Abstract
PKA contributes to many physiological processes, including glucose homeostasis and cell migration. The substrate specificity of PKA is low compared with other kinases; thus, complex formation with A-kinase-anchoring proteins is important for the localization of PKA in specific subcellular regions and the phosphorylation of specific substrates. Here, we show that PKA forms a complex with WAVE2 (Wiskott-Aldrich syndrome protein family verprolin-homologous protein 2) in MDA-MB-231 breast cancer cells and mouse brain extracts. Two separate regions of WAVE2 are involved in WAVE2-PKA complex formation. This complex localizes to the leading edge of MDA-MB-231 cells. PKA activation results in enlargement of the membrane protrusion. WAVE2 depletion impairs PKA localization at membrane protrusions and the enlargement of membrane protrusion induced by PKA activation. Together, these results suggest that WAVE2 works as an A-kinase-anchoring protein that recruits PKA at membrane protrusions and plays a role in the enlargement of membrane protrusions induced by PKA activation.
Keywords: Actin, Adaptor Proteins, Cell Migration, Cyclic AMP (cAMP), Protein Kinase A (PKA), Protein Targeting, A-kinase-anchoring Protein
Introduction
Cell migration is essential for many physiological and pathological phenomena, including development, inflammation, wound healing, angiogenesis, and cancer metastasis. Cell migration is a complex process that can be divided into several steps: protrusion of the leading edge, adhesion of the protruded leading edge to the extracellular matrix, translocation of the cell body forward, and detachment/retraction of the tail. The protrusion of the leading edge, called filopodia and lamellipodia, involves marked actin cytoskeletal remodeling mediated by WASP (Wiskott-Aldrich syndrome protein) family proteins (1).
WASP family proteins link various upstream signals to the stimulation of Arp2/3-mediated actin polymerization. The WASP family consists of WASP, N-WASP, and WAVE (WASP family verprolin-homologous protein) family proteins (WAVE1, WAVE2, and WAVE3) (1–3). Among them, WAVE2 is ubiquitously expressed in mammals (4) and plays crucial roles in lamellipodial formation and cell-cell contact organization under a Rho family small GTPase Rac1 signal (5, 6). WAVE2 contains conserved protein regions interacting with other molecules. For example, the WAVE homology domain (WHD)2 interacts with the Abi (Abl-interacting protein)-Sra1-Nap1 complex (7–10). The basic region binds to phosphatidylinositol 3,5-triphosphate (PIP3) (11), and the proline-rich region is associated with IRSp53 (12) as well as vinexin-β (13). Sra1 and IRSp53 connect Rac1 signaling to WAVE2 by interacting with Rac1. The verprolin homology region interacts with G-actin, and the cofilin homology region and acidic region binds to Arp2/3 complexes, resulting in enhanced Arp2/3-mediated actin polymerization (14, 15). Complex formation also contributes to WAVE2 localization and stability. The depletion of any components of the Abi-Sra1-Nap1 complex induces the mislocalization of WAVE2 and decrease in the WAVE2 protein level (7, 8, 16). Vinexin-β interaction with WAVE2 through the proline-rich region stabilizes WAVE2 (13).
PKA is a serine/threonine kinase that contributes to a variety of physiological phenomena, including glucose homeostasis, as well as cell proliferation, differentiation, and apoptosis (17, 18). PKA is a heterotetrameric enzyme containing two regulatory (R) and two catalytic (C) subunits. The R subunit inhibits the kinase activity of the C subunit in the complex; thus, the PKA holoenzyme is inactive. After cAMP binding, the R subunit dissociates from the C subunit, and then the released C subunits become active (18). Increasing evidence indicates the importance of PKA in cell migration (19). PKA is activated in response to growth factors, and PKA activity is required for growth factor-induced chemotaxis (20–22). Furthermore, the front-to-back gradient of cAMP in migrating cells, as well as PKA activation at the leading edge of cells, has been reported (20–24).
PKA phosphorylates a number of substrates, including channels, transcription factors, and cytoskeletal proteins, and shows broad substrate specificity (17). The association of PKA with A-kinase-anchoring proteins (AKAPs) enables PKA to localize in specific subcellular regions and phosphorylate the appropriate substrates (25–27). More than 50 AKAPs have been reported, but AKAPs that contribute to PKA-mediated cell migration are not known well. In this study, we demonstrate that WAVE2 interacts with PKA through its WHD and VCA region. This complex localizes to the leading edge of MDA-MB-231 breast cancer cells. Depletion of WAVE2 expression impairs PKA localization at membrane protrusions and enlargement of membrane protrusions induced by PKA activation. These results suggest that WAVE2 works as an AKAP that recruits PKA at membrane protrusions and plays a role in the enlargement of membrane protrusions induced by PKA activation.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Mouse anti-FLAG monoclonal antibody M2 was obtained from Sigma. High affinity rat anti-HA monoclonal antibody was obtained from Roche Applied Science. Rabbit (H-110) and goat (D-16) anti-WAVE2 polyclonal antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-PKA C subunit monoclonal antibody was obtained from BD Transduction Laboratories. Duolink, a proximity ligation assay (PLA) reagent, was obtained from Olink Bioscience (Uppsala, Sweden). Type I collagen was from Nitta Zeratin (Osaka, Japan). Forskolin (Fsk) was from Calbiochem. H-89, 3-isobutyl-1-methylxanthine (IBMX; a phosphodiesterase inhibitor), and 8-bromo-cAMP were obtained from Sigma.
