Abstract
CDK9/cyclin T1, a key enzyme in HIV-1 transcription, is negatively regulated by 7SK RNA and the HEXIM1 protein. Dephosphorylation of CDK9 on Thr186 by protein phosphatase 1 (PP1) in stress-induced cells or by protein phosphatase M1A in normally growing cells activates CDK9. Our previous studies showed that HIV-1 Tat protein binds to PP1 through the Tat Q35VCF38 sequence, which is similar to the PP1-binding RVXF motif and that this interaction facilitates HIV-1 transcription. In the present study, we analyzed the effect of expression of the central domain of nuclear inhibitor of PP1 (cdNIPP1) in an engineered cell line and also when cdNIPP1 was expressed as part of HIV-1 pNL4-3 in place of nef. Stable expression of cdNIPP1 increased CDK9 phosphorylation on Thr186 and the association of CDK9 with 7SK RNA. The stable expression of cdNIPP1 disrupted the interaction of Tat and PP1 and inhibited HIV-1 transcription. Expression of cdNIPP1 as a part of the HIV-1 genome inhibited HIV-1 replication. Our study provides a proof-of-concept for the future development of PP1-targeting compounds as inhibitors of HIV-1 replication.
Keywords: CDK (Cyclin-dependent Kinase), HIV, Protein Phosphatase, RNA Polymerase II, Transcription Regulation, HIV-1 Tat
Introduction
Cyclin-dependent kinase-9 (CDK9)3/cyclin T1 is a protein kinase that phosphorylates the C-terminal domain of the largest subunit of RNA polymerase II during transcription elongation (1). CDK9/cyclin T1 is part of positive transcription elongation complex (P-TEFb) and present in the cells within large and small complexes. The large complex contains 7SK RNA (2, 3) and the hexamethylene bisacetamide-induced protein (HEXIM1) (4, 5). The activity of CDK9 in the large complex is inhibited by the interaction with 7SK RNA and HEXIM1 (2–5). In stress-induced cells, CDK9/cyclin T1 dissociates from the 7SK RNA and HEXIM1 protein and then binds to the bromodomain protein 4 (Brd4) (6, 7), a member of bromodomain-containing extraterminal domain protein family that interact with acetylated histones. CDK9/cyclin T1 bound to Brd4 forms the transcriptionally active P-TEFb (7). Brd4 recruits P-TEFb to the promoters of cellular genes that are expressed at the end of mitosis (8). HIV-1 Tat protein can also recruit P-TEFb by binding directly to CDK9/cyclin T1 and targeting it to HIV-1 TAR RNA (7). Hence, Tat induces HIV-1 transcription by recruiting CDK9/cyclin T1 (9–11) and histone acetyltransferases to the HIV-1 promoter (7, 12–14). The HIV-1 Tat prevents the formation of the large complex and promotes the dissociation of CDK9/cyclin T1 from 7SK RNA/HEXIM1 complex (15). The mechanism of this Tat-mediated dissociation of CDK9/cyclin T1 and 7SK RNA/HEXIM1 involves the dephosphorylation of CDK9 on Thr186 (16, 17). Protein phosphatase 1 (PP1) dephosphorylates Thr186 of CDK9 in vitro, and the dephosphorylated CDK9/cyclin T1 does not associate with 7SK RNA (16). In stress-induced cells, PP1α dephosphorylates Thr186 of CDK9 and in cooperation with protein phosphatase 2B (PP2B) disrupts the interaction between CDK9/cyclin T1 and 7SK RNA/HEXIM1 (18). In normally growing cells, protein phosphatase M1A (PPM1A) dephosphorylates CDK9 on Thr186 (19).
Our previous studies showed that PP1 stimulates Tat-dependent HIV-1 transcription in vitro (20) and in cultured cells (21). PP1 holoenzymes consist of a constant catalytic subunit (PP1α, PP1β/δ, or PP1γ) and a variable regulatory subunit that determines the localization, activity, and substrate specificity of the phosphatase (22, 23). One of the major nuclear regulators of PP1 is NIPP1 (nuclear inhibitor of PP1), which inhibits the dephosphorylation of a wide range of PP1 substrates (22, 23). Tat-mediated HIV-1 transcription is blocked by the expression of NIPP1, and the inhibition is reversed by the co-expression of PP1γ (21). Tat contains a PP1-binding motif and potentially functions as a PP1 regulatory subunit (24). Mutation of residues in the PP1-binding motif (V36A and F38A) prevents Tat from inducing HIV-1 transcription and the translocation of PP1 to the nucleus (24). We showed that in cultured cells, PP1, but not PP2A, likely dephosphorylates CDK9 and contributes to the regulation of activated HIV-1 transcription (25).
In the present paper, we analyzed the effect of the expression of cdNIPP1, an inhibitory 82-residue PP1-binding fragment of the central domain of NIPP1 (cdNIPP1), on the CDK9 phosphorylation and activity and HIV-1 transcription. We further analyzed the effect of cdNIPP1 expression on HIV-1 replication in a physiologically relevant manner by expressing cdNIPP1 as part of the HIV-1 genome in place of nef. Our results indicate that the expression of PP1 inhibitor suppresses HIV-1 replication.
