Abstract
Chondroadherin is a leucine-rich repeat protein known to mediate adhesion of isolated cells via the integrin α2β1 and to interact with collagen. In this work, we show that cell adhesion to chondroadherin leads to activation of MAPKs but does not result in cell spreading and division. This is in contrast to the spreading and dividing of cells grown on collagen, although the binding is mediated via the same α2β1 receptor. We identified a cell binding motif, CQLRGLRRWLEAK318 by mass spectrometry after protease digestion of chondroadherin. Cells adhering to the synthetic peptide CQLRGLRRWLEAK318 remained round, as was observed when they bound to the intact protein. The peptide added in solution was able to inhibit cell adhesion to the intact protein in a dose-dependent manner and was also verified to bind to the α2β1 integrin. A cyclic peptide, CQLRGLRRWLEAKASRPDATC326, mimicking the structural constraints of this sequence in the intact protein, showed similar efficiency in inhibiting binding to chondroadherin. The unique peptide motif responsible for cellular binding is primarily located in the octamer sequence LRRWLEAK318. Binding of cells to the active peptide or to chondroadherin immobilized on cell culture plates rapidly induces intracellular signaling (i.e. ERK phosphorylation). Thus, chondroadherin interaction with cells may be central for maintaining the adult chondrocyte phenotype and cartilage homeostasis. The peptides, particularly the more stable cyclic peptide, open new opportunities to modulate cell behavior in situations of tissue pathology.
Keywords: Cell Surface Receptor, Cellular Regulation, Collagen, Connective Tissue, Extracellular Matrix Proteins, Integrin, Cell Matrix Interaction, Chondroadherin, Leucine-rich Repeat Protein, Cartilage
Introduction
Cartilage contains one specific type of cell, the chondrocyte, surrounded by an extensive extracellular matrix accounting for ∼90% of the tissue volume. This matrix provides the structure for mechanical stability and resistance to load. A key element of the matrix is the fibrillar network, with collagen type II as a major constituent, which provides the tensile properties important for distributing load and retaining the extremely anionic proteoglycan aggrecan (1, 2). Aggrecan binds specifically to hyaluronan via its N-terminal domain, thereby forming multimolecular aggregates consisting of 100 or more molecules. This creates a very high fixed charge density, allowing for water retention and resistance to deformation (3).
Cells orchestrate and regulate the formation and maintenance of the extracellular matrix by interacting with their surroundings via a number of different receptors, such as integrins (4), heparan sulfate proteoglycans (5), hyaluronan receptors (6), and discoidin receptors (7). These interactions elicit signals that lead to degradation and removal of matrix elements, as well as to the production of new constituents. Many of the components found in the matrix have the ability to interact both with other matrix components and cell surface receptors. Hyaluronan is an archetypal example that can bind to specific receptors, such as CD44, whereas other portions of this extended molecule can bind to aggrecan molecules (8, 9). Other matrix components include collagens, which bind to integrins, particularly α1β1, α2β1, α10β1, and α11β1 (for a review, see Ref. 4). The small leucine-rich repeat protein PRELP (10) binds to both collagen and the heparan sulfate chains of proteoglycans (11), such as syndecans and glypicans, present at the cell surface.5 Fibronectin, quantitatively a minor component of articular cartilage, interacts with integrin α5β1 as well as heparan sulfate and collagen (12).
Chondroadherin is a cartilage matrix protein that is primarily found close to the cells. It is particularly prominent in the cartilage of the growth plate, primarily between the proliferative and hypertrophic zones (13). The protein is a member of the leucine-rich repeat family but distinguishes itself by having a double disulfide loop close to its C terminus, whereas all of the other members have a single loop in this region. It does not contain the N-terminal extension common to the other LRR proteins. Chondroadherin is also unique among these proteins in having no posttranslational glycosylation (14). The protein can bind with high affinity at two distinct sites of triple helical collagens (15), a process that, in analogy to that of other LRR proteins, may be mediated by motifs in the LRR repeat region. Chondroadherin can also bind to the α2β1 integrin (16) at the cell surface of chondrocytes. The region of chondroadherin interacting with the integrin is not known.
Cells adhering via α2β1 to chondroadherin remain round, as do cells adhering via α5β1 to the integrin binding domain of fibronectin (12). In the case of fibronectin, treatment with a heparin-binding fragment of the molecule (HepII), a peptide from this domain, or phorbol esters to activate protein kinase C is necessary to provide sufficient signals inducing spreading, formation of focal adhesions, and stress fibers. This process involves both integrin and syndecan receptors at the cell surface. Similarly, cell spreading can be induced on chondroadherin as a consequence of phorbol ester treatment (16).
The unique effects of chondroadherin on cellular morphology suggest a role in modulating chondrocyte activities and matrix composition. Therefore, this study was undertaken to identify the specific peptide motif of the chondroadherin molecule that mediates the interaction with the α2β1 integrin. Identification of a specific peptide mediating the binding may provide an opening for the design of compounds promoting cartilage repair.
MATERIALS AND METHODS
Proteins
Recombinant human chondroadherin was expressed in (U293) EBNA cells (15) and purified as described elsewhere. Collagen type II was isolated from bovine nasal cartilage by pepsin digestion (17). Matrilin-1 was isolated from bovine tracheal cartilage as described (18). Fibronectin and laminin were from Invitrogen, and vitronectin was a kind gift from Prof. Björn Dahlbäck (Lund University).
Expression of Unfolded Recombinant Chondroadherin in Bacteria
Recombinant human chondroadherin, including the endogenous signal peptide, was ligated into the pQE8 vector (Qiagen) containing a 6-histidine residue stretch and expressed in Escherichia coli M15 (pREP4) (Qiagen). The cells were harvested by centrifugation and resuspended in 8 ml of 6 m guanidine HCl, 0.1 m NaH2PO4, 0.5 m NaCl, 5 mm N-ethylmaleimide, pH 8. The cell suspension was passed 10 times through a 0.8-mm injection needle to ensure complete lysis, and the supernatant was collected by centrifugation. Recombinant protein was purified using a 1-ml HiTrap chelating column (GE Healthcare) preloaded with NiCl2 equilibrated in 6 m guanidine HCl, 0.1 m NaH2PO4, 0.5 m NaCl, pH 8. The column was washed with equilibration buffer followed by the same buffer adjusted to pH 6.3. The bound protein was eluted by stepwise lowering of the pH (from pH 4 to pH 3.2). Eluted fractions were neutralized by the addition of 1 m Tris-HCl, pH 9. The purity and identity of the protein was confirmed by SDS-PAGE and Western blotting using antibodies raised against the C-terminal fragment of chondroadherin (15). Fractions containing the recombinant protein were dialyzed against water and lyophilized.
