Abstract
The CYP3A subfamily of hepatic cytochromes P450, being engaged in the metabolism and clearance of >50% of clinically relevant drugs, can significantly influence therapeutics and drug-drug interactions. Our characterization of CYP3A degradation has indicated that CYPs 3A incur ubiquitin-dependent proteasomal degradation (UPD) in an endoplasmic reticulum (ER)-associated degradation (ERAD) process. Cytochromes P450 are monotopic hemoproteins N-terminally anchored to the ER membrane with their protein bulk readily accessible to the cytosolic proteasome. Given this topology, it was unclear whether they would require the AAA-ATPase p97 chaperone complex that retrotranslocates/dislocates ubiquitinated ER-integral and luminal proteins into the cytosol for proteasomal delivery. To assess the in vivo relevance of this p97-CYP3A association, we used lentiviral shRNAs to silence p97 (80% mRNA and 90% protein knockdown relative to controls) in sandwich-cultured rat hepatocytes. This extensive hepatic p97 knockdown remarkably had no effect on cellular morphology, ER stress, and/or apoptosis, despite the well recognized strategic p97 roles in multiple important cellular processes. However, such hepatic p97 knockdown almost completely abrogated CYP3A extraction into the cytosol, resulting in a significant accumulation of parent and ubiquitinated CYP3A species that were firmly ER-tethered. Little detectable CYP3A accumulated in the cytosol, even after concomitant inhibition of proteasomal degradation, thereby documenting a major role of p97 in CYP3A extraction and delivery to the 26 S proteasome during its UPD/ERAD. Intriguingly, the accumulated parent CYP3A was functionally active, indicating that p97 can regulate physiological CYP3A content and thus influence its clinically relevant function.
Keywords: ATPases, Cytochrome P450, E3 Ubiquitin Ligase, Endoplasmic Reticulum (ER), Proteasome, Protein Degradation, Ubiquitination, ERAD, Intracellular Trafficking, p97/VCP AAA ATPase
Introduction
Hepatic cytochrome P450 hemoproteins (P450s)2 of the CYP3A subfamily include CYP3A4, the major human liver drug metabolizing P450 enzyme engaged in the metabolism of over 50% of clinically relevant drugs and other xenobiotics (1). In common with other microsomal P450s (2–10), CYPs 3A are integral endoplasmic reticulum (ER) membrane-anchored monotopic proteins, with their N terminus embedded in the ER and their catalytic domain exposed to the cytosol. Our findings in various in vivo and in vitro reconstituted eukaryotic systems have documented that both native3 and structurally inactivated CYPs 3A incur ubiquitin (Ub)-dependent proteasomal degradation (UPD) (11–20), in a typical ER-associated degradation (ERAD) process (21–28). Indeed, CYPs 3A qualify as bona fide ERAD-C substrates following mechanism-based inactivation by certain agents (29–31) (see below).
Thus, consistent with a typical ERAD process, we have found that CYP3A ERAD involves posttranslational phosphorylation by cytosolic kinases (15, 32, 33); ubiquitination by the ER-integral “glycoprotein” 78/autocrine motility factor receptor (gp78/AMFR) and the cytosolic C terminus of Hsp70-interacting protein (CHIP) E3 Ub ligases along with their respective cognate Ub-conjugating enzymes UBC7/Ube2g2 and UbcH5a (33–35); and subsequent degradation by the 26 S proteasome (12–17). As a monotopic ER protein, the bulk of the P450 molecule is exposed to the cytosol and thus amply accessible to the 26 S proteasome, a fraction of which is thought to be intimately associated with the ER membrane (36, 37). Thus, it was unclear whether ER membrane extraction into the cytosol would be at all necessary for P450 proteasomal processing, as in the case of polytopic transmembrane or luminal ER proteins (21, 38–40, 42–55). The latter require the p97 AAA ATPase-Npl4-Ufd1 chaperone complex for their retrotranslocation from the ER into the cytosol, a process that is fueled by ATP hydrolysis (21, 38–40, 42–56).
p97, also known, albeit erroneously, as VCP (valosin-containing protein) or Cdc48p (in yeast), is an abundant cytosolic AAA ATPase (ATPase associated with various cellular activities) involved in an ever growing number of cellular functions and processes (55–61). Perhaps its best characterized functional role is its ER to cytosol retrotranslocation of ER integral and luminal proteins, a prerequisite for their subsequent UPD (21, 38–40, 42–61). p97 has a homohexameric barrel structure with each subunit composed of two ATPase domains stacked on top of each other (62–64). It functions as a heterotrimeric complex with two additional heterodimeric adapters, Ufd1p and Npl4p, which bind to its N-terminal domain (49–57). Ufd1p and Npl4p are thought to assist in the recruitment of polyubiquitinated target substrates to the p97 complex by engaging the poly-Ub chains decorating an ERAD target substrate (49–51, 55–57).
Because Saccharomyces cerevisiae yeast strains with defects in each of the three individual components of the homologous Cdc48p-Npl4p-Ufd1p chaperone complex are available (21, 40, 42), we examined CYP3A4 ERAD in each of these defective strains along with the corresponding wild type strains upon heterologous expression of CYP3A4 (19). Indeed, findings of our stationary chase analyses revealed that CYP3A4 was markedly stabilized in the cdc48-2 yeast and even more strikingly so in the npl4-1 and ufd1-1 strains, relative to that in their wild type counterparts (19). These results provided the initial clue that despite its ER topology, strategically poised for imminent access to the 26 S proteasome, CYP3A4 ERAD in S. cerevisiae requires the yeast Cdc48p-Npl4p-Ufd1p chaperone complex. These findings led us to examine the role of p97 in cultured rat hepatocytes (14). Collectively, these findings provided evidence of a strong association between the native CYP3A and p97 that was significantly enhanced after mechanism-based CYP3A inactivation by the suicide inactivator, 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine (DDEP). DDEP, in the course of its CYP3A-catalyzed oxidation, results in the oxidative fragmentation of the CYP3A prosthetic heme into mono- and dipyrrolic products that irreversibly bind to the active site of the enzyme (11, 65), fatally damaging it both structurally and functionally. In vivo, this structurally damaged protein with active site lesions within its cytosolic domain is marked for rapid clearance via ERAD/UPD in a typical ERAD-C process.
Given on one hand the imminent accessibility of CYPs 3A to the 26 S proteasome that renders p97 potentially dispensable in its ERAD and, on the other, this collective evidence in yeast and rat hepatocytes supporting a role for the p97 complex in CYP3A ERAD, we sought to determine the relative importance of the p97 complex to CYP3A ERAD/UPD through RNA interference (RNAi) targeted against p97. Our findings of p97 knockdown reveal that despite the predominantly cytosol-oriented ER topology of CYP3A, p97 plays a major role in CYP3A extraction into the cytosol and its subsequent delivery to the 26 S proteasome. Accordingly, p97 knockdown results in a strikingly marked accumulation of both the native and inactivated parent and ubiquitinated CYP3A species that remain firmly tethered to the ER membrane. Surprisingly, the native non-ubiquitinated CYP3A species accumulating in the ER on p97 knockdown is functionally active. This suggests that the p97 complex can regulate the hepatic content of the parent non-ubiquitinated CYP3A species and thus may critically influence its physiological function. Given the vast array of clinically relevant drugs, carcinogens, and other xenobiotics that depend on these particular P450s for their metabolic clearance (1), our findings, if extrapolated to the human liver, would be clinically and/or toxicologically relevant.
EXPERIMENTAL PROCEDURES
Materials
The sources of common cell culture media, supplements, and culture plastic ware and the commercial sources of protease inhibitors, dexamethasone (Dex), β-glucuronidase/arylsulfatase mixture, 7-hydroxy-4-trifluoromethylcoumarin (HFC), and 7-benzyloxy-4-trifluoromethylcoumarin (BFC) have been reported previously (35, 66, 67). Goat or rabbit polyclonal IgGs were raised commercially against purified recombinant rat hepatic CYP3A23 and purified by Hi-Trap® Protein A-Sepharose affinity chromatography. Peptides (tagged either N-terminally with keyhole limpet hemocyanin or C-terminally with 8-MAP) corresponding to (keyhole limpet hemocyanin)-1MDLLSALTLET11 in the signal-anchor, 20VLLYGFGTRTHGLF33-(8-MAP) in the basic sequence, 35KQGIPGPKPLPFFG48-(8-MAP) in the proline-rich segment of the N-terminal domain, and (keyhole limpet hemocyanin)-486KPIILKVVPRDEIITGS502 in the C-terminal domain were synthesized, and polyclonal antibodies were generated commercially in rabbits (ResGen/Invitrogen). These antibodies have been referred to as α-N-terminal (NT), α-Middle, α-Internal, and α-C-terminal. The Proteasome-GloTM cell-based assay system was purchased from Promega (Madison, WI).
Isolation of Microsomes and Cytosol from DDEP-treated Rats
Male Sprague-Dawley rats (6–8 weeks old) purchased from Simonsen Laboratories (Gilroy, CA) were fed and given water ad libitum and handled according to institutional animal care and use committee guidelines. After acclimation for ∼5–7 days, they were pretreated with the CYP3A inducer Dex (100 mg/kg/day; i.p.) in corn oil for 4 days. Twenty-four h after the last dose, they were given a single i.p. injection of DDEP (125 mg/kg) and killed 2 h later. Livers were perfused and isolated, and homogenates were prepared and subjected to ultracentrifugation at 100,000 × g at 4 °C for 1 h; and microsomes (100,000 × g pellet) and/or cytosol (100,000 × g supernatant) were prepared as described previously (11–13). Dex-pretreated rats not given DDEP were killed at time 0 h and used as the corresponding controls. Microsomes were washed free of cytosol by two sequential rounds of resedimentation at 100,000 × g at 4 °C for 30 min and resuspension, and stored as “pellets” overlaid with Tris buffer, pH 7.4, 20% (v/v) glycerol, EDTA (1 mm) at −80 °C until use.
Construction of Recombinant p97 Lentiviral shRNA Vectors
The following oligonucleotides targeting three regions of p97/VCP mRNA were designed and synthesized: p97-shRNA 1 targeting exon 6 of the rat p97 sequence (GenBankTM accession number NM053864) (nt 812–832), 5′-aaaactgcagaaaaaGTAGGCTATGATGACATCGGTGGTTtctcttgaaAACCACCGATGTCATCATAGCCTACggtgtttcgtcctttccacaag-3′ (reverse primer); p97-shRNA 2 targeting p97 exon 3 (nt 346–366), 5′-aaaactgcagaaaaaGATGGATGAACTACAGTTGTTCAGAtctcttgaaTCTGAACAACTGTAGTTCATCCATCggtgtttcgtcctttccacaag-3′ (reverse primer); and p97-shRNA 3 targeting p97 exon 3 (nt 454–474), 5′-aaaactgcagaaaaaGATTCGAATGAATAGAGTTGTTCGGtctcttgaaCCGAACAACTCTATTCATTCGAATCggtgtttcgtcctttccacaag-3′ (reverse primer). Using plasmid pTZU6+1 as the template, the individual oligonucleotide (which also served as reverse primer) was inserted downstream of the U6+1 promoter by PCR using a forward primer (5′-AAAAACTAGTAAGGTCGGGCAGGAAGAGGGC-3′) and the corresponding reverse primer. SpeI and PstI restriction enzyme sites were introduced into the forward and reverse primers, respectively. The PCR fragments were cloned into pGEM-T Easy TA vectors and confirmed by DNA sequencing. The SpeI/PstI-digested PCR fragments of pGEM vectors were cloned into pHR CMV PURO Wsin18 vector, resulting in plasmids containing the oligonucleotides encoding shRNAs. The control shRNA sequence has been recently described (35).