EXPERIMENTAL PROCEDURES
Materials
293T cells were purchased from American Type Cultue Collection (Manassas, VA). The 84-31 line is a subclone of 293 cells that stably expresses E4 of Ad and that supports growth of adeno-associated virus (26). The catalytic subunit of PP1 (PP1C) was purified from rabbit skeletal muscle as previously described (27). WT Tat was expressed in Escherichia coli from pGEM2 Tat vector obtained from the NIH AIDS Research and Reference Reagent Program and purified on an Aquapore RP-300 column (Applied Biosystems) by reverse-phase chromatography as we described previously (28). Rabbit polyclonal antibodies to CDK9, rabbit polyclonal antibodies to cyclin T1, and rabbit polyclonal antibodies to enhanced green fluorescent protein (EGFP) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-Tat rabbit polyclonal antibodies were received from AIDS Research and Reference Reagents Program (National Institutes of Health). Anti-α-tubulin and anti-FLAG monoclonal antibodies were purchased from Sigma. Rabbit anti-PP1γ antibodies were from Calbiochem. Anti-CDK9 phospho-Thr186, anti-Brd4, and anti-HEXIM1 polyclonal antibodies were kind gifts from Dr. Qiang Zhou (University of California, Berkeley). Monoclonal antibodies specific to the unphosphorylated form of the C-terminal domain (8WG16) or C-terminal domain phosphorylated on serine 2 (H5) were purchased from BabCo (Richmond, CA). Rabbit polyclonal anti-NIPP1 antibodies were described in Ref. 29.
293T-cdNIPP1 Cells
293T cells stably expressing the central domain of NIPP1 (amino acids 143–224) were generated by transfection of NIPP1-(143–224)-EGFP and limiting dilution cloning. Genomic DNA was isolated from parental 293T cells and three different clones that were stably transformed with NIPP1-(143–224)-EGFP. About 500 ng of these genomic DNAs were used as a template in a PCR with the sense primer (5′-ATGGGTGGAGAGGATGATGAACTC-3′), corresponding to nucleotides 436–459 of bovine NIPP1 cDNA (accession number Z50748) and the antisense primer (5′-CGTCGCCGTCCAGCTCGACCAG-3′), corresponding to the inverse complement of nucleotides 724–745 of pEGFP-N1 vector (Clontech). The amplified PCR products were subcloned in the pGEM-T Easy vector (Promega) and sequenced by T7 and SP6 primer.
Plasmids
The expression vectors for EGFP-fused central domain of NIPP1, NIPP1-(143–224), or NIPP1-(143–224) V201A/F203A (RATA mutant) were described previously (30). NIPP1 mutated within its RVXF motif has low binding affinity for PP1 (30). 7SK expression vector was a gift from Dr. Shona Murphy (Oxford University, UK). FLAG-CDK9 expression vectors were kind gifts from Dr. Qiang Zhou. Expression vectors for PP1γ-EGFP and for PP1γ-D64N-EGFP are described (31).
RT-PCR to Detect 7SK RNA
RNA was isolated from complexes that co-immunoprecipitated with anti-CDK9 antibodies using TRIzol reagent according the Invitrogen protocol. RNA was reverse-transcribed using a SuperScript II kit (Invitrogen) with the 7SK reverse primer. The following primer sequences were used: forward, 3′-GGATGTGAGGCGATCTGGCTG-5′ and reverse, 3′-TAAAGAAAGGCAGACTGCCAC-5′. These primers were used in a PCR that followed the RT reaction. In semiquantitative PCR, products were resolved on 2% agarose gel and photographed. In real time PCR, PCRs were run with Syber Green on a MioQ PCR machine and quantified using cdNIPP1 expression vector as control for the determination of copy number.
Fractionation of Cellular Lysates on Glycerol Gradient
293T and 293T-cdNIPP1 cells were grown in 100-mm plates and then lysed with 0.5 ml of whole cell lysis buffer (50 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 1% Nonidet P-40, 0.1% SDS) supplemented with protease inhibitors mixture (Sigma). Cell lysates were clarified by centrifugation for 30 min at 10,000 × g and loaded on top of a 10–30% glycerol (9-ml) gradient. Glycerol gradient buffer contained 20 mm HEPES-KOH, pH 7.9, 150 mm KCl, 200 μm EDTA. The gradient was spun in SW 41Ti rotor (Beckman) at 38,000 rpm for 18 h. Fractions (0.5 ml) were collected through a needle inserted into the bottom of the tube and analyzed by immunoblotting using the indicated antibodies. The x-ray films with the immunoblots for NIPP1 and CDK9 phosphorylated on Thr186 were scanned and quantified using OptiQuant software from Packard Cyclone PhosphorImager (Perkin-Elmer). The pixels were adjusted to have the same total amount for the 293T and the cdNIPP1 extracts, and the normalized results were graphed.
Effect of cdNIPP1-derived Peptides on HIV-1 Transcription in CEM-GFP Cell Activated by Recombinant Tat
CEM cells containing integrated HIV-1 LTR EGFP (CEM-GFP obtained from the NIH AIDS Research and Reference Reagent Program) were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1% glutamine, 1% antibiotic solution (penicillin and streptomycin), and 500 μg/ml G418. CEM-GFP cells were treated in a 96-well plate with purified recombinant Tat added at 0.8 μg/300,000 cells per well in the presence of 100 μm chloroquine and, where indicated supplemented with 50 μm KNSRVTFSED or KNSRATASED peptides. After an 18-h incubation, the cells were precipitated by centrifugation and lysed for 20 min at room temperature in 50 μl of lysis buffer, containing 20 mm HEPES at pH 7.9, 0.1% Nonidet P-40, and 5 mm EDTA and transferred into 150 μl of PBS. The fluorescence was measured with 480-nm excitation and 510-nm emission on a Luminescence Spectrometer LS50B (Perkin-Elmer Life Sciences) equipped with a robotic 96-well scanner.
