-B | Intrahomodimer | 2DH4 | 3402.7g | −51.7g |
| Ph-GGPPsf | Homodimer | A-A | Intrahomodimer | 1WY0 (dimerized A) | 1064.1 | −16.0 |
| Sa-GGPPsf | Homodimer | A-B | Intrahomodimer | 2J1P | 1534.4 | −36.8 |
| Hs-GGPPsf | Homohexamer | A-B | Intrahomodimer | 2Q80 | 1579.8 | −30.1 |
| C-D | 1586.9 | −30.0 | ||||
| E-F | 1570.4 | −30.1 | ||||
| AB-CD | Interhomodimer | 1221.2 | −7.7 | |||
| CD-EF | 1197.8 | −8.9 | ||||
| EF-AB | 1240.5 | −10.0 | ||||
| Ss-HexPPsf | Homodimer | A-B | Intrahomodimer | 2AZJ | 1774.8 | −28.4 |
| Tm-OPPsf | Homodimer | A-B | Intrahomodimer | 1V4E | 1690.7 | −25.6 |
b Interface area was calculated as difference in total accessible surface areas of isolated and interfacing structures divided by 2.
c ΔiG indicates the solvation free energy gain upon formation of the interface. The value was calculated as difference in total solvation energies of isolated and interfacing structures.
d The pairs of subunits S1 plus L1 and S2 plus L2 form the functional heterodimers. The subunit names S1, L1, S2, and L2 are assigned as chains A, B, C, and D in PDB coordinate 3AQB and as chains B, A, C, and D in 3KRP, respectively.
e The intra- and interheterodimer interface areas are consistent with those (1980 and 380 Å2, respectively) previously reported by Chang et al. (59).
f The subunit names are the same as the chain IDs in each PDB coordinate.
g The N-terminal helix of each subunit protrudes into the other subunit, resulting in the very large intrahomodimer interface area and stabilization energy (12).
The large subunit of Ml-HexPPs, HexB, is composed of 17 antiparallel α-helices (A–Q) joined by connecting loops. The other small subunit HexA is also composed of seven antiparallel α-helices (R–X) and loops. No β-strand is found in either subunit. This α-helix-rich folding is also observed in all known trans-prenyltransferases (8–20, 22, 59) and several isoprenoid biosynthesis-related enzymes (74–79). Despite the low sequence identity between HexA and HexB and their distinct polypeptide chain lengths, these two heterodimeric subunits are related by a pseudo-2-fold axis at the center of the heterodimer (Fig. 2, A and B), as observed in the dimeric units of trans-prenyltransferases (8–20, 22, 59). The topology diagram showed that HexA corresponds well with the first half of HexB (helices A–I; Fig. 2D). Superposition of the HexA composed of 134 Cα atoms onto the corresponding part of HexB (321 Cα atoms) showed that the r.m.s. deviation value was of 2.22 Å for 104 Cα atom pairs (Fig. 2E).
HexB is well superposed onto the structures of all known trans-prenyltransferases. The r.m.s. deviation values are in the range of 1.90–3.50 Å for 233–314 Cα pairs. The best-superposed coordinates are homodimeric GGPPs from Pyrococcus horikoshii Ot3 (Ph-GGPPs) (PDB code 1WY0) (r.m.s. deviation 2.70 Å for 314 Cα of 321 HexB Cα atoms) (Fig. 2F). One of the two subunits (chain A) of homodimeric HexPPs from Sulfolobus solfataricus (Ss-HexPPs) (PDB code 2AZJ), which synthesizes the same product with heterodimeric Ml-HexPPs, is superposed onto HexB with an r.m.s. deviation value of 2.29 Å for 233 Cα of 276 Ss-HexPPs Cα atoms (Fig. 2F). The LSU of heterotetrameric Mp-GPPs is also well superposed onto HexB (r.m.s. deviation 1.98 Å for 249 Cα of 284 LSU Cα atoms) (Fig. 2G). The overall structure similarity and the catalytic motif conservation between HexB and the functional subunits of known trans-prenyltransferases imply that HexB catalyzes the condensation reactions with the similar mechanisms to typical trans-prenyltransferases.
The structure of HexA is also similar to that of all known trans-prenyltransferases, but the size of this subunit (composed of 7 helices) is much smaller than that of the HexB subunit (17 helices), any subunits of the homooligomeric enzymes (typically over 10 helices), and both the small and the large subunits of heterotetrameric Mp-GPPs (11 and 14 helices). Because HexA lacks several conserved residues dominant in the catalysis of trans-prenyltransferase, the role of this subunit is deduced to be different from the roles of HexB or any of the catalytic subunits of trans-prenyltransferases. In addition, it is very hard to estimate the function of HexA from that of the SSU of Mp-GPPs. The suggested role of SSU consists of simply lining the cleft wall in the large subunit using a long loop (the purple loop indicated by the black arrow in Fig. 2H). However, the corresponding loop in HexA is much shorter than the loop in SSU. This implies that the small subunit HexA plays an unrevealed role(s) in the function of heterooligomeric trans-prenyltransferases. Thus, we determined the structure of the ligand complex of Ml-HexPPs in order to more deeply understand the molecular mechanisms of the HexA subunit as well as the HexB subunit.