Generation of Chondroadherin Peptides
Intact recombinant protein (1 mg, expressed in EBNA cells) (19) was digested with endoproteinase Lys-C (Roche Applied Science) at an enzyme/substrate ratio of 1:50 for 16 h at 37 °C, according to the manufacturer's instructions. Generated peptides were separated by reversed phase chromatography on a Sephasil C8 column (GE Healthcare) with a gradient of 0–70% acetonitrile, 0.1% TFA (0.1 ml/min over 50 min) using the SMART System (GE Healthcare). The effluent was monitored for absorbance at 215 and 280 nm. Fractions were pooled as indicated, lyophilized, and resuspended in PBS (137 mm NaCl, 2.7 mm KCl, 9.6 mm sodium phosphate, pH 7.4). These pools were used to block cell adhesion to intact chondroadherin coated onto a plastic surface.
Mass Spectrometry
Mass spectrometry was performed using a Bruker Scout 384 Reflex III matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer. The instrument was used in the positive ion mode with delayed extraction and an acceleration voltage of 26 kV. Peptide samples were primarily analyzed using the reflector mode, and 50–150 single-shot spectra were accumulated for improved signal/noise ratio. The ProFound software was used to identify the peptides.
Synthetic Peptides
The candidate peptides were synthesized and purified by Schafer-N (Copenhagen, Denmark). Their primary structures were verified by MALDI-TOF mass spectrometry.
Further Purification of Synthetic Peptides
The synthetic peptides (1 mg) were dissolved in 4 m guanidine HCl, 20 mm Tris, pH 8.0, at 1 mg/ml and purified using a Source 5 RPC column (polystyrene/divinylbenzene beads, GE Healthcare) coupled to an ÄKTA basic HPLC-system (GE Healthcare). Bound material was eluted with a gradient of 0–50% acetonitrile in 0.1% TFA using 20 column volumes. The effluent was monitored for absorbance at 215 and 280 nm. Content and identity of peptides in the fractions were analyzed by mass spectrometry as described above. In parallel, a sample of the cyclic peptide was reduced in 10 mm dithiothreitol for 1 h at 56 °C in 4 m guanidine HCl, 20 mm Tris-HCl, pH 8, followed by alkylation with a 2.5-fold molar excess of N-ethylmaleimide for 0.5 h at room temperature. Purification and demonstration of complete reduction and alkylation were done by HPLC and mass spectrometry as described. Defined fractions were pooled, lyophilized, and repeatedly redissolved in water. Finally, the peptides were weighed, dissolved in PBS, and adjusted to the same molar concentration according to the absorbance at 280 nm, which primarily reflects the content of the single tryptophan in the peptides.
Cells and Cell Culture
Bovine articular cartilage chondrocytes were isolated by collagenase (CLS1, Worthington) digestion of articular cartilage from 4–6-month-old calves as described (20). Human chondrocytes were prepared from cartilage obtained at autopsy from a 2-year-old individual. Cells were isolated as described previously (21) and stored frozen after passage 1. For the work described here, frozen chondrocytes were expanded and used at passage level 5. Cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 100 units/ml penicillin plus 100 μg/ml streptomycin (Invitrogen).
Human cells (K9) originating from a human chondrosarcoma were a kind gift from Drs. Sven Inerot and Anders Lindahl (Sahlgrenska University Hospital, Gothenburg, Sweden). The K9 cells were cultured in DMEM/nutrient mixture F-12 (DMEM/F-12, 1:1) with GLUTAMAX I (Invitrogen), supplemented with 10% fetal calf serum, 25 μg/ml ascorbic acid, 50 IU of penicillin, and 50 μg/ml streptomycin. A different human cell line (105KC) originating from another human chondrosarcoma was a kind gift from Dr. Sven Inerot (Sahlgrenska University Hospital). The 105KC cells were cultured in a mix of 40% DMEM, 40% α-minimum essential medium (Invitrogen), 10% nutrient mixture F-12 I (Invitrogen) supplemented with 100 nm hydrocortisone (Sigma), 100 ng/ml insulin (Sigma), 10% fetal bovine serum, 25 μg/ml ascorbic acid, 50 IU/ml penicillin, and 50 μg/ml streptomycin. Human mammary tumor cells (T47D) from the American Type Culture Collection (Manassas, VA) were cultured in RPMI 1640 (Invitrogen), 10% FCS, 0.2 units/ml insulin.
To harvest cells, culture dishes were rinsed three times with Ca2+/Mg2+-free PBS, and the cells were incubated with 0.5% trypsin and 1 mm EDTA for 30 s. Detached cells were suspended in PBS, Ca2+/Mg2+, containing 1 mg/ml soybean trypsin inhibitor (Sigma-Aldrich) and then washed in PBS. To remove cell surface collagens, the cells were treated with 100 units/ml purified collagenase (CLSPA, Worthington) in PBS for 30 min at 37 °C, followed by a wash with PBS. Primary bovine chondrocytes were treated with collagenase as described and in addition treated with 10 units/ml hyaluronidase (Sigma-Aldrich) before direct binding to the peptide and its inhibition of cell attachment.
Cell Signaling
Tissue culture grade 6-well dishes (Corning Life Sciences) were coated overnight with 5 μg/ml bacterially expressed chondroadherin in 4 m guanidine HCl, 50 mm sodium acetate, pH 5.8; native chondroadherin expressed in EBNA cells; or a 100 μm concentration of the peptide CQLRGLRRWLEAK318 in PBS. The dishes were blocked for nonspecific binding with 0.5% BSA in PBS for 3 h. To test ERK phosphorylation, the human chondrocytes were serum-starved for 48 h prior to experiments. Cells were harvested as indicated and washed and suspended in PBS (with 1 mm CaCl2 and MgCl2) containing 0.1% BSA. Cells were added to the wells coated with chondroadherin or BSA (500,000 cells/well) and allowed to adhere for 1 h. All cells (bound and unbound) were collected and mixed with sample buffer for SDS-PAGE. Cell lysates were analyzed by SDS-PAGE followed by Western blotting. Blots were exposed to the phosphorylation-specific antibody (phospho-p44/42MAPK, New England Biolabs) at dilutions recommended by the supplier. Following visualization with chemiluminescence detection using the Pierce SuperSignal® West Dura extended duration substrate (Pierce), they were stripped by incubation in a 0.2 m Tris/glycine buffer, pH 2.8 (containing 0.1% SDS and 0.1% Tween 20), and reprobed with an antibody to determine total ERK1/ERK2 (p44/42MAPK, New England Biolabs). The relative abundance of phospho-ERK in relation to total ERK of scanned blots was assessed using Gel-Pro Analyzer® software (Media Cybernetics).