Quantitative RT-PCR to Analyze NIPP1 Expression
RT was performed using the SuperscriptTM RT-PCR kit (Invitrogen). Random hexamers were used for RT-PCR. Primers for PCR were designed for β-actin (forward, GCGGGAAATCGTGCGTGCGTGACATT; reverse, GATGGAGTTGAAGGTAGTTTCGTG) and NIPP1 (forward, AGAATTCAACACTGCCACA; reverse, CACCCGCTTC TTCTTGACTG). Real-time PCR was performed with SYBR Green Super Mix (Bio-Rad) at 95 °C for 15 s, annealing at 55 °C for 30 s, and extension at 72 °C for 45 s for 45 cycles on a MyiQ real time PCR detection system (Bio-Rad). The obtained Ct numbers were converted to the cDNA copy numbers using as a calibration, amplification of NIPP1-expressing plasmid, and primers for NIPP1.
Cloning cdNIPP1 into pNL4-3
We used a 398-6 shuttle vector which was created by subcloning of BamHI-NcoI fragment from pNL4-3 containing nef sequence into a pUC vector and introducing CTCGAGTCTAGAGCGGCCGCTTCGA linker with XhoI, XbaI, and NotI sites into the XhoI site of the nef sequence. Then 398-6 shuttle vector was digested with BamHI and NcoI. The 2-kb fragment containing the linker was subcloned back into the Nco-BamH1 sites of pNL4-3 vector. The insert into the nef gene leads to the deletion of the first 36 amino acids of nef, which are critical for its function (32). The pNL4-3 398 vector was used as positive control in the infection experiment. To subclone cdNIPP1, pNL4-3 398 vector was digested with XhoI and NotI restriction enzymes, and the backbone was used to subclone the NIPP1-EGFP fragment that was obtained by digestion with XhoI and NotI from the WT and mutant NIPP1-(143–224)-EGFP expression vector. Correct cloning was verified by sequencing. Nuclear EGFP expression was verified by fluorescence in transfected 293 cells, and cdNIPP1 expression was verified by Western blotting using anti-NIPP1 antibodies.
Lactate Dehydrogenase (LDH) Cytotoxicity Assay
LDH release, used as an indicator of cell membrane damage, was measured in the culture medium using an LDH assay kit (LDH Cytotoxicity Assay kit; BioVision, Mountain View, CA). Briefly, the medium was collected and mixed with 100 μl of reaction mixture provided by the manufacturer and incubated for 30 min at room temperature protected from light. High control was generated by treating cells with 1% Triton X-100 for 2 h prior to the beginning of the assay. The colorimetric change was measured with a microtiter reader (Labsystem Multiskan) at 450 nm.
Analysis of HIV-1 Replication
HIV-1 viruses were prepared by transfecting HeLa cells with genomic pNL4-3, pNL4-3 398, pNL4-3-cdNIPP1-EGFP, and pNL4-3-cdNIPP1mut-EGFP and collecting media at 72 h after transfection. Viral RT activity was determined and adjusted in the collected media. RT was measured in 10 μl incubated in a 96-well plate with RT reaction mixture containing 1× RT buffer (50 mm Tris-HCl, 1 mm DTT, 5 mm MgCl2, 20 mm KCl), 0.1% Triton X-100, 10−2 units poly(A), 10−2 units poly(dT), and [3H]TTP. The mixture was incubated overnight at 37 °C, and 5 μl of the reaction mix was spotted on a DEAE Filtermat paper, washed four times with 5% Na2HPO4 and three times with water, and then dried completely. RT activity was measured in a Betaplate counter (PerkinElmer Life Sciences). MT4 cells were grown to 70% confluence, inoculated with pNL4-3 viruses, and medium was collected at the indicated time points. The RT activity was determined as described above.
RESULTS
Expression of cdNIPP1 Redistributes Cellular Endogenous NIPP1 and Increases CDK9 Phosphorylation and Its Association with 7SK RNA
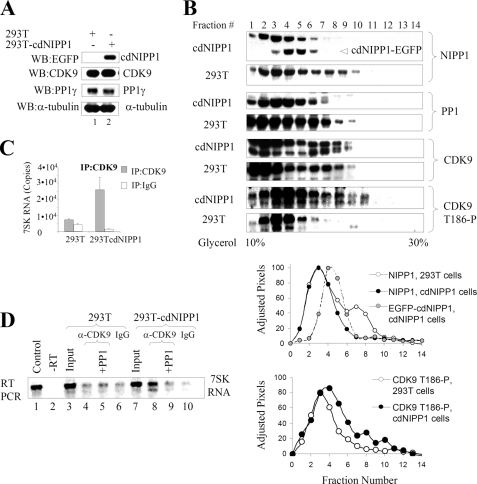
We analyzed the effect of the expression of a PP1 inhibitor in an engineered cell line that stably expressed the central, inhibitory domain of NIPP1 (residues 143–224) fused to EGFP (Fig. 1A, lane 2). By Western blotting analyses, the expression of CDK9 or PP1γ was not affected in these cells (Fig. 1A, lanes 1 and 2). Next, the distribution of NIPP1 and CDK9 phosphorylated on Thr186 between small and large molecular weight complexes was analyzed in whole cell extracts prepared from 293T or 293T-cdNIPP1 cells after fractionation on glycerol gradients (Fig. 1B). In 293T-cdNIPP1 cells, cdNIPP1-EGFP expression was detected, and the WT NIPP1 was shifted toward the lower molecular weight complexes, compared with 293T cells (Fig. 1B, NIPP1 panels). Quantification of the band densities of NIPP1 immunoblots after adjusting the pixels to equalize the total amount of NIPP1 confirmed this observation (Fig. 1B). By comparison, there was no difference in the distribution of CDK9 (Fig. 1B, CDK9 panel, quantification not shown) and a slight difference in PP1 distribution (Fig. 1B, PP1 panel). The amount of CDK9 phosphorylated on Thr186 was increased in the higher molecular weight fractions (fractions 7–10) of 293T-cdNIPP1 cells (Fig. 1B, CDK9-T186P panel). These results suggest that endogenous NIPP1 is redistributed in 293T-cdNIPP1 cells toward the smaller molecule fractions and that CDK9 phosphorylated on Thr186 is present in higher molecular weight fractions. There were no changes in the distribution of HEXIM1, cyclin T1, or Brd4 in 293T-cdNIPP1 cells versus 293T cells (data not shown).