Two Substrate-binding Sites and the Condensation Reaction
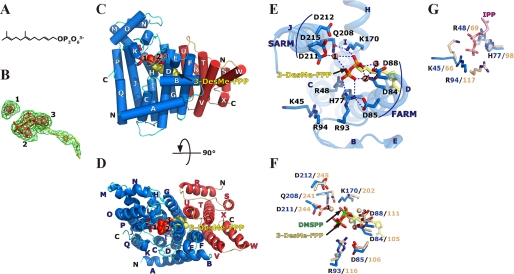
An FPP analog, 3-DesMe-FPP (Fig. 3A), was used to prepare an allylic substrate complex of Ml-HexPPs. This compound is not reactive with IPP by prenyltransferase family enzymes, because removing the methyl group from the C3 position of FPP destabilizes its allyl cation intermediate, which is essential for the condensation reaction (80). The complex structure was determined at 2.6 Å resolution (Table 1). The Fo − Fc omit electron density map clearly showed the presence of 3-DesMe-FPP and coordinated magnesium ions (Fig. 3B) in both of the heterodimers (S1L1 and S2L2) in a crystallographic asymmetric unit. The binding site is shown in Fig. 3, C and D. No significant difference was found between the backbone structure of the substrate-free enzyme and that of the 3-DesMe-FPP-bound complex. The r.m.s. deviation values are of 0.24 Å (456 Cα atom pairs) and 0.21 Å (459 Cα) for heterodimers S1L1 and S2L2, respectively. The diphosphate moiety of 3-DesMe-FPP is bound on two catalytically essential aspartate-rich motifs, FARM and SARM, located at the top of helices D and J, respectively (Fig. 3E). B-Asp84 (Asp84 of HexB) and B-Asp88 in FARM coordinate two magnesium ions (Mg2 and Mg3), and the two cations interact with the diphosphate part of 3-DesMe-FPP (Fig. 3E). Another magnesium ion (Mg1) bridges B-Asp211 in SARM and the diphosphate moiety of the ligand. Mg1 and Mg3 are also found in the substrate-free structure, whereas Mg2 can be seen only in the complex. Such features of the magnesium ions are the same as those in yeast GGPPs (12, 18). Other conserved motifs, GKXXR (B-Lys45 and B-Arg48), RRG (B-Arg93 and B-Arg94), G(Q/E) (B-Glu146), KT (B-Lys170), and (F/Y)Q (B-Gln208), are located around FARM and SARM. Positive groups of B-Arg93 in RRG and B-Lys170 in KT directly interact with the diphosphate moiety (Fig. 3E). This binding mode is similar to that seen in the “A-site” in all known trans-prenyltransferases (13, 14, 16–20, 22, 26, 59). As shown in Fig. 3F, the superposed structures of Ml-HexPPs on Ec-FPPs (PDB code 1RQI) (10) show that the binding modes of the diphosphate part of allylic substrates (DMSPP and 3-DesMe-FPP for Ec-FPPs and Ml-HexPPs, respectively) are essentially the same in both enzymes. Similar binding modes are also observed in other enzymes, such as homodimeric Sc-GGPPs (PDB codes 2E8T, 2E8X, and 2E90) (18), FPPs from Gallus gallus (Gg-FPPs) (PDB codes 1UBW, 1UBX, and 1UBY) (26), and heterotetrameric Mp-GPPs (PDB code 3KRF and 3KRO) (59).
FIGURE 3.
Substrate binding on Ml-HexPPs. A, chemical structure of 3-demethyl farnesyl diphosphate (3-DesMe-FPP), an analog of FPP. B, 3-DesMe-FPP and magnesium ions superposed on the final Fo − Fc omit electron density map contoured at the 2.5 σ (green) and 5.0 σ levels (red). The molecules are shown by a stick model and pink spheres, respectively. Carbons, oxygens, and phosphoruses are shown in yellow, red, and orange, respectively. The pink spheres labeled with 1, 2, and 3 correspond to Mg1, Mg2, and Mg3 under “Results and Discussion,” respectively. C, front view of the Ml-HexPPs structure bound with 3-DesMe-FPP. Red and blue subunits represent the HexA and HexB subunits, respectively. The first and the second aspartate-rich motifs in HexB, FARM (DDXX2–4D), and SARM (DDXXD) are shown by stick models as in Fig. 2, A and B. 3-DesMe-FPP and magnesium ions are shown by sphere models using the same color code as in B. D, top view of the Ml-HexPPs structure bound with 3-DesMe-FPP. The same colors are used as in C. E, close-up view around the substrate-binding sites on Ml-HexPPs bound with 3-DesMe-FPP. The first and the second aspartate-rich motifs in HexB, FARM and SARM, are shown. All residues shown in this panel are HexB residues. F, A-site of HexB superposed on a subunit (chain A) of Ec-FPPs in complex with DMSPP, an analog of DMAPP, and IPP (PDB code 1RQI). Ml-HexPPs and Ec-FPPs are colored in blue and wheat, respectively. DMSPP is shown by a stick model, and its phosphorus and carbon atoms are colored in green. Magnesium ions of Ml-HexPPs and Ec-FPPs are shown by pink and wheat spheres, respectively. G, I-site. The superposition was performed as in F. IPP bound on Ec-FPPs is colored in pink.