Lentiviral Packaging in HEK 293T Cells
This was carried out exactly as described (35). A 5–10-ml aliquot of the filtered viral stock was used for total RNA extraction. The extracted RNA concentration was determined by the nanodrop technique (average concentration is 100 ng/μl). The samples were stored at −80 °C until the reverse transcription step to generate cDNA. The lentiviral RNA concentration was also determined by real-time quantitative PCR (qRT-PCR) analyses (68) using the following: forward primer (HIV1-LTR-FO), 5′-TGTGTGCCCGTCTGTTGTGT-3′; reverse primer (HIV1-LTR-RE), 5′-GAGTCCTGCGTCGAGAGAGC-3′; and the Taqman probe (HIV1-LTR-TaqProbe), 5′-FAM-CAGTGGCGCCCGAACAGGGA-TAMRA-3′ (FAM, 6-carboxyfluorescein; TAMRA, tetramethyl rhodamine carboxylic acid derivative). Based on the RNA concentration of this 5–10-ml aliquot, the entire viral stock was pelleted and resuspended in PBS/hepatocyte culture medium to a final concentration of 1 μg of viral RNA/ml for infection of hepatocytes as described (35).
Rat Hepatocyte Viral Infection and Culture
Hepatocytes were isolated from rats (4–6 weeks old) by in situ liver perfusion with collagenase (liver digest medium) and cultured as described (14, 66, 67, 69). Cells were maintained for 2 days with a daily change of medium to enable recovery and hepatic function restoration (70). At 72 h of culture, puromycin (5 μg/ml) was added to the medium for selection of cells that were successfully infected by the shRNA-containing virus. The medium was replaced daily, the Dex concentration was increased to 10 μm to induce CYPs 3A, and cells were cultured for an additional 5–6 days, with daily light microscopic examination for any signs of cell death or cytotoxicity. In some cases, cells were treated with vehicle (DMSO), Dex (10 μm), or Dex (10 μm) plus DDEP (100 μm) for 4 h.
In preliminary experiments, the optimal time for ≥80% knockdown of p97 was determined to be >7 days. At this time, cells were harvested in lysis buffer (66), and clarified lysate supernatants were subjected to Western immunoblotting analyses. In parallel, some shRNA-infected cell cultures were also harvested in RNA-stabilizing reagent (Qiagen) and used for total RNA extraction and qRT-PCR analyses (see below).
Preparation of Microsomal and Cytosolic Fractions
Microsomes were isolated from rat hepatocytes as described (13, 69). The supernatant obtained after the first 100,000 × g ultracentrifugation was used as the cytosolic fraction and subjected to CYP3A immunoprecipitation as described below. To determine the content of CYP3A normally ER-integrated relative to that dislocated but still loosely associated with the external ER surface, microsomes from 35S-pulse-chase experiments were subjected to a 0.1 m Na2CO3 wash exactly as detailed (35). The 0.1 m Na2CO3-washed microsomes representing firmly integrated proteins were sedimented at 180,000 × g, and the pellet was used for CYP3A immunoprecipitation. The resulting supernatant was precipitated with 100% trichloroacetic acid, and the protein pellet was washed with 100% acetone and referred to as the “Na2CO3-wash.” This Na2CO3-wash was subjected to CYP3A immunoprecipitation as described below. It represents any proteins that were dislocated from the ER but remained loosely associated with the ER membranes.
CYP3A Immunoprecipitation
CYP3A was routinely immunoprecipitated before the extent of its ubiquitination or 35S-incorporation was determined as described previously (14). For CYP3A immunoprecipitation, 1 mg of lysate protein or 2 mg of cytosolic protein was immunoprecipitated with 2 mg of goat anti-CYP3A antibody, whereas 500 μg of microsomal protein was immunoprecipitated with 2 mg of goat anti-CYP3A antibody. The Na2CO3-washes (200 μg of protein) were resuspended in immunoprecipitation buffer, boiled, and used for CYP3A immunoprecipitation with 500 μg of goat anti-CYP3A antibody.
Immunoblotting Analyses
CYP3A and ubiquitinated CYP3A immunoprecipitates and ubiquitinated hepatic protein were immunoblotted as described except that 5% (w/v) nonfat milk in 0.1% (v/v) Tween TBS (TTBS) was used for blocking (17, 66). Because the CYP3A antigenicity of the peptide-derived antibodies varied, the protein amounts loaded in each immunoblotting analyses also varied. Thus, with polyclonal α-Internal, α-C-terminal, and α-CYP3A IgGs, 20, 10, and 5 μg of microsomal protein were loaded, respectively. For immunoblotting analyses of the cytosolic fraction, 20 μg (α-Internal IgG) and 10 μg (polyclonal α-C-terminal and α-CYP3A IgGs) of protein were loaded. For p97 immunoblotting analyses, lysate protein (20 μg) was subjected to 7.5% SDS-PAGE and electroblotted onto a nitrocellulose membrane, and the p97 protein was immunoblotted with a primary rabbit polyclonal antibody (sc-20799, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) followed by goat anti-rabbit HRP-conjugated secondary antibody (catalog no. 170-6515) from Bio-Rad. All immunoblots were developed with the SuperSignal West maximum sensitivity Femto or Pico chemiluminescent substrate from Pierce. Actin immunoblotting analyses were routinely conducted with each lysate (10 μg of protein) to ensure equivalent protein loading.
Immunoaffinity Capture
Polyclonal rabbit α-NT antibody was dialyzed against 0.1 m phosphate, 0.15 m NaCl, pH 7.2, to remove traces of Tris and then irreversibly cross-linked to Protein A-Sepharose beads (∼8 mg of antibody/ml of bead) using disuccinimidyl suberate. The following buffers were used: antibody binding/wash buffer (50 mm sodium borate, pH 8.2); blocking buffer (0.1 m ethanolamine, pH 8.2) (to block remaining active sites); IgG elution buffer (0.1 m glycine, pH 2.8) (to elute non-cross-linked IgGs).
These α-NT antibody-cross-linked beads were used for immunoaffinity capture of CYP3A from microsomes or cytosol prepared from hepatocytes. Microsomal pellets were resuspended in immunoprecipitation buffer containing fresh protease inhibitors (final 2% (v/v) Triton, 1% (w/v) deoxycholate (DOC)). Fresh protease inhibitors were added to the cytosolic fraction and 1% (w/v) final DOC. Microsomal protein (1 mg) or cytosolic protein (2 mg) was used with 200 μl of beads. The mix was gently rolled in tightly capped tubes overnight at 4 °C. Beads were spun down in a microcentrifuge, and the supernatant was removed. The pellet was used to calculate how much protein was actually bound to the beads. In order to elute the immunoaffinity-captured CYP3A, the beads were sequentially washed with (i) 0.5 m NaCl, 0.05 m Tris-HCl, pH 8.2, 1 mm EDTA, 0.5% (v/v) Nonidet P-40; (ii) 0.15 m NaCl, 0.05 m Tris-HCl, pH 8.2, 1 mm EDTA, 0.5% (v/v) Nonidet P-40, 0.1% (w/v) SDS; (iii) 0.15 m NaCl, 0.5% (w/v) DOC (twice), as described (71).
The beads were packed into small columns onto an Eppendorf tube containing 100 μl of 1 m Tris-HCl, pH 6.8, for collection of the eluate. Bound antigen was eluted with 50 μl of 0.1 m diethylamine, pH 11.5, containing 0.5% (w/v) DOC and 10% (v/v) glycerol. This elution step was repeated with an additional 50 μl. Contents of the tube were mixed vigorously to adjust the pH to 7.4. Following this, the beads were repeatedly washed with borate buffer and stored in 0.1 m borate buffer with 0.02% (w/v) sodium azide at 4 °C. After elution, these samples were used for Western immunoblotting analyses with goat anti-CYP3A23 antibody.
Densitometric Quantification
Direct quantification of the immunoblots was performed by ImageJ (National Institutes of Health) analyses as described (67). Densitometrically derived arbitrary units of individual immunoblots were normalized against corresponding values of the actin loading controls and then expressed as a percentage of the control/basal values. When [35S]CYP3A immunoprecipitates were quantified, gels were dried, exposed to PhosphorImager screens, visualized using a Typhoon scanner, and quantified using ImageQuant software.
qRT-PCR Analyses
Total RNA was extracted and reverse transcribed to cDNA exactly as described (67, 69). Universal PCR master mix (catalog no. 430447) and Taqman primer-probe mixes were purchased from Applied Biosystems Inc. (Foster City, CA) for the detection of rat mRNA sequences for p97 (catalog no. Rn00587865_m1), cytochrome b5 (catalog no. Rn01483963_m1), CYP3A23 (catalog no. Rn01412959_g1), β-glucuronidase (catalog no. Rn00566655_m1), Grp78 (catalog no. Rn00565250_m1), Grp94 (catalog no. Rn01760569_m1), gp78, CHIP, and GAPDH (catalog no. Rn01775763_g1). The gp78 primers were ACCTCATGCACCACATTCACATGC (forward) and CAAGACACCTCTTGTCCAACATGC (reverse). The CHIP primers were ACCCGGAACCCACTTGTGGCAGTG (forward) and CTGGATGGGCAGTCTGTGAAGGCG (reverse). qRT-PCR was carried out as described (35). The Ct (cycle number at which the fluorescent signal reaches the threshold level) value reflecting the expression of each gene was normalized to that of the endogenous control glucuronidase gene. Relative gene expression was calculated as 2−ΔCt, where ΔCt is defined as Ct for the gene of interest minus Ct for glucuronidase. All values are expressed as percentage increase/decrease with respect to the corresponding RNA value in control shRNA-infected hepatocytes.
Immunofluorescence Microscopy
Immunochemically detectable CYP3A content of shRNA-infected and control cells was detected with a Zeiss Axiovert 200M, LSM 510 Meta confocal microscope as described previously (35). The relative immunofluorescence intensity was quantitated as the mean intensity of a cytoplasmic area (4 μm × 5 μm) excluding the nucleus within each cell using the dedicated Zeiss confocal software. Intensity values (mean ± S.D.; n = 50 cells) ranged from 1 to 256, corresponding to the fluorescence dynamic range of 1 (no signal) to 256 maximal (saturation) immunofluorescence signal, respectively, of the 20 μm2 area. The relative nuclear diameters of control shRNA-infected and p97 shRNA 1+2-infected hepatocytes (n = 33 each) were determined as an index of apoptotic shrinkage induced by p97 knockdown.
CYP3A Functional Assay
A fluorescence-based P450 assay (72) was performed as described (35) by direct incubation of rat hepatocytes cultured in 60-mm Permanox plates and infected with p97 shRNA 1+2 or control shRNA for 7 days. Results were expressed as mean ± S.D. μmol of HFC metabolite formed/3.5 × 106 hepatocytes/h of three individual cultures.