FIGURE 1.
Effect of cdNIPP1 expression on NIPP1 distribution, CDK9 phosphorylation, and association with 7SK RNA. A, analysis of 293T-cdNIPP1 cells is shown. Western blot analysis of 293T and 293T-cdNIPP1 cells with antibodies for EGFP (to check for the expression of cdNIPP1-EGFP), CDK9, PP1γ, and α-tubulin is shown. B, expression of cdNIPP1 changes NIPP1 distribution and increases CDK9 phosphorylation on Thr186. Whole cell extracts were prepared from 293T and 293T-cdNIPP1 cells and separated on glycerol gradients by ultracentrifugation. The gradients were fractionated and analyzed by Western blotting for NIPP1, PP1, CDK9, and CDK9 phosphorylated on Thr186. The NIPP1 and CDK9 phosphorylated on Thr186 immunoblots were scanned and quantified using PhosphorImager OptiQuant software. The pixels were adjusted to have the same total amount for the 293T and the cdNIPP1 extracts, and the normalized results are graphed (lower panels). C, expression of cdNIPP1 increases the association of CDK9 with 7SK RNA. RNA was isolated from material that co-precipitated with CDK9 from 239T or 293T-cdNIPP1 whole cell lysates. The RNA was reverse transcribed and analyzed by real time PCR. Results are presented as numbers of copies of 7SK RNA. D, PP1 removes CDK9-associated 7SK RNA. RNA was isolated from material that co-immunoprecipitated with CDK9 from 239T or 293T-cdNIPP1 cells, reverse-transcribed, and analyzed by semiquantitative PCR (26 cycles) using 7SK-specific primers. Lane 1, control using 7SK expression vector. Lane 2, control that was not reverse-transcribed. Lanes 3 and 7 are inputs. Lanes 4 and 8, 7SK RNA associated with CDK9 in 293T or 293T-cdNIPP1 whole cell lysates. Lanes 5 and 9, immunoprecipitated CDK9 was treated with purified rabbit muscular PP1C prior to the RNA extraction. Lanes 6 and 10, controls in which the immunoprecipitations were carried out with nonspecific rabbit IgG.
Because the phosphorylation of CDK9 on Thr186 promotes the association of CDK9/cyclin T1 with 7SK RNA (16, 17), we analyzed whether the increased CDK9 phosphorylation affected the association of CDK9 with 7SK RNA. Analysis by quantitative real time PCR showed an increase in the amount of 7SK RNA that precipitated with CDK9 from 293T-cdNIPP1 cells compared with 293T cells (Fig. 1C). These results were replicated in a semiquantitative RT-PCR, which showed 7SK RNA co-precipitating with CDK9 from 293T-cdNIPP1 cell lysate (Fig. 1D, lane 8) in contrast to a little co-precipitation of 7SK RNA with CDK9 from 293T cells (Fig. 1D, lane 4). Pretreatment of the CDK9/7SK complex immunoprecipitated from 293T-cdNIPP1 cells with purified PP1c removed 7SK RNA (Fig. 1D, compare lanes 8 and 9), suggesting that association of 7SK RNA with CDK9 is phosphorylation-dependent and sensitive to dephosphorylation by PP1. Taken together, these results indicate that CDK9 is additionally phosphorylated on Thr186 in 293T-cdNIPP1 cells and that phosphorylated CDK9 is associated with 7SK RNA in high molecular weight fractions.
Interaction of Tat with PP1 Is Altered in 293T-cdNIPP1 Cells
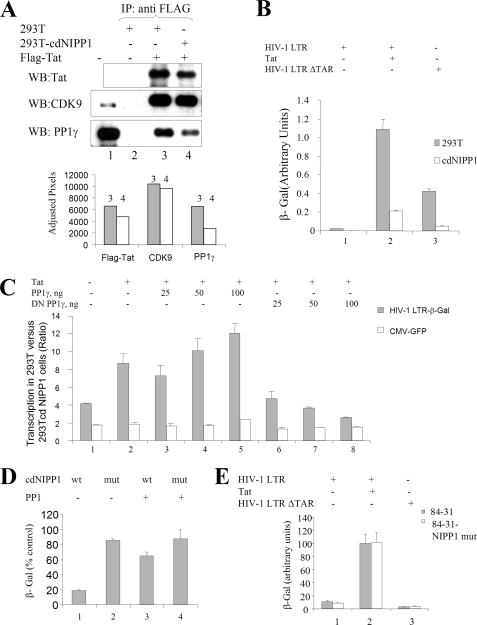
To investigate the interaction of Tat with PP1, FLAG-tagged Tat was expressed in 293T and 293T-cdNIPP1 cells and then immunoprecipitated from cellular extracts with anti-FLAG antibodies (Fig. 2A). The amount of FLAG-Tat expressed in 293T-cdNIPP1 cells was less than in 293T, and less Tat protein was immunoprecipitated from mutant cell line whole cell lysate (Fig. 2A, compare lanes 3 and 4). The amount of CDK9 co-precipitated with Tat was slightly reduced in 293T-cdNIPP1 cells (Fig. 2A, lanes 3 and 4). In contrast, the amount of PP1 co-precipitated with Tat was reduced substantially in 293T-cdNIP1 cells (Fig. 2A, lanes 3 and 4), even comparing with a reduced amount of Tat precipitated from the mutated cell line. Thus, the expression of cdNIPP1 reduced the amount of PP1 available to bind Tat.