Cell Adhesion
Tissue culture grade 48-well dishes (Corning Life Sciences) were coated overnight with 5 μg/ml chondroadherin or matrilin-1 in 4 m guanidine HCl, 50 mm sodium acetate, pH 5.8, whereas native proteins were coated in PBS. Alternatively, coating was with collagen type II (5 μg/ml) diluted in PBS from a stock solution (2.1 mg/ml in 0.5 m acetic acid). In other experiments, wells were coated with collagen, fibronectin, laminin, vitronectin, or chondroadherin (5 μg/ml) or with peptides (20 μm) in PBS overnight at room temperature. The dishes were blocked for nonspecific binding with 0.5% BSA in PBS for 3 h.
An assay to test the inhibition by peptides derived from chondroadherin of cell binding to the immobilized protein was applied. The cells (usually 105KC) were suspended in PBS (with 20 mm HEPES, 1 mm CaCl2 and MgCl2) containing 0.1% BSA and added to the wells coated with chondroadherin or other cell-binding proteins (50,000 cells/well) in the absence or presence of various peptide pools corresponding to an identical aliquot of the chondroadherin digest. Alternatively, synthetic peptides were added at different concentrations. Non-adherent cells were removed after 1 h, and adhesion was determined by lysing the bound cells and measuring lysosomal N-acetylglucosaminidase (22). In studies to establish the time course of the adhesion and spreading, 105KC cells were allowed to adhere for 30 min, unbound cells were removed, and serum-free medium supplemented with or without peptides (250 μg/ml) was added. Cell morphology was visualized by light microscopy after 1, 3, and 20 h.
Affinity Purification of the Cell Surface Receptor on the CQLRGLRRWLEAK318 Peptide
The synthetic peptide CQLRGLRRWLEAK318 was coupled to Ultra Link-agarose (Pierce) via its N-terminal cysteine residue at 1 mg/ml matrix suspension according to the manufacturer's instructions. Control matrix was treated in the same manner but omitting the peptide.
105KC cells were harvested by brief incubation with trypsin-EDTA as described above. Isolated cells were homogenized in 25 mm Tris, pH 7.4, 0.15 m NaCl, 1% Triton X-100, 1 mm MgCl2, 1 mm CaCl2, and 1 mm MnCl2 supplemented with the COMPLETE® protease inhibitor mixture followed by end-over-end incubation at 4 °C for 1 h to further solubilize membrane proteins. The control and peptide affinity matrices (1 ml) were packed in separate minicolumns (Bio-Rad) and equilibrated with 20 volumes of running buffer (10 mm Tris, pH 7.4, 0.1% Triton X-100, 1 mm MgCl2, 1 mm CaCl2, 1 mm MnCl2), supplemented with COMPLETE® protease inhibitor. The cleared cell extract was passed twice through the control column, and the flow-through was subsequently passed four times through the peptide affinity column. Both columns were washed with 15 volumes of running buffer and eluted (0.5-ml fractions) with 10 mm Tris, pH 7.4, 0.1% Triton X-100, 20 mm EDTA supplemented with COMPLETE® protease inhibitor. Eluted proteins were precipitated with methanol and materials were separated by 10% SDS-PAGE under non-reducing conditions and electrotransferred to a PVDF membrane (Pall Corp.). The membrane was blocked with 3% BSA in 10 mm Tris-HCl, 0.15 m NaCl, 1% Tween, pH 7.4, and incubated with an integrin α2-specific antibody (H00003673-M01, Abnova Corp., Taipei, Taiwan) or an integrin β1 antibody (Ab SM2118P, Acris Antibodies GmbH). Bound antibodies were detected by a rabbit anti-mouse HRP-conjugated antibody (Dako A/S) and visualized by chemiluminescence detection using Pierce SuperSignal® West Dura extended duration substrate (Pierce).
RESULTS
Adhesion of 105KC Cells to Chondroadherin and Matrilin-1 via Transmembrane Collagens
Many cells are known to contain transmembrane collagens that may mediate binding to matrix protein. To discern integrin interaction, we needed to develop approaches where we could eliminate effects of the surface collagens.
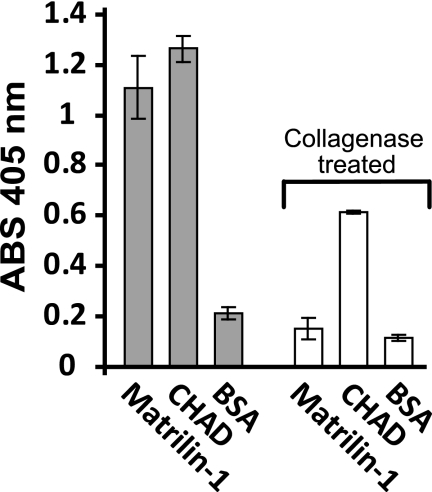
Chondroadherin (15) and matrilin-1 (23) are known to interact with collagen. To identify any binding due to interaction with cell surface collagen (e.g. collagen XIII) (24), we seeded cells (105KC) on matrilin-1 or chondroadherin-coated surfaces with or without pretreatment with collagenase. The proteins were immobilized on 48-well culture dishes, and remaining sites were blocked with BSA. We found that adhesion to matrilin-1 decreased after collagenase treatment to background levels seen with BSA (Fig. 1), indicating that cell surface collagens mediate adhesion to matrilin-1. Binding of cells to chondroadherin was partially decreased upon collagenase treatment (Fig. 1), suggesting that this molecule may bind to cell surface collagens in addition to integrins. To focus on binding to plasma membrane receptors other than collagens, cells were treated with collagenase in all subsequent experiments.
FIGURE 1.