Assay of Proteasomal Function
The Proteasome-Glo cell-based assay system was used to monitor the chymotrypsin-like, trypsin-like, and postglutamyl peptide hydrolytic (caspase-like) 20 S proteasomal core proteolytic activities (73) in intact rat hepatocytes with and without p97 knockdown to assess whether p97 RNAi had concomitantly affected hepatic proteasomal function. In brief, Suc-LLVY aminoluciferin, Suc-benzyloxycarbonyl-LRR aminoluciferin, or Suc-benzyloxycarbonyl-nLPnLD (where nL represents norleucine) aminoluciferin substrate probe dissolved in a cell-permeabilizing buffer was incubated with 20,000 intact rat hepatocytes for 10 min at room temperature. The aminoluciferin generated from each of these substrates following its release by the proteasomal chymotrypsin-like, trypsin-like, or caspase-like proteolytic activities, respectively, was monitored by an indirect coupled assay, wherein a recombinant luciferase utilizes the aminoluciferin produced to generate a luminescent signal (73). The luminescence was monitored using a Spectramax M5E plate reader in a 96-well plate assay. In parallel, some cells were incubated with the proteasomal inhibitor epoxomycin (10 μm) or MG132 (50 μm) and the lysosomal inhibitor NH4Cl (20 mm) for 3 h in order to verify the relative proteasomal versus lysosomal specificity of these proteolytic activities (supplemental Fig. S1).
Statistical Analyses
Experiments were performed in triplicate. The Kolmogorov-Smirnov test was performed on the data sets to ensure that the data were normally distributed. Data were compared by analysis of variance, and p values <0.05 were considered statistically significant.
RESULTS
CYP3A Extraction into the Cytosol of DDEP-treated Rats
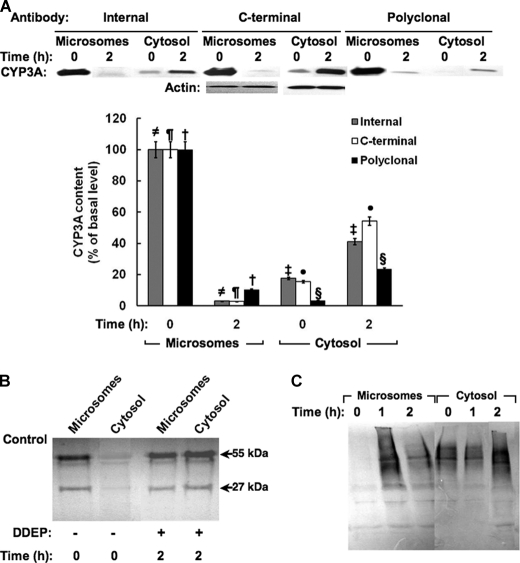
In preliminary experiments in rats treated with the CYP3A suicide inactivator DDEP for 0, 0.5, 1, and 2 h, we sought to define the time course of the ER extraction into the cytosol (data not shown). These initial findings along with our previously published observations in DDEP-treated rats and DDEP-incubated rat hepatocytes in suspension (12, 13) revealed that, as expected of an ER membrane-anchored monotopic protein, a major fraction of hepatic CYP3A (CYP3A23/CYP3A2) was initially (0 h) in the microsomal fraction, with a small amount in the cytosol. However, within 2 h of DDEP inactivation, CYP3A was largely lost from the ER and could be retrieved as the parent protein (55 kDa) and/or its high molecular mass (HMM; 65–250 kDa) ubiquitinated species in the cytosol. These cytosolic CYP3A species accumulated in the presence of proteasomal inhibitors (12–14). For these reasons, we used this 2-h period to scrutinize the process of CYP3A extraction from the ER. Was CYP3A extracted out as the intact protein, or was it cleaved from its ER membrane signal anchor and then delivered to the cytosolic 26 S proteasome?
Immunoblotting analyses with antibodies to the N-terminal proline-rich domain peptide (α-Internal) and to the C-terminal peptide (α-C-terminal) revealed that the protein spanning residues 35–502 initially existed largely in the liver ER membrane (microsomes) but was removed intact from the ER membrane into the cytosol over the 2 h of DDEP treatment (Fig. 1A). Similar findings were obtained in cultured rat hepatocytes treated with DDEP (100 μm) for 4 h (data not shown). Parallel immunoblotting analyses of liver microsomal and cytosolic subfractions from these DDEP-treated rats with antibodies to the signal anchor (α-NT) and basic sequence (α-Middle) were unsuccessful because the antibodies recognized several N termini from other microsomal proteins and were nonspecific. We therefore resorted to immunoaffinity capture, wherein the α-NT was cross-linked to Protein A-Sepharose and then exposed to solubilized liver microsomes or cytosol from rats treated with DDEP for 0–2 h. The α-NT immunoaffinity-captured fractions were then subjected to immunoblotting analyses with goat polyclonal anti-CYP3A antibody (Fig. 1B). These findings revealed that the CYP3A23 N terminus was indeed initially in the liver microsomes but progressively moved into the cytosol along with the rest of the protein over the 2-h period of DDEP treatment (Fig. 1B).
FIGURE 1.
Extraction of DDEP-inactivated CYP3A into cytosol. A, microsomes or cytosol from Dex-pretreated rats (n = 3) treated with DDEP for 0 or 2 h were subjected to immunoblotting analyses with polyclonal antibody against the proline-rich (Internal), C-terminal, or intact CYP3A23 domains as the primary antibody, and the CYP3A content was densitometrically quantified as described (see “Experimental Procedures”). Corresponding actin loading controls are also included. A representative CYP3A immunoblot obtained with each antibody is shown at the top, and the corresponding densitometric quantification of three individual immunoblots is shown at the bottom. Values are mean ± S.D. (error bars) of immunochemically detected CYP3A content in liver microsomes or cytosol from three individual rats, expressed as a percentage of their initial (0 h) values. Statistically significant differences in CYP3A content were observed between the two mean ± S.D. values, each marked with the same symbol as follows. ≠, p < 0.0001; ¶, p < 0.0001; †, p < 0.0001; §, p < 0.005; ‡, p < 0.001; ●, p < 0.001. B, a typical CYP3A immunoblotting analysis with a goat polyclonal antibody against the intact protein is shown of CYP3A immunoaffinity-captured with an antibody against the extreme N terminus of the CYP3A23 signal anchor using the corresponding microsomes and cytosol shown in A. C, Ub-immunoblotting analyses of CYP3A immunoprecipitates from liver microsomes and cytosol obtained from rats treated with DDEP for 0, 1, or 2 h. A typical immunoblot developed with alkaline phosphatase-conjugated secondary antibody is shown.
Corresponding Ub-immunoblotting analyses of CYP3A immunoprecipitates from liver microsomal and cytosolic subfractions from these same rats with goat polyclonal anti-CYP3A IgGs revealed CYP3A ubiquitination at 1 h, while the CYP3A was apparently largely in the ER, with the gradual loss of this ER-bound ubiquitinated species into the cytosol over the next 1 h (Fig. 1C). These findings are entirely consistent with our previous observations that upon inactivation, CYP3A is ubiquitinated while still in the ER, and subsequently extracted into the cytosol for proteasomal degradation (12–14). Our present findings additionally reveal that CYP3A is indeed dislocated intact from the ER membrane along with its extreme N-terminal signal anchor.
RNAi Evidence for an in Vivo Role of p97 AAA-ATPase Complex in Hepatic CYP3A Extraction
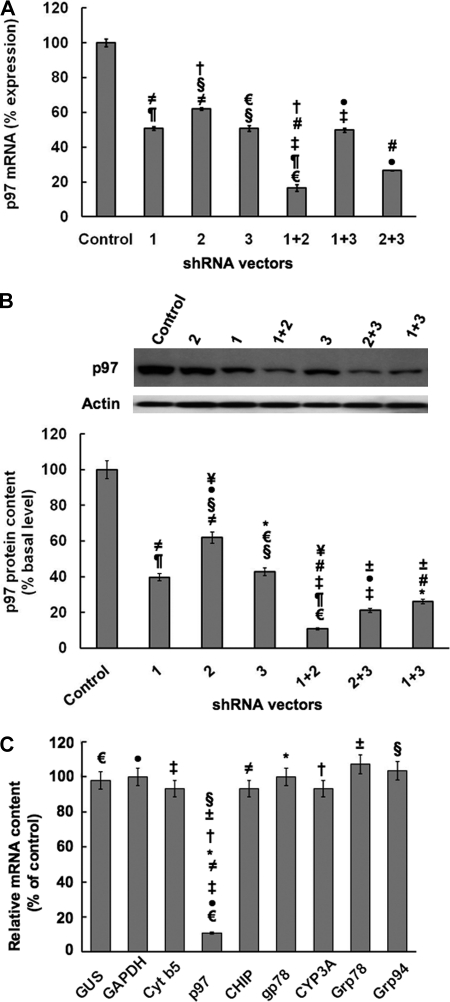
Given our previous findings both in yeast and cultured rat hepatocytes suggesting a plausible role for the p97 complex, we examined whether p97 was indeed involved in CYP3A extraction into the cytosol through RNAi directed against p97 (Fig. 2). Lentivirus-mediated delivery of three shRNAs targeted against the rat p97 gene were used singly or in combination in cultured rat hepatocytes pretreated with Dex to induce CYP3A content and thus improve its detection (Fig. 2A). A “control” shRNA was used in parallel as the corresponding vector control. qRT-PCR analyses of total RNA from hepatocytes infected with the control or p97-targeted shRNAs indicated that although all three shRNAs were capable of knocking down p97 mRNA and protein expression, the combination of shRNA 1+2 seemed to be the most effective, with a knockdown of >85% p97 mRNA expression (Fig. 2A), and >90% p97 protein expression (Fig. 2B). This particular shRNA 1+2 combination was therefore used in all subsequent studies. Parallel qRT-PCR analyses of shRNA 1+2-infected hepatocytes indicated no corresponding mRNA knockdown of CYP3A23, cytochrome b5, and gp78 (three integral ER proteins); Grp78/BiP and Grp94 (two ER-associated chaperones); CHIP (a cytosolic E3 Ub ligase cochaperone); or GAPDH and glucuronidase (two housekeeping genes), thereby attesting to the specificity of the p97 knockdown (Fig. 2C). Additionally, the findings of essentially unaltered hepatic Grp78/BiP and Grp94 mRNA levels, sensitive indices of ER stress induction/unfolded protein response, also attest to no such intracellular perturbation upon such an extensive p97 knockdown.
FIGURE 2.