Plasmid Construction
FLAG-tagged WAVE2 mutants (N-terminal half, amino acids 1–269; C-terminal half, 270–498; WHD, 1–170; Pro-rich region, 270–406; VCA region, 407–498; ΔWHD, 171–498; and ΔVCA, 1–406) (see Fig. 3A) were generated by PCR using p401-WAVE2 (13) as a template and subcloned into the p401 expression vector (28). cDNAs coding for PKA subunits were amplified by PCR using human PKA Cα, RI, and RII cDNAs obtained from the National Institute of Technology and Evaluation (Tokyo, Japan) as templates and subcloned into the pcDNA3.1-HA expression vector and pColdI expression vector. Vinculin cDNA (29) was also subcloned into the pColdI expression vector.
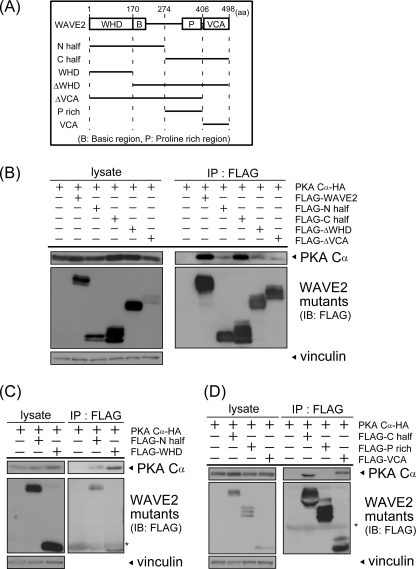
FIGURE 3.
The WHD and VCA region are involved in WAVE2-PKA complex formation. A, illustrations of WAVE2 deletion mutants are shown. Numbers indicate the amino acid number of full-length WAVE2. N half, N-terminal half; C half, C-terminal half; P rich, proline-rich region. B–D, WAVE2 deletion mutants were transfected with HA-PKA Cα into COS-7 cells. Cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG antibody. Precipitated proteins were visualized by immunoblotting (IB) using the indicated antibodies. Asterisks indicate IgG. Vinculin was used as a loading control.
Cell Culture and Transfection
COS-7 and MDA-MB-231 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen) at 37 °C under a humidified atmosphere (5% CO2). COS-7 cells were transfected with expression plasmids using Lipofectamine Plus (Invitrogen). For knockdown experiments, MDA-MB-231 cells were transfected with 10 nm ON-TARGETplus SMARTpool against human WASF2 (Dharmacon) or 20 nm Stealth Select RNAiTM siRNA against WASF2 (Invitrogen) with Lipofectamine RNAi Max (Invitrogen).
Immunoprecipitation and Immunoblotting
Cells and mouse brain were lysed with cell lysis buffer (1% Nonidet P-40, 0.1 mg/ml p-amidinophenylmethanesulfonyl fluoride, 5 μg/ml leupeptin, and 10 μg/ml aprotinin in PBS). Mouse brain was homogenized using a Polytron homogenizer. Equal amounts of cell lysates were incubated with antibodies for 1 h or overnight at 4 °C, followed by incubation with protein G-Sepharose beads for 1 h. Precipitates were washed three times with cell lysis buffer and resolved by SDS-PAGE. Proteins were transferred onto PVDF membranes (Millipore). After blocking with Blocking One (Nacalai Tesque, Kyoto, Japan), the membrane was incubated with primary antibodies and then with secondary antibodies. The complex was detected using an ECL kit (GE Healthcare).
Immunostaining and PLA
MDA-MB-231 cells were cultured on coverslips coated with type I collagen for 24 h. The cells were preincubated with or without 20 μm H-89 for 15 min and then treated with 10 μm Fsk and 100 μm IBMX for 30 min. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.2% Triton X-100 in PBS for 5 min, and blocked with 10% goat serum in PBS for 30 min. They were then incubated with each antibody or phalloidin for 1 h at room temperature or overnight at 4 °C. For immunostaining analysis, they were incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG and Alexa Fluor 543-labeled goat anti-goat IgG (Molecular Probes) for 1 h. For the PLA (30), cells were incubated with PLA probes (anti-mouse MINUS and anti-rabbit PLUS reagents), which are antibodies conjugated with oligonucleotides. These oligonucleotides are combined to form a circle when the MINUS and PLUS probes, i.e. two target proteins, are in close proximity, resulting in a rolling circle amplification of the specific repeated sequence. PLA signals were detected by hybridizing fluorescently labeled probes with the amplified sequences following the manufacturer's instructions. Thus, the PLA assay showed amplified signals only when two target proteins were in close proximity in cells (30). Fluorescence images of immunostaining and PLA were taken with a Pascal confocal microscopy system (Carl Zeiss Co., Ltd.).
PKA-positive membrane protrusion was quantitated as follows. After immunostaining with anti-PKA antibody and phalloidin, phalloidin-positive peripheral regions were postulated as a membrane protrusion, and then the protrusion that co-immunostained with anti-PKA antibody were counted by eye. Scoring was done blindly as to which cell was being analyzed. The lengths of membrane protrusions were measured as illustrated in supplemental Fig. 1.
Protein Purification
GST-WAVE2 (kindly provided by Drs. Shiro Suetsugu and Tadaomi Takenawa) (11) was expressed in Sf9 cells. Cells were lysed with cell lysis buffer, and GST-WAVE2 was precipitated by GST tag affinity using Sepharose 4B (GE Healthcare). GST-WAVE2 was eluted from the resin by 25 mm glutathione-containing buffer (0.1% Nonidet P-40, protease inhibitor mixture (Roche Applied Science), and 10% glycerol in PBS).
