Abstract
Nonvisual arrestins are regulated by direct post-translational modifications, such as phosphorylation, ubiquitination, and nitrosylation. However, whether arrestins are regulated by other post-translational modifications remains unknown. Here we show that nonvisual arrestins are modified by small ubiquitin-like modifier 1 (SUMO-1) upon activation of β2-adrenergic receptor (β2AR). Lysine residues 295 and 400 in arrestin-3 fall within canonical SUMO consensus sites, and mutagenic analysis reveals that Lys-400 represents the main SUMOylation site. Depletion of the SUMO E2 modifying enzyme Ubc9 blocks arrestin-3 SUMOylation and attenuates β2AR internalization, suggesting that arrestin SUMOylation mediates G protein-coupled receptor endocytosis. Consistent with this, expression of a SUMO-deficient arrestin mutant failed to promote β2AR internalization as compared with wild-type arrestin-3. Our data reveal an unprecedented role for SUMOylation in mediating GPCR endocytosis and provide novel mechanistic insight into arrestin function and regulation.
Keywords: Adaptor Proteins, Adrenergic Receptor, Endocytosis, G Protein-coupled Receptors (GPCR), Signal Transduction, SUMOylation, Ubc9, Arrestin
Introduction
Nonvisual arrestins play an important role in the regulation of G protein-coupled receptor (GPCR)3 desensitization, trafficking, and signaling. Arrestins bind to agonist-activated and GPCR kinase phosphorylated receptors to uncouple the associated heterotrimeric G protein from the receptor through a process likely to involve steric hindrance, culminating in the termination of further signaling (1). Arrestins also promote GPCR internalization by virtue of their ability to bind to components of the trafficking machinery, such as clathrin and AP2, thus enabling the recruitment of receptors to clathrin-coated pits where they are internalized (2, 3). In addition, arrestins interact with and activate several downstream signaling molecules to initiate G protein-independent signaling events (4–7).
Arrestins are regulated by post-translational modifications, including ubiquitination, phosphorylation, and nitrosylation (8–10). Ubiquitination of arrestin has been linked to its ability to promote internalization of β2-adrenergic receptor (β2AR) (10). It has also been linked to G protein-independent and arrestin-dependent GPCR signaling whereby the arrestin ubiquitination status correlates with its ability to stably associate with GPCRs upon internalization onto endosomes and to activate downstream signaling cascades (11). Dephosphorylation and nitrosylation facilitate the ability of arrestins to promote GPCR trafficking, possibly by facilitating interactions with the internalization machinery (8, 9). Whether arrestins are regulated by other post-translational modifications remains unknown.
To our knowledge, other than a few studies, the role of SUMO, another common post-translational modifier, in GPCR signaling remains relatively unexplored (12–14). Proteins that are modified by SUMO typically encode a SUMO consensus motif, defined as ΨKX(D/E), where Ψ represents an aliphatic amino acid followed by an acceptor lysine residue, and X represents any amino acid adjacent to an acidic residue (Asp/Glu), although there are exceptions (15). SUMO is a member of the ubiquitin-like molecules family of proteins (16, 17). It is structurally related to ubiquitin, although at the primary amino acid level they share very little amino acid identity (18). SUMO is attached to proteins via an enzymatic cascade, reminiscent of ubiquitination reactions but uses dedicated SUMO E1, E2 and E3 enzymes. Unlike the ubiquitin system, which includes ∼40 E2s and ∼700 E3s, the SUMO system has only a single E2 (Ubc9) and approximately a dozen E3s (15, 19, 20). SUMO is attached to proteins via its C-terminal glycine residue, which forms an isopeptide bond with the epsilon amine group of the acceptor lysine residue on the target protein. SUMO modification of proteins has been linked to several processes such as DNA repair, chromatin remodeling, and signal transduction (21–23). SUMO has also been linked to modulation of ion channel activity, possibly by regulating ion channel surface levels, but mechanistic insight is lacking (24, 25).
Here we show for the first time that nonvisual arrestins are subject to stimulus-dependent SUMOylation. We identify lysine residue 400 of arrestin-3 as the main SUMOylation site, and interestingly, SUMOylation of this site is required for arrestin-3 to promote GPCR endocytosis. Our study reveals for the first time a role for SUMO in GPCR trafficking.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Reagents
Human embryonic kidney (HEK) 293 cells, obtained from Microbix (Toronto, Canada), and COS-1 and HeLa cells, obtained from American Tissue Type Culture Collection (Manassas, VA), were maintained in DMEM (Media Tech, Manassas, VA) supplemented with 10% FBS (Hyclone (Logan, UT)). The cells were transfected using TransIT-LT1 transfection reagent according to the manufacturer's recommendations (Mirus, Madison, WI). Lipofectamine 2000 transfection reagent was from Invitrogen and was used to transfect siRNA alone or co-transfected with DNA. The His antibody was from Qiagen (Valencia, CA). The FLAG M2 horseradish peroxidase and alkaline phosphatase-conjugated antibodies were from Sigma. Monoclonal and polyclonal HA antibodies were from Covance (Emeryville, CA). The GST antibody was from Rockland (Gilbertsville, PA). The SUMO-1 and Ubc9 polyclonal antibodies were from Boston Biochem (Cambridge, MA) and Enzo Life Sciences (Plymouth Meeting, PA). The arrestin (21-B1) monoclonal antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The arrestin polyclonal antibody was kindly provided by Dr. Jeffrey Benovic (Thomas Jefferson University, Philadelphia, PA). The GAPD and Ubc9 (siGenome Smart pool M-004910) siRNA were from Dharmacon RNAi Technologies (Lafayette, CO). Alexa Fluor-conjugated secondary antibodies and transferrin were from Invitrogen.