RNAi-mediated p97 knockdown; Effects on p97 mRNA and protein expression in cultured rat hepatocytes and specificity of the p97 knockdown. A, effects of shRNA 1, shRNA 2, and shRNA 3 targeted against hepatic p97 individually or in combination or a control shRNA (Control) on hepatic p97 mRNA content derived from qRT-PCR analyses of total RNA isolated from each shRNA-infected cell culture. Values are mean ± S.D. (error bars) of three individually treated hepatocyte cultures. Statistically significant differences in hepatic p97 mRNA content were observed between the control values and values from shRNA 1+2- and shRNA 2+3-infected cells at p < 0.0001 and p < 0.0005, respectively, and all other shRNA-infected cells at p < 0.01. Similar statistically significant differences between the two mean ± S.D. values each marked with the same symbol were as follows. ≠, p < 0.01; ¶, p < 0.001; †, p < 0.001; §, p < 0.01; €, p < 0.001; #, p < 0.01; ‡, p < 0.001; ●, p < 0.005. B, effects of p97 shRNAs individually or in combination on hepatic p97 protein. A representative example of p97 Western immunoblotting analyses of these hepatocyte lysates (50 μg of protein) is shown at the top, with corresponding aliquots used for actin immunoblotting analyses as loading controls. Densitometric quantification (mean ± S.D.) of hepatic p97 content from three individual experiments is shown at the bottom. Shown are statistically significant differences in hepatic p97 protein content (mean ± S.D.) between control shRNA-infected cells and those infected with shRNA 1 or shRNA 3 at p < 0.001, those infected with shRNA 2 at p < 0.005, those infected with shRNA 1+3 or shRNA 2+3 at p < 0.0005, and those infected with shRNA 1 + 2 at p < 0.0001. Statistically significant differences in hepatic p97 protein content were observed between the control values and values from shRNA 1+2-, shRNA 1+3-, and shRNA 2+3-infected cells at p < 0.0001, p < 0.0005, and p < 0.0005, respectively; shRNA 1- and shRNA 3-infected cells at p < 0.001; and shRNA 2-infected cells at p < 0.005. Similar statistically significant differences between the two mean ± S.D. values each marked with the same symbol were as follows. ≠, p < 0.01; ¶, p < 0.005; ¥, p < 0.001; ±, p < 0.05; #, p < 0.01; §, p < 0.01; €, p < 0.005; *, p < 0.01; ‡, p < 0.01; ●, p < 0.002. C, evidence of the relative target specificity of p97 shRNA 1+2 against hepatic p97 mRNA by qRT-PCR analyses is documented. Statistically significant differences in hepatic p97 mRNA content were observed between the two mean ± S.D. values (n = 3 individual cultures), each marked with the same symbol as follows. ≠, p < 0.0001; ±, p < 0.0001; †, p < 0.0001; §, p < 0.0001; €, p < 0.0001; *, p < 0.0001; ‡, p < 0.0001; ●, p < 0.0001.
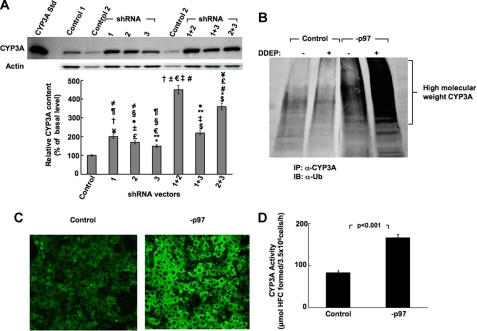
Corresponding immunoblotting analyses of hepatocyte lysates also revealed a marked >4.5-fold stabilization of the native parent CYP3A protein (55 kDa) over the control levels in hepatocytes infected with shRNA 1+2, versus any other combination or any of the individual shRNAs (Fig. 3A). However, the individual shRNAs also significantly stabilized CYP3A content over the control levels, and the shRNA 2+3 combination led to a respectable and statistically significant 3.5-fold stabilization of the native CYP3A protein over the control levels (Fig. 3A). Ub-immunoblotting analyses of CYP3A immunoprecipitates from lysates of cells infected with shRNA 1+2 showed a marked accumulation of HMM ubiquitinated CYP3A species, which was further enhanced upon DDEP-induced CYP3A inactivation (Fig. 3B). These findings unequivocally indicated that p97 knockdown indeed stabilized the hepatic content of both the parent and ubiquitinated CYP3A species.
FIGURE 3.
Effects of RNAi-mediated p97 knockdown on hepatic CYP3A content. A, effects of shRNA 1, shRNA 2, and shRNA 3 targeted against hepatic p97 individually or in combination or a control shRNA (Control) on hepatic CYP3A content of cell lysates derived from such shRNA-infected cell cultures. Hepatocytes were infected with each shRNA individually or in combination and then treated with the CYP3A inducer Dex. Cells were harvested on the 8th day of knockdown. A representative example of CYP3A Western immunoblotting analyses of these hepatocyte lysates (50 μg of protein) is shown at the top, with corresponding aliquots used for actin immunoblotting analyses as loading controls. Densitometric quantification of hepatic CYP3A content from three individual experiments (mean ± S.D. (error bars)) is shown at the bottom. Statistical analyses revealed significant differences in hepatic CYP3A content (mean ± S.D.) between control shRNA-infected cells and cells infected with shRNA 1, shRNA 2, or shRNA 1+3 at p < 0.01; shRNA 3 at p < 0.05; shRNA 2+3 at p < 0.001; and shRNA 1+2 at p < 0.0001. Statistically significant differences in hepatic CYP3A protein content observed between the two mean ± S.D. values each marked with the same symbol were as follows. ≠, p < 0.05; ¶, p < 0.01; †, p < 0.0002; ¥, p < 0.001; ±, p < 0.0001; #, p < 0.005; §, p < 0.05; €, p < 0.0001; **, p < 0.01; ‡, p < 0.0002; $, p < 0.001; #, p < 0.005; £, p < 0.001; *, p < 0.01; ●, p < 0.02. B, effects of p97 shRNA 1+2-mediated knockdown (−p97) on hepatic CYP3A ubiquitination monitored on the 8th day post-p97 knockdown relative to control shRNA-infected cells (Control). Some control and −p97 cells were treated with the CYP3A suicide inactivator DDEP for 4 h. CYP3A immunoprecipitates (IP) from lysates derived from DDEP-treated control and −p97 cells at 0 and 4 h were subjected to Ub-immunoblotting analyses (IB) as detailed (see “Experimental Procedures”). A representative immunoblot is shown. C, in situ verification of CYP3A stabilization in cultured rat hepatocytes with confocal immunofluorescence microscopy. Shown are rat hepatocyte cultures infected with the control shRNA (Control), or p97 shRNA 1+2 (−p97) for 7 days. On the 8th day, shRNA-infected rat hepatocyte cultures were fixed and stained with antibodies to CYP3A (green). Data from a representative experiment showing relative CYP3A accumulation are shown. D, functional relevance of hepatic CYP3A stabilization after p97 knockdown (−p97). Rat hepatocyte cultures were infected with the control shRNA or p97 shRNA 1+2 (−p97) for 7 days. On the 8th day, CYP3A functional activity was assayed in intact hepatocytes by assessing their ability to catalyze the 7-O-debenzylation of BFC, a diagnostic CYP3A functional probe, to HFC. The relative HFC formation (μmol of HFC formed/3.5 × 106 cells/h) in the medium was assayed (as detailed under “Experimental Procedures”). Experimental values (mean ± S.D.) from three individual experiments are shown. Statistical analyses revealed significant differences in hepatic CYP3A function between control and −p97 cells at p < 0.01.
In situ confocal immunofluorescence microscopic analyses of hepatocytes infected with the control shRNA or p97 shRNA 1+2 showed no significant effects on cellular morphology following 7 days of infection. Thus, no deleterious cellular effects and no evidence of apoptotic morphology or nuclear hypertrophy and/or other perturbations characteristic of apoptosis upon such an extensive (>90%) endogenous p97 protein knockdown were detected. In fact, the nuclear diameter (14.6 ± 1.44 μm; n = 33 cells) of the p97 knocked down hepatocytes was found to be comparable to that of the corresponding control shRNA-infected cells (14.0 ± 1.41 μm; n = 33 cells). By contrast, the immunofluorescent CYP3A content, consistent with the immunoblotting analyses, was visibly increased in p97 shRNA 1+2-infected cells over the controls (Fig. 3C). Accordingly, quantification of the relative immunofluorescence intensity of a 20-μm2 cytoplasmic area (excluding the nucleus) of control and p97 shRNA 1+2-infected cells yielded values of 47.2 ± 4.8 and 116 ± 6.3 (mean ± S.D.; n = 50 cells), respectively.
To determine whether the stabilized hepatic CYP3A fraction represented a functional CYP3A species or a structurally inactive species already marked for cellular disposal, we examined the functional activity of the CYP3A stabilized after p97 knockdown in cultured rat hepatocytes “in vivo” (Fig. 3D). We therefore monitored the CYP3A-dependent O-debenzylation of BFC, a relatively selective diagnostic probe for CYP3A function, in a fluorescence-based assay (72). A significant increase of HFC, the O-debenzylated BFC metabolite, was detected in the extracellular medium after p97 knockdown relative to corresponding controls at 1 h (Fig. 3D). No corresponding increase in HFC formation was detected in the cell lysates, thereby revealing that most of the HFC formed was exported into the extracellular medium. These findings in intact non-DDEP-treated hepatocytes indicate that the CYP3A stabilization observed upon p97 knockdown is functionally relevant.
A Definitive Role for p97 in the ER Extraction of Hepatic CYP3A
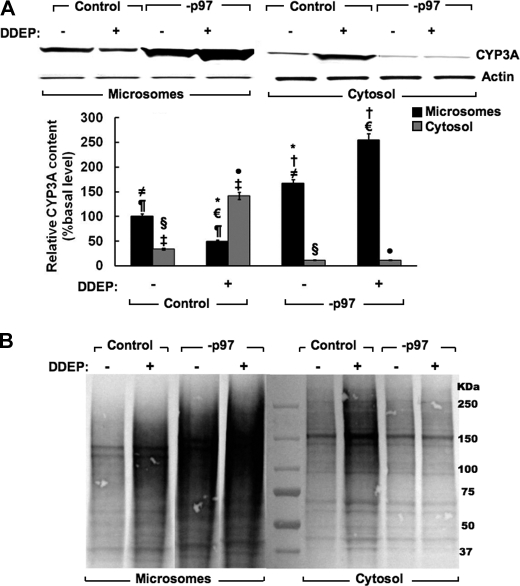
A considerable fraction of the 26 S proteasome is associated with the ER membranes (36, 37), and the relative extent to which the p97 complex and the 26 S proteasome are involved in the extraction of a monotopic ERAD-C substrate, such as hepatic CYP3A, was unclear. Liver microsomal and cytosolic subfractions from control shRNA and shRNA 1+2-infected hepatocytes in culture were therefore examined with or without DDEP treatment (Fig. 4A). In the absence of DDEP treatment, immunoblotting analyses revealed that most of the native CYP3A is in the microsomes, with a small fraction in the cytosol. However, following DDEP treatment for 4 h, CYP3A immunoblotting analyses of microsomes derived from these cells documented the characteristic loss of parent CYP3A protein (∼55 kDa), stemming from its DDEP-induced UPD/ERAD (Fig. 4A). CYP3A immunoblotting analyses of the cytosolic subfraction isolated from these same cells showed a corresponding increase of CYP3A protein, consistent with the loss of ER-bound CYP3A protein species upon its dislocation/extraction from the ER membrane into the cytosol (Fig. 4A). p97 knockdown markedly increased not only the native CYP3A microsomal content (Fig. 4A; −DDEP, lane 3) but also that of the DDEP-inactivated CYP3A species (Fig. 4A; +DDEP, lane 4). By contrast, p97 knockdown significantly abrogated the appearance of any native or DDEP-inactivated parent CYP3A ∼55-kDa species into the cytosol (Fig. 4A, lanes 7 and 8). Corresponding densitometric quantification of the relative CYP3A content after immunoblotting analyses is shown (Fig. 4A, bottom) and supports this assessment. This provided the first clear evidence that p97 was indeed crucial for CYP3A dislocation from the ER.