Effect of collagenase treatment on cellular adhesion to chondroadherin. 105KC cells were pretreated in the presence or absence of collagenase to remove cell surface-bound collagen, resuspended in PBS containing 0.1% BSA, and added to wells coated with human chondroadherin (CHAD) expressed in E. coli M15, collagen type II or bovine matrilin-1 (50,000 cells/well). The cells were allowed to adhere for 1 h at 37 °C. Non-adherent cells were removed by washing. Spreading was visualized by light microscopy, and adhesion was determined by analysis of lysosomal N-acetylglucosaminidase. The data represent the average of two wells. Error bars, S.D. in one experiment. Several independent experiments showed similar results.
Adhesion of K9 Cells to Chondroadherin and Collagen Type II
To determine the requirement for a native structure of the chondroadherin molecule in the adhesion process, preparations of different origins were used to analyze adhesion of human chondrosarcoma cells (K9). Chondroadherin of tissue origin that would contain any unique posttranslational modifications of the molecule was extracted with 4 m guanidine HCl from bovine cartilage, whereas recombinant chondroadherin expressed in E. coli M15 represents the protein without disulfide bonds, expected to assume a random coil structure due to the lack of disulfide bond formation. Chondroadherin expressed by EBNA cells represents a native, functional form of the protein (e.g. with collagen binding). Collagen type II was used for comparison because it interacts with the same α2β1 integrin on the cell surface. All proteins were immobilized on 48-well culture dishes. Similar results (below) were obtained with 105KC and K9 cells, showing that the binding is not unique to one cell type. 105KC cells expand in culture much more rapidly than K9; therefore, for convenience, we used this cell type for the bulk of the experiments.
The cells adhered well to the various substrates (Fig. 2). Notably, the cells bound as efficiently to the bacterially expressed chondroadherin. The bacterial protein was most likely unfolded because coating was performed under denaturing conditions in 4 m guanidine HCl, and formation of disulfide bonds had been blocked during isolation by N-ethylmaleimide. Cell binding thus appeared to be mediated by a linear peptide sequence.
FIGURE 2.
Adhesion of K9 cells to native and denatured chondroadherin. 48-well tissue culture dishes were coated with 5 μg/ml tissue-extracted bovine chondroadherin, human chondroadherin expressed in EBNA cells, and human chondroadherin expressed in E. coli (M15) or collagen type II and blocked for nonspecific binding with BSA. The wells were seeded with K9 cells at 50,000 cells/well, and cells were allowed to adhere for 1 h at 37 °C. Non-adherent cells were removed by washing, and adhesion/cell content was determined by analyzing lysosomal N-acetylglucosaminidase. Adhesion is expressed as number of cells. Binding to type II collagen is shown for comparison. Data represent the average of two wells. Error bars, S.D. in one experiment. Independent experiments showed similar results.
Isolation of Chondroadherin Fragments and Identification of Peptide Pools with Binding Activity
Endoproteinase Lys-C-digested recombinant chondroadherin (from EBNA cells) was separated using reversed phase chromatography, and fractions were pooled as indicated in Fig. 3A. Cells were preincubated with the different pools and allowed to adhere to intact bacterially expressed protein (Fig. 3B). The ability of the isolated peptide pools to abolish cell binding was determined. We found that Pool P7 was most potent in inhibiting cell adhesion to chondroadherin. Adhesion to collagen type II was used as a positive control for binding, and adhesion to BSA-coated wells was used as a negative control.
FIGURE 3.
Identification of active chondroadherin peptides. A, human recombinant chondroadherin expressed in EBNA cells was digested with Lys-C and fractionated on a Sephasil C8 column (GE Healthcare). Elution was with a gradient of 0–70% acetonitrile, 0.1% TFA (0.1 ml/min over 50 min) using the SMART® system (GE Healthcare). The absorbance profile of eluted fractions at 280 nm is shown. A similar profile was obtained for absorbance at 215 nm. Pools used for binding and inhibition studies are marked P1–P11. B, inhibition of chondrocyte adhesion to chondroadherin by peptides derived by Lys-C digestion. 48-well tissue culture dishes were coated with 5 μg/ml human chondroadherin (CHAD) expressed in E. coli and blocked for nonspecific binding with BSA. K9 cells were suspended in PBS containing 0.1% BSA and seeded at 50,000 cells/well in the absence (representing adhesion to chondroadherin or collagen type II as a control, without any additions) or in the presence of various peptide pools at amounts corresponding to an equivalent aliquot of the chondroadherin digest. Cells were allowed to adhere for 1 h at 37 °C. Non-adherent cells were removed by washing. Adhesion was determined by analyzing lysosomal N-acetylglucosaminidase. Data represent the average of two wells. Error bars, S.D. in one experiment. Independent experiments showed similar results.
Chondroadherin peptides in Pool P7 were identified by MALDI-TOF MS and are listed in Table 1. By comparing efficiency in inhibiting cell binding by adhesion to different partly separated peptide pools generated by digestion with endoproteinase Glu-C-digested samples (data not shown), a common denominator for inhibition was implicated (i.e. peptide 1947.01 (residues 306–318) was identified as the most likely candidate for mediating cell adhesion to chondroadherin).
TABLE 1.
Identification of peptide fragments in P7
MALDI-TOF MS was used to determine the molecular masses and identify the chondroadherin fragments in Pool 7 (Fig. 3) of a digest separated on a C8 reverse phase column.
| Mass | Amino acid sequence |
|---|---|
| Da | |
| 1029.63 | 46IPKVSEKTK52 |
| 1355.72 | 258FSDGAFLGVTTLK271 |
| 1775.05 | 217LRVVEELKLSHNPLK232 |
| 1947.01 | 304CTCQLRGLRRWLEAK318 |
Effects of Peptide CQLRGLRRWLEAK318 on Cell Adhesion to Chondroadherin and Collagen Type II
A 12-amino acid peptide, CQLRGLRRWLEAK318, representing the candidate sequence identified by mass spectrometry, was synthesized, and its ability to inhibit cell adhesion was determined. We found that adhesion of 105KC cells to immobilized chondroadherin decreased in a dose-dependent manner in the presence of increasing concentrations of the peptide (Fig. 7, CQLRGLRRWLEAK318, ♢). As shown in Fig. 4A, this peptide at 250 μg/ml (150 μm) abolished cell binding to chondroadherin, whereas it had only a minor effect on the adhesion of the cells to collagen type II. To specifically focus on the role of the α2β1 integrin, binding of the T47D cells to the active structure was investigated. These cells only express α2β1 of the collagen-binding integrins (α1β1, α2β1, α10β1, and α11β1) (25). The peptide inhibited the binding to chondroadherin and had only a minor effect on the binding to collagen type II (Fig. 4C). In corroborating experiments, primary bovine chondrocytes adhered to native chondroadherin expressed in EBNA cells as well as to chondroadherin expressed in E. coli (Fig. 4D). The peptide inhibited the binding to both native and denaturated chondroadherin and had a minor effect on the binding to collagen type II. This confirms that the same structure is used by other cell types in the binding.
