Abstract
Ube2g2 is a human ubiquitin conjugating (E2) enzyme involved in the endoplasmic reticulum-associated degradation pathway, which is responsible for the identification and degradation of unfolded and misfolded proteins in the endoplasmic reticulum compartment. The Ube2g2-specific role is the assembly of Lys-48-linked polyubiquitin chains, which constitutes a signal for proteasomal degradation when attached to a substrate protein. NMR chemical shift perturbation and paramagnetic relaxation enhancement approaches were employed to characterize the binding interaction between Ube2g2 and ubiquitin, Lys-48-linked diubiquitin, and Lys-63-linked diubiquitin. Results demonstrate that ubiquitin binds to Ube2g2 with an affinity of 90 μm in two different orientations that are rotated by 180° in models generated by the RosettaDock modeling suite. The binding of Ube2g2 to Lys-48- and Lys-63-linked diubiquitin is primarily driven by interactions with individual ubiquitin subunits, with a clear preference for the subunit containing the free Lys-48 or Lys-63 side chain (i.e. the distal subunit). This preference is particularly striking in the case of Lys-48-linked diubiquitin, which exhibits an ∼3-fold difference in affinities between the two ubiquitin subunits. This difference can be attributed to the partial steric occlusion of the subunit whose Lys-48 side chain is involved in the isopeptide linkage. As such, these results suggest that Lys-48-linked polyubiquitin chains may be designed to bind certain proteins like Ube2g2 such that the terminal ubiquitin subunit carrying the reactive Lys-48 side chain can be positioned properly for chain elongation regardless of chain length.
Keywords: Biophysics, Enzyme Processing, NMR, Ubiquitin, Ubiquitin Conjugating Enzyme (Ubc), Ubiquitination
Introduction
Proteins to which Lys-48-linked polyubiquitin chains have been attached are destined for degradation by the proteasome (1). The specific polyubiquitylation of protein substrates is accomplished by a cascade of enzymatic steps starting with the ATP-dependent activation of ubiquitin by an E1 protein and culminating in the attachment to substrate of polyubiquitin chains linked together by isopeptide bonds formed between the C terminus of one ubiquitin and the Lys-48 side chain of another ubiquitin (2, 3). Exquisite substrate specificity is achieved by the concerted action of specific pairs of ubiquitin conjugating (E2) and ubiquitin ligase (E3) proteins, of which there are tens and many hundreds found in the cell, respectively.
One such E2:E3 pair is the ubiquitin conjugating enzyme Ube2g2 and the ubiquitin ligase enzyme gp78, which function together as part of the endoplasmic reticulum-associated degradation pathway (4). The endoplasmic reticulum-associated degradation pathway is responsible for the degradation of misfolded, unassembled, and tightly regulated proteins within the endoplasmic reticulum compartment (5–7). Proteins targeted for degradation by endoplasmic reticulum-associated degradation must be retro-translocated to the cytosol to be polyubiquitylated and ultimately degraded by the ubiquitin-proteasome machinery (8, 9). Proper function of the endoplasmic reticulum-associated degradation pathway is essential for cell homeostasis, and its dysfunction is implicated in several diseases including Parkinson (10), cystic fibrosis (11), and cancer metastasis (12, 13).
Although few details are known, some general features of Ube2g2·gp78-mediated polyubiquitylation are emerging. It has been shown that Ube2g2 preassembles Lys-48-specific polyubiquitin chains on its active site before en bloc transfer to substrate in a gp78-mediated process (14). Apparently, polyubiquitin chain assembly is carried out by the concerted action of different Ube2g2 molecules, each bearing active site-linked ubiquitin molecules. But the rate of assembly of Lys-48-linked polyubiquitin chains by Ube2g2 depends strongly on the presence of gp78 (14–17). Recent studies have shown that Ube2g2 binds the G2BR helix of gp78 with a 5–20 nm affinity with the subsequent allosteric activation of Ube2g2 binding to the RING domain of gp78 (15, 16).