FIGURE 4.
Effects of hepatic p97 knockdown on CYP3A extraction from the ER into the cytosol. A, cells were infected with control shRNA or p97 shRNA 1+2 (−p97) for 7 days. On the 8th day, cells were treated with DDEP (100 μm; +) or equivalent volume of the vehicle (DMSO; −) for 4 h. Cells were harvested, and microsomes (5 μg of protein) and cytosol (10 μg) derived from cell homogenates were subjected to CYP3A immunoblotting analyses as described (see “Experimental Procedures”). A representative CYP3A immunoblot is shown at the top, and the corresponding densitometric quantification of three individual immunoblots is shown at the bottom. Values are mean ± S.D. (error bars) of immunochemically detected CYP3A content in liver microsomes or cytosol from three individual cell cultures, expressed as a percentage of their corresponding vehicle-treated basal (−DDEP) values. Statistically significant differences in hepatic CYP3A protein content observed between the two mean ± S.D. values each marked with the same symbol were as follows. ≠, p < 0.001; ¶, p < 0.002; †, p < 0.001; ¥, p < 0.001; §, p < 0.01; €, p < 0.0001; ‡, p < 0.001; *, p < 0.001; ●, p < 0.0005. B, corresponding effects of p97 shRNA 1+2-mediated knockdown (−p97) on hepatic CYP3A ubiquitination are shown. Some control and −p97 cells were treated with the CYP3A suicide inactivator DDEP for 4 h. CYP3A immunoprecipitates from microsomes and cytosol derived from DDEP-treated control and −p97 cells at 0 and 4 h were subjected to Ub-immunoblotting analyses as detailed (see “Experimental Procedures”). A representative immunoblot is shown.
However, it was plausible that the CYP3A species accumulating in the cytosol following p97 knockdown was largely ubiquitinated and thus at higher molecular masses not reflected by the parent CYP3A 55 kDa band. We therefore subjected CYP3A immunoprecipitates from the same microsomal and cytosolic subfractions examined in Fig. 4A to Ub-immunoblotting analyses (Fig. 4B). These findings revealed that indeed, in cells infected just with the control shRNA and no other treatment, a considerable fraction of CYP3A was found ubiquitinated both in the microsomes and cytosol (Fig. 4B). DDEP treatment, as expected, further increased the extent of this ubiquitination in both cellular compartments (Fig. 4B). However, upon p97 knockdown, the fraction of both the ubiquitinated native and DDEP-inactivated HMM CYP3A species dramatically increased in the microsomes while visibly declining in the cytosol (Fig. 4B). Together, these findings are entirely consistent not only with a major role for p97 in the dislocation of the parent as well as the ubiquitinated CYP3A species into the cytosol, but also with the ubiquitination of CYP3A occurring to a substantial degree while the protein is still in the ER by the ER-integral Ub ligase gp78 and/or cytosolic CHIP (33–35).
p97 Rather than the 26 S Proteasome Plays a Major Role in the ER Extraction of Hepatic CYP3A
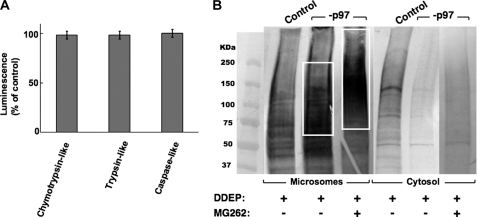
The above findings were surprising because hepatocytes with p97 specifically knocked down still retained their full 26 S proteasomal complement. Indeed, a cell-based bioluminescent assay monitoring three proteasomal ATPase-independent proteolytic activities (trypsin-like, chymotrypsin-like, and caspase-like, corresponding to 20 S proteasomal β2, β5, and β1 subunits (73)) indicated no significant differences in proteasomal function between the control shRNA-infected cells and p97 shRNA 1+2-infected cells (Fig. 5A and supplemental Fig. S1). Thus, the functionally intact 26 S proteasome could have compensated for such a major loss of p97 function by both dislocating the ubiquitinated CYP3A and degrading it. As such, if all of the HMM ubiquitinated CYP3A species extracted by the 26 S proteasome into the cytosol were to be promptly degraded by it, it would be difficult to assess the specific contribution of p97 relative to that of the 26 S proteasome to the ER extraction of ubiquitinated CYP3A. We therefore examined the effect of blocking such proteasomal degradation were it to occur, by concomitant treatment of the cells with MG262, an effective inhibitor of 20 S proteasomal Thr proteases (74). Cultured hepatocytes were infected with either the control shRNA or with p97 shRNA 1+2 for 7 days, as detailed above. On the 8th day, they were treated at time 0 with MG262 (10 μm), followed 1 h later by DDEP. After 4 h of DDEP treatment, control and MG262-treated cells were harvested, and microsomal and cytosolic subfractions were subjected to CYP3A immunoprecipitation (see “Experimental Procedures”) (Fig. 5B). Ub-immunoblotting analyses of CYP3A immunoprecipitates derived from the microsomal and cytosolic subfractions from DDEP-treated control cells once again exhibited the pronounced CYP3A ubiquitination seen in Fig. 4B, consistent with DDEP-mediated enhancement of CYP3A ERAD/UPD (Fig. 5B). Furthermore, as also documented in Fig. 4B, the dislocation of HMM ubiquitinated CYP3A protein species into the cytosol was markedly diminished upon p97 knockdown (Fig. 5B). However, when the hepatic proteasomal (proteolytic) function was concomitantly inhibited by MG262 treatment of hepatocytes, the level of ubiquitinated CYP3A species in the cytosol upon p97 knockdown did not significantly increase. In fact, it remained considerably lower than that in the cytosol of corresponding DDEP-treated control cells containing a functional p97 (Fig. 5B). On the other hand, the level of HMM ubiquitinated CYP3A species in the microsomal fraction not only increased upon p97 knockdown, consistent with the findings depicted in Fig. 4B, but CYP3A ubiquitination also was found to extend to an even higher molecular mass range with the concomitant MG262-mediated proteasomal inhibition (Fig. 5B). These findings thus reveal not only that p97 rather than the 26 S proteasome plays a major role in CYP3A dislocation but also that, upon concomitant inhibition of the proteasomal function, the CYP3A species accumulating in the ER, undergoes further ubiquitination to higher molecular mass species.
FIGURE 5.
Relative contribution of p97 versus the 26 S proteasome to the extraction of ubiquitinated CYP3A from the ER. A, assessment of the effects of p97 knockdown on hepatic proteasomal function. Cells were infected with control shRNA or p97 shRNA 1+2 (−p97) for 7 days, and on the 8th day they were examined with the specific probes for the proteasomal chymotrypsin-like, trypsin-like, and caspase-like activities in a cell-based bioluminescent assay as detailed (see “Experimental Procedures”). Values are mean ± S.D. (error bars) relative luminescence units derived from three separate −p97 cultures and expressed as a percentage of the values for corresponding activities in control shRNA-treated cells. No statistically significant differences were observed between activities from control and −p97 cells. B, cells were infected with control shRNA or p97 shRNA 1+2 (−p97) for 7 days and then on the 8th day treated with or without DDEP for 4 h as described in the legend to Fig. 4. Some DDEP-treated cells were also treated concomitantly with the proteasomal inhibitor, MG262. Microsomes or cytosol were isolated and subjected to CYP3A immunoprecipitation. The corresponding CYP3A immunoprecipitates from microsomes or cytosol were subjected to Ub-immunoblotting analyses and development with alkaline phosphatase-conjugated secondary antibody as detailed (see “Experimental Procedures”). A representative immunoblot is shown.
CYP3A Species Accumulating in the ER upon p97 Knockdown Is Largely Anchored to the ER
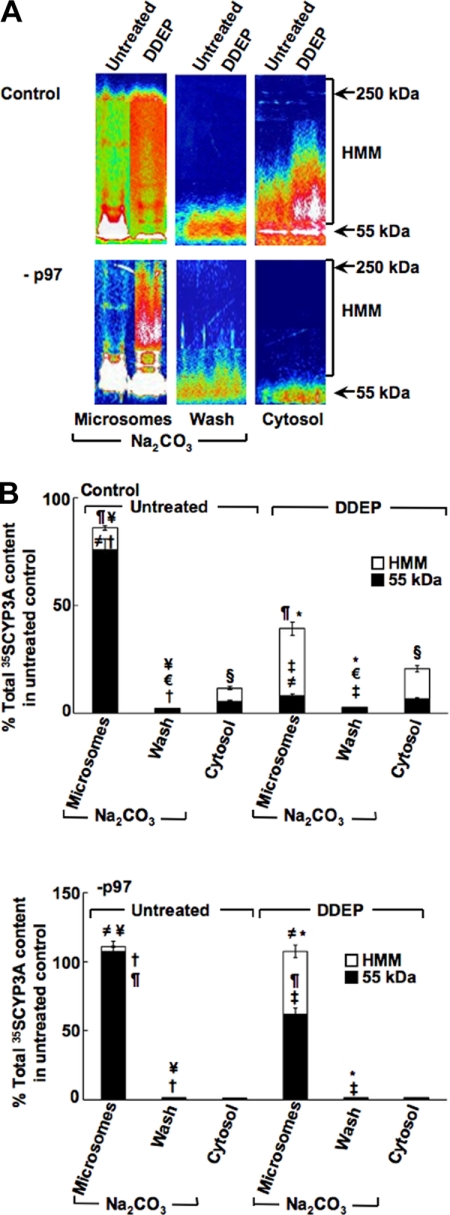
CYPs 3A, in common with most hepatic microsomal P450s, are highly hydrophobic proteins that, even after cleavage from their ER membrane anchor, tend to partition into the microsomal subfraction upon ultracentrifugation (4, 5). We therefore examined the relative extent of CYP3A (i) truly anchored to the ER membrane and thus not removed by a 0.1 m Na2CO3 wash of the liver microsomes; (ii) solubilized by the 0.1 m Na2CO3 wash of the liver microsomes and thus representing the CYP3A species dislocated from the ER membrane but still loosely associated with its external surface; and (iii) in the cytosolic subfraction representing the CYP3A species dislocated from the ER membrane and truly extracted into the cytosol (Fig. 6A). To enhance CYP3A detection, the control shRNA-infected and shRNA 1+2-infected cells were first pulse-chased with [35S]Met/Cys for 1 h and then, following chase with unlabeled Met/Cys, treated for 4 h with DDEP. Cells were then harvested, and the various organellar subfractions were prepared. [35S]CYP3A immunoprecipitated from each of these subfractions was subjected to SDS-PAGE and PhosphorImager scanning and ImageQuant analyses of the dried gels as detailed (see “Experimental Procedures”) (Fig. 6A).
FIGURE 6.