FIGURE 2.
Expression of cdNIPP1 inhibits HIV-1 transcription. A, expression of cdNIPP1 disrupts the interaction of Tat and PP1. 293T and 293T-cdNIPP1 cells were transfected with FLAG-Tat expression vectors. Tat and associated proteins were immunoprecipitated with anti-FLAG antibodies, resolved on SDS-PAGE, and probed with antibodies against Tat, CDK9, and PP1γ. Lane 1, inputs. Lane 2, 293T cells without FLAG-Tat expressed. Lanes 3 and 4, 293T and 293T-cdNIPP cells, respectively, transfected with FLAG-Tat. B, expression of cdNIPP1 inhibits Tat-induced HIV-1 transcription. 293T and 293T-cdNIPP1 cells were transiently transfected with HIV-1 LTR-LacZ without (lane 1) or with co-transfection of Tat expressing plasmid (lane 2); or a TAR deleted construct of HIV-1 LTR-LacZ (HIV-1 LTRΔTAR) (lane 3). At 24 h after the transfection, the cells were lysed and analyzed for β-galactosidase activity. HIV-1 LTR-expression plasmid was analyzed by real time PCR and used for normalization. C, co-expression of cdNIPP1 and PP1γ or PP1γ-D64N-EGFP (DN PP1γ-EGFP) affects basal and Tat-induced HIV-1 transcription. 293T and 293T-cdNIPP1 cells were transfected with HIV-1 LTR-LacZ (lane 1) or in combination with Tat expression vector (lanes 2–8) and the indicated amounts of PP1γ (lanes 3–5) or DN PP1γ (lanes 6–8) expression vectors and co-transfected with CMV-EGFP expression vector. At 24 h the cells were lysed and analyzed for β-galactosidase and EGFP fluorescence. D, expression of PP1γ rescues cdNIPP1-mediated inhibition of Tat-induced HIV-1 transcription. 293T cells were transfected with HIV-1 LTR-LacZ and Tat expression vectors and with WT cdNIPP1 (lanes 1 and 3) or mutant cdNIPP1 (lanes 2 and 4) expression vectors and with PP11γ expression vector (lanes 3 and 4). At 48 h after transfection, the cells were lysed and analyzed for β-galactosidase activity. E, expression of mutant NIPP1 does not affect HIV-1 transcription. 84-31 cells and 84-31 cells stably expressing mutant NIPP1 were transfected with HIV-1 LTR-LacZ without (lane 1) or with co-transfection of Tat-expressing plasmid (lane 2); or HIV-1 LTRΔTAR (lane 3). At 24 h the cells were lysed and analyzed for β-galactosidase activity.
HIV-1 Transcription Is Inhibited in 293T-cdNIPP1 Cells
To investigate the effect of the stable expression of cdNIPP1 on HIV-1 transcription, 293T and 293T-cdNIPP1 cells were transiently transfected with HIV-1 LTR-Lac Z expression vector alone or in combination with Tat (Fig. 2B), and the β-galactosidase results were normalized to the amount of plasmid DNA in the cells, which was determined by real time PCR. Tat-induced HIV-1 transcription was reduced significantly in 293T-cdNIPP1 cells compared with parental 293T cells (Fig. 2B, lane 2). Basal HIV-1 transcription from a mutant HIV-1 LTR with a deletion of the TAR RNA sequence (HIV-1 LTRΔTAR) was also reduced in 293T-cdNIPP1 (Fig. 2B, lane 3). Although the CMV-LacZ transcription was also reduced, the effect on HIV-1 promoter was much stronger (factor 4–10 compared with factor 2 for CMV promoter, Fig. 2C) and exceeded the effect on CMV promoter. Overexpression of PP1γ increased HIV-1 transcription in 293T cells over cdNIPP1 cells, and overexpression of PP1γ-D64N-EGFP (DN PP1γ) decreased the transcription (Fig. 2C, lanes 3–5 and lanes 6–8), suggesting the involvement of PP1. Co-expression of PP1γ reduced the inhibitory effect of the cdNIPP1 expression on Tat-induced HIV-1 transcription (Fig. 2D). Stable expression of mutant NIPP1 that does not bind PP1 in 84-31 cells did not have an effect on basal or Tat-induced HIV-1 transcription (Fig. 2E). To rule out the possibility that expression of cdNIPP1 causes general cellular toxicity, we analyzed the effect of cdNIPP1 expression on the release of LDH, which is an indicator of cell membrane damage. Measurement of LDH in the medium of 239T cells transfected with EGFP alone was similar to the LDH levels released from cells transfected with EGFP-fused WT or mutant NIPP1, cdNIPP1 or WT or DN PP1γ (supplemental Fig. S1A). The effect of chemically synthesized KNSRVTFSED peptide derived from the central domain of NIPP1 was analyzed in CEM-GFP T cells containing integrated HIV-1 LTR-EGFP reported which was activated by the addition of purified Tat protein to the culture medium (33). Addition of purified peptides KNSRVTFSED had small inhibitory and KNSRATASED, slightly stimulatory effect on LTR-driven EGFP expression (supplemental Fig. S1B), confirming the results obtained above with stable expression of cdNIPP1.