His-PKA subunits were expressed in Escherichia coli BL21. Cell lysates were subjected to His tag affinity purification with TALON metal affinity resin (Clontech). Precipitated proteins were eluted with PBS containing 100 mm imidazole, protease inhibitor mixture, and 10% glycerol. Eluted proteins were separated by size exclusion chromatography using Buffer A (50 mm NaCl, protease inhibitor mixture, and 10% glycerol in 10 mm HEPES, pH 7.0) with Superdex 200 10/300 GL on an ÅKTA purifier system (GE Healthcare).
PKA Holoenzyme Formation
PKA holoenzymes were generated as reported with a slight modification (31). Briefly, RI or RII subunits were mixed with the C subunit at a 1:1 molar ratio and preserved overnight at 4 °C in buffer containing 50 mm NaCl, 0.8 mm AMP-PNP (Sigma), 4 mm MnCl2, protease inhibitor mixture, and 10% glycerol in 10 mm HEPES, pH 7.0. Holoenzymes were separated from each subunit using size exclusion chromatography as described above.
In Vitro Binding Assay
Purified GST or GST-WAVE2 was incubated with glutathione beads for 1 h at 4 °C and washed once with Buffer A, followed by incubation with purified PKA subunits or holoenzyme overnight at 4 °C. The precipitates were washed twice with Buffer A and resolved by SDS-PAGE. The gels were subjected to Coomassie Brilliant Blue staining.
Statistical Analysis
Statistical analysis was performed using Student's paired t test.
RESULTS
PKA Holoenzyme Forms a Complex with WAVE2
We have previously reported that PKA can be coprecipitated with overexpressed WAVE2 (13). To characterize the interaction further, we first performed a reciprocal co-immunoprecipitation assay in COS-7 cells. After FLAG-WAVE2 and HA-PKA Cα were transfected into COS-7 cells, cell lysates were immunoprecipitated using anti-FLAG (Fig. 1A) or anti-HA (Fig. 1B) antibody. HA-PKA Cα was clearly detected in immunoprecipitates prepared using anti-FLAG antibody. FLAG-WAVE2 was also detected in immunoprecipitates prepared using anti-HA antibody. To further confirm the association, endogenously expressed WAVE2 was immunoprecipitated from MDA-MB-231 breast cancer cells with anti-WAVE2 antibody, and coprecipitated PKA was examined by immunoblotting with anti-PKA C subunit antibody. As shown in Fig. 1C, the PKA C subunit was significantly coprecipitated with WAVE2 in MDA-MB-231 cells. The co-immunoprecipitation of endogenously expressed WAVE2 with the PKA C subunit was also observed in mouse brain extracts (Fig. 1D). These results suggest that the PKA C subunit associates with WAVE2 under physiological conditions.
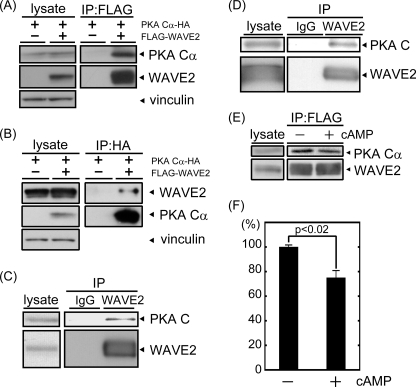
FIGURE 1.
PKA is co-immunoprecipitated with WAVE2. A and B, FLAG-WAVE2 and HA-PKA Cα were transfected into COS-7 cells. The cell lysate was subjected to immunoprecipitation (IP) with anti-FLAG (A) or anti-HA (B) antibody. Coprecipitated proteins were visualized by immunoblotting using anti-FLAG and anti-HA antibodies. Vinculin was used as a loading control. C and D, endogenous WAVE2 was immunoprecipitated from MDA-MB-231 cells (C) or mouse brain extracts (D) with anti-WAVE2 antibody. Coprecipitated PKA was immunoblotted with anti-PKA C subunit antibody. E, cell lysate was isolated from COS-7 cells transfected with FLAG-WAVE2 and HA-PKA Cα. Following the addition of 8-bromo-cAMP (10−5 m), lysates were subjected to immunoprecipitation with anti-FLAG antibody. Proteins were visualized by immunoblotting using the indicated antibodies. F, the amount of coprecipitated PKA Cα subunit with WAVE2 (PKA Cα/WAVE2) was quantified by capturing film images and processing with ImageJ software (n = 3).
The PKA holoenzyme contains two R and two C subunits. cAMP binding to R subunits induces the dissociation of R subunits from C subunits. We next examined the effect of cAMP treatment on WAVE2-PKA complexes to determine whether the PKA holoenzyme is included in this complex. The addition of 10 μm 8-bromo-cAMP to cell lysates induced the dissociation of C and R subunits (data not shown). This treatment released a significant amount of PKA C subunits from the complex (Fig. 1, E and F), suggesting that PKA holoenzymes are included in the WAVE2-PKA complex.