The binding mode of the other substrate IPP can be deduced from the complex structure of Ec-FPPs with its two substrates, including IPP (10). The IPP molecule was bound to the other site, called the “I-site” (Fig. 3G) (15). The positive groups on Lys66 and Arg69 in GKXXR, His98, as well as Arg117 in RRG in Ec-FPPs, are bound to the diphosphate moiety of IPP with electrostatic interactions. Similar IPP binding modes are also found in homodimeric Sc-GGPPs (PDB codes 2E8T and 2E8U) (18), Hs-FPPs (14, 17) and FPPs from Trypanosoma cruzi (13) (PDB codes 1ZW5 and 1YHM, respectively), and heterotetrameric Mp-GPPs (PDB codes 3KRC, 3KRF, and 3KRO) (59). As illustrated in Fig. 3G, the important residues for the recognition of IPP are conserved in Ml-HexPPs. The side chain conformations of B-Lys45 and B-Arg94 of Ml-HexPPs are different from those of the corresponding residues of Ec-FPPs. They probably take a different conformation when IPP binds on this enzyme.
As shown above, A- and I-sites on the large subunit HexB in Ml-HexPPs are structurally very similar to those on known trans-prenyltransferases. This similarity suggests that HexB catalyzes the condensation reaction using a mechanism similar to that of known trans-prenyltransferases (10, 15). In contrast, the other small subunit HexA does not seem to be directly involved in the condensation reaction because no dominant residues in catalysis are conserved in both the primary and three-dimensional structures. The specific role of HexA is discussed below.
Hydrophobic Cleft at the Heterodimer Interface; the Small Subunit Is Directly Involved in the Product Chain Length Regulation Together with the Large Subunit
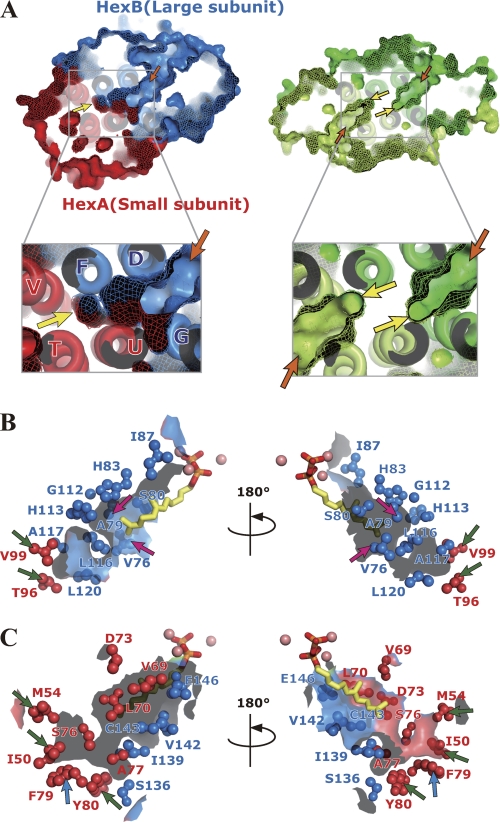
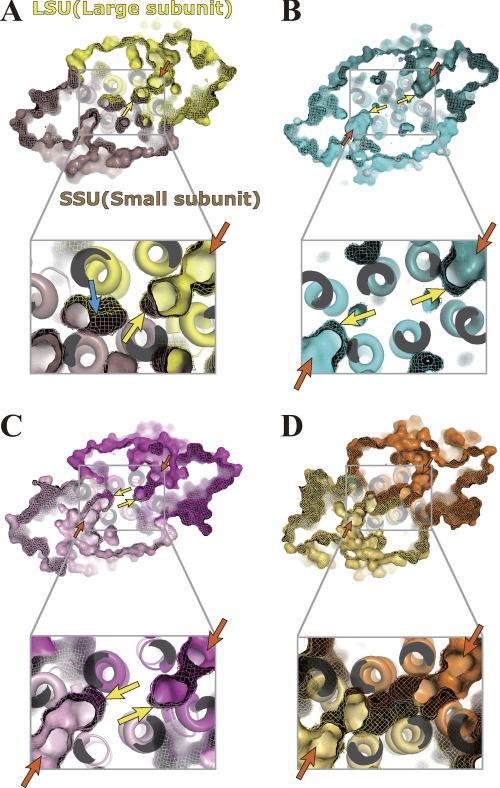
A large hydrophobic cleft is found at the heterodimer interface of the Ml-HexPPs (Fig. 4A, left). The size of the cleft is about 10 × 15 × 25 Å3 in width, height, and depth, respectively. The cleft entrance starts from the two substrate-binding sites (A- and I-sites) on the HexB surface (orange arrows in Fig. 4A, left), and the cleft bottom contacts the helices V and T in the other subunit HexA (yellow arrows in Fig. 4A, left). The cleft is sandwiched by a pair of helix bundles (helices D, F, and V as well as helices G, U, and T). Residues in both subunits participate in forming the wall of this cleft (Fig. 4, B and C). Hydrophobic clefts themselves are also found in various trans-prenyltransferases (8–12, 15, 18, 22, 59), but their aspects are discriminated from that for Ml-HexPPs as follows. Two independent clefts are separately located at the center of each subunit in most of the homodimeric enzymes, as illustrated in Fig. 4A (right) and in Fig. 5, B and C. The cleft architecture of the heterotetrameric Mp-GPPs is essentially the same as that of the homodimeric enzymes in that the cleft does not penetrate from the entrance at the large subunit LSU to inside the small subunit SSU (Fig. 5A). In contrast, the cleft of Ml-HexPPs starts from the large subunit HexB and extends to the inside of the small subunit HexA (Fig. 4A, left).