Analysis by real time PCR showed no difference in the expression of endogenous NIPP1 in the WT and mutant cdNIPP1-expressing cells (supplemental Fig. S1C), suggesting that the expression of NIPP1 was not altered. Taken together, these results indicate that the expression of cdNIPP1 inhibited HIV-1 transcription and that this inhibition was not due to generalized cellular toxicity of the PP1 inhibitor.
HIV-1 Replication Is Inhibited When cdNIPP1 Is Expressed as Part of the Viral Genome
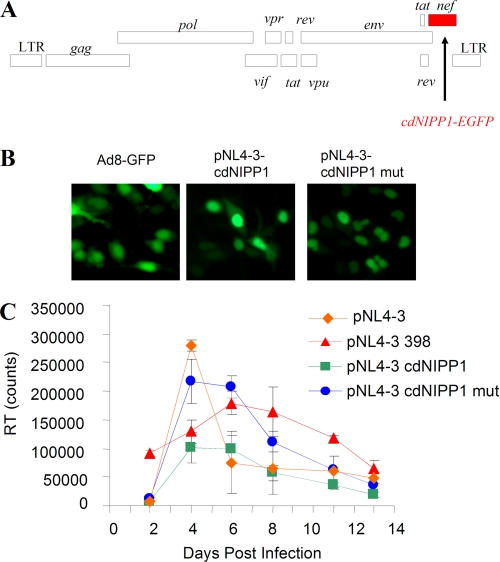
To determine directly the effect of cdNIPP1 on HIV-1 viral replication in a more physiologically relevant system, we subcloned the cdNIPP1-EGFP sequence in place of nef into the pNL4-3 vector (Fig. 3A). The expression of cdNIPP1-expressing molecular clones was verified by the nuclear fluorescence of cdNIPP1-EGFP in 293T cells transfected with pNL4-3 cdNIPP1 WT and mutant and compared with the expression of EGPF-AD8 molecular clone (34) (Fig. 3B). The recombinant HIV-1 viruses were created by transfecting HeLa cells with pNL4-3 cdNIPP1 (WT or mutant) and collecting culture supernatants at 48 h after transfection. The viruses were equalized for RT activities and used to infect cultured MT4 cells. As positive controls, we used WT pNL4-3 virus and also pNL4-3 398 virus that contained inactivating insert in the nef gene that deletes 36 N-terminal amino acids of nef critical for the nef function (32) (see “Experimental Procedures”). The expression of NIPP1 in the infected MT4 cells was monitored by Western blotting (not shown). The RT activity was analyzed over the course of 13 days to determine the kinetics of viral replication (Fig. 3C). Compared with the WT pNL4-3, the viruses with the nef inserts showed delayed replication kinetics, likely due to the impairment of the nef function. Expression of cdNIPP1 in the context of an HIV-1 genome significantly inhibited viral replication compared with the expression of a control mutant cdNIPP1 in the same genome (Fig. 3B) or expression of WT pNL4-3 or pNL4-3 398 virus with the inactivating insert in the nef gene. These results verified that productive HIV-1 replication is inhibited when cdNIPP1 is expressed during the course of the viral infection in T cells.
FIGURE 3.
Expression of cdNIPP1 inhibits HIV-1 replication. A, cloning strategy. Diagram of the HIV-1 genome indicates the insertion of cdNIPP1-EGFP into nef. B, expression of cdNIPP1. EGFP-fused WT cdNIPP1 and mutant RATA cdNIPP1 were expressed in 293T cells transfected with recombinant pNL4-3 constructs or Ad8-GFP expression vector as control. C, replication of recombinant pNL4-3 cdNIPP1. Replication kinetics of WT pNL4-3 and control pNL4-3 398 and recombinant pNL4-3 expressing cdNIPP1 and mutant RATA cdNIPP1 in MT4 cells determined by RT measurement in cell media are shown. No new cells were added into the infected cultures.
DISCUSSION
Here, we show that expression of a relatively short, 82 residues PP1-binding peptide, derived from NIPP1 redistributed cellular PP1, modified CDK9 phosphorylation and association with 7SK RNA, and inhibited HIV-1 replication. Although the PP1 targets relevant to HIV-1 transcription are still not conclusively understood, one possibility is that PP1 dephosphorylates CDK9 (25). Thr186 of CDK9 can be dephosphorylated by PP1α in vivo (18). Our data are in agreement with this previous work because we documented an increased phosphorylation of CDK9 Thr186 and increased association of CDK9 with 7SK RNA when PP1 inhibitor was overexpressed. We previously hypothesized that during HIV-1 infection, the viral Tat protein serves as a PP1-targeting subunit. Here, we found that the expression of cdNIPP1 disrupts the association of Tat with PP1; this disruption could be a mechanism for inhibiting HIV-1 transcription, similar to the previously reported HIV-1 inhibitory effect of Tat mutations that prevented binding of Tat to PP1 (24). Accordingly, we observed the inhibition of HIV-1 transcription by a short peptide containing the RVTF sequence, although the effect was relatively small likely due to the problem with cell permeability and inefficient delivery.