The noncovalent interaction of ubiquitin and Ube2g2 has not been reported but may be anticipated due to the already known details of its function and in analogy to other E2 proteins. For some E2 proteins, the interaction with ubiquitin is mediated by outlying ubiquitin associated domains contained within C- or N-terminal extensions to the E2 core (18). Yet ubiquitin has also been observed to bind to the E2 core, as in the case of HsUbc2b (19) and UbcH5c (20) (although not for UbcH7). The NMR structure of UbcH5c·Ub complex shows ubiquitin bound to the β-sheet region of UbcH5c, a region that is remote from the active site cysteine yet necessary for BRCA1-mediated ubiquitin chain elongation in vitro (20). These results suggest the involvement of multiple E2 proteins as well as raising the question as to whether E2 proteins might interact with polyubiquitin chains differently than with ubiquitin alone. Furthermore, one might expect that any interaction might be linkage-dependent given that polyubiquitin chains with different linkages assume different conformations in solution (21, 22).
We have probed the interaction of Ube2g2 with ubiquitin and diubiquitin using NMR chemical shift perturbation (CSP)4 and paramagnetic relaxation enhancement (PRE) experiments. Rosetta-based conformational searches were subsequently employed to produce models of the corresponding complexes. Our results indicate that ubiquitin binds to Ube2g2 at the same site in two different orientations, one of which agrees very closely with that observed for the UbcH5c·Ub complex (20). In addition, we have found that both Lys-48- and Lys-63-linked diubiquitin molecules bind to this same site of Ube2g2 in a fashion that appears to be dominated by interactions between Ube2g2 and a single Ub subunit. In the case of Lys-63-linked diubiquitin, Ube2g2 binds independently to both subunits with similar but not identical affinity. On the other hand, we find that Ube2g2 exhibits a clear preference for binding to the distal subunit of the Lys-48-linked diubiquitin molecule (containing the free Lys-48 side chain) presumably due to steric hindrance of proximal subunit binding by the Lys-48—Gly-76 isopeptide linkage connecting the two ubiquitin molecules. These results suggest that Ube2g2 will in general preferentially bind to the terminal ubiquitin subunit of a Lys-48-linked polyubiquitin chain.
EXPERIMENTAL PROCEDURES
Protein Preparation
Ube2g2 was expressed and purified using a pET28b vector modified to contain a tobacco etch virus cleavage site as previously described (23). Wild type Ub-pET3a plasmid was the gift of Prof. Cecile Pickart. With the exception of the G75CΔ ubiquitin mutant, all other ubiquitin mutants, E2–25K, Ubc13, and MMS2 plasmids were gifts of Prof. David Fushman. G75CΔ-Ub-pET3a was prepared by PCR mutagenesis using oligonucleotide primers from Invitrogen using Pfu Turbo (Stratagene) at an annealing temperature of 40 °C. Plasmid sequence was verified by sequencing at the Johns Hopkins Medical Institute Synthesis and Sequencing Facility. Ubiquitin was expressed and purified as described previously (24). Uniform 15N-labeling of proteins was achieved by first growing transformed BL21(DE3) Escherichia coli in LB media followed by replacement just before induction with M9 minimal medium containing 1.0 g/liter 15NH4Cl as the sole nitrogen source (25). Lys-48-linked diubiquitin was synthesized from 10 mg/ml ubiquitin-D77 (C terminus-blocked) and 10 mg/ml ubiquitin-K48R (lysine side chain replaced) in 50 mm Tris-HCl buffer (pH 8.3) with 5 mm MgCl2, 1 mm tris(2-carboxyethyl)phosphine, 4 mm ATP, 10 mm phosphocreatine, 0.6 units/ml creatine phosphokinase, 0.6 units/ml inorganic phosphatase, 20 μm E2–25K, and 0.1 μm E1 (Boston Biochem) as previously described (26). Lys-63-linked diubiquitin was synthesized from 10 mg/ml ubiquitin-D77 and 10 mg/ml ubiquitin-K63R in 50 mm Tris-HCl buffer (pH 7.5) with 5 mm MgCl2, 1 mm tris(2-carboxyethyl)phosphine, 4 mm ATP, 10 mm phosphocreatine, 0.6 units/ml creatine phosphokinase, 0.6 units/ml inorganic pyrophosphatase, 20 μm Ubc13, 20 μm MMS2, and 0.1 μm E1 as previously described (27). Both diubiquitin molecules were purified by Superdex-75 (GE Healthcare) gel filtration using 10 mm sodium phosphate (pH 7.5), 1 mm tris(2-carboxyethyl)phosphine, 0.1 mm EDTA, and 0.05% NaN3 as a column buffer, with final purity was assayed by SDS-PAGE.