Relative intracellular localization of parent and ubiquitinated HMM CYP3A species after p97 knockdown in cultured hepatocytes. Rat hepatocyte cultures were infected with control shRNA or p97 shRNA 1+2 (−p97) for 7 days. On the 8th day, they were subjected to 35S-pulse-chase analyses. Two h after cold chase, some cells were treated with or without DDEP for 4 h as described in the legend to Fig. 4. Cells were harvested, and homogenates were subfractionated into cytosol and microsomes. CYP3A immunoprecipitates (45 μl) from cytosol (Cytosol), Na2CO3-washed microsomes (Microsomes), and wash corresponding to the Na2CO3-solubilized material (Wash) were obtained as described under “Experimental Procedures” and subjected to SDS-PAGE analyses. The gels were dried and then exposed to PhosphorImager screens and visualized using a Typhoon scanner. A, a typical SDS-PAGE gel is shown as a representative of corresponding CYP3A immunoprecipitates from pooled hepatocyte cultures. The color wheel intensity code is as follows: white > magenta > red > orange > yellow > green > light blue > dark blue > black. B, the corresponding relative [35S]CYP3A intensity of the parent CYP3A (55–60 kDa), Na2CO3-solubilized (dislocated), and the HMM ubiquitinated CYP3A species between 65 and 250 kDa in each lane were quantified using ImageQuant software. Values are mean ± S.D. (error bars) of three individual determinations and are expressed as a percentage of the total (parent, HMM, and dislocated) [35S]CYP3A protein content in untreated control cells. Statistically significant differences in hepatic CYP3A protein content observed between the two mean ± S.D. values each marked with the same symbol were as follows. Top, ≠, p < 0.0005; ¶, p < 0.001; †, p < 0.0001; ¥, p < 0.001; ‡, p < 0.01; §, p < 0.01; *, p < 0.0005; €, not significant. Bottom, ≠, p < 0.0005; ¶, p < 0.005; †, p < 0.0001; *, p < 0.0001; ‡, p < 0.0005; ¥, not significant.
These combined analyses of [35S]CYP3A immunoprecipitates revealed that, in untreated control shRNA-infected hepatocytes, [35S]CYP3A is initially largely present as an integral 55-kDa ER protein with minimal ubiquitination detectable as the HMM species ranging between 65 and 250 kDa (Fig. 6A, top). A smaller fraction that includes both parent (55 kDa) and HMM ubiquitinated [35S]CYP3A species was found in the cytosol, and an even smaller fraction was found to be dislocated from the ER but loosely bound to it. The latter fraction recovered in the 0.1 m Na2CO3 microsomal wash was trichloroacetic acid-precipitable and immunoprecipitable. However, after 4 h of DDEP treatment, the ER-integral parent [35S]CYP3A species rapidly declined, whereas the corresponding HMM [35S]CYP3A species increased, consistent with DDEP-mediated structural inactivation and subsequent ubiquitination of the protein while still anchored to the ER membrane (Fig. 6A, top). CYP3A immunoprecipitation analyses of the corresponding cytosolic subfraction also revealed some parent species but an increase of the ubiquitinated [35S]CYP3A species, consistent with a substantial fraction of the DDEP-inactivated P450 protein being extracted into the cytosol in the presence of a functional p97 (Fig. 6A, top). The Na2CO3 wash after 4 h of DDEP treatment revealed no appreciable increase in [35S]CYP3A fraction that remained ER-associated upon dislocation (Fig. 6A, top). These findings confirm our previous findings that DDEP-mediated CYP3A inactivation enhances its ubiquitination and dislocation from the ER membrane into the cytosol, wherein it is normally degraded by the 26 S proteasome (13).
On the other hand, following p97 knockdown of untreated hepatocytes, the parent [35S]CYP3A species firmly anchored to the ER membrane markedly increased, whereas the corresponding [35S]CYP3A species in the cytosol and that dislocated from the ER membrane into the Na2CO3 wash significantly declined (Fig. 6A, bottom). After a 4-h treatment of these cells with DDEP, CYP3A inactivation once again resulted in protein ubiquitination, as documented by the HMM [35S]CYP3A species, but these ubiquitinated [35S]CYP3A species remained firmly bound to the ER. Furthermore, unlike the findings in control cells with their full p97 complement, following p97 knockdown, almost no parent and/or HMM ubiquitinated [35S]CYP3A species were dislocated and escaped into the cytosol (Fig. 6A, bottom). The relative quantitative differences in [35S]CYP3A trafficking as parent and ubiquitinated species from the ER into the cytosol with and without p97 knockdown were verified by our PhosphorImager/ImageQuant analyses (Fig. 6B). Collectively, these findings attest to p97 indeed playing an obligatory role in the ER extraction of the monotopic CYP3A protein. The corresponding ubiquitination profiles of hepatic protein in microsomes, cytosol, and Na2CO3 wash reveal a similar abrogation of ubiquitinated hepatic protein extraction into the cytosol with a corresponding accumulation of ubiquitinated hepatic proteins in the ER, thereby confirming the key role of p97 in the ERAD of other hepatic ER proteins (supplemental Fig. S2).
DISCUSSION
Collectively, our data suggest that despite the topological accessibility of ER-anchored CYPs 3A to the cytosolic 26 S proteasome, a functional p97 is essential for the extraction of both the native and DDEP-inactivated CYPs 3A from the ER membrane into the cytosol. This extraction entails the dislocation of the intact CYP3A proteins, including their N-terminal ER membrane signal anchor, into the cytosol. However, not all ERAD substrates require p97 for their extraction/retrotranslocation/delivery to the cytosol for their cellular turnover because some are efficiently extracted by the functionally similar AAA ATPases of the PA700 (19 S) proteasomal cap (75–82). Our findings (Fig. 5B) reveal that this is clearly not the case with CYPs 3A, which require p97 in a nearly obligatory manner. Upon p97 knockdown and inhibition of the 20 S proteasomal proteases by MG262, parent and/or ubiquitinated CYP3A would be expected to accumulate in the cytosol if PA700 AAA ATPases were significantly involved in their extraction, which did not occur (Fig. 5B). Thus, despite a functional 26 S proteasomal complement, unaffected by the p97 knockdown (Fig. 5A and supplemental Fig. S1), and its documented role in the extraction of other integral/luminal ER proteins (75–82), p97 knockdown virtually shuts down CYP3A extraction from the ER membrane and their subsequent proteasomal degradation.
p97 is an abundant cytosolic AAA ATPase chaperone that works in concert with its cochaperone adapters Npl4 and Ufd1 as a heterotrimeric complex in the retrotranslocation/extraction of integral/luminal ER proteins (21, 39, 40, 42–56). This p97 complex-mediated ERAD function is essentially energy-driven by ATP hydrolysis (45, 49). Although we have not directly addressed the specific roles of its Npl4 and Ufd1 adapters in the present study, our previous findings have revealed that, along with p97, both of these adapters were also critically required for the ERAD of heterologously expressed CYP3A4 in yeast (19). We therefore infer by analogy to yeast that in rat hepatocytes, an intact functional p97-Npl4-Ufd1 complex is similarly involved in the delivery of ER-anchored CYP3A to the cytosolic 26 S proteasome for its subsequent degradation and that this process too is also critically dependent on ATP-derived energy. Taken together with our previously published findings (11–17), the present findings underscore that both the native and DDEP-inactivated CYP3A species are genuine ERAD-C substrates, subscribing to all of the requisite criteria of an ERAD-C process (29–31).
The findings above also reveal that CYP3A ubiquitination occurs to a substantial degree in the ER while the P450 protein is still tethered to the ER membrane. CYP3A ubiquitination thus precedes and to a large extent can be independent of CYP3A extraction into the cytosol. In vitro CYP3A4 ubiquitination analyses with functionally reconstituted recombinant mammalian enzymes have identified two effective E2-E3 systems: (i) UBC7 (an ERAD-associated Ub-conjugating enzyme) and gp78 (a polytopic ER-integral E3 Ub ligase) and (ii) UbcH5a (a Ub-conjugating enzyme) and CHIP (a cytosolic E3 Ub ligase) that acts in concert with Hsp70/Hsp40 chaperones (33–35). Our in vivo RNAi analyses targeted against either of these Ub ligases in cultured rat hepatocytes not only have confirmed this dual involvement but have also revealed that, physiologically, a substantial cellular fraction of native (non-DDEP-inactivated) CYP3A that is targeted by both of these E2-E3 systems is functionally active (35). These findings are fully congruent with our present findings of p97 RNAi, wherein a substantial fraction of hepatic CYP3A content is also found to be functionally active upon p97 knockdown (Fig. 3D). This is consistent with the relative increase in the content of the unmodified parent [35S]CYP3A (55-kDa species) observed upon p97 knockdown in microsomes (Fig. 6A).
Parallel assessment of the relative immunodetectable CYP3A content in cultured hepatocytes following RNAi targeted against hepatic p97, gp78, or CHIP, through quantification of the relative immunofluorescence intensity of a 20-μm2 cytoplasmic area by confocal immunofluorescence microscopic analyses revealed values (mean ± S.D.) of 116 ± 6.3, 84.5 ± 4.3, and 69.4 ± 3.6 versus 47.2 ± 4.8 for corresponding control shRNA-infected hepatocytes (Fig. 3C) (35). These findings suggest that CYP3A accumulation in hepatocytes was considerably higher after p97 knockdown than after knockdown of either gp78 or CHIP Ub ligase. They are consistent with the finding that p97 knockdown led to the ER accumulation of both the parent and ubiquitinated CYP3A species (Fig. 4). However, despite an otherwise effective ER protein ubiquitination process and no significant impairment of either gp78 or CHIP Ub ligase (Fig. 2C), this fraction of functionally active CYP3A protein accumulating after p97 knockdown was apparently comparable to that stabilized by the RNAi knockdown of either gp78 or CHIP (35). This suggests that either (i) the p97 complex can directly target both the parent non-ubiquitinated and ubiquitinated CYP3A species, consistent with its known targeting of both the ubiquitinated and the non-ubiquitinated domains of ERAD substrates (45, 47), or (ii) functionally active parent CYP3A accumulates upon p97 knockdown because it is possibly spared Ub modification due to exhaustion of available free Ub stores as follows: p97 knockdown impairs the extraction/retrotranslocation into the cytosol of all ubiquitinated ER proteins (including CYPs 3A) that are p97-dependent (supplemental Fig. S2). This ER accumulation of ubiquitinated proteins coupled with their impaired 26 S proteasomal processing would concurrently reduce recycling of poly-Ub chains required for fresh cycles of protein ubiquitination. In turn, this would spare a fraction of ER proteins, including CYPs 3A, from being normally de novo ubiquitinated, thereby retaining this population in the native functionally active form. Although in the case of other ER-integral proteins, such as the inositol 1,4,5-triphosphate receptors and Ste6p*, evidence has been provided that p97 targets only the ubiquitinated species (26, 50) and thus acts postubiquitination but before proteasomal degradation of these ERAD substrates, this remains to be conclusively established in the case of p97 targeting of CYPs 3A. Our findings (Fig. 6A) revealing the dislocation of the unmodified parent [35S]CYP3A (55-kDa) species into the cytosol and to the external surface of the ER membrane (Na2CO3-wash) in the presence of a fully functional p97 complement suggest that protein ubiquitination may not be absolutely essential for the extraction of all CYP3A molecules.4
We also find it noteworthy that, upon p97 knockdown, a substantial fraction of DDEP-inactivated CYP3A species not only is similarly ubiquitinated while firmly ER-anchored and accumulates in this form, but also its ubiquitination is considerably enhanced and extends to an even higher molecular mass range upon MG262-mediated proteasomal inhibition (Fig. 5B). Such enhanced CYP3A ubiquitination may result from the delay in CYP3A dislocation, thereby providing additional time for the elaboration of longer Ub chains and/or the appendage of Ub chains on additional CYP3A residues.