Another possible biological target for PP1 dephosphorylation in HIV-1 transcription could be the transcription factor Sp1 because it is known to associate with PP1 at many cellular promoters (35). Sp1 is required by HIV-1 for basal transcription (36) and can target cyclin T1 to the LTR in the absence of Tat or TAR RNA (37). An increase in Sp1 phosphorylation induced by Tat and DNA-PK can enhance HIV-1 transcription (38). RNA polymerase II could be a third PP1 substrate; we have previously shown that the C-terminal domain of RNA polymerase II is indeed dephosphorylated by PP1 (39). Our recent study showed that NIPP1 can serve as an RNA polymerase II-targeting subunit of PP1 (40), and thus altered RNA polymerase II dephosphorylation could also be considered as a potential inhibitory mechanism. Interestingly, the results presented here indicate that despite the expression of cdNIPP1, cells remained viable even though the association of CDK9 with 7SK RNA was increased. Also, a transient expression of cdNIPP1 was not toxic. Thus, our observations indicate a future possibility for the design of the HIV-1 inhibitory compounds that affect the interaction of Tat with PP1.
Here, we utilized an RVXF-binding peptide (i.e. cdNIPP1) to disrupt the interaction of PP1 with a subset of cellular regulatory subunits and inhibit HIV-1. Recently, a combined approach of in silico screening and a multistep biochemical validation procedure identified many novel PP1 interactors that contained novel PP1 binding motifs, “SILK” and “MyPhoNE” (41). Thus, small molecule compounds that disrupt either of these PP1-binding motifs or an RVXF motif might deregulate subsets of cellular PP1 holoenzymes and selectively target viral or cellular pathways. Taken together, previous studies and our findings here indicate that PP1 is involved in HIV-1 transcription. Efficient inhibition of HIV-1 transcription and replication through the redistribution of PP1 indicate that PP1 can be a future target for antiviral therapeutics.
Supplementary Material
Acknowledgments
We thank members of Dr. Victor Gordeuk's Center for Sickle Cell Disease, Howard University, for valuable discussions, and Qiang Zhou (University of California, Berkeley) for the gift of Thr186 phospho-specific antibodies and antibodies for Brd4 and HEXIM1.
This work was supported by National Institutes of Health Research Grant 2 R25 HL003679-12 funded by the NHLBI. This work was also supported by the Office of Research on Minority, Grant 1SC1GM082325-03, funded by the National Institute for General Medical Sciences; by a Howard University Seed Grant (to S. N.); and by Grant RCMI-NIH 2G12RR003048-22 from the Research Centers in Minority Institutions Program, Division of Research Infrastructure, National Center for Research Resources, National Institutes of Health.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CDK9
- cyclin-dependent kinase-9
- Brd4
- bromodomain protein 4
- cdNIPP1
- central domain NIPP1
- EGFP
- enhanced green fluorescent protein
- HEXIM1
- hexamethylene bisacetamide-induced protein
- NIPP1
- nuclear inhibitor of PP1
- P-TEFb
- positive transcription elongation complex
- PP1
- protein phosphatase 1
- PP1C
- catalytic subunit of PP1
- PP2B
- protein phosphatase 2B
- PPM1A
- protein phosphatase M1A
- TAR RNA
- transactivation response RNA.
REFERENCES
- 1. Nekhai S., Jeang K. T. (2006) Future Microbiol. 1, 417–426 [DOI] [PubMed] [Google Scholar]
- 2. Yang Z., Zhu Q., Luo K., Zhou Q. (2001) Nature 414, 317–322 [DOI] [PubMed] [Google Scholar]
- 3. Nguyen V. T., Kiss T., Michels A. A., Bensaude O. (2001) Nature 414, 322–325 [DOI] [PubMed] [Google Scholar]
- 4. Michels A. A., Nguyen V. T., Fraldi A., Labas V., Edwards M., Bonnet F., Lania L., Bensaude O. (2003) Mol. Cell. Biol. 23, 4859–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yik J. H., Chen R., Nishimura R., Jennings J. L., Link A. J., Zhou Q. (2003) Mol. Cell 12, 971–982 [DOI] [PubMed] [Google Scholar]
- 6. Jang M. K., Mochizuki K., Zhou M., Jeong H. S., Brady J. N., Ozato K. (2005) Mol. Cell 19, 523–534 [DOI] [PubMed] [Google Scholar]
- 7. Yang Z., Yik J. H., Chen R., He N., Jang M. K., Ozato K., Zhou Q. (2005) Mol. Cell 19, 535–545 [DOI] [PubMed] [Google Scholar]
- 8. Dey A., Nishiyama A., Karpova T., McNally J., Ozato K. (2009) Mol. Biol. Cell 20, 4899–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garber M. E., Wei P., Jones K. A. (1998) Cold Spring Harbor Symp. Quant. Biol. 63, 371–380 [DOI] [PubMed] [Google Scholar]
- 10. Zhu Y., Pe'ery T., Peng J., Ramanathan Y., Marshall N., Marshall T., Amendt B., Mathews M. B., Price D. H. (1997) Genes Dev. 11, 2622–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X., Gold M. O., Tang D. N., Lewis D. E., Aguilar-Cordova E., Rice A. P., Herrmann C. H. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 12331–12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ott M., Schnölzer M., Garnica J., Fischle W., Emiliani S., Rackwitz H. R., Verdin E. (1999) Curr. Biol. 9, 1489–1492 [DOI] [PubMed] [Google Scholar]