NMR Spectroscopy
All NMR experiments were acquired at 295 K on a Bruker Avance II 600 MHz spectrometer equipped with a single axis pulsed field gradient TCI cryoprobe. For two-dimensional 1H,15N HSQC spectroscopy, 639 and 94 complex data points were acquired with spectral widths of 10000 and 1820 Hz in the 1H and 15N dimensions, respectively. Total experimental acquisition times were typically 45 min per two-dimensional dataset. Data were multiplied by 63°-shifted (1H) and 90°-shifted (15N) sinebell apodization functions, zero-filled, and then Fourier-transformed using NMRPipe software (28). The resulting data matrices were 4096 by 1024 points.
Chemical Shift Perturbation Measurements
Samples were prepared using ∼100 μm 15N isotopically labeled protein in 300 μl NMR buffer (10% D2O, 10 mm NaPi (pH 7.5), 1 mm tris(2-carboxyethyl)phosphine, 0.1 mm EDTA, 0.05% NaN3) in a susceptibility matched NMR tube (Shigemi, Inc.). A stock solution of the titrant protein was prepared in the identical NMR buffer and then added incrementally with two-dimensional 1H,15N HSQC spectra acquired at each point. Peak positions were quantified using the program Sparky (29), with changes in chemical shifts Δδ determined according to,
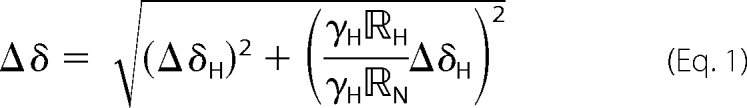 |
with ΔδH, and ΔδN corresponding to the chemical shift changes (in ppm) in the 1H and 15N dimensions, γH and γN corresponding to the gyromagnetic ratios for 1H and 15N, and RH and RN corresponding to the digital resolutions of the 1H and 15N dimensions (respectively 7.82 and 9.68 Hz).
Determination of Binding Affinities
Measured values for Δδ were fit to a two-state binding model simultaneously for the 15–20 most perturbed residues from all associated titrations (either 2 or 3 datasets) using the equation,
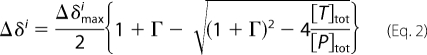 |
with Γ = ([T]Tot + Kd)/[P]Tot. Δi denotes the measured chemical shift change for the ith residue as a function of the concentrations of total titrant protein, [T]tot, and total 15N-labeled protein, [P]tot. Dilution effects were explicitly corrected based on the known added volumes of titrant protein solution. The floating parameters during fitting were the dissociation constant, Kd, the maximum chemical shift changes for each residue, Δδimax, and a multiplicative correction factor, cUb, to correct the concentration of the ubiquitin containing solution (whether titrant or 15N-labeled protein). Concentrations of Ube2g2 were determined by use of the bicinchoninic acid method (30) (Thermo Scientific) using a BSA standard calibrated by amino acid analysis (University of California, Davis, CA). Concentrations for ubiquitin were also determined by the bicinchoninic acid method, with the obtained value used as the initial guess in the fitting procedure. The best fit-optimized ubiquitin concentrations were found to range from 40 to 70% percent of the bicinchoninic acid estimated concentrations. All concentrations for ubiquitin reported in the text have been corrected based on the values resulting from the best fit to the data.