FIGURE 7.
A cyclic peptide representing a native disulfide loop shows the same efficient inhibitory activity as the linear peptide. A, 48-well tissue culture plates were coated with human chondroadherin expressed in E. coli. 105 KC cells were suspended in PBS containing 0.1% BSA and added to the wells at 50,000 cells/well in the presence of increasing concentrations (0, 1.25, 2.5, 5, 10, 20, and 40 μm) of either the linear peptides QLRGLRRWLEAK318 (○), CQLRGLRRWLEAK318 (♢), and CQLRGLRRWLEAKASRPDATC326 with the cysteine residues alkylated with N-ethylmaleimide (♦) or the cyclic peptides CQLRGLRRWLEAKASRPDATC326 (●). The cyclic peptide CQLRGLRRWEKLAASRPDATC326 (▴) was scrambled at the putative active site. The cells were allowed to adhere for 1 h at 37 °C. Non-adherent cells were removed by washing, and adhesion was quantified by analyzing lysosomal N-acetylglucosaminidase. Adhesion is expressed as percentage adhesion, where adhesion to chondroadherin without the addition of peptides represents 100%. Data represent the average of two wells. Independent experiments showed similar results. B, schematic diagram of chondroadherin structure with the sequence of the peptides studied indicated. The identified peptide is located in one of the C-terminal disulfide loops.
FIGURE 4.
Inhibition of adhesion to human chondroadherin by the peptide CQLRGLRRWLEAK318. 105KC cells (A and B) and T47D cells (C) pretreated with 100 units/ml collagenase or primary bovine chondrocytes (D) pretreated with 100 units/ml collagenase and 10 units/ml hyaluronidase were added to 48-well plates coated with human chondroadherin expressed in E. coli, human chondroadherin expressed in EBNA cells, the CQLRGLRRWLEAK318 peptide (striped bars) or collagen type II at 50,000 cells/well in the presence (white bars)or absence (gray bars) of the synthetic peptide (250 μg/ml, 150 μm for 105 kc cells and T47D cells, 33 μg/ml, 20 μm for primary bovine chondrocytes). The cells were allowed to adhere for 1 h at 37 °C, and non-adherent cells were removed by washing. Adhesion was quantified by analyzing lysosomal N-acetylglucosaminidase. Adhesion is expressed as number of cells, where the data represent the average of two wells. Error bars, S.D. in one experiment. Independent experiments showed similar results.
In addition to inhibiting adhesion to chondroadherin, the peptide itself promoted attachment of chondrocytes. 105KC cells as well as primary bovine chondrocytes plated on immobilized peptide (CQLRGLRRWLEAK318) adhered and remained round (Fig. 4, B and D). This adhesion was prevented in the presence of the same peptide in solution. These results show that the synthetic peptide contains the motif mediating binding of cells to chondroadherin and that there appears to be only one region in the chondroadherin molecule responsible for cell attachment that is present within the tested synthetic peptide.
Affinity Purification of Integrin α2β1 as a CQLRGLRRWLEAK318 Binding Cell Surface Protein
Cell adhesion to intact chondroadherin is known to be mediated via the integrin α2β1 (16). To verify that the identified peptide binds to this integrin, Triton X-100-solubilized membrane proteins were affinity-purified on a CQLRGLRRWLEAK318 column. Any bound integrins were eluted with EDTA because removal of divalent cations is known to disrupt integrin ligand interactions (26). Eluted proteins were separated by SDS-PAGE and transferred to a PVDF membrane. Staining by Western blotting with an antibody specific for the α2 integrin chain showed a reaction with a protein with an approximate molecular size of 145 kDa, as expected for the integrin α2 subunit, which was not present in material eluted from the control column (Fig. 5A). Affinity-purified material was also stained with an antibody specific for the β1 subunit. This antibody did react with a protein of the expected molecular size in material eluted from both columns, indicating the nonspecific adsorption of other β1-containing integrin forms (Fig. 5B). In parallel, a gel lane representing the material eluted with EDTA from the affinity column and stained with Coomassie Blue was analyzed by proteomic approaches using trypsin digestion of the bands observed followed by reversed phase separation of peptides on line with an ion trap mass spectrometer for their identification using standard protocols. A number of intracellular proteins were found as well as Annexins 1 and 2. None of these are candidates for mediating cell adhesion (data not shown). These results demonstrate that the CQLRGLRRWLEAK318 peptide interacts with α2β1 specifically in accordance with our earlier demonstration that chondroadherin mediates cell adhesion through this integrin.
FIGURE 5.
Affinity purification of CQLRGLRRWLEAK318 binding integrins. Membrane proteins were extracted from 105KC cells using Triton X-100. Cleared lysate was passed first over a control column followed by incubation with a peptide affinity column (CQLRGLRRWLEAK318). Bound proteins were eluted with EDTA from the control (ctrl) and peptide columns (pept). Eluted material was separated by SDS-PAGE and transferred to a PVDF membrane. The membrane was incubated with antibodies specific to the α2 (A) and β1 (B) integrin subunits, respectively.
Identification of the Minimal Sequence with Retained Cell Binding
Two overlapping peptides covering the peptide sequence were produced, QLRGLRRW314 and LRRWLEAK318 (outlined in Fig. 7B). They were found to be inefficient at blocking the initial binding to chondroadherin (data not shown). This does not exclude weaker but still functional cell interactions. We therefore decided to test their long term effect on the behavior of cells bound to chondroadherin. Human chondrosarcoma cells (105KC) were allowed to adhere to immobilized chondroadherin, and non-adherent cells were removed after 30 min by washing. Medium without serum was added, with or without peptides representing the whole or overlapping segments of the fragment isolated from chondroadherin. Bound cells were visualized by light microscopy after 1, 3, and 20 h (Fig. 6, A–D). After long incubation times, some spreading was observed on chondroadherin (Fig. 6D). Peptides QLRGLRRWLEAK318 and the truncated version LRRWLEAK318 both prevented spreading and induced cells to form aggregates (Fig. 6, A and B). The N-terminal part of the large peptide QLRGLRRW314 had no effect on cell spreading and migration (Fig. 6C). The sequence active in cell binding and subsequent behavior thus appears to be located in the C-terminal eight residues of the originally identified peptide. In the same type of assay, the addition of antibodies blocking the β1 integrin showed effects similar to those of the active peptides on cell adhesion and behavior. The anti-β1 integrin antibody prevented spreading, and the cells formed aggregates (data not shown). In controls, the addition of EDTA, known to dissociate integrin interactions, lead to cell detachment (data not shown).