To determine whether WAVE2 binds to PKA directly, we performed the in vitro binding assay. First, His-tagged PKA RI and RII subunits and the PKA C subunit, as well as GST-tagged WAVE2, were expressed in and purified from E. coli or Sf9 cells (Fig. 2, A and B). GST-WAVE2 or GST immobilized on glutathione beads was incubated with these subunits and then precipitated. As shown in Fig. 2B, the PKA C subunit and both R subunits (RI and RII) were clearly coprecipitated with GST-WAVE2 but not with GST. Vinculin, another actin-binding protein, was not coprecipitated with either GST or GST-WAVE2. We also investigated the direct interaction of WAVE2 with PKA holoenzymes. PKA type I and II holoenzymes formed in vitro were also coprecipitated with purified GST-WAVE2 (Fig. 2C). The observation that holoenzymes, as well as both C and R subunits, can associate with WAVE2 directly is consistent with the results of Fig. 1 (E and F), in which WAVE2 partially dissociated from the PKA C subunit after treatment with 8-bromo-cAMP. These results suggest that WAVE2 can directly interact with PKA subunits as well as PKA holoenzymes.
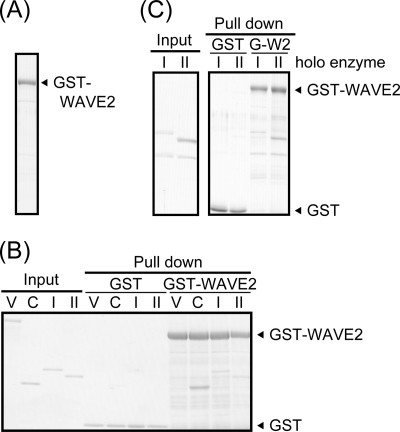
FIGURE 2.
WAVE2 directly binds to PKA. A, purified GST-WAVE2 protein was confirmed by Coomassie Brilliant Blue staining. B and C, purified PKA subunits (B) or the PKA holoenzyme (C) was subjected to in vitro binding assay with purified GST or GST-WAVE2 as described under “Experimental Procedures.” Precipitates were separated by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. Vinculin (V) was used as a negative control. C, I, and II, PKA Cα, RI, and RII subunits, respectively.
The WHD and VCA Region of WAVE2 Mediate the Formation of WAVE2-PKA Complexes
WAVE family proteins have conserved regions, including the WHD and basic, proline-rich, and VCA regions. To elucidate which regions of WAVE2 are important for complex formation, deletion mutants containing either the N- or C-terminal half of WAVE2 were constructed (Fig. 3A) and subjected to the co-immunoprecipitation assay. Interestingly, PKA was coprecipitated with both the N- and C-terminal halves (Fig. 3B). The amount of PKA coprecipitated with the C-terminal half was similar to that of wild-type WAVE2, whereas a smaller amount of PKA was coprecipitated with the N-terminal half. This observation suggests that PKA associates with WAVE2 through at least two regions of WAVE2.
To analyze the interacting regions in detail, we constructed WAVE2 mutants containing or lacking the conserved regions, as shown in Fig. 3A. Focusing on the N-terminal half, which contains the WHD and basic region, we first examined the coprecipitation of PKA with the WHD (Fig. 3C). PKA was substantially coprecipitated with the WHD. Deletion of the WHD (ΔWHD) reduced the amount of coprecipitated PKA (Fig. 3B), indicating that the WHD is involved in the association of WAVE2 with PKA. The C-terminal half is composed of the VCA and proline-rich regions. PKA was coprecipitated with the VCA region but not with the proline-rich region (Fig. 3D). Furthermore, PKA was coprecipitated slightly with a deletion mutant lacking the VCA region (ΔVCA) (Fig. 3B), indicating that the VCA region is required for the association of the C-terminal half with PKA. Together, these results suggest that both the WHD and VCA region are critical for the formation of WAVE2-PKA complexes.
PKA Colocalizes with WAVE2 at the Leading Edge of Membrane Protrusions
WAVE2 regulates membrane protrusions and cell migration by promoting Arp2/3 complex-dependent actin reorganization. Recently, it was reported that PKA activation occurs at the leading edge of migrating cells (23). To test whether the WAVE2-PKA complex is at the leading edge, we first analyzed the subcellular localization of WAVE2 and PKA in MDA-MB-231 breast cancer cells. Immunostaining analysis using confocal microscopy showed that WAVE2 was localized at the leading edge of membrane protrusions, where F-actin was accumulated (Fig. 4, A and B). PKA was also localized at the leading edge of membrane protrusions and colocalized with WAVE2. To show that these two molecules are in the complex, we performed the PLA, in which signals can be detected only when two proteins are in close proximity (30). We counted the PLA signals at membrane protrusions and found 10-fold more PLA signals when anti-WAVE2 and anti-PKA antibodies were used as probes compared with control IgG (Fig. 4, C and D), indicating the close localization of WAVE2 and PKA at membrane protrusions. Together, these data suggest that the WAVE2-PKA complex is localized at membrane protrusions.
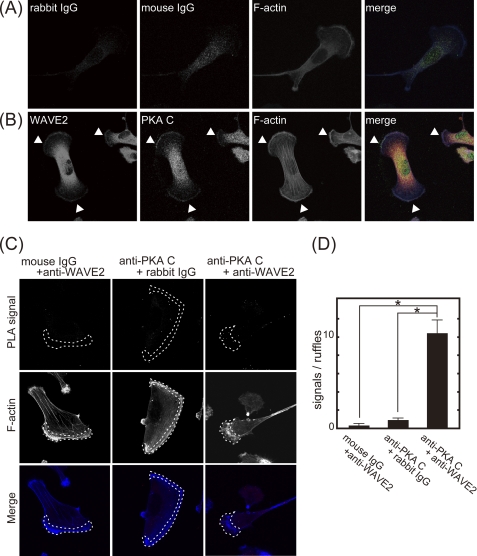
FIGURE 4.