Constructs, Cloning, and Mutagenesis
HA-tagged bovine arrestin-2 and arrestin-3 were as described previously (26). DNA encoding SUMO-1 was from Dr. Frauke Melchior (University of Heidelberg, Heidelberg, Germany) and was used to make FLAG-tagged SUMO-1. Site-directed mutagenesis by sequential PCR steps was employed to generate the single and multiple point mutants described in this study. The sequences of all of the constructs were verified by dideoxy sequencing.
SUMOylation Assay
To detect SUMOylation of arrestin-2 and arrestin-3, HEK293 cells were transiently transfected with HA-tagged arrestin-2 and arrestin-3 and empty vector (pcDNA) plus FLAG-tagged SUMO-1 or empty vector (pCMV) for 48 h. The cells were lysed in immunoprecipitation buffer 1 (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, 0.5% (w/v) sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, 20 mm N-ethylmaleimide, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin), sonicated, and clarified by centrifugation. The lysates were incubated with an anti-HA polyclonal antibody to immunoprecipitate tagged arrestins followed by 7% SDS-PAGE and immunoblotting with the FLAG M2 HRP-conjugated antibody to detect the incorporated FLAG-tagged SUMO-1. To detect endogenous arrestin SUMOylation, HEK293 cells grown to confluency in 10-cm dishes were lysed in 1 ml of immunoprecipitation buffer, and clarified lysates were divided into equal aliquots and incubated with IgG (Santa Cruz) control and an arrestin monoclonal antibody followed by immunoblotting with a polyclonal SUMO-1 antibody. To detect arrestin-3 SUMOylation in COS-1 cells, cells grown in six-well plates were co-transfected with 1 μg of HA-arrestin-3 (WT; K400R,K295R; 2K/R; and 4K/R) plus FLAG-Ubc9 (1 μg), His-SUMO-1 (1 μg) and pcDNA alone, or with FLAG-Ubc9 and His-SUMO-1 together. For siRNA transfections, HeLa cells grown to ∼80% confluency in six-well plates were co-transfected with 1 μg of HA-arrestin-3, His-SUMO-1, or empty vector plus 200 pmol of GAPDH or Ubc9 siRNA using Lipofectamine 2000. The cells were grown for 24 h and lysed directly in 2× sample buffer (0.0375 m Tris-HCl, pH 6.5, 8% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.003% bromphenol blue), and equal amounts were analyzed by immunoblotting. For agonist-dependent SUMOylation, COS-1 cells grown in six-well plates were co-transfected with DNA encoding β2-adrenergic receptor (1 μg), HA-arrestins (Arr2; Arr3; K400R; K2/R; 1 μg), and His-SUMO-1 (1 μg). Twenty-four hours later, the cells were washed once in warm DMEM and treated with DMEM supplemented with 20 mm HEPES, pH 7.0, containing either vehicle (0.01 mm ascorbic acid) or isoproterenol (10 μm) for 5 min. The cells were washed once on ice with cold PBS and collected in immunoprecipitation buffer 2 (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100, 0.1% SDS, 20 mm N-ethylmaleimide, and 10 μg/ml each of leupeptin, aprotonin, and pepstatin A), followed by sonication on ice and centrifugation at 21,000 × g, for 20 min at 4 °C. Clarified samples were incubated with an anti-HA polyclonal antibody and protein A-agarose to immunoprecipitate HA-tagged arrestins, followed by SDS-PAGE and immunoblotting using an anti-His monoclonal antibody to detect incorporated His-tagged SUMO-1. Lysates were also subject to immunoblotting to detect expression of the various constructs.
Co-immunoprecipitation Assay
HEK293 cells were transiently transfected with HA-tagged wild-type arrestin-3, arrestin-3–4K/R, and empty vector (pcDNA3) plus His-SUMO-1 or pcDNA3 using TransIT-LT1 transfection reagent. Twenty-four hours later, the cells were lysed in immunoprecipitation buffer 3 (20 mm Na2PO4, pH 6.5, 150 mm NaCl, 1% (v/v) Triton X-100, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin A) and incubated at 4 °C for 30 min. The cells were sonicated and centrifuged, and clarified lysates were incubated with an anti-HA polyclonal antibody to immunoprecipitate HA-tagged arrestin followed by immunoblotting to detect bound β2-adaptin.
Receptor Internalization Assay
To measure receptor internalization, we employed an enzyme-linked immunosorbant assay, as described previously (26). COS-1 cells grown in six-well plates to ∼70–80% confluency were transfected with FLAG-β2AR (1 μg) or FLAG-AT1aR plus 6–12 ng of HA-tagged wild-type arrestin-3 and the 4K/R or 2K/R arrestin-3 SUMO-deficient mutants, respectively. For internalization measured in HEK293 cells, the cells grown in six-well plates were co-transfected with 250 ng of FLAG-β2AR DNA plus 50 pmol of GAPDH or Ubc9 siRNA using Lipofectamine 2000, according to the manufacturer's recommendations (Invitrogen). The following day, the cells were passaged onto 24-well plates coated with poly-l-lysine (0.1 mg/ml) and grown for an additional 24 h. On the day of the experiment, the cells were washed once in warm DMEM and were treated with vehicle and 10 μm isoproterenol for 15 min in DMEM containing 20 mm HEPES and 0.01 mm ascorbic acid, after which receptor internalization was measured using the M2-alkaline phosphatase-conjugated FLAG antibody (Sigma), using a protocol we have described previously (27).