Together, these findings are entirely consistent not only with a major role for p97 in the dislocation of the parent as well as the ubiquitinated CYP3A species into the cytosol, but also with CYP3A ubiquitination by the ERAD-associated Ub ligases gp78 and CHIP occurring while the proteins are still tethered to the ER membrane (35). The functional involvement of both gp78 and p97 in CYP3A UPD/ERAD is fully consistent with the intimate structural-functional association of these two proteins that effectively couples the ubiquitination of numerous integral and luminal ER proteins to their ER retrotranslocation/dislocation during their ERAD (49, 58, 83–86). Accordingly, the ER polytopic gp78 Ub ligase protein is proposed to contain a p97-docking domain, the VIM, in its cytosolic C terminus that serves to recruit p97 to its ER-bound ubiquitinated substrate5 (58, 83–85). Whether a similar direct functional association exists between p97 and CHIP remains to be established. Some evidence exists in support of such a functional p97-CHIP association (87, 88).
The ER also contains the monotopic VCP-interacting membrane protein (VIMP), which is proposed to recruit p97 to the ER membrane and, together with p97, to function in a hetero-oligomeric complex with Derlin-1, another polytopic homo-oligomeric ER membrane protein (46, 49). Together, this p97-VIMP-Derlin-1 complex is reportedly involved in the retrotranslocation of misfolded and luminal ER proteins across the ER membrane into the cytosol for delivery to the 26 S proteasome (49). Whether Derlin-1 and/or VIMP are also involved in CYP3A ERAD remains to be determined. The direct interaction of p97 with gp78 as well as other ERAD-associated Ub ligases (i.e. Hrd1 and CHIP) purportedly enables p97 recruitment of these E3s to the ER substrate in question within the Derlin-1 translocation channel in the ER membrane. However, our findings indicate that this is not absolutely essential for CYP3A ERAD because CYPs 3A are quite efficiently ubiquitinated in the ER following such an extensive p97 knockdown (Figs. 4B, 5B, and 6A).
p97 is known to function in diverse cellular processes, including homotypic membrane fusion, vesicular transport, ERAD, autophagic lysosomal degradation and UFD-mediated degradation of cytosolic proteins, such as IκBα (55–61). As such, it is thought to interact with >30 different cellular proteins (61). Thus, its knockdown is expected to affect a myriad of physiologically relevant cellular processes.
To our knowledge, this is perhaps the most extensive (>90%) cellular knockdown of this abundant cytosolic p97 protein reported in the literature. Given the vast array of cellular functions dependent on p97 (55–61), we find it both surprising and remarkable that our confocal immunofluorescence microscopic analyses attested to relatively healthy viable hepatocytes with no disruption of their cellular morphology or any detectable cell necrosis after 7 days of such an extensive p97 knockdown (Fig. 3C). No evidence of extensive cytoplasmic vacuolization, ER stress and expansion, impaired cell function, cell death, and/or apoptosis was found (Fig. 3C), as reported in neuronal PC12 cells expressing a dominant negative p97 (89) or HeLa cells transiently transfected with synthetic siRNAs that knocked down p97 by ∼80% (60, 61). For comparison, corresponding analyses of similarly cultured hepatocytes treated with the proteasomal inhibitor MG-132 (300 μm), a concentration known to induce ER stress as determined by the activation and/or induction of hepatic ER stress markers PERK and GCN2 eIF2α kinases as well as the elevation of hepatic Grp78/Bip protein levels (66), are included (supplemental Fig. S3). qRT-PCR analyses of total RNA from control and p97-knocked down hepatocytes revealed no significant increases in hepatic Grp78 and Grp94 mRNA content (Fig. 2C), sensitive indices of a full-blown unfolded protein response/ER stress response (90). Furthermore, none of the prototypic morphologic hallmarks of apoptosis, such as chromatin condensation, collapse, or budding and/or significant nuclear shrinkage or fragmentation were detected at this time (Fig. 3C), in contrast to those observed in cultured hepatocytes treated with high MG132 concentrations (300 μm; supplemental Fig. S3) (66). Indeed, not only were the nuclei morphologically similar to control nuclei, but also the nuclear size of the p97-knocked down hepatocytes was comparable to that of the corresponding control shRNA-treated hepatocytes (Fig. 3C). In addition, cellular processes, such as protein ubiquitination (including that of CYPs 3A) (supplemental Fig. S2), CYP3A synthesis,6 and functional activity (Fig. 3D), and proteasomal function (Fig. 5A) were unaffected following this extensive 7-day p97 knockdown. Thus, it appears that such an extensive knockdown, albeit impairing hepatic ERAD/UPD of CYP3A proteins and resulting in the accumulation of ubiquitinated proteins, is relatively well tolerated by the hepatocyte. Accordingly, just ∼10% of functional hepatic p97 protein content apparently is sufficient and compatible with cellular survival and health of sandwich-cultured primary rat hepatocytes at least for a day or so after the 7-day p97 knockdown. Thus, primary hepatocytes appear to be strikingly more tolerant of extensive p97 knockdown than cell lines, and whether this tolerance extends to other primary cells remains to be determined. Alternatively, the lentiviral approach of shRNA delivery that entails integration of the silencing sequence into the hepatocyte chromosome, albeit slower in producing the desired knockdown, may enable the cells to adjust more gradually to the changes in their intracellular milieu and thus to better withstand the cellular stress of p97 knockdown.
p97 knockdown of this extent clearly impairs CYP3A ERAD, resulting in the stabilization of a functionally active CYP3A, which in the human liver could lead to clinically relevant drug-drug interactions. However, it remains to be determined whether a less severe p97 impairment due to genetic polymorphisms or faulty expression of allelic variants is at all encountered clinically and would influence P450-dependent function and consequent drug-drug interactions. The expression of a p97 mutant with dominant negative mutations in the catalytic domains of both of its ATPase subunits is indeed capable of impairing UPD (45, 89). Thus, impaired CYP3A ERAD may be of clinical relevance in individuals with p97 proteins with functional defects in their ATPase domains and/or gp78/CHIP-interacting domains if such defects were not embryonically lethal.
In summary, our findings reveal that p97 plays a critical physiological role in CYP3A ERAD by extracting the CYPs 3A from the ER membrane into the cytosol and subsequently delivering them to the 26 S proteasome for destruction. p97 knockdown via RNAi abrogates this process and results in the accumulation of functionally active CYP3A species as well as extensively ubiquitinated species tethered to the ER membrane.
Supplementary Material
Acknowledgments
We gratefully thank Chris Her (University of California San Francisco (UCSF) Liver Center Cell and Tissue Biology Core Facility) (Dr. J. J. Maher, Director) for hepatocyte isolation and Ahalya Sriskandarajan (UCSF) for able technical assistance in some of these studies. We also thank Dr. Alex Y. So (Keith Yamamoto laboratory, UCSF) for generous advice and training in the use of lentiviral shRNAi technology and Dr. Michelle Flanniken (Raul Andino laboratory, UCSF) for technical advice. We also acknowledge the UCSF Liver Center Core on Cell and Tissue Biology (supported by NIDDK, National Institutes of Health, Grant P30DK26743).
This work was supported, in whole or in part, by National Institutes of Health Grants DK26506 and GM44037.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
By “native” we mean the CYP3A/P450 enzyme that has not been structurally and/or functionally inactivated by any exogenous mechanism-based inactivator, such as DDEP.
Although our data are consistent with both ubiquitinated and the parent species being extracted out of the ER (microsomes) by p97, our recent findings (35) reveal that such extraction of the parent species requires CHIP E3 Ub ligase as well. Knockdown of CHIP but not gp78, even in the presence of a fully functional p97, virtually abolishes such parent CYP3A extraction into the cytosol (35). The inclusion of N-ethylmaleimide (5 mm), an inhibitor of deubiquitinating enzymes, in our harvesting/lysis buffer should exclude the possibility that this cytosolic “parent” CYP3A species stems from CYP3A deubiquitination following lysis of the cells.
It is noteworthy, nevertheless, that mutation of gp78 residue 611 in this domain that reportedly abolishes gp78 binding to p97 (58) failed to abolish the degradation of gp78 itself and of its heterologous substrate CD3-δ (41).
This is deduced from our qRT-PCR analyses indicating that p97 knockdown failed to affect hepatic CYP3A mRNA levels (Fig. 2B) and our observation that the [35S]radioactivity (cpm/mg of lysate protein/h) incorporated into CYP3A immunoprecipitates (Fig. 6) at time 0 h following pulse-chase was equivalent: 77,434 ± 2560 (control) versus 77,000 ± 2554 (−p97).
- P450
- cytochrome P450
- CYPs 3A
- P450s of the hepatic CYP3A subfamily
- AMFR
- autocrine motility factor receptor
- CHIP
- C terminus of Hsp70-interacting protein
- BFC
- 7-benzyloxy-4-trifluoromethylcoumarin
- DDEP
- 3,5-dicarbethoxy-2,6-dimethyl-4-ethyl-1,4-dihydropyridine
- Dex
- dexamethasone
- DOC
- deoxycholate
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- HFC
- 7-hydroxy-4-trifluoromethylcoumarin
- HMM
- high molecular mass
- Ub
- ubiquitin
- UPD
- Ub-dependent proteasomal degradation
- VIMP
- VCP-interacting membrane protein
- −p97
- p97 knockdown
- NT
- N-terminal
- qRT-PCR
- quantitative RT-PCR
- 8-MAP
- multiple antigen octapeptide.