PKA colocalizes with WAVE2 at the leading edge of membrane protrusions. MDA-MB-231 cells were seeded on coverslips coated with type I collagen (6 μg/ml). A and B, cells were immunostained with control rabbit IgG (red), mouse IgG (green), and phalloidin (blue) (A) or with anti-WAVE2 antibody (red), anti-PKA C subunit antibody (green), and phalloidin (blue) (B). Arrowheads indicate WAVE2 and PKA colocalization at membrane protrusions. Note that signals detected by anti-WAVE2 antibody in the central region of the cell are nonspecific because pre-adsorption of anti-WAVE2 antibody with purified GST-WAVE2 removed the signal in peripheral region but not in the central region (supplemental Fig. 2). C, cells were subjected to the PLA using control IgG or anti-WAVE2 and anti-PKA C subunit antibodies. PLA signals and phalloidin staining are shown in the upper and middle panels, respectively. Merged images are shown in the lower panels (PLA signal, red; and F-actin, blue). Phalloidin-positive protrusions are indicated by dashed lines (middle panels). The central region of the cells also showed some PLA signals even with control IgG, suggesting that signals in the central region may be nonspecific signals. D, shown is the quantification of PLA signals. PLA signals localized at phalloidin-positive protrusions were counted.
WAVE2 Is Required for PKA Localization at the Leading Edge
Some groups have reported that the phosphorylation of WAVE2 is important for regulating its function (9, 32, 33). We also reported that vinexin regulates WAVE2 expression in a PKA-dependent manner (13). To examine whether WAVE2 is directly phosphorylated by PKA, COS-7 cells transfected with FLAG-WAVE2 and HA-PKA or MDA-MB-231 cells were treated with and without Fsk. Exogenous or endogenous WAVE2 was immunoprecipitated and immunoblotted using anti-phospho-PKA substrate antibody. Although several phosphorylated proteins were detected in Fsk-stimulated lysates, no signal was detected on immunoprecipitated WAVE2 even after Fsk stimulation (data not shown), suggesting that WAVE2 is not directly phosphorylated by PKA under this condition.
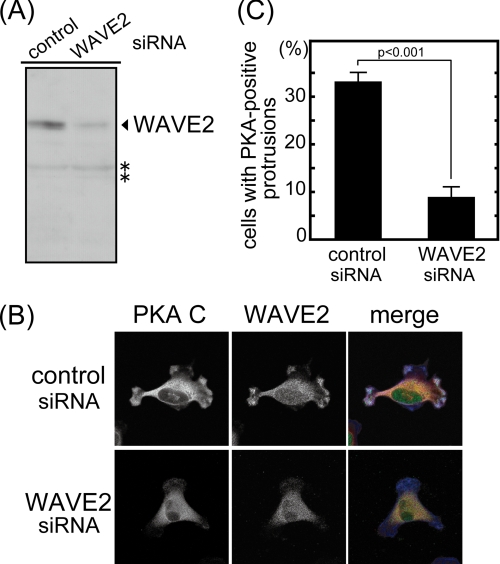
We next performed WAVE2 knockdown by siRNA treatment to test whether WAVE2 has a role in the subcellular localization of PKA. The siRNA treatment reduced WAVE2 protein expression by 70% (Fig. 5A). MDA-MB-231 cells treated with siRNA were then immunostained with anti-WAVE2 and anti-PKA C subunit antibodies. Knockdown of WAVE2 expression did not affect membrane protrusion under quiescent conditions (see below and Fig. 7). WAVE2 and PKA were localized at the leading edge of membrane protrusions in cells transfected with control siRNA. In contrast, WAVE2 and PKA disappeared from the leading edge of membrane protrusions in WAVE2 knockdown cells (Fig. 5B). Thirty-three percent of control siRNA-transfected cells contained PKA-positive membrane protrusions, whereas only 9% of WAVE2 siRNA-transfected cells did (Fig. 5C). These results indicate that PKA requires WAVE2 to be localized at the leading edge of protrusions.
FIGURE 5.
WAVE2 recruits PKA at membrane protrusions. MDA-MB-231 cells were transfected with siRNA against WAVE2. A, the expression level of WAVE2 was detected by immunoblotting. Vinculin was used as a loading control. Signals marked with asterisks seem to be nonspecific because WAVE2 siRNA did not affect these signals. B, MDA-MB-231 cells transfected with siRNA against WAVE2 were seeded on type I collagen and immunostained with anti-WAVE2 and anti-PKA C subunit antibodies. Diffused signals observed in WAVE2 immunostaining of knockdown cells may include nonspecific signals because the expression of WAVE2 estimated by immunoblotting was decreased to 30% in these cells. Merged images (PKA C subunit, red; WAVE2, green; and F-actin, blue) are also shown. C, >65 cells were counted and are denoted as the percentage of cells having PKA-positive membrane protrusions. The graph shows the average of three independent experiments.
FIGURE 7.