Confocal Fluorescence Microscopy
HeLa cells transiently transfected with FLAG-β2AR and HA-arrestin-3 wild-type or HA-arrestin-3–4K/R were passaged onto poly-l-lysine-coated coverslips and grown for 24 h. The cells were serum-starved by incubating with warm DMEM containing 20 mm HEPES, pH 7.5, for 3–4 h at 37 °C followed by treatment with 10 μm isoproterenol or vehicle (0.01 mm ascorbic acid) for 5 min. The cells were fixed with 3.7% paraformaldehyde and permeabilized with 0.05% (w/v) saponin for 10 min, similar to a protocol that we have described previously (26). Briefly, after permeabilization and fixation, the cells were incubated with 1% BSA in 0.01% saponin-PBS for 30 min at 37 °C, followed by immunostaining with FLAG polyclonal antibody used at 1:100 dilution. For HA-arrestin-3–4K/R, the cells were also immunostained with anti-HA monoclonal antibody. The cells were washed five times with 0.01% saponin-PBS, followed by incubating with Alexa Fluor 594- or 488-conjugated secondary antibodies for 30 min at 37 °C. Finally, the cells were washed with PBS and fixed again in 3.7% formaldehyde-PBS and mounted onto glass slides using mounting medium containing DAPI (Vector Laboratories, Burlingame, CA). The samples were analyzed using a Zeiss LSM 510 laser scanning confocal microscope equipped with a Plan-Apo 63×/1.4 oil lens objective. The images were acquired using a 1.4 megapixel cooled extended spectra range RGB digital camera set at 512 × 512 resolution. Acquired images were analyzed using Image J software (version 1.41o).
To determine a role for Ubc9 in internalization of FLAG-β2AR and transferrin receptor in HeLa cells, the cells grown in 10-cm dishes were co-transfected with DNA encoding FLAG-β2AR (3 μg) and Ubc9 or GAPDH siRNA (600 pmol) using Lipofectamine 2000. The next day, the cells were passaged onto poly-l-lysine-coated coverslips and grown for an additional 24 h. The cells were serum-starved, followed by treatment in the absence and presence of 10 μm isoproterenol for 30 min. During the final 5 min of the incubation, the cells were treated with 25 μg/μl transferrin conjugated to Alexa Fluor 594 (Invitrogen). The cells were processed as described above. FLAG-β2AR was labeled with a FLAG polyclonal antibody (1:100 dilution) and an Alexa Fluor 483-conjugated anti-rabbit IgG secondary antibody (1:200 dilution). The cells expressing FLAG-β2AR were counted from 10 random fields containing 5–10 cells/field, and the amount of receptor internalized was calculated as the percentage of FLAG-β2AR-expressing cells showing only punctate staining. The cells that showed staining on the surface of cells were indicative of cells in which receptor internalization had not occurred or was incomplete. Transferrin receptor internalization was determined in the same cells used to calculate FLAG-β2AR internalization. The fluorescence intensity of Alex Fluor 594-conjugated transferrin was calculated using the histogram feature of Zeiss LSM 510 image analysis software (4.2 SP1).
RESULTS
Nonvisual Arrestins Are Modified by SUMO
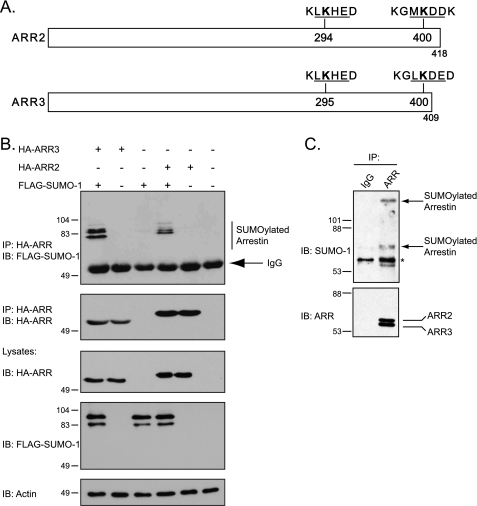
As part of our efforts to understand how ubiquitin-like molecules govern GPCR regulation, we noticed the presence of two SUMO consensus sequences within nonvisual arrestin-2 and arrestin-3 (Fig. 1A), suggesting that they are modified by SUMO. To determine whether arrestins are indeed SUMOylated, HEK293 cells were transiently transfected with DNA encoding HA-tagged arrestin-2, arrestin-3, or empty vector (pcDNA3) with FLAG-tagged SUMO-1 or empty vector (pCMV-10). HA-tagged arrestins were immunoprecipitated with an anti-HA antibody followed by SDS-PAGE and immunoblotting to detect incorporation of FLAG tagged-SUMO-1. As shown in Fig. 1B, several migrating species were detected in cells transfected with HA-arrestin-2 or HA-arrestin-3 and FLAG-SUMO-1 but not in cells transfected with either arrestin or SUMO-1 alone, suggesting that these migrating species represent SUMOylated arrestins. The presence of multiple species suggests that arrestins are modified by SUMO-1 on multiple lysine residues, consistent with the presence of two SUMO consensus sites in both arrestins (Fig. 1A). We also observed that endogenous arrestins are modified by endogenous SUMO-1 in HEK293 cells (Fig. 1C). The migration of endogenous SUMOylated arrestin is faster than that of exogenous arrestin, likely because of the absence of the HA and FLAG epitope tags on endogenous arrestin and SUMO, respectively.
FIGURE 1.
Arrestins are modified by SUMO. A, schematic representation of arrestin-2 and arrestin-3 depicting the putative SUMO consensus sites (underlined) and the surrounding amino acid residues. The SUMO acceptor lysine residues are in bold type. B, SUMOylation analysis of nonvisual arrestins. HEK293 cells were transiently transfected with HA-tagged arrestin-2, HA-arrestin-3, empty vector (pcDNA) plus FLAG-tagged SUMO-1, or empty vector (pCMV) for 48 h. Transfections with empty vector are indicated with a minus sign. HA-tagged arrestins were immunoprecipitated (IP) and followed by immunoblotting (IB) to detect the incorporated FLAG-SUMO-1. The top blot was stripped and reprobed with an anti-HA monoclonal antibody to determine the level of tagged arrestin in the IP. Cross-reactivity to IgG is indicated. The lysates were analyzed by IB to detect the expression of the various constructs. Representative blots from one of three independent experiments are shown. The positions of the molecular mass markers in kDa are shown. C, SUMOylation of endogenous arrestins. Endogenous arrestins were IP from HEK293 cells, followed by IB for endogenous SUMO-1. SUMOylated arrestin is indicated with arrows. The asterisk represents possible cross-reactive bands present in samples incubated with arrestin and IgG antibodies. Representative blots from one of three independent experiments are shown. The positions of the molecular mass markers in kDa are shown.