FIGURE 6.
Identification of the minimal peptide sequence interfering with chondrocyte adhesion to chondroadherin. The wells of 48-well tissue culture dishes were coated with 5 μg/ml chondroadherin expressed in E. coli and blocked for nonspecific binding with BSA. 105KC cells were suspended in PBS containing 0.1% BSA, and 50,000 cells were dispensed into each well. Non-adherent cells were removed after a 30-min incubation at 37 °C by washing. Peptides as indicated (250 μg/ml (A), 150 μm (B), and 230 μm (C)) were added in serum-free medium (A–C). Control cells were incubated without peptides (D). Cell morphology was recorded by light microscopy after 1, 3, and 20 h of incubation.
Activity of a Cyclic Peptide Corresponding to the Loop Containing the Active Peptide
Because the cell binding motif is present in a disulfide-linked loop in the C-terminal part of chondroadherin (14) (illustrated in Fig. 7B), we synthesized this structure containing the active peptide, CQLRGLRRWLEAKASRPDATC326, where the cysteine residues were linked to form a cyclic peptide. We then compared the blocking activity of this cyclic peptide with the activity of the linear peptide CQLRGLRRWLEAKASRPDATC326 derived from the cyclic peptide by reduction and alkylation as well as the linear peptide QLRGLRRWLEAK318 and the peptide CQLRGLRRWLEAK318. In preliminary experiments, we found that the activity of various batches of individual peptides varied. We found by reverse phase chromatography followed by MALDI-TOF mass spectrometry of individual fractions that there were contaminants lacking single amino acids. Therefore, all preparations of peptides were purified by chromatography on a reverse phase column, and fractions were analyzed by mass spectrometry to identify those containing the correct and pure peptide. These were pooled, and the peptide was recovered by cycles of freeze drying and dissolution in water. To obtain the same molar amount of peptide inhibitor in the inhibition assay, the individual preparations were dissolved at the appropriate concentration, which was verified by absorbance at 280 nm, representing the single tryptophan present in the peptides.
Cell adhesion to immobilized chondroadherin decreased in a dose-dependent manner in the presence of increasing concentrations of the cyclic and linear peptides (Fig. 7A). The cyclic peptide unfolded by reduction and alkylation with N-ethylmaleimide before purification, and the linear cysteine-containing peptide showed a somewhat (∼2 times) higher efficiency in the dose-dependant inhibition of cell binding compared with the cyclic peptide and the shorter variant of the peptide, indicating some contribution by surrounding amino acids. We synthesized and purified a peptide, CQLRGLRRWEKLAASRPDATC326, where we shifted the order of amino acids in the apparent active WLEAK sequence found in the natural peptide. This peptide (Fig. 7, scrambled) did not inhibit cell adhesion to chondroadherin. Another set of peptides where the lysine residue in the cyclic peptide or the short QLRGLRRWLEAK peptide was replaced with an arginine residue showed the same inhibition as the peptides containing lysine (data not shown).
Peptide Specificity for the α2β1 Integrin Interaction with Chondroadherin
The specificity of the cyclic peptide CQLRGLRRWLEAKASRPDATC326 in blocking cell adhesion was determined. Human chondrosarcoma cells (105KC) were allowed to adhere to a number of matrix proteins that bind cells via different integrins. These represent collagen type II (adhering to the α1β1, α2β1, α10β1, and α11β1 integrins), fibronectin (adhering primarily to the α5β1 integrin), laminin (adhering to the α6β1 integrin), vitronectin (adhering to the αVβ3 integrin), and chondroadherin (interacting only with the α2β1 integrin). The cells adhered equally well to all of the proteins. Only adhesion to chondroadherin was inhibited by the cyclic peptide (Fig. 8).
FIGURE 8.
Specificity of the cyclic peptide CQLRGLRRWLEAKASRPDATC326 for blocking adhesion of chondrocytes to chondroadherin. Tissue culture dishes were coated with 5 μg/ml collagen type II (CII), fibronectin (FN), laminin (LN), vitronectin (VN), or human chondroadherin expressed in E. coli (CHAD) and blocked for nonspecific binding with BSA. 105KC cells were suspended in PBS containing 0.1% BSA and added to the wells at 50,000 cells/well in the absence (dark gray bars) or presence (light gray bars) of synthetic peptide (20 μm) and allowed to adhere for 1 h at 37 °C. Non-adherent cells were removed by washing. Adhesion was determined by analyzing lysosomal N-acetylglucosaminidase and expressed as number of cells, where the data represent the average of two wells, and error bars indicate S.D. in one experiment. Independent experiments showed similar results.
Cell Signaling in Response to Chondroadherin Adhesion
To determine if adhesion to chondroadherin and engagement of cell surface integrins lead to the activation of signaling cascades, ERK1/2 phosphorylation was analyzed because integrin-mediated activation of this signaling pathway is well documented (27). In contrast to the chondrosarcoma cell lines, human chondrocytes show a low background level of ERK1/2 phosphorylation and were therefore used to study activation of cell surface receptors. These cells display similar adhesion characteristics to chondroadherin as all other cell lines tested. The level of ERK1/2 phosphorylation relative to total ERK was detected by Western blotting using a phospho-ERK-specific antibody or a phosphorylation-independent antibody (total ERK). Adhesion to bacterially expressed chondroadherin resulted in a 3-fold increase in phosphorylated ERK. Cells adhered to the linear peptide CQLRGLRRWLEAK318 resulted in a 2.8-fold increase in phosphorylated ERK, and adhesion to native chondroadherin results in a 5.1-fold increase without changes in total ERK levels (Fig. 9). This signaling response is in line with a direct interaction of chondroadherin with cell surface integrins.