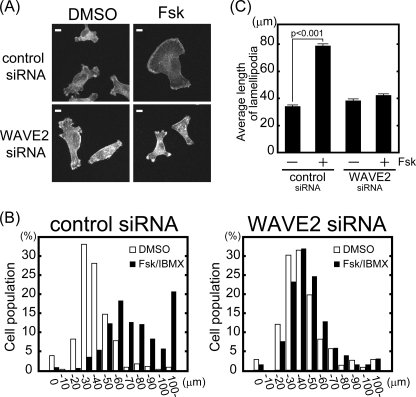
WAVE2 knockdown inhibits membrane protrusions induced by PKA activation. MDA-MB-231 cells were transfected with siRNA against WAVE2. A, cells were seeded on type I collagen, followed by stimulation with and without Fsk and IBMX. F-actin was visualized with phalloidin. Scale bars = 10 μm. B and C, cells were classified according to the length of their longest protrusion (n > 300, combination of three independent experiments). White and black bars indicate dimethyl sulfoxide (DMSO)- and Fsk/IBMX-treated cells, respectively (B). The average length of protrusions from three independent experiments is shown (C).
WAVE2 Is Required for Membrane Protrusion Enlargement Induced by PKA
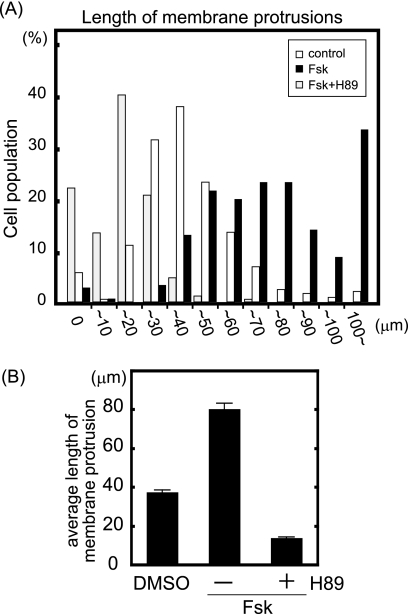
Several recent reports suggest the involvement of PKA in cell migration (19–24). To investigate the effect of PKA activity on cell migration, we examined membrane protrusions, the first step of cell migration, and cell morphology of MDA-MB-231 breast cancer cells under PKA-activated and PKA-inhibited conditions (Fig. 6, A and B). Cells were incubated with Fsk and IBMX, which increases the cAMP concentration in cells, with and without pretreatment with H-89, a PKA-specific inhibitor. Control cells had 37.4 ± 1.0-μm membrane protrusions. Fsk with IBMX treatment enlarged membrane protrusions and increased the length of protrusions 2-fold (80.0 ± 3.2 μm). H-89 pretreatment inhibited the enlargement of protrusions induced by Fsk with IBMX and decreased the length of protrusions to one-third (13.8 ± 0.6 μm). These results indicate that PKA activation stimulates the enlargement of membrane protrusions and that basal PKA activity is required for basal membrane protrusion in quiescent MDA-MB-231 cells.
FIGURE 6.
PKA activation enlarges membrane protrusions. MDA-MB-231 cells were seeded on coverslips coated with type I collagen. Cells were stimulated by Fsk (10 μm) and IBMX (100 μm) for 30 min with and without preincubation with H-89 (20 μm) for 15 min and then stained with phalloidin. The length of phalloidin-positive protrusions was quantified using ImageJ software. A, cells were classified according to the length of their largest protrusion (n > 250, combination of three independent experiments). White and black bars indicate cells treated with dimethyl sulfoxide (DMSO) and Fsk, respectively. Gray bars indicate H-89-preincubated cells treated with Fsk. B, shown is the average length of protrusions from three independent experiments.
We next performed WAVE2 knockdown to test whether WAVE2 is involved in membrane protrusion induced by PKA activation. Under quiescent conditions, i.e. without Fsk treatment, WAVE2 knockdown did not affect the length of membrane protrusions (Fig. 7). Control and WAVE2 knockdown cells had 34.1 ± 0.9-μm and 38.3 ± 1.2-μm membrane protrusions, respectively. This result indicates that WAVE2 is not necessary for membrane protrusions under quiescent conditions. Fsk with IBMX treatment enlarged membrane protrusions ∼2-fold in control knockdown cells (78.7 ± 2.5 μm). In contrast, Fsk with IBMX treatment did not increase membrane protrusions in WAVE2 knockdown cells (42.2 ± 1.1 μm). Similar results were also obtained using cells transfected with two different siRNAs against WAVE2 (supplemental Fig. 3). These results suggest that WAVE2 is involved in membrane protrusions induced by PKA activation.
DISCUSSION
An increasing body of evidence has suggested that PKA activation regulates cell migration as well as many other physiological phenomena. PKA has broad substrate specificity (17). Thus, protein-protein interaction forming a complex with AKAPs is considered to be necessary for the localization of PKA in specific subcellular regions and PKA phosphorylation of specific substrates. However, AKAPs, as well as other proteins associated with PKA that contribute to PKA-mediated cell migration, are not fully understood. In this study, we have demonstrated that WAVE2 associates with PKA in MDA-MB-231 cells and mouse brain extracts. The PKA holoenzyme and both C and R subunits bind directly to WAVE2 in vitro. Both WAVE2 and PKA were localized at membrane protrusions, and WAVE2 was necessary for the localization of PKA at membrane protrusions. From these observations, we propose that WAVE2 works as an AKAP involved in membrane protrusions.