FIGURE 4.
Hydrophobic clefts of heterodimeric and homodimeric trans-prenyltransferases. A, surface representation of the heterodimeric Ml-HexPPs (C30 synthesis) (left) and homodimeric Sc-GGPPs (C20) (PDB code 2E8V) (right). The bottom two panels represent enlarged drawings of the cleft around the subunit interface for each enzyme. HexA and HexB of Ml-HexPPs as well as each subunit of Sc-GGPPs are drawn in red, blue, green and light green, respectively. Orange and yellow arrows indicate the two substrate-binding sites (cleft entrance) and the bottom of the cleft, respectively. B and C, residues of HexA and HexB forming the hydrophobic cleft shown in the ball-and-stick model. 3-DesMe-FPP is shown by a stick model. Magnesium ions are shown by pink spheres. Magenta, blue, and green arrows are as described under “Results and Discussion.”
FIGURE 5.
Hydrophobic clefts of various trans-prenyltransferases in increasing order of the product chain lengths. Surface representations of the heterotetrameric Mp-GPPs (C10 synthesis) (PDB code 3KRP) (A), homodimeric Gg-FPPs (C15) (PDB code 1FPS) (B), Sa-GGPPs (C20) (PDB code 2J1P) (C), and Tm-OPPs (C40) (PDB code 1V4E) (D) are shown. The bottom panels show an enlarged drawing of the cleft around the subunit interface for each enzyme. Orange and yellow arrows indicate the two substrate-binding sites (cleft entrance) and the bottom of the cleft, respectively. The third cleft indicated by the blue arrow in A is separated from the “main” and “functional” cleft indicated by the yellow and orange arrows.
The aliphatic tail moiety of 3-DesMe-FPP is accommodated in this hydrophobic cleft. The ω-end of the compound contacts the residues B-Val76 and B-Ala79 (magenta arrows in Fig. 4B) located at the middle position of the cleft. Mutation of B-Ala79 into Leu or Phe is known to change the product chain length into C25 or C20, respectively (60). This replacement probably decreases the whole size of the cleft and prevents the enzyme from elongating the product chain length longer than C20/C25, which is shorter than the original length C30. In contrast, mutation of B-Val76 into a smaller residue, Gly, changes the ultimate product chain length into C40 (60). The longer product chain length is consistent with the increase of the total cleft size by the mutation. Similar product chain length investigations were performed with homologous heterodimeric trans-prenyltransferases, HepPP synthases from B. subtilis and from B. stearothermophilus (60–62). Mutation of I-Tyr103 of the enzyme from B. subtilis3 into a smaller residue, Ser, elongates the chain length of its final product into C50 prenyl diphosphate, which is C15 longer than the original length C35 (62). This residue corresponds to A-Phe79 of Ml-HexPPs (blue arrows in Fig. 4C and supplemental Fig. 4A) located at the bottom of the cleft. The cleft probably accommodates the elongated prenyl chain using the bottom space surrounded by residues A-Ile50 and A-Met54 on helix T, A-Thr96 and A-Val99 on helix V, and A-Phe79 and A-Tyr80 on helix U (green and blue arrows in Fig. 4, B and C). The structurally bulky and rigid residues, A-Phe79 and A-Tyr80, form the bottom end of the cleft and seem to block the further prenyl chain elongation from exceeding the appropriate length (C30). These residues forming the cleft wall in HexA are highly conserved in HepPPs from B. subtilis and B. stearothermophilus (supplemental Fig. 4A), as are those forming the cleft wall in HexB (supplemental Fig. 4B). In addition, the cleft volume of Ml-HexPPs (∼1400 Å3) is reasonable compared with other trans-prenyltransferases (Table 3). Enzymes synthesizing longer products have larger cleft size. Also, the cleft volume of homodimeric Ss-HexPPs (∼1400 Å3) is almost the same as that of Ml-HexPPs. These structural and mutational analyses imply that the hydrophobic cleft regulates the product chain length by utilizing the whole size of the cleft formed by both of the two heteromeric subunits as a ruler and that the small subunit HexA is also directly involved in the product chain length regulation in cooperation with the large subunit HexB beyond the heterodimer interface.
TABLE 3.
Calculated cleft volume
| Enzyme | Oligomeric type | Product chain length | PDB code and chains | Cleft volumea |
|---|---|---|---|---|
| Å3 | ||||
| Mp-GPPs | Heterotetramer | C10 | 3KRP (A, B) | 422.1 |
| Gg-FPPs | Homodimer | C15 | 1FPS (dimerized A) | 549.6,549.8 |
| Sc-GGPPs | Homodimer | C20 | 2DH4 (A, B) | 774.0,786.0 |
| Ph-GGPPs | Homodimer | C20 | 1WY0 (dimerized A) | 748.5,748.4 |
| Sa-GGPPs | Homodimer | C20 | 2J1P (A, B) | 903.1,b 519.5b |
| Ss-HexPPs | Homodimer | C30 | 2AZJ (A, B) | 1411.7,c 670.3c |
| Ml-HexPPs | Heterodimer | C30 | 3AQB (A, B) | 1419.0 |
| Tm-OPPs | Homodimer | C40 | 1V4E (A, B) | 2000.6 |
b Chain A accommodates the product GGPP, whereas the other chain is vacant. The ligand may enlarge the cleft size with the shrinkage of the other vacant cleft. The averaged volume of the two chains is ∼710 Å3, which is comparable with those of Sc- and Ph-GGPPs.