Arrestin-3 Is Modified by SUMO on Lysine Residue 400
The SUMO consensus site represents a binding site for the SUMO-specific E2 enzyme Ubc9 (28) and to facilitate the detection of arrestin SUMOylation, we co-expressed HA-tagged arrestin-3 with exogenous FLAG-tagged Ubc9 alone or together with His-tagged SUMO-1 in COS-1 cells and analyzed whole cell lysates for the presence of SUMOylated arrestin by immunoblotting. As shown in Fig. 2A, two main species were observed that migrated more slowly than unmodified arrestin in cells transfected with His-SUMO-1 alone and were more abundant in cells that were co-transfected with His-SUMO-1 and FLAG-Ubc9, suggesting that these species represented SUMOylated arrestin-3. These species were not present in cells transfected with vector, HA-arrestin-3, or FLAG-Ubc9 alone, further suggesting that they represent SUMOylated arrestin-3. We observed similar results from three independent experiments. It is important to note that SUMOylated arrestin represents a small percentage of the total cellular complement of arrestin. This is not unusual, because for nearly all proteins that have been shown to be SUMOylated, the SUMOylated form represents a small percentage of the total cellular complement of that protein (15, 29, 30).
FIGURE 2.
Identification of the SUMO acceptor sites on arrestin-3. A, SUMOylation analysis of arrestin-3 by co-expressing SUMO-1 and Ubc9 in COS-1 cells. COS-1 cells grown in six-well plates were transfected either with empty vector (pcDNA) or HA-arrestin-3, plus either empty vector (pcDNA), FLAG-Ubc9 alone, His-SUMO-1 alone, or together with FLAG-Ubc9. Twenty-fours hours later, the cells were lysed directly in 2× sample buffer, and equal amounts were analyzed by IB. Unmodified and SUMOylated arrestins are indicated. Shown are representative IBs from one of three independent experiments. B, SUMOylation analysis of arrestin-3 lysine mutants. Lysine residues 295 and 400, both of which reside within SUMOylation consensus sites, were changed individually to arginine residues. Lysine residues 398 and 400 were changed simultaneously to create the 2K/R mutant, and lysine residues 293 and 295 were changed simultaneously to arginine residues in the 2K/R background to create the 4K/R mutant. The SUMOylation status of these mutants was assessed as described in A. Shown are representative IBs from one of three independent experiments.
We next set out to identify the lysine residues that are modified by SUMO. As noted in Fig. 1A, arrestin-3 has two consensus sites for SUMOylation and thus two possible lysine acceptor sites: Lys-295 and Lys-400. To determine whether these residues are SUMO acceptor sites, they were changed to conserved arginine residues, and the SUMOylation status of these arrestin mutants (K295R and K400R) was assessed by immunoblot analysis, as described above. As shown in Fig. 2B, the SUMOylation status of the K295R mutant was indistinguishable from wild-type arrestin-3; however, in sharp contrast, the K400R mutant was not modified by SUMO-1, suggesting that Lys-400 is the primary SUMO-1 site. However, upon longer exposure the presence of additional species, albeit less abundant, were evident, suggesting that additional lysine residues are modified by SUMO-1. To rule out the possibility that neighboring lysine residues to Lys-400 and Lys-295 are SUMOylated, we created two additional arrestin-3 lysine residue mutants. In one mutant, referred to as the 2K/R mutant, Lys-400 and Lys-398 were changed simultaneously to arginine residues, whereas in another mutant, referred to as 4K/R, Lys-295 and Lys-293 were changed simultaneously to arginine residues in the 2K/R background. As shown in Fig. 2B, SUMOylation of the 2K/R mutant was observed only upon a long exposure of the immunoblot and was indistinguishable from that of the K400R mutant, suggesting that other lysine residues, possibly Lys-295 and/or Lys-293, were modified by SUMO-1 in the 2K/R mutant. Accordingly, when these residues were changed to arginine residues in the 2K/R background, SUMOylation of this mutant (i.e. 4K/R) was not observed. Taken together, these data suggest that Lys-400 is the predominant SUMOylation site, whereas Lys-295 and other lysine residues (Lys-293 and Lys-398) may also be modified by SUMO-1, although to a lesser degree.
Receptor Activation Promotes Arrestin SUMOylation
Upon binding to ligand activated and phosphorylated receptors, arrestins are thought to undergo several conformational changes, exposing the C-tail that ultimately facilitates downstream interactions (31, 32). Because Lys-400 resides within the C-tail of arrestin-3, it is possible that arrestin SUMOylation is regulated by receptor activation. To examine this, we used β2AR, a prototypical GPCR used to study many aspects of arrestin function on GPCR signaling and trafficking (33). As shown in Fig. 3A, treatment of COS-1 cells expressing FLAG-β2AR and His-SUMO-1 with the nonselective β-adrenergic agonist isoproterenol (10 μm) for 5 min enhanced SUMOylation of both arrestin-2 and arrestin-3, as compared with vehicle-treated cells. In contrast, the K400R and 2K/R arrestin mutants were not modified by SUMO-1, as compared with wild-type arrestin-3 (Fig. 3B). Densitometric analysis of data from three independent experiments reveals that receptor activation significantly enhances arrestin-3 SUMOylation on Lys-400 (Fig. 3C).
FIGURE 3.