WAVE family proteins have been shown to stimulate Arp2/3-mediated actin polymerization. In addition to this function, one WAVE family protein, WAVE1, has been reported to associate with PKA (34) and works as an AKAP for recruiting PKA to mitochondria (35, 36). WAVE1 forms a complex containing PKA as well as Bad (Bcl-2 antagonist of cell death) and glucokinase and integrates the glycolytic and apoptotic pathways. Our observations in this study suggest that WAVE2 functions as an AKAP recruiting PKA to membrane protrusions. WAVE2 associated directly with PKA subunits as well as PKA holoenzymes. Immunocytochemical analysis showed that endogenous WAVE2 and PKA were colocalized at membrane protrusions in MDA-MB-231 cells. PLA analysis indicated that they were in close proximity at membrane protrusions. There are at least two possibilities for the function of the WAVE2-PKA complex. One possibility is that WAVE2 is a PKA substrate and that complex formation enhances PKA phosphorylation of WAVE2 and WAVE2-mediated Arp2/3 activation; however, we could not detect any increase in phosphorylation by PKA activation. Another possibility is that the association recruits PKA to membrane protrusions where WAVE2 is localized. Indeed, we have clearly succeeded in showing that WAVE2 knockdown reduced PKA localization at membrane protrusions without affecting membrane protrusions under quiescent conditions. These results suggest that WAVE2 makes a complex with PKA and acts as AKAP that recruits PKA at membrane protrusions. In addition to WAVE2, integrin α4 (37) and AKAP-lbc (38) have been reported to form a complex with PKA and recruit PKA to protrusions in colon carcinoma cells and CHO cells. Interestingly, AKAP-lbc ablation reduces but does not completely disrupt PKA localization at protrusions (38). Thus, it is quite possible that several AKAPs, including WAVE2, integrin α4, and AKAP-lbc, work cooperatively to recruit and anchor PKA at protrusions in migrating cells.
We have also shown that PKA activation induces the enlargement of membrane protrusions of MDA-MB-231 cells. WAVE2 was required for this enlargement as well as the localization of PKA at membrane protrusions. PKA activation at the leading edge in response to PDGF stimulation and the front-to-back gradient of cAMP in migrating cells have been reported (23). Membrane ruffles induced by PDGF are reported to involve polarized PKA activation (20). These observations are consistent with our findings. Several proteins that regulate cell adhesion and cytoskeleton, including integrin α4, VASP, and Rho GTPase, are phosphorylated by PKA at the leading edge of cells (19, 39–41). PKA at the membrane protrusions recruited by WAVE2 could phosphorylate these substrates in a spatially restricted manner.
Mutational analysis of WAVE2 in this study revealed that both the WHD and VCA region are involved in WAVE2-PKA complex formation. Westphal et al. (34) reported that WAVE1 forms a complex with PKA through its VCA region. They also reported that WAVE2 and WAVE3 do not form a complex with PKA in HEK293 cells. This apparent inconsistent observation may imply that different mechanisms of binding of WAVE proteins to PKA, which is mediated by both the WHD and VCA region or the VCA region alone, affect the affinity of each cell type or that the different expressions of binding partners of WAVE proteins in each cell type affect the affinity. The observation that PKA preferred WAVE1 to WAVE2 in NIH3T3 cells but preferred WAVE2 to WAVE1 in COS-7 cells supports this explanation (supplemental Fig. 4).
PKA activity is reported to be required for PIP3 accumulation at membrane protrusions after PDGF stimulation (20). PIP3 binds to WAVE2 and directs the localization of WAVE2 at membrane protrusions. In this study, we have demonstrated that WAVE2 is necessary for the localization of PKA under quiescent conditions as well as for membrane protrusions induced by PKA activation. From these observations, it can easily be speculated that WAVE2 first recruits PKA at membrane protrusions, where recruited PKA induces the accumulation of PIP3 after growth factor stimulation. Enriched PIP3 would further recruit WAVE2 protein at membrane protrusions and stimulate Arp2/3-mediated actin polymerization as a positive feedback, leading to further membrane protrusions. Clarifying how each molecule contributes to membrane protrusions in combination with other molecules will be an important issue for future studies.
Supplementary Material
Acknowledgments
We thank Dr. S. Suetsugu (Tokyo University) and Dr. T. Takenawa (Kobe University) for providing valuable material.
This work was supported in part by The Naito Foundation, a Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, research fellowships from the Japan Society for the Promotion of Science for Young Scientists, and a grant from the Bio-oriented Technology Research Advancement Institution.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- WHD
- WAVE homology domain
- PIP3
- phosphatidylinositol 3,5-triphosphate
- R
- regulatory
- C
- catalytic
- AKAP
- A-kinase-anchoring protein
- VCA
- verprolin homology/cofilin homology/acidic
- PLA
- proximity ligation assay
- Fsk
- forskolin
- IBMX
- 3-isobutyl-1-methylxanthine