- 13. Kiernan R. E., Vanhulle C., Schiltz L., Adam E., Xiao H., Maudoux F., Calomme C., Burny A., Nakatani Y., Jeang K. T., Benkirane M., Van Lint C. (1999) EMBO J. 18, 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng L., de la Fuente C., Fu P., Wang L., Donnelly R., Wade J. D., Lambert P., Li H., Lee C. G., Kashanchi F. (2000) Virology 277, 278–295 [DOI] [PubMed] [Google Scholar]
- 15. Barboric M., Yik J. H., Czudnochowski N., Yang Z., Chen R., Contreras X., Geyer M., Matija Peterlin B., Zhou Q. (2007) Nucleic Acids Res. 35, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen R., Yang Z., Zhou Q. (2004) J. Biol. Chem. 279, 4153–4160 [DOI] [PubMed] [Google Scholar]
- 17. Li Q., Price J. P., Byers S. A., Cheng D., Peng J., Price D. H. (2005) J. Biol. Chem. 280, 28819–28826 [DOI] [PubMed] [Google Scholar]
- 18. Chen R., Liu M., Li H., Xue Y., Ramney W. N., He N., Ai N., Luo H., Zhu Y., Zhou N., Zhou Q. (2008) Genes Dev. 22, 1356–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y., Dow E. C., Liang Y. Y., Ramakrishnan R., Liu H., Sung T. L., Lin X., Rice A. P. (2008) J. Biol. Chem. 283, 33578–33584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bharucha D. C., Zhou M., Nekhai S., Brady J. N., Shukla R. R., Kumar A. (2002) Virology 296, 6–16 [DOI] [PubMed] [Google Scholar]
- 21. Ammosova T., Jerebtsova M., Beullens M., Voloshin Y., Ray P. E., Kumar A., Bollen M., Nekhai S. (2003) J. Biol. Chem. 278, 32189–32194 [DOI] [PubMed] [Google Scholar]
- 22. Bollen M., Beullens M. (2002) Trends Cell Biol. 12, 138–145 [DOI] [PubMed] [Google Scholar]
- 23. Bollen M. (2001) Trends Biochem. Sci. 26, 426–431 [DOI] [PubMed] [Google Scholar]
- 24. Ammosova T., Jerebtsova M., Beullens M., Lesage B., Jackson A., Kashanchi F., Southerland W., Gordeuk V. R., Bollen M., Nekhai S. (2005) J. Biol. Chem. 280, 36364–36371 [DOI] [PubMed] [Google Scholar]
- 25. Ammosova T., Washington K., Debebe Z., Brady J., Nekhai S. (2005) Retrovirology 2, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chirmule N., Propert K., Magosin S., Qian Y., Qian R., Wilson J. (1999) Gene Ther. 6, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 27. DeGuzman A., Lee E. Y. (1988) Methods Enzymol. 159, 356–368 [DOI] [PubMed] [Google Scholar]
- 28. Deng L., Ammosova T., Pumfery A., Kashanchi F., Nekhai S. (2002) J. Biol. Chem. 277, 33922–33929 [DOI] [PubMed] [Google Scholar]
- 29. Lesage B., Beullens M., Nuytten M., Van Eynde A., Keppens S., Himpens B., Bollen M. (2004) J. Biol. Chem. 279, 55978–55984 [DOI] [PubMed] [Google Scholar]
- 30. Beullens M., Vulsteke V., Van Eynde A., Jagiello I., Stalmans W., Bollen M. (2000) Biochem. J. 352, 651–658 [PMC free article] [PubMed] [Google Scholar]
- 31. Ceulemans H., Vulsteke V., De Maeyer M., Tatchell K., Stalmans W., Bollen M. (2002) J. Biol. Chem. 277, 47331–47337 [DOI] [PubMed] [Google Scholar]
- 32. Ali S. A., Huang M. B., Campbell P. E., Roth W. W., Campbell T., Khan M., Newman G., Villinger F., Powell M. D., Bond V. C. (2010) AIDS Res. Hum. Retroviruses 26, 173–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nekhai S., Bhat U. G., Ammosova T., Radhakrishnan S. K., Jerebtsova M., Niu X., Foster A., Layden T. J., Gartel A. L. (2007) Oncogene 26, 3899–3903 [DOI] [PubMed] [Google Scholar]
- 34. Vanham G., Penne L., Allemeersch H., Kestens L., Willems B., van der Groen G., Jeang K. T., Toossi Z., Rich E. (2000) Aids 14, 2299–2311 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y., Liao M., Dufau M. L. (2008) J. Biol. Chem. 283, 24039–24046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jochmann R., Thurau M., Jung S., Hofmann C., Naschberger E., Kremmer E., Harrer T., Miller M., Schaft N., Stürzl M. (2009) J. Virol. 83, 3704–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yedavalli V. S., Benkirane M., Jeang K. T. (2003) J. Biol. Chem. 278, 6404–6410 [DOI] [PubMed] [Google Scholar]
- 38. Chun R. F., Semmes O. J., Neuveut C., Jeang K. T. (1998) J. Virol. 72, 2615–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Washington K., Ammosova T., Beullens M., Jerebtsova M., Kumar A., Bollen M., Nekhai S. (2002) J. Biol. Chem. 277, 40442–40448 [DOI] [PubMed] [Google Scholar]
- 40. Jerebtsova M., Klotchenko S. A., Artamonova T. O., Ammosova T., Washington K., Egorov V. V., Shaldzhyan A. A., Sergeeva M. V., Zatulovskiy E. A., Temkina O. A., Petukhov M. G., Vasin A. V., Khodorkovskii M. A., Orlov Y. N., Nekhai S. (2010) Mol. Cell Biochem. 347, 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. (2009) Chem. Biol. 16, 365–371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.