FIGURE 9.
Adhesion of human chondrocytes to chondroadherin or peptide induces phosphorylation of ERK1/2. Human chondrocytes were added to 6-well plates coated with human chondroadherin expressed in E. coli, native chondroadherin expressed in EBNA cells, synthetic peptide CQLRGLRRWLEAK318 (M15 CHAD) or BSA at 500,000 cells/well, and the cells were allowed to adhere for 1 h at 37 °C. Bound and unbound cells were collected, lysed in SDS-PAGE sample buffer, and separated on a linear (10%) SDS-polyacrylamide gel. The level of ERK1/2-phosphorylation (pERK) relative to total ERK (totERK) was analyzed by Western blotting using a phospho-ERK-specific antibody and a panspecific antibody (total ERK). The relative abundance of phospho-ERK in relation to total ERK of scanned blots was assessed using Gel-Pro Analyzer® software (Media Cybernetics). The data set shown represents one of three similar experiments.
DISCUSSION
The present study describes the identification of a cell-binding motif in chondroadherin located in one of the C-terminal disulfide loops. The amino acid sequence, WLEAK318 contained in this loop appears to be essential for cell interaction with chondroadherin.
Chondroadherin interacts with the α2β1 integrin (16) but also with triple helical collagen (15). Therefore, membrane-bound collagen could contribute to cellular binding by the protein. Indeed, upon collagenase treatment, the cells showed somewhat lower adhesion to chondroadherin. Adhesion of the chondrosarcoma cells used in this study to matrilin-1, also capable of collagen binding, was completely abolished by collagenase treatment, indicating that cell surface collagens have the capacity to interact with the immobilized proteins but that this could be removed. Matrilin-1 has previously been implicated in mediating cell adhesion via the integrin α1β1 (28), but the cells we used appear to bind to matrillin-1 via a membrane collagen rather than via an integrin. Supporting data were obtained in a previous study (9) using primary chondrocytes that were maintained in suspension culture using serum- and ascorbate-free medium. These cells produced little or no collagen and did not bind to matrilin-1 (then named the 148-kDa protein). The same cells bound efficiently to chondroadherin (then called the 36-kDa protein), showing that membrane-associated collagen is not an obligate partner for cell binding to chondroadherin.
Although the cells do not spread on a chondroadherin surface, adhesion to the protein was found to induce ERK phosphorylation in human articular cartilage chondrocytes. These cells bound to chondroadherin with an efficiency similar to that observed with the chondrosarcoma cell lines. However, the chondrosarcoma cells, due to their transformed nature, displayed very high background levels of MAPK activation, making them unsuitable for these studies. Because chondroadherin has previously been shown to bind to integrin α2β1 (16), its capacity to induce a signaling response is similar to that observed via the interaction of other integrin ligands (27). In pilot experiments, we found that human chondrosarcoma cells (K9) adhered equally well to native and unfolded chondroadherin, suggesting that a linear amino acid segment of the chondroadherin molecule was responsible for the binding. The use of bacterially expressed chondroadherin lacking post-translational modifications provided further support for the notion that binding is mediated via a straight peptide sequence not involving posttranslational modifications.
Knowing that a linear peptide sequence can mediate binding, we devised a strategy to search for the binding sequence based on proteolytic cleavage of chondroadherin expressed in EBNA cells. We chose this recombinant protein produced in a mammalian expression system because the bacterially expressed chondroadherin contained a range of fragments, which yielded very complex peptide patterns upon cleavage, hampering their identification. The enzyme proteinase Lys-C was used to cleave the protein into fragments sufficiently large to be likely to retain an active structure. Peptides generated were fractionated and used to inhibit binding of cells to intact protein.
Our data show that the peptide sequence CQLRGLRRWLEAK318 mediates the binding of chondrocytes to chondroadherin. Cells adhering to this peptide remained round, as is also observed when they adhere to the intact protein. Adhesion to other matrix proteins was not significantly affected. Affinity purification verified that the α2β1 integrin complex is responsible for mediating adhesion to the linear sequence QLRGLRRWLEAK located in one of the two cysteine loops in the C terminus of chondroadherin.
To identify a shorter active sequence, we synthesized two overlapping peptides, CQLRGLRR313 and LRRWLEAK318 contained in the originally identified sequence. Neither of these peptides prevented cells from binding to coated chondroadherin. However, the LRRWLEAK peptide retained some effect, because its presence induced cells to round up and form aggregates, as did the larger CQLRGLRRWLEAK318 peptide. The N-terminal part, CQLRGLRR313 peptide, did not show any detectable effects. It thus appears that the C-terminal eight residues of the longer peptide, most likely involving the WLEAK sequence, indeed do mediate binding to the integrin and interfere with the multipoint attachment to the chondroadherin substratum. Short peptides are known to be less active and in some cases show a reduced specificity. A good example are the short RGD peptides that bind to several different integrins, whereas the intact proteins containing this sequence, such as fibronectin, vitronectin, and osteopontin, show more specific receptor preferences (29).
Changing the order of the amino acids in the cyclic peptide CQLRGLRRWLEAKASRPDATC326 from the natural WLEAK sequence to WEKLA demonstrated that the WLEAK sequence is critical for the binding of this peptide. Extending the peptide beyond the lysine did not interfere with cell binding, as would be expected for an internal motif. The active sequence is located in one of the two loop structures found in the C-terminal part of chondroadherin (14). Interestingly, a cyclic peptide corresponding to the whole loop showed similar efficiency in blocking cell adhesion. We also noted that further purification of synthetic peptides is necessary to obtain a reproducible activity. It is possible that the variant peptides interacted in some manner to interfere with the activity of the correct peptide. To ensure that the cyclic peptide was not binding directly to immobilized chondroadherin and thereby blocking cell binding, we used a solid phase assay to demonstrate that the cyclic peptide tagged at its N-terminal with carboxyfluorescein or with biotin showed no binding to chondroadherin (data not shown). The latter peptide was further tested and shown to bind cells similar to the non-biotinylated structure (data not shown). The cyclic peptide CQLRGLRRWLEAKASRPDATC326 and the linear peptide CQLRGLRRWLEAK318 showed specificity for blocking the binding to chondroadherin but were unable to affect cell adhesion to the matrix proteins collagen type II, fibronectin, laminin, and vitronectin, all binding integrins with different specificity (30).