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1. Takenawa T., Suetsugu S. (2007) Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 2. Pollitt A. Y., Insall R. H. (2009) J. Cell Sci. 122, 2575–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stradal T. E., Rottner K., Disanza A., Confalonieri S., Innocenti M., Scita G. (2004) Trends Cell Biol. 14, 303–311 [DOI] [PubMed] [Google Scholar]
- 4. Suetsugu S., Miki H., Takenawa T. (1999) Biochem. Biophys. Res. Commun. 260, 296–302 [DOI] [PubMed] [Google Scholar]
- 5. Miki H., Suetsugu S., Takenawa T. (1998) EMBO J. 17, 6932–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamazaki D., Oikawa T., Takenawa T. (2007) J. Cell Sci. 120, 86–100 [DOI] [PubMed] [Google Scholar]
- 7. Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L. B., Steffen A., Stradal T. E., Di Fiore P. P., Carlier M. F., Scita G. (2004) Nat. Cell Biol. 6, 319–327 [DOI] [PubMed] [Google Scholar]
- 8. Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T. E. (2004) EMBO J. 23, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. (2002) Nature 418, 790–793 [DOI] [PubMed] [Google Scholar]
- 11. Oikawa T., Yamaguchi H., Itoh T., Kato M., Ijuin T., Yamazaki D., Suetsugu S., Takenawa T. (2004) Nat. Cell Biol. 6, 420–426 [DOI] [PubMed] [Google Scholar]
- 12. Miki H., Yamaguchi H., Suetsugu S., Takenawa T. (2000) Nature 408, 732–735 [DOI] [PubMed] [Google Scholar]
- 13. Mitsushima M., Sezaki T., Akahane R., Ueda K., Suetsugu S., Takenawa T., Kioka N. (2006) Genes Cells 11, 281–292 [DOI] [PubMed] [Google Scholar]
- 14. Machesky L. M., Insall R. H. (1998) Curr. Biol. 8, 1347–1356 [DOI] [PubMed] [Google Scholar]
- 15. Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E., Pollard T. D. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kunda P., Craig G., Dominguez V., Baum B. (2003) Curr. Biol. 13, 1867–1875 [DOI] [PubMed] [Google Scholar]
- 17. Shabb J. B. (2001) Chem. Rev. 101, 2381–2411 [DOI] [PubMed] [Google Scholar]
- 18. Taylor S. S., Kim C., Vigil D., Haste N. M., Yang J., Wu J., Anand G. S. (2005) Biochim. Biophys. Acta 1754, 25–37 [DOI] [PubMed] [Google Scholar]
- 19. Howe A. K. (2004) Biochim. Biophys. Acta 1692, 159–174 [DOI] [PubMed] [Google Scholar]
- 20. Deming P. B., Campbell S. L., Baldor L. C., Howe A. K. (2008) J. Biol. Chem. 283, 35199–35211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edin M. L., Howe A. K., Juliano R. L. (2001) Exp. Cell Res. 270, 214–222 [DOI] [PubMed] [Google Scholar]
- 22. O'Connor K. L., Mercurio A. M. (2001) J. Biol. Chem. 276, 47895–47900 [DOI] [PubMed] [Google Scholar]
- 23. Lim C. J., Kain K. H., Tkachenko E., Goldfinger L. E., Gutierrez E., Allen M. D., Groisman A., Zhang J., Ginsberg M. H. (2008) Mol. Biol. Cell 19, 4930–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Howe A. K., Baldor L. C., Hogan B. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14320–14325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beene D. L., Scott J. D. (2007) Curr. Opin. Cell Biol. 19, 192–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Langeberg L. K., Scott J. D. (2005) J. Cell Sci. 118, 3217–3220 [DOI] [PubMed] [Google Scholar]
- 27. Wong W., Scott J. D. (2004) Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 28. Kioka N., Sakata S., Kawauchi T., Amachi T., Akiyama S. K., Okazaki K., Yaen C., Yamada K. M., Aota S. (1999) J. Cell Biol. 144, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Takahashi H., Mitsushima M., Okada N., Ito T., Aizawa S., Akahane R., Umemoto T., Ueda K., Kioka N. (2005) Biochem. Biophys. Res. Commun. 336, 239–246 [DOI] [PubMed] [Google Scholar]
- 30. Gustafsdottir S. M., Schallmeiner E., Fredriksson S., Gullberg M., Söderberg O., Jarvius M., Jarvius J., Howell M., Landegren U. (2005) Anal. Biochem. 345, 2–9 [DOI] [PubMed] [Google Scholar]
- 31. Kim C., Cheng C. Y., Saldanha S. A., Taylor S. S. (2007) Cell 130, 1032–1043 [DOI] [PubMed] [Google Scholar]
- 32. Danson C. M., Pocha S. M., Bloomberg G. B., Cory G. O. (2007) J. Cell Sci. 120, 4144–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pocha S. M., Cory G. O. (2009) Cell Motil. Cytoskeleton 66, 36–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Westphal R. S., Soderling S. H., Alto N. M., Langeberg L. K., Scott J. D. (2000) EMBO J. 19, 4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Danial N. N., Gramm C. F., Scorrano L., Zhang C. Y., Krauss S., Ranger A. M., Datta S. R., Greenberg M. E., Licklider L. J., Lowell B. B., Gygi S. P., Korsmeyer S. J. (2003) Nature 424, 952–956 [DOI] [PubMed] [Google Scholar]
- 36. Rawe V. Y., Ramalho-Santos J., Payne C., Chemes H. E., Schatten G. (2004) Hum. Reprod. 19, 2594–2604 [DOI] [PubMed] [Google Scholar]
- 37. Lim C. J., Han J., Yousefi N., Ma Y., Amieux P. S., McKnight G. S., Taylor S. S., Ginsberg M. H. (2007) Nat. Cell Biol. 9, 415–421 [DOI] [PubMed] [Google Scholar]
- 38. Paulucci-Holthauzen A. A., Vergara L. A., Bellot L. J., Canton D., Scott J. D., O'Connor K. L. (2009) J. Biol. Chem. 284, 5956–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goldfinger L. E., Han J., Kiosses W. B., Howe A. K., Ginsberg M. H. (2003) J. Cell Biol. 162, 731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Butt E., Abel K., Krieger M., Palm D., Hoppe V., Hoppe J., Walter U. (1994) J. Biol. Chem. 269, 14509–14517 [PubMed] [Google Scholar]
- 41. Howe A. K., Hogan B. P., Juliano R. L. (2002) J. Biol. Chem. 277, 38121–38126 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.