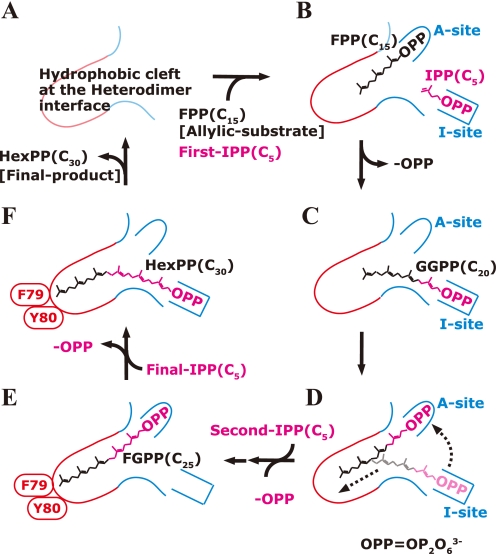
The chain elongation cycle of Ml-HexPPs is hypothesized as illustrated in Fig. 6. First, FPP(C15) and IPP(C5) bind at the A- and I-sites, respectively, and couple to form a C20 intermediate with the release of an inorganic pyrophosphate (Fig. 6, A–C). Second, the diphosphate head of the C20 intermediate moves from the I-site to the A-site with the movement of the C20 prenyl tail toward the bottom of the cleft (Fig. 6D). Another condensation elongates the chain length to C25, and the cleft from the A-site to the bottom is fully occupied by the elongated chain (Fig. 6E). Finally, the ultimate product HexPP (C30) is formed by the final IPP (C5) condensation onto the C25 intermediate (Fig. 6F). The bulky side chains of A-Phe79 and A-Tyr80 block any further chain elongation reaction at the bottom of the cleft. The synthesized C30 product is released, and the enzyme is recycled for the next reaction cycle. Similar reaction steps have been proposed for known trans-prenyltransferases (13, 59). The crystal structure of homodimeric Sc-GGPPs revealed that Tyr and His residues located at the bottom of the cleft contacted the ω-end of the product GGPP and obstructed further chain elongation (PDB code 2E8V) (12). It is widely believed that each subunit of the homodimer in homooligomeric enzymes independently synthesizes the elongated product with a desired chain length (9, 11, 12, 33, 37, 43). In the case of heterotetrameric Mp-GPPs, the condensation reaction and the C10 chain length control are essentially dominated only by the large subunit LSU (59). The small subunit SSU just remotely regulates the conformation of a loop on the LSU. In contrast, the product chain length of heterodimeric Ml-HexPPs may be cooperatively controlled by both the HexA and HexB subunits, whereas the condensation reaction is dominated by only the HexB subunit.
FIGURE 6.
Hypothetical chain length determination mechanism of Ml-HexPPs. The red and blue lines represent the cleft wall formed by the small subunit HexA and the large subunit HexB, respectively. The A- and I-sites are the allylic substrate-binding site and IPP-binding site, respectively. One cycle of the elongation reaction is shown.
This intersubunit product chain length control seems to apply to some homooligomeric enzymes. Mutational and crystallographic analyses of homodimeric type II GGPPs (C20 synthesis) from Pantoea ananatis and from Sinapis alba (Sa-GGPPs) (PDB code 2J1P), respectively, have suggested that the bottoms of the clefts were located at the subunit interface (15, 34) (Fig. 5C). Homodimeric octaprenyl diphosphate synthase (OPPs; C40 synthesis) from Thermotoga maritima (Tm-OPPs) has a “tunnel” connecting the two condensation reaction sites on the two subunits (Fig. 5D) in addition to the two deep clefts parallel to the bundle of the helices (PDB code 1V4E) (9, 33). The cooperative regulation of the product chain length might be widely seen in both homo- and heterooligomeric trans-prenyltransferases synthesizing medium or long prenyl chain.
In summary, we have determined the crystal structure of a heterodimeric Ml-HexPPs and proposed its molecular mechanism. The overall structure of this enzyme closely resembles that of all known trans-prenyltransferases, despite the low amino acid sequence homology and the distinct polypeptide chain lengths between the small subunit HexA and typical catalytic subunits of trans-prenyltransferases as well as the large subunit HexB. Ml-HexPPs probably catalyzes the condensation reaction using the HexB subunit. The structural similarity of the two substrate-binding sites in HexB suggests that the condensation reaction mechanism of this enzyme is the same as that of trans-prenyltransferases. It is implied that the final product chain length is cooperatively regulated by both the HexA and HexB subunits using the whole size of the hydrophobic cleft as a ruler. Such cooperation of the two subunits has not been widely investigated in trans-prenyltransferases but might be a common mechanism of both homo- and heterooligomeric trans-prenyltransferases. The dominant roles of the small subunit seem to consist of stabilizing the functional heterodimeric unit of the enzyme through the large hydrophobic interface and directly regulating the product chain length together with the large catalytic subunit HexB.