Role of receptor activation on arrestin SUMOylation. A, examination of nonvisual arrestin SUMOylation by GPCR activation. Stimulus-dependent SUMOylation of arrestin-2 and arrestin-3 was examined in COS-1 cells co-transfected with FLAG-tagged β2AR, His-tagged SUMO-1, and HA-tagged arrestin-2, arrestin-3, or empty vector (pcDNA). The cells were treated with vehicle (0.01 mm ascorbic acid) and 10 μm isoproterenol (ISO) for 5 min, followed by IP of tagged arrestin and IB to detect incorporated His-SUMO-1. Shown are IBs from one of three independent experiments. B, role of lysine residues in arrestin-3 SUMOylation by GPCR activation. Stimulus-dependent SUMOylation was examined in COS-1 cells transfected with FLAG-tagged β2AR and His-tagged SUMO-1 plus HA-tagged arrestin-3, wild-type, K400R, 2K/R, or empty vector (pcDNA). Treatment and analysis of arrestin SUMOylation was as described in A. Shown are IBs from one of three independent experiments. C, arrestin SUMOylation is represented graphically. The data represent the average results from the densitometric analysis of immunoblots from three independent experiments. The fold increase upon isoproterenol (ISO) treatment as compared with vehicle (VEH) treatment was normalized to HA-arrestin-3 levels present in the immunoprecipitates. The error bars represent the standard error of the mean. The data were analyzed by an unpaired Student's t test (*, p < 0.05).
Arrestin SUMOylation Regulates GPCR Internalization
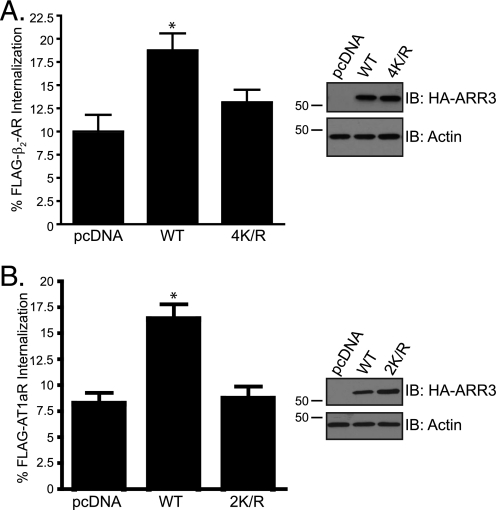
Because nonvisual arrestins mediate GPCR endocytosis, it is possible that SUMOylation modulates the role of arrestin in GPCR trafficking. To confirm a role for SUMOylation in GPCR endocytosis, we examined arrestin SUMOylation and internalization of β2AR in cells depleted of the SUMO E2 enzyme Ubc9. We first determined whether depletion of Ubc9 modulated arrestin-3 SUMOylation. As shown in Fig. 4A, siRNA-mediated depletion of Ubc9 attenuated arrestin SUMOylation as compared with control siRNA-transfected cells. We next examined the role of Ubc9 on agonist-induced internalization of β2AR. HeLa cells co-transfected with FLAG-β2AR and GAPDH or Ubc9 siRNA were treated with 10 μm isoproterenol for 30 min, and receptor internalization was assessed by confocal immunofluorescene microscopy, as described under “Experimental Procedures.” As shown in Fig. 4B, upon isoproterenol treatment in cells transfected with GAPDH siRNA, FLAG-β2AR staining was mostly punctate, indicating that the receptor had internalized into endocytic vesicles. In contrast, upon agonist treatment of cells transfected with Ubc9 siRNA, most FLAG-β2AR staining was restricted to the cell surface, indicating that the receptor had failed to internalize (Fig. 4B, bottom panels). Quantification of FLAG-β2AR internalization in multiple cells from 10 randomly selected fields from three independent experiments revealed that agonist-induced internalization of FLAG-β2AR was significantly inhibited in cells transfected with Ubc9 siRNA as compared with GAPDH siRNA (Fig. 4C). To rule out the possibility that Ubc9 siRNA has a global effect on endocytosis, we also examined internalization of the transferrin receptor in the same cells expressing FLAG-β2AR. As shown in Fig. 4 (B and D), internalization of the transferrin receptor was not impaired in cells transfected with Ubc9 siRNA as compared with GAPDH siRNA-transfected cells. The extent of Ubc9 depletion in these experiments was greater than 90% as revealed by immunoblotting of whole cell lysates (Fig. 4E). To rule out the possibility that Ubc9 may modulate GPCR endocytosis in a cell type-selective manner, we also examined the role of Ubc9 on β2AR internalization in HEK293 cells. Similar to HeLa cells, agonist-induced internalization of β2AR was significantly (Student's t test, p < 0.05) attenuated in HEK293 cells transfected with Ubc9 siRNA as compared with GAPDH siRNA-transfected cells (Fig. 4F). Taken together, these data suggest that SUMOylation is important for arrestin function in promoting GPCR internalization. To further link arrestin SUMOylation to β2AR internalization, we examined internalization of β2AR in COS-1 cells co-expressing wild-type HA-arrestin-3 and the SUMOylation-deficient 4K/R mutant. As shown in Fig. 5A, when expressed to equal levels, the SUMOylation-deficient 4K/R mutant failed to enhance β2AR internalization as compared with wild-type arrestin-3, further suggesting that arrestin SUMOylation mediates β2AR endocytosis. To examine the possibility that arrestin SUMOylation may modulate the internalization of other GPCRs, we also examined the role of arrestin SUMOylation on internalization of the angiotension 1A receptor (AT1aR). As shown in Fig. 5B, the SUMOylation-deficient 2K/R mutant failed to enhance AT1aR internalization as compared with wild-type arrestin-3, suggesting that arrestin SUMOylation regulates internalization of AT1aR and that it may play a broad role in mediating GPCR endocytosis.
FIGURE 4.