REFERENCES
- 1. Guengerich F. P. (2005) Cytochrome P450: Structure, Mechanism and Biochemistry (Ortiz de Montellano P. ed) pp. 377–530, Kluwer-Academic/Plenum Press, New York [Google Scholar]
- 2. Sakaguchi M., Mihara K., Sato R. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 3361–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Lemos-Chiarandini C., Frey A. B., Sabatini D. D., Kreibich G. (1987) J. Cell Biol. 104, 209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vergères G., Winterhalter K. H., Richter C. (1989) Biochemistry 28, 3650–3655 [DOI] [PubMed] [Google Scholar]
- 5. Brown C. A., Black S. D. (1989) J. Biol. Chem. 264, 4442–4449 [PubMed] [Google Scholar]
- 6. Sato T., Sakaguchi M., Mihara K., Omura T. (1990) EMBO J. 9, 2391–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards R. J., Murray B. P., Singleton A. M., Boobis A. R. (1991) Biochemistry 30, 71–76 [DOI] [PubMed] [Google Scholar]
- 8. Kemper B., Szczesna-Skorupa E. (1989) Drug Metab. Rev. 20, 811–820 [DOI] [PubMed] [Google Scholar]
- 9. Sakaguchi M., Omura T. (1993) Front. Biotransform. 8, 59–72 [Google Scholar]
- 10. Szczesna-Skorupa E., Kemper B. (2008) Expert Opin. Drug Metab. Toxicol. 4, 123–136 [DOI] [PubMed] [Google Scholar]
- 11. Correia M. A., Decker C., Sugiyama K., Caldera P., Bornheim L., Wrighton S. A., Rettie A. E., Trager W. F. (1987) Arch. Biochem. Biophys. 258, 436–451 [DOI] [PubMed] [Google Scholar]
- 12. Correia M. A., Davoll S. H., Wrighton S. A., Thomas P. E. (1992) Arch. Biochem. Biophys. 297, 228–238 [DOI] [PubMed] [Google Scholar]
- 13. Wang H. F., Figueiredo Pereira M. E., Correia M. A. (1999) Arch. Biochem. Biophys. 365, 45–53 [DOI] [PubMed] [Google Scholar]
- 14. Faouzi S., Medzihradszky K. F., Hefner C., Maher J. J., Correia M. A. (2007) Biochemistry 46, 7793–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Korsmeyer K. K., Davoll S., Figueiredo-Pereira M. E., Correia M. A. (1999) Arch. Biochem. Biophys. 365, 31–44 [DOI] [PubMed] [Google Scholar]
- 16. Correia M. A. (2003) Drug Metab. Rev. 35, 107–143 [DOI] [PubMed] [Google Scholar]
- 17. Correia M. A., Sadeghi S., Mundo-Paredes E. (2005) Annu. Rev. Pharmacol. Toxicol. 45, 439–464 [DOI] [PubMed] [Google Scholar]
- 18. Murray B. P., Correia M. A. (2001) Arch. Biochem. Biophys. 393, 106–116 [DOI] [PubMed] [Google Scholar]
- 19. Liao M., Faouzi S., Karyakin A., Correia M. A. (2006) Mol. Pharmacol. 69, 1897–1904 [DOI] [PubMed] [Google Scholar]
- 20. Correia M. A., Liao M. (2007) Expert Opin. Drug Metab. Toxicol. 3, 33–49 [DOI] [PubMed] [Google Scholar]
- 21. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 22. Hampton R. Y. (2002) Curr. Opin. Cell Biol. 14, 476–482 [DOI] [PubMed] [Google Scholar]
- 23. Römisch K. (2005) Annu. Rev. Cell Dev. Biol. 21, 435–456 [DOI] [PubMed] [Google Scholar]
- 24. Raasi S., Wolf D. H. (2007) Semin. Cell Dev. Biol. 18, 780–791 [DOI] [PubMed] [Google Scholar]
- 25. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. (2008) Cell 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirsch C., Gauss R., Horn S. C., Neuber O., Sommer T. (2009) Nature 458, 453–460 [DOI] [PubMed] [Google Scholar]
- 29. Taxis C., Hitt R., Park S. H., Deak P. M., Kostova Z., Wolf D. H. (2003) J. Biol. Chem. 278, 35903–35913 [DOI] [PubMed] [Google Scholar]
- 30. Vashist S., Ng D. T. (2004) J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahner A., Brodsky J. L. (2004) Trends Cell Biol. 14, 474–478 [DOI] [PubMed] [Google Scholar]
- 32. Wang X., Medzihradszky K. F., Maltby D., Correia M. A. (2001) Biochemistry 40, 11318–11326 [DOI] [PubMed] [Google Scholar]
- 33. Wang Y., Liao M., Hoe N., Acharya P., Deng C., Krutchinsky A. N., Correia M. A. (2009) J. Biol. Chem. 284, 5671–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pabarcus M. K., Hoe N., Sadeghi S., Patterson C., Wiertz E., Correia M. A. (2009) Arch. Biochem. Biophys. 483, 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim S. M., Acharya P., Engel J. C., Correia M. A. (2010) J. Biol. Chem. 285, 35866–35877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rivett A. J., Palmer A., Knecht E. (1992) J. Histochem. Cytochem. 40, 1165–1172 [DOI] [PubMed] [Google Scholar]
- 37. Enenkel C., Lehmann A., Kloetzel P. M. (1998) EMBO J. 17, 6144–6154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dai R. M., Li C. C. (2001) Nat. Cell Biol. 3, 740–744 [DOI] [PubMed] [Google Scholar]
- 39. Ye Y., Meyer H. H., Rapoport T. A. (2001) Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 40. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bays N. W., Hampton R. Y. (2002) Curr. Biol. 12, R366–R371 [DOI] [PubMed] [Google Scholar]
- 43. Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 44. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ye Y., Meyer H. H., Rapoport T. A. (2003) J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 47. Elkabetz Y., Shapira I., Rabinovich E., Bar-Nun S. (2004) J. Biol. Chem. 279, 3980–3989 [DOI] [PubMed] [Google Scholar]
- 48. Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 49. Ye Y., Shibata Y., Kikkert M., van Voorden S., Wiertz E., Rapoport T. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14132–14138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alzayady K. J., Panning M. M., Kelley G. G., Wojcikiewicz R. J. (2005) J. Biol. Chem. 280, 34530–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bar-Nun S. (2005) Curr. Top. Microbiol. Immunol. 300, 95–125 [DOI] [PubMed] [Google Scholar]
- 52. Elsasser S., Finley D. (2005) Nat. Cell Biol. 7, 742–749 [DOI] [PubMed] [Google Scholar]
- 53. Leichner G. S., Avner R., Harats D., Roitelman J. (2009) Mol. Biol. Cell 20, 3330–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ikeda Y., Demartino G. N., Brown M. S., Lee J. N., Goldstein J. L., Ye J. (2009) J. Biol. Chem. 284, 34889–34900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jentsch S., Rumpf S. (2007) Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 57. Woodman P. G. (2003) J. Cell Sci. 116, 4283–4290 [DOI] [PubMed] [Google Scholar]
- 58. Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 59. Dreveny I., Pye V. E., Beuron F., Briggs L. C., Isaacson R. L., Matthews S. J., McKeown C., Yuan X., Zhang X., Freemont P. S. (2004) Biochem. Soc. Trans. 32, 715–720 [DOI] [PubMed] [Google Scholar]
- 60. Wójcik C., Yano M., DeMartino G. N. (2004) J. Cell Sci. 117, 281–292 [DOI] [PubMed] [Google Scholar]
- 61. Wójcik C., Rowicka M., Kudlicki A., Nowis D., McConnell E., Kujawa M., DeMartino G. N. (2006) Mol. Biol. Cell 17, 4606–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huyton T., Pye V. E., Briggs L. C., Flynn T. C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P. S. (2003) J. Struct. Biol. 144, 337–348 [DOI] [PubMed] [Google Scholar]
- 63. Isaacson R. L., Pye V. E., Simpson P., Meyer H. H., Zhang X., Freemont P. S., Matthews S. (2007) J. Biol. Chem. 282, 21361–21369 [DOI] [PubMed] [Google Scholar]
- 64. Davies J. M., Brunger A. T., Weis W. I. (2008) Structure 16, 715–726 [DOI] [PubMed] [Google Scholar]
- 65. He K., Bornheim L. M., Falick A. M., Maltby D., Yin H., Correia M. A. (1998) Biochemistry 37, 17448–17457 [DOI] [PubMed] [Google Scholar]
- 66. Acharya P., Engel J. C., Correia M. A. (2009) Mol. Pharmacol. 76, 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67. Acharya P., Chen J. J., Correia M. A. (2010) Mol. Pharmacol. 77, 575–592 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Geraerts M., Willems S., Baekelandt V., Debyser Z., Gijsbers R. (2006) BMC Biotechnol. 6, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Han X. M., Lee G., Hefner C., Maher J. J., Correia M. A. (2005) J. Pharmacol. Exp. Ther. 314, 128–138 [DOI] [PubMed] [Google Scholar]
- 70. LeCluyse E., Bullock P., Madan A., Carroll K., Parkinson A. (1999) Drug Metab. Dispos. 27, 909–915 [PubMed] [Google Scholar]
- 71. Chefalo P. J., Oh J., Rafie-Kolpin M., Kan B., Chen J. J. (1998) Eur. J. Biochem. 258, 820–830 [DOI] [PubMed] [Google Scholar]
- 72. Donato M. T., Jiménez N., Castell J. V., Gómez-Lechón M. J. (2004) Drug Metab. Dispos. 32, 699–706 [DOI] [PubMed] [Google Scholar]
- 73. Moravec R. A., O'Brien M. A., Daily W. J., Scurria M. A., Bernad L., Riss T. L. (2009) Anal. Biochem. 387, 294–302 [DOI] [PubMed] [Google Scholar]
- 74. Mc Cormack T., Baumeister W., Grenier L., Moomaw C., Plamondon L., Pramanik B., Slaughter C., Soucy F., Stein R., Zühl F., Dick L. (1997) J. Biol. Chem. 272, 26103–26109 [DOI] [PubMed] [Google Scholar]
- 75. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 76. Rubin D. M., Glickman M. H., Larsen C. N., Dhruvakumar S., Finley D. (1998) EMBO J. 17, 4909–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mayer T. U., Braun T., Jentsch S. (1998) EMBO J. 17, 3251–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xiong X., Chong E., Skach W. R. (1999) J. Biol. Chem. 274, 2616–2624 [DOI] [PubMed] [Google Scholar]
- 79. Lee R. J., Liu C. W., Harty C., McCracken A. A., Latterich M., Römisch K., DeMartino G. N., Thomas P. J., Brodsky J. L. (2004) EMBO J. 23, 2206–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Carlson E. J., Pitonzo D., Skach W. R. (2006) EMBO J. 25, 4557–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lipson C., Alalouf G., Bajorek M., Rabinovich E., Atir-Lande A., Glickman M., Bar-Nun S. (2008) J. Biol. Chem. 283, 7166–7175 [DOI] [PubMed] [Google Scholar]
- 82. Zhao M., Zhang N. Y., Zurawel A., Hansen K. C., Liu C. W. (2010) J. Biol. Chem. 285, 4771–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ballar P., Shen Y., Yang H., Fang S. (2006) J. Biol. Chem. 281, 35359–35368 [DOI] [PubMed] [Google Scholar]
- 84. Kostova Z., Tsai Y. C., Weissman A. M. (2007) Semin. Cell Dev. Biol. 18, 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ballar P., Fang S. (2008) Biochem. Soc. Trans. 36, 818–822 [DOI] [PubMed] [Google Scholar]
- 86. Jo Y., Debose-Boyd R. A. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Janiesch P. C., Kim J., Mouysset J., Barikbin R., Lochmüller H., Cassata G., Krause S., Hoppe T. (2007) Nat. Cell Biol. 9, 379–390 [DOI] [PubMed] [Google Scholar]
- 88. Custer S. K., Neumann M., Lu H., Wright A. C., Taylor J. P. (2010) Hum. Mol. Genet. 19, 1741–1755 [DOI] [PubMed] [Google Scholar]
- 89. Kobayashi T., Tanaka K., Inoue K., Kakizuka A. (2002) J. Biol. Chem. 277, 47358–47365 [DOI] [PubMed] [Google Scholar]
- 90. Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.