Supplementary Material
Acknowledgments
We are grateful for the special gift of 3-DesMe-FPP from Prof. Yuji Maki (Yamagata University, Japan). We also thank the beamline scientists of the Photon Factory, KEK, Japan.
This work was partly supported by Global Centers of Excellence program “International Center for Integrated Research and Advanced Education in Materials Science” of the Ministry of Education, Culture, Sports, Science and Technology of Japan Grant B-024.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
The atomic coordinates and structure factors (codes 3AQB and 3AQC) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The heterodimeric subunits of HepPPs from B. subtilis are named HepPPs-I and HepPPs-II. These subunits correspond to HexA and HexB, respectively.
- IPP
- isopentenyl diphosphate(s)
- 3-DesMe-FPP
- 7,11-dimethyl-2,6,10-dodecatrien-1-yl diphosphate ammonium salt
- DMAPP
- dimethylallyl diphosphate
- DMSPP
- dimethylallyl S-thiolodiphosphate
- FARM
- first aspartate-rich motif
- FPP
- farnesyl diphosphate
- FPPs
- farnesyl diphosphate synthase
- GGPP
- geranylgeranyl diphosphate
- GGPPs
- geranylgeranyl diphosphate synthase
- GPP
- geranyl diphosphate
- GPPs
- geranyl diphosphate synthase
- HepPP
- heptaprenyl diphosphate
- HepPPs
- heptaprenyl diphosphate synthase
- HexA and HexB
- hexaprenyl diphosphate synthase A and B subunit, respectively
- HexPP
- hexaprenyl diphosphate
- HexPPs
- hexaprenyl diphosphate synthase
- LSU
- large subunit of GPPs from M. piperita
- OPPs
- octaprenyl diphosphate synthase
- PDB
- Protein Data Bank
- r.m.s.
- root mean square
- SARM
- second aspartate-rich motif
- SSU
- small subunit of GPPs from M. piperita
- aa
- amino acids.
REFERENCES
- 1. Christianson D. W. (2007) Science 316, 60–61 [DOI] [PubMed] [Google Scholar]
- 2. Ogura K., Koyama T. (1998) Chem. Rev. 98, 1263–1276 [DOI] [PubMed] [Google Scholar]
- 3. Koyama T. (1999) Biosci. Biotechnol. Biochem. 63, 1671–1676 [DOI] [PubMed] [Google Scholar]
- 4. Ito M., Kobayashi M., Koyama T., Ogura K. (1987) Biochemistry 26, 4745–4750 [DOI] [PubMed] [Google Scholar]
- 5. Wang W., Dong C., McNeil M., Kaur D., Mahapatra S., Crick D. C., Naismith J. H. (2008) J. Mol. Biol. 381, 129–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fujihashi M., Zhang Y. W., Higuchi Y., Li X. Y., Koyama T., Miki K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo R. T., Ko T. P., Chen A. P., Kuo C. J., Wang A. H., Liang P. H. (2005) J. Biol. Chem. 280, 20762–20774 [DOI] [PubMed] [Google Scholar]
- 8. Tarshis L. C., Yan M., Poulter C. D., Sacchettini J. C. (1994) Biochemistry 33, 10871–10877 [DOI] [PubMed] [Google Scholar]
- 9. Guo R. T., Kuo C. J., Chou C. C., Ko T. P., Shr H. L., Liang P. H., Wang A. H. (2004) J. Biol. Chem. 279, 4903–4912 [DOI] [PubMed] [Google Scholar]
- 10. Hosfield D. J., Zhang Y., Dougan D. R., Broun A., Tari L. W., Swanson R. V., Finn J. (2004) J. Biol. Chem. 279, 8526–8529 [DOI] [PubMed] [Google Scholar]
- 11. Sun H. Y., Ko T. P., Kuo C. J., Guo R. T., Chou C. C., Liang P. H., Wang A. H. (2005) J. Bacteriol. 187, 8137–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang T. H., Guo R. T., Ko T. P., Wang A. H., Liang P. H. (2006) J. Biol. Chem. 281, 14991–15000 [DOI] [PubMed] [Google Scholar]
- 13. Gabelli S. B., McLellan J. S., Montalvetti A., Oldfield E., Docampo R., Amzel L. M. (2006) Proteins 62, 80–88 [DOI] [PubMed] [Google Scholar]
- 14. Kavanagh K. L., Guo K., Dunford J. E., Wu X., Knapp S., Ebetino F. H., Rogers M. J., Russell R. G., Oppermann U. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 7829–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kloer D. P., Welsch R., Beyer P., Schulz G. E. (2006) Biochemistry 45, 15197–15204 [DOI] [PubMed] [Google Scholar]
- 16. Mao J., Mukherjee S., Zhang Y., Cao R., Sanders J. M., Song Y., Zhang Y., Meints G. A., Gao Y. G., Mukkamala D., Hudock M. P., Oldfield E. (2006) J. Am. Chem. Soc. 128, 14485–14497 [DOI] [PubMed] [Google Scholar]
- 17. Rondeau J. M., Bitsch F., Bourgier E., Geiser M., Hemmig R., Kroemer M., Lehmann S., Ramage P., Rieffel S., Strauss A., Green J. R., Jahnke W. (2006) Chem. Med. Chem. 1, 267–273 [DOI] [PubMed] [Google Scholar]