The role of Ubc9 on arrestin SUMOylation and receptor trafficking. A, HeLa cells transfected with control (CON, GAPDH) and Ubc9 siRNA plus His-tagged SUMO or empty vector (pcDNA) were solubilized in 2× sample buffer and subjected to IB to detect HA-tagged arrestin-3. SUMOylated arrestin is indicated. Shown are IBs from one of three independent experiments. B, the role of Ubc9 on GPCR internalization was examined in HeLa cells co-transfected with DNA encoding FLAG-β2AR and control (Ctrl, GAPDH) or Ubc9 siRNA. The cells were treated with 10 μm isoproterenol for 30 min, and during the final 5 min of the incubation, the cells were treated with 25 μg/μl Alexa Fluor 594-conjugated transferrin (Tfn). The cells were processed as described under “Experimental Procedures.” Shown are representative confocal fluorescence microscopy images of cells analyzed from three independent experiments. Bar, 20 μm. C, shown is a graph representing FLAG-β2AR internalization calculated in control (GAPDH) and Ubc9 siRNA-transfected cells shown in B. The percentage of receptor internalization was determined by counting cells showing punctate staining and no membrane staining in 10 randomly selected fields containing 5–10 cells/field. The data represent the averages from three independent experiments, and the error bars represent the standard error of the mean. The data were analyzed by a Student's t test (*, p < 0.0001). D, internalization of the transferrin receptor was determined in the same cells used for the analysis performed in C. Internalization of the transferrin receptor was calculated by measuring the fluorescence intensity of transferrin Alexa Fluor 594 using LSM 510 image analysis software, as described under “Experimental Procedures.” The percentage of fluorescence intensity in Ubc9 transfected cells was normalized to control (GAPDH) transfected cells. The error bars represent the standard error of the mean. E, levels of Ubc9 and actin were detected by immunoblotting whole cell lysates from control and Ubc9 siRNA-transfected cells. F, FLAG-β2AR internalization was examined in HEK293 cells co-transfected with DNA encoding FLAG-β2AR and control (GAPDH) or Ubc9 siRNA, as described under “Experimental Procedures.” The cells were treated with vehicle (0.01 mm ascorbic acid) and 10 μm isoproterenol for 15 min, and receptor internalization was measured by cell surface ELISA. The data represent the averages from three or four independent experiments, and the error bars represent the standard error of the mean. The data were analyzed by a Student's t test (*, p < 0.05). Shown in the inset are representative IBs to detect Ubc9 and actin levels.
FIGURE 5.
Role of arrestin-3 SUMOylation on GPCR internalization. FLAG-β2AR (A) and FLAG-AT1aR (B) internalization was assessed in COS-1 cells co-transfected with HA-arrestin-3 WT, 4K/R, 2K/R, or empty vector (pcDNA) upon treatment with vehicle (0.01 mm ascorbic acid) and 10 μm isoproterenol for 30 min by ELISA. The data represent the averages from three independent experiments, and the error bars represent the standard error of the mean. The data were analyzed by a one-way analysis of variance followed by Bonferroni's post-hoc test. Wild-type arrestin-3, but not the SUMO-deficient mutants, was significantly different from pcDNA (p < 0.05). Shown in the insets are representative IBs to detect expression of HA-tagged arrestins.
To rule out the possibility that the SUMOylation-deficient arrestin-3 mutants may be impaired in their ability to be recruited to the receptor upon agonist treatment, we examined their distribution in cells upon receptor activation by confocal immunofluoresence microscopy. As shown in Fig. 6A, in vehicle-treated cells, wild-type HA-arrestin-3 was diffusely distributed throughout the cytoplasm; however, upon treatment with isoproterenol for 5 min, HA-arrestin-3 rapidly translocated to the plasma membrane where it co-localized with β2AR. Similarly, the SUMOylation-deficient 4K/R mutant rapidly translocated to the plasma membrane upon agonist treatment where it co-localized with β2AR (Fig. 6B). In addition, we also examined the role of SUMOylation in the distribution of arrestin-3 in response to angiotensin II treatment. In cells co-expressing HA-AT1aR and wild-type arrestin-3-yellow fluorescent protein, angiotensin II treatment for 30 min promoted co-internalization of both the receptor and arrestin-3, as assessed by confocal fluorescence microscopy (supplemental Fig. S1A). Similarly, the SUMO-deficient K400R mutant co-internalized with HA-AT1aR (supplemental Fig. S1B). Quantification of the percentage of co-localization between receptor and arrestin puncta revealed no difference between wild-type and the SUMO-deficient arrestin (supplemental Fig. S1C), which is consistent with the idea that SUMOylation is not involved in arrestin binding to the receptor. Taken together, these data suggest that SUMOylation is not involved in arrestin recruitment to GPCRs but rather is likely involved in mediating interactions with factors of the internalization machinery to promote GPCR endocytosis.
FIGURE 6.
Confocal microscopy analysis of arrestin distribution upon agonist activation. HeLa cells co-transfected with FLAG-β2AR and HA-arrestin-3-WT (A) or HA-Arrestin-3–4K/R (B) were serum-starved before being treated with vehicle (0.01 mm ascorbic acid) and 10 μm isoproterenol (ISO) for 5 min. The cells were then fixed, permeabilized, and immunostained with anti-FLAG and anti-HA antibodies. Yellow in the merged images represent co-localization between FLAG-β2AR and HA-arrestin-3. The differential interference contrast (DIC) image is shown. Shown are representative images from cells analyzed from three independent experiments in which at least 10 cells were analyzed per experiment. Bar, 20 μm.