Chondroadherin interaction with cells has rather specific effects on the cellular phenotype with modulation of matrix protein synthesis (data not shown) but an apparent lack of stimulatory effects on cell division and migration. This phenotype differs from that induced by collagen type II binding and could be central for maintaining the chondrocyte phenotype in adult cartilage and its role in matrix homeostasis. Such a role would primarily involve modulation of production of growth factors, proteases, and molecules for matrix building. This is essential for the repair and adaptation of the extracellular matrix, which is continuously exposed to different loads, sometimes in the form of minor trauma, which needs to be dealt with efficiently. A role for the protein in cell division is implicated by its narrow distribution in the growth plate between the major zones of proliferation and hypertrophy (13). To further unravel the role of chondrocyte-matrix interaction in cartilage homeostasis, the use of short active peptides with a distinct specificity for disturbing the binding of matrix components, like the one identified in the present work, will allow us to selectively manipulate the function of chondroadherin in cartilage and identify cellular reactions important for cartilage homeostasis.
Footnotes
This work was supported by the Swedish Research Council, King Gustaf V: 80 Years Fund, the Crafoord Foundation for Research in Rheumatology, Greta and Johan Kock's Foundations, the Royal Physiographic Society, the Österlund Foundation, and ANAMAR. Funding of mass spectrometers was through a grant from the IngaBritt and Arne Lundbergs Research Foundation. A patent application has been submitted for use of the identified active peptides to modulate cell activity.
E. Bengtsson, D. Heinegård, and A. Aspberg, unpublished data.
REFERENCES
- 1. Hardingham T. E., Fosang A. J., Dudhia J. (1992) in Articular Cartilage and Osteoarthritis (Kuettner K. E., Schleyerbach R., Peyron J. G., Hascall V. C. eds) pp. 5–20, Raven Press, New York [Google Scholar]
- 2. Heinegård D., Wieslander J., Sheehan J., Paulsson M., Sommarin Y. (1985) Biochem. J. 225, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maroudas A. (1968) Biophys. J. 8, 575–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heino J. (2000) Matrix Biol. 19, 319–323 [DOI] [PubMed] [Google Scholar]
- 5. Beauvais D. M., Rapraeger A. C. (2004) Reprod. Biol. Endocrinol. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Knudson C. B., Knudson W., Smith R. L. (2004) Clin. Orthop. 427, (suppl.) S152–S162 [PubMed] [Google Scholar]
- 7. Xu L., Peng H., Wu D., Hu K., Goldring M. B., Olsen B. R., Li Y. (2005) J. Biol. Chem. 280, 548–555 [DOI] [PubMed] [Google Scholar]
- 8. Knudson W., Chow G., Knudson C. B. (2002) Matrix Biol. 21, 15–23 [DOI] [PubMed] [Google Scholar]
- 9. Sommarin Y., Larsson T., Heinegård D. (1989) Exp. Cell Res. 184, 181–192 [DOI] [PubMed] [Google Scholar]
- 10. Bengtsson E., Neame P. J., Heinegård D., Sommarin Y. (1995) J. Biol. Chem. 270, 25639–25644 [DOI] [PubMed] [Google Scholar]
- 11. Bengtsson E., Mörgelin M., Sasaki T., Timpl R., Heinegård D., Aspberg A. (2002) J. Biol. Chem. 277, 15061–15068 [DOI] [PubMed] [Google Scholar]
- 12. Woods A., Longley R. L., Tumova S., Couchman J. R. (2000) Arch. Biochem. Biophys. 374, 66–72 [DOI] [PubMed] [Google Scholar]
- 13. Shen Z., Gantcheva S., Mânsson B., Heinegârd D., Sommarin Y. (1998) Biochem. J. 330, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neame P. J., Sommarin Y., Boynton R. E., Heinegård D. (1994) J. Biol. Chem. 269, 21547–21554 [PubMed] [Google Scholar]
- 15. Månsson B., Wenglén C., Mörgelin M., Saxne T., Heinegård D. (2001) J. Biol. Chem. 276, 32883–32888 [DOI] [PubMed] [Google Scholar]
- 16. Camper L., Heinegârd D., Lundgren-Åkerlund E. (1997) J. Cell Biol. 138, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller E. J. (1972) Biochemistry 11, 4903–4909 [DOI] [PubMed] [Google Scholar]
- 18. Paulsson M., Heinegård D. (1981) Biochem. J. 197, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mânsson B., Gülfe A., Geborek P., Heinegård D., Saxne T. (2001) Ann. Rheum. Dis. 60, 27–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sommarin Y., Heinegård D. (1983) Biochem. J. 214, 777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Recklies A. D., Golds E. E. (1992) Arthritis Rheum. 35, 1510–1519 [DOI] [PubMed] [Google Scholar]
- 22. Landegren U. (1984) J. Immunol. Methods 67, 379–388 [DOI] [PubMed] [Google Scholar]
- 23. Hägg P., Rehn M., Huhtala P., Väisänen T., Tamminen M., Pihlajaniemi T. (1998) J. Biol. Chem. 273, 15590–15597 [DOI] [PubMed] [Google Scholar]
- 24. Chen Q., Johnson D. M., Haudenschild D. R., Tondravi M. M., Goetinck P. F. (1995) Mol. Biol. Cell 6, 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sonnenberg A., Linders C. J., Modderman P. W., Damsky C. H., Aumailley M., Timpl R. (1990) J. Cell Biol. 110, 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tuckwell D., Calderwood D. A., Green L. J., Humphries M. J. (1995) J. Cell Sci. 108, 1629–1637 [DOI] [PubMed] [Google Scholar]
- 27. Howe A. K., Aplin A. E., Juliano R. L. (2002) Curr. Opin. Genet. Dev. 12, 30–35 [DOI] [PubMed] [Google Scholar]
- 28. Makihira S., Yan W., Ohno S., Kawamoto T., Fujimoto K., Okimura A., Yoshida E., Noshiro M., Hamada T., Kato Y. (1999) J. Biol. Chem. 274, 11417–11423 [DOI] [PubMed] [Google Scholar]
- 29. Ruoslahti E. (1996) Annu. Rev. Cell Dev. Biol. 12, 697–715 [DOI] [PubMed] [Google Scholar]
- 30. Buck C. A., Horwitz A. F. (1987) Annu. Rev. Cell Biol. 3, 179–205 [DOI] [PubMed] [Google Scholar]