- 18. Guo R. T., Cao R., Liang P. H., Ko T. P., Chang T. H., Hudock M. P., Jeng W. Y., Chen C. K., Zhang Y., Song Y., Kuo C. J., Yin F., Oldfield E., Wang A. H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10022–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Artz J. D., Dunford J. E., Arrowood M. J., Dong A., Chruszcz M., Kavanagh K. L., Minor W., Russell R. G., Ebetino F. H., Oppermann U., Hui R. (2008) Chem. Biol. 15, 1296–1306 [DOI] [PubMed] [Google Scholar]
- 20. Cao R., Chen C. K., Guo R. T., Wang A. H., Oldfield E. (2008) Proteins 73, 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sagami H., Morita Y., Ogura K. (1994) J. Biol. Chem. 269, 20561–20566 [PubMed] [Google Scholar]
- 22. Kavanagh K. L., Dunford J. E., Bunkoczi G., Russell R. G., Oppermann U. (2006) J. Biol. Chem. 281, 22004–22012 [DOI] [PubMed] [Google Scholar]
- 23. Miyagi Y., Matsumura Y., Sagami H. (2007) J. Biochem. 142, 377–381 [DOI] [PubMed] [Google Scholar]
- 24. Sagami H., Korenaga T., Ogura K., Steiger A., Pyun H. J., Coates R. M. (1992) Arch. Biochem. Biophys. 297, 314–320 [DOI] [PubMed] [Google Scholar]
- 25. Sagami H., Ishii K., Ogura K. (1981) Biochem. Int. 3, 669–675 [Google Scholar]
- 26. Tarshis L. C., Proteau P. J., Kellogg B. A., Sacchettini J. C., Poulter C. D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15018–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koyama T., Tajima M., Sano H., Doi T., Koike-Takeshita A., Obata S., Nishino T., Ogura K. (1996) Biochemistry 35, 9533–9538 [DOI] [PubMed] [Google Scholar]
- 28. Koyama T., Tajima M., Nishino T., Ogura K. (1995) Biochem. Biophys. Res. Commun. 212, 681–686 [DOI] [PubMed] [Google Scholar]
- 29. Song L., Poulter C. D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3044–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Joly A., Edwards P. A. (1993) J. Biol. Chem. 268, 26983–26989 [PubMed] [Google Scholar]
- 31. Blanchard L., Karst F. (1993) Gene 125, 185–189 [DOI] [PubMed] [Google Scholar]
- 32. Marrero P. F., Poulter C. D., Edwards P. A. (1992) J. Biol. Chem. 267, 21873–21878 [PubMed] [Google Scholar]
- 33. Guo R. T., Kuo C. J., Ko T. P., Chou C. C., Liang P. H., Wang A. H. (2004) Biochemistry 43, 7678–7686 [DOI] [PubMed] [Google Scholar]
- 34. Noike M., Katagiri T., Nakayama T., Koyama T., Nishino T., Hemmi H. (2008) FEBS J. 275, 3921–3933 [DOI] [PubMed] [Google Scholar]
- 35. Hemmi H., Noike M., Nakayama T., Nishino T. (2003) Eur. J. Biochem. 270, 2186–2194 [DOI] [PubMed] [Google Scholar]
- 36. Hemmi H., Noike M., Nakayama T., Nishino T. (2002) Biochem. Biophys. Res. Commun. 297, 1096–1101 [DOI] [PubMed] [Google Scholar]
- 37. Wang K., Ohnuma S. (1999) Trends Biochem. Sci. 24, 445–451 [DOI] [PubMed] [Google Scholar]
- 38. Ohnuma S., Narita K., Nakazawa T., Ishida C., Takeuchi Y., Ohto C., Nishino T. (1996) J. Biol. Chem. 271, 30748–30754 [DOI] [PubMed] [Google Scholar]
- 39. Ohnuma S., Hirooka K., Hemmi H., Ishida C., Ohto C., Nishino T. (1996) J. Biol. Chem. 271, 18831–18837 [DOI] [PubMed] [Google Scholar]
- 40. Rogers M. J. (2003) Curr. Pharm. Des. 9, 2643–2658 [DOI] [PubMed] [Google Scholar]
- 41. Rodan G. A., Reszka A. A. (2002) Curr. Mol. Med. 2, 571–577 [DOI] [PubMed] [Google Scholar]
- 42. Russell R. G., Rogers M. J. (1999) Bone 25, 97–106 [DOI] [PubMed] [Google Scholar]
- 43. Ohnuma S., Hirooka K., Tsuruoka N., Yano M., Ohto C., Nakane H., Nishino T. (1998) J. Biol. Chem. 273, 26705–26713 [DOI] [PubMed] [Google Scholar]
- 44. Saiki R., Nagata A., Kainou T., Matsuda H., Kawamukai M. (2005) FEBS J. 272, 5606–5622 [DOI] [PubMed] [Google Scholar]
- 45. Burke C., Klettke K., Croteau R. (2004) Arch. Biochem. Biophys. 422, 52–60 [DOI] [PubMed] [Google Scholar]
- 46. Saiki R., Nagata A., Uchida N., Kainou T., Matsuda H., Kawamukai M. (2003) Eur. J. Biochem. 270, 4113–4121 [DOI] [PubMed] [Google Scholar]
- 47. Burke C., Croteau R. (2002) J. Biol. Chem. 277, 3141–3149 [DOI] [PubMed] [Google Scholar]