To explore this possibility, we examined the role of arrestin SUMOylation on binding to β2-adaptin by co-immunoprecipitation. The β2-adaptin subunit of AP2 binds to nonvisual arrestins, and this interaction is important for promoting GPCR endocytosis. HA-tagged wild-type arrestin-3 and the 4K/R mutant transiently transfected in HEK293 cells were subject to immunoprecipitation followed by immunoblotting to detect bound endogenous β2-adaptin. As shown in Fig. 7, both wild-type arrestin-3 and the 4K/R mutant bound to β2-adaptin to equal levels. However, when His-tagged SUMO-1 was co-expressed, binding of β2-adaptin to wild-type arrestin-3 was markedly enhanced, as compared with the 4K/R mutant, suggesting that arrestin SUMOylation promotes binding to β2-adaptin.
FIGURE 7.
Role of arrestin SUMOylation on binding to AP2. HEK293 cells were transfected with HA-tagged wild-type arrestin-3 and the 4K/R mutant plus His-SUMO-1 or empty vector (pcDNA). The next day, HA-tagged arrestins were subjected to IP followed by IB to detect bounds β2-adaptin, as described under “Experimental Procedures.” Shown are representative IBs from one of three independent experiments.
DISCUSSION
Our data reveal for the first time that arrestins are subject to stimulus-dependent SUMOylation. Lysine residue 400 in arrestin-3 is the main SUMOylation site, and the arrestin SUMOylation status correlates with its ability to mediate GPCR endocytosis, possibly by promoting binding to AP2. Whether SUMOylation also modulates the role of arrestin in GPCR desensitization and/or signaling remains to be determined. Our data reveal an unprecedented function for protein SUMOylation in GPCR endocytosis and extends our knowledge about the molecular mechanisms by which arrestins govern GPCR trafficking.
We identified lysine residues 295 and 400 as the primary SUMOylation sites in arrestin-3 (Fig. 2A). Each of these residues falls within a canonical SUMO consensus motif, and although it is possible that other lysine residues may be modified by SUMO, Lys-400 and Lys-295 likely represent the main sites. Arrestin SUMOylation is regulated by GPCR activation because SUMOylation of Lys-400 is enhanced by agonist activation of the β2AR (Fig. 3). Upon agonist exposure and binding to phosphorylated receptor, arrestin is thought to undergo a conformational change that exposes elements in the C-tail. Because the SUMO site resides within the C-tail, it is likely that agonist activation and a subsequent conformational change in arrestin facilitates Ubc9 binding and subsequent SUMO modification of Lys-400. SUMO modification of this site enables arrestin to regulate GPCR trafficking. This is consistent with our data showing that 1) overexpression of a SUMO-deficient arrestin fails to promote β2AR and AT1aR internalization (Fig. 5); and 2) siRNA-mediated depletion of Ubc9 inhibits arrestin SUMOylation and β2AR internalization (Fig. 4, A–C). To our knowledge, these data link SUMOylation for the first time to endocytic trafficking of GPCRs. It is possible that SUMOylation has a broad role in regulating endocytosis. Dynamin, a key component of the internalization machinery, may be regulated by the SUMO machinery (34). Therefore, it is possible that Ubc9 siRNA may also affect other factors of the trafficking machinery, in addition to arrestin-3, to modulate GPCR endocytosis. However, depletion of Ubc9 siRNA did not have a global effect on endocytosis, because transferrin receptor internalization was not affected by depletion of Ubc9 (Fig. 4, B and D). The fact that the arrestin SUMOylation-deficient mutants are unable to support GPCR endocytosis indicates that arrestin SUMOylation is required for this process (Fig. 5).
Arrestins are also regulated by other post-translational modifications including phosphorylation, nitrosylation, and ubiquitination. Stimulus-dependent dephosphorylation of arrestin-3 (also called β-arrestin-2) on serine (Ser-361) and threonine (Thr-383) residues is required for internalization of β2AR, likely by facilitating arrestins association with clathrin (8). Ubiquitination of arrestin-3 is required to support internalization of β2AR, although mechanistic insight is lacking (10). Nitrosylation of the C-terminal cysteine residue of arrestin-3 has been linked to its ability to regulate GPCR internalization through a mechanism that may involve release of the buried C-tail, thus facilitating binding to clathrin and β2-adaptin, thereby promoting GPCR endocytosis (9). It is possible that SUMOylation of Lys-400 may favor the release of the buried C-tail, thereby facilitating these interactions to promote GPCR endocytosis. Our data in Fig. 7 are consistent with this idea, because expression of His-SUMO-1 enhanced β2-adaptin binding to wild-type arrestin-3 but not to the SUMO-deficient 4K/R mutant. It is unlikely that this mutant does not interact with the receptor, because the 4K/R mutant was not impaired in its ability to be recruited to β2AR at the plasma membrane (Fig. 6), suggesting that SUMOylation is not involved in the ability of arrestins to bind to the receptor.
It is possible that the attached SUMO moiety may play a more direct role in GPCR endocytosis. SUMO often interacts with SUMO interaction motifs. SUMO interaction motifs are found in several proteins and are characterized by a short stretch of hydrophobic amino acids near an acidic cluster of residues that interact noncovalently with the SUMO moiety while it is attached to its target protein (35). It is possible that the SUMO moiety on arrestin interacts with a SUMO interaction motif-harboring protein to execute its function in GPCR endocytosis. Alternatively, SUMOylation may be linked to arrestin ubiquitination. For example, SUMO modification has been shown to prevent a protein from being ubiquitinated, because some ubiquitin acceptor sites may also serve as SUMO acceptor sites (36). As mentioned above, ubiquitination of arrestin facilitates its role in promoting GPCR internalization (10), as we observed with SUMOylation in this study. Interestingly, the SUMO acceptor site Lys-400 in arrestin-3 may not be a ubiquitination site (11). Therefore, it is possible that ubiquitination and SUMOylation have distinct roles in mediating arrestin-dependent GPCR internalization.