- 48. Burke C. C., Wildung M. R., Croteau R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13062–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y. W., Koyama T., Marecak D. M., Prestwich G. D., Maki Y., Ogura K. (1998) Biochemistry 37, 13411–13420 [DOI] [PubMed] [Google Scholar]
- 50. Shimizu N., Koyama T., Ogura K. (1998) J. Bacteriol. 180, 1578–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y. W., Koyama T., Ogura K. (1997) J. Bacteriol. 179, 1417–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koike-Takeshita A., Koyama T., Obata S., Ogura K. (1995) J. Biol. Chem. 270, 18396–18400 [DOI] [PubMed] [Google Scholar]
- 53. Yoshida I., Koyama T., Ogura K. (1989) Biochim. Biophys. Acta 995, 138–143 [DOI] [PubMed] [Google Scholar]
- 54. Yoshida I., Koyama T., Ogura K. (1989) Biochem. Biophys. Res. Commun. 160, 448–452 [DOI] [PubMed] [Google Scholar]
- 55. Fujii H., Koyama T., Ogura K. (1983) FEBS Lett. 161, 257–260 [DOI] [PubMed] [Google Scholar]
- 56. Fujii H., Koyama T., Ogura K. (1982) J. Biol. Chem. 257, 14610–14612 [PubMed] [Google Scholar]
- 57. Takahashi I., Ogura K., Seto S. (1980) J. Biol. Chem. 255, 4539–4543 [PubMed] [Google Scholar]
- 58. Yoshida I., Koyama T., Ogura K. (1987) Biochemistry 26, 6840–6845 [DOI] [PubMed] [Google Scholar]
- 59. Chang T. H., Hsieh F. L., Ko T. P., Teng K. H., Liang P. H., Wang A. H. (2010) Plant Cell 22, 454–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y. W., Li X. Y., Koyama T. (2000) Biochemistry 39, 12717–12722 [DOI] [PubMed] [Google Scholar]
- 61. Hirooka K., Ohnuma S., Koike-Takeshita A., Koyama T., Nishino T. (2000) Eur. J. Biochem. 267, 4520–4528 [DOI] [PubMed] [Google Scholar]
- 62. Zhang Y. W., Li X. Y., Sugawara H., Koyama T. (1999) Biochemistry 38, 14638–14643 [DOI] [PubMed] [Google Scholar]
- 63. Zhang D., Poulter C. D. (1993) Anal. Biochem. 213, 356–361 [DOI] [PubMed] [Google Scholar]
- 64. Fujii H., Koyama T., Ogura K. (1982) Biochim. Biophys. Acta 712, 716–718 [PubMed] [Google Scholar]
- 65. Fujikura K., Maki Y., Ohya N., Satoh M., Koyama T. (2008) Biosci. Biotechnol. Biochem. 72, 851–855 [DOI] [PubMed] [Google Scholar]
- 66. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 67. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1937–1940 [DOI] [PubMed] [Google Scholar]
- 69. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 70. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 71. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 72. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 74. Zhang H., Seabra M. C., Deisenhofer J. (2000) Structure 8, 241–251 [DOI] [PubMed] [Google Scholar]
- 75. Pandit J., Danley D. E., Schulte G. K., Mazzalupo S., Pauly T. A., Hayward C. M., Hamanaka E. S., Thompson J. F., Harwood H. J., Jr. (2000) J. Biol. Chem. 275, 30610–30617 [DOI] [PubMed] [Google Scholar]
- 76. Long S. B., Casey P. J., Beese L. S. (1998) Biochemistry 37, 9612–9618 [DOI] [PubMed] [Google Scholar]
- 77. Wendt K. U., Poralla K., Schulz G. E. (1997) Science 277, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 78. Starks C. M., Back K., Chappell J., Noel J. P. (1997) Science 277, 1815–1820 [DOI] [PubMed] [Google Scholar]
- 79. Lesburg C. A., Zhai G., Cane D. E., Christianson D. W. (1997) Science 277, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 80. Nagaki M., Kimura K., Kimura H., Maki Y., Goto E., Nishino T., Koyama T. (2001) Bioorg. Med. Chem. Lett. 11, 2157–2159 [DOI] [PubMed] [Google Scholar]
- 81. Weiss M. S. (2001) J. Appl. Crystallogr. 34, 130–135 [Google Scholar]
- 82. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 83. Krissinel E. (2010) J. Comput. Chem. 31, 133–143 [DOI] [PubMed] [Google Scholar]
- 84. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 85. Liang J., Edelsbrunner H., Woodward C. (1998) Protein Sci. 7, 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liang J., Edelsbrunner H., Fu P., Sudhakar P. V., Subramaniam S. (1998) Proteins 33, 1–17 [PubMed] [Google Scholar]
- 87. Liang J., Edelsbrunner H., Fu P., Sudhakar P. V., Subramaniam S. (1998) Proteins 33, 18–29 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.