We observed that only a small percentage of the total cellular pool of arrestin-3 is SUMOylated. This is not uncommon, because with most proteins that are SUMOylated, only a small fraction of the total cellular pool of a protein is in fact SUMOylated at any given time (15, 30). Although the reason remains poorly understood, it may be because only a small fraction of a protein is in the appropriate location to be modified by SUMO, and once it performs its action it may be rapidly de-SUMOylated to return the protein to its basal state. This may be true for arrestin because SUMOylation may occur before or once arrestin is recruited to clathrin-coated pits, where we propose it facilitates interactions with factors of the internalization machinery. De-SUMOylation would antagonize these interactions and subserve to terminate SUMOylated arrestin activity. We anticipate that the identification of the machinery, such as the SUMO E3 ligase and the de-SUMOylase, that regulates the arrestin SUMOylation status will provide significant insight into these events.
In summary, we provide evidence that SUMOylation of arrestin-3 on lysine 400 promotes internalization of β2AR. This study reveals for the first time a role for SUMOylation in promoting GPCR endocytosis. Whether SUMOylation plays a role in other aspects of GPCR signaling remains to be examined.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants GM75159 and DA026040 (to A. M.). This work was also supported by Predoctoral Fellowship 0910098G from the American Heart Association (to R. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- GPCR
- G protein-coupled receptor
- SUMO
- small ubiquitin-like modifier
- β2AR
- β2-adrenergic receptor
- IP
- immunoprecipitated/immunoprecipitation
- IB
- immunoblot/immunoblotting
- HEK
- human embryonic kidney.
REFERENCES
- 1. Lohse M. J., Benovic J. L., Codina J., Caron M. G., Lefkowitz R. J. (1990) Science 248, 1547–1550 [DOI] [PubMed] [Google Scholar]
- 2. Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 3. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 5. DeFea K. A., Vaughn Z. D., O'Bryan E. M., Nishijima D., Déry O., Bunnett N. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luttrell L. M., Roudabush F. L., Choy E. W., Miller W. E., Field M. E., Pierce K. L., Lefkowitz R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2449–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beaulieu J. M., Sotnikova T. D., Marion S., Lefkowitz R. J., Gainetdinov R. R., Caron M. G. (2005) Cell 122, 261–273 [DOI] [PubMed] [Google Scholar]
- 8. Lin F. T., Chen W., Shenoy S., Cong M., Exum S. T., Lefkowitz R. J. (2002) Biochemistry 41, 10692–10699 [DOI] [PubMed] [Google Scholar]
- 9. Ozawa K., Whalen E. J., Nelson C. D., Mu Y., Hess D. T., Lefkowitz R. J., Stamler J. S. (2008) Mol. Cell 31, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shenoy S. K., McDonald P. H., Kohout T. A., Lefkowitz R. J. (2001) Science 294, 1307–1313 [DOI] [PubMed] [Google Scholar]
- 11. Shenoy S. K., Lefkowitz R. J. (2005) J. Biol. Chem. 280, 15315–15324 [DOI] [PubMed] [Google Scholar]
- 12. Rodríguez-Muñoz M., Bermúdez D., Sánchez-Blázquez P., Garzón J. (2007) Neuropsychopharmacology 32, 842–850 [DOI] [PubMed] [Google Scholar]
- 13. Tang Z., El Far O., Betz H., Scheschonka A. (2005) J. Biol. Chem. 280, 38153–38159 [DOI] [PubMed] [Google Scholar]
- 14. Klenk C., Humrich J., Quitterer U., Lohse M. J. (2006) J. Biol. Chem. 281, 8357–8364 [DOI] [PubMed] [Google Scholar]
- 15. Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 16. Su H. L., Li S. S. (2002) Gene 296, 65–73 [DOI] [PubMed] [Google Scholar]
- 17. Hochstrasser M. (2009) Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. (1998) J. Mol. Biol. 280, 275–286 [DOI] [PubMed] [Google Scholar]
- 19. Ye Y., Rape M. (2009) Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) PLoS One 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009) Nature 462, 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Girdwood D., Bumpass D., Vaughan O. A., Thain A., Anderson L. A., Snowden A. W., Garcia-Wilson E., Perkins N. D., Hay R. T. (2003) Mol. Cell. 11, 1043–1054 [DOI] [PubMed] [Google Scholar]
- 23. Kang J. S., Saunier E. F., Akhurst R. J., Derynck R. (2008) Nat. Cell Biol. 10, 654–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin S., Nishimune A., Mellor J. R., Henley J. M. (2007) Nature 447, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benson M. D., Li Q. J., Kieckhafer K., Dudek D., Whorton M. R., Sunahara R. K., Iñiguez-Lluhí J. A., Martens J. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1805–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhandari D., Trejo J., Benovic J. L., Marchese A. (2007) J. Biol. Chem. 282, 36971–36979 [DOI] [PubMed] [Google Scholar]
- 27. Malik R., Marchese A. (2010) Mol. Biol. Cell 21, 2529–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernier-Villamor V., Sampson D. A., Matunis M. J., Lima C. D. (2002) Cell 108, 345–356 [DOI] [PubMed] [Google Scholar]
- 29. Hay R. T. (2007) Trends Cell Biol. 17, 370–376 [DOI] [PubMed] [Google Scholar]
- 30. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 31. Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shenoy S. K., Lefkowitz R. J. (2005) Sci. STKE, cm10. [DOI] [PubMed] [Google Scholar]
- 33. Pierce K. L., Premont R. T., Lefkowitz R. J. (2002) Nat. Rev. Mol. Cell Biol. 308, 639–650 [DOI] [PubMed] [Google Scholar]
- 34. Mishra R. K., Jatiani S. S., Kumar A., Simhadri V. R., Hosur R. V., Mittal R. (2004) J. Biol. Chem. 279, 31445–31454 [DOI] [PubMed] [Google Scholar]
- 35. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) Mol. Cell 2, 233–239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.