Attachment of Spin Labels
The paramagnetic spin label, S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL), was purchased from Toronto Research Chemicals, Inc. and then reacted with the appropriate single cysteine-containing ubiquitin or diubiquitin mutant according to the following procedure. Lyophilized protein was resuspended in 10 mm DTT, 10 mm NaPi (pH 7.5), 0.1 mm EDTA, and 0.05% NaN3 followed by removal of DTT over a 225-ml Superdex-75 FPLC column (GE Healthcare) using 10 mm NaPi (pH 7.5), 0.1 mm EDTA, and 0.05% NaN3 as a column buffer. Immediately, protein was added to a 5-fold molar excess of MTSL dissolved in acetonitrile, and incubated in darkness at room temperature for 72 h. The solution was then reapplied to the Superdex-75 FPLC column with the same buffer to remove any undesirable disulfide-linked products as well as any unreacted spin label. The proteins, which were spin-labeled in this fashion, were K48C-ubiquitin, G75CΔ-ubiquitin, distal-K48C-ubiquitin in Lys-48-linked diubiquitin, and proximal-G75CΔ-ubiquitin in Lys-48-linked diubiquitin.
Measurement of PREs
Samples were prepared to ∼150 μm 15N-labeled Ube2g2 in 300 μl of NMR buffer (10 mm NaPi (pH 7.5), 10% D2O, 0.1 mm EDTA, 0.05% NaN3) in a Shigemi NMR tube. MTSL-labeled ubiquitin species were then added stepwise to final concentrations ranging from 150 to 500 μm. All protein concentrations were estimated based on the measured A280 in the presence of 6 m guanidine hydrochloride using the method of Gill and von Hippel (31). Two-dimensional 1H,15N HSQC spectra were acquired before and after each addition of the spin-labeled titrant protein. Once the desired final titrant concentration was attained, the spin label was quenched by the addition of 10 mm sodium dithionite (from a 250 mm stock), and a final reference two-dimensional 1H,15N HSQC spectrum was recorded. Peak heights were quantified using the program Sparky (29) with the reported PRE values calculated as the ratio of peak heights measured with active spin label to those measured with reduced spin label (32).
Free Spin Label Binding Assay
The possibility of direct binding of MTSL to Ube2g2 was tested using l-2-amino-3-[thiomethyl-1-(1-oxyl-2,2,5,5-tetramethyl-3 -pyrrolin-3-yl)]propanoic acid dihydrochloride. This compound was selected to avoid the cysteine reactivity of MTSL. A solution containing 150 μm 15N-labeled Ube2g2 in 300 μl of NMR buffer (10 mm NaPi (pH 7.5), 10% D2O, 0.1 mm EDTA, 0.05% NaN3) was placed into a Shigemi NMR tube, and then the free paramagnetic species was added to a final concentration of 150 μm. Two-dimensional 1H,15N HSQC spectroscopy was before and after the addition of spin label to quantify any PRE effects.
RosettaDock Modeling of Ube2g2·Ub and Ube2g2·Lys-48-diUb
The Ube2g2·Ub and Ube2g2·Lys-48-diUb complexes were modeled using the Rosetta 3.2 biomolecular modeling suite (33) with experimentally observed CSP and PRE measurements included to bias the simulations toward physically realistic models. Local docking was performed with a version of the RosettaDock rigid body docking protocol modified to allow backbone flexibility of the four C-terminal ubiquitin residues (34–36). Furthermore, the score functions normally used in RosettaDock were modified to award a bonus to models satisfying the observed CSPs and a penalty for interfacial contacts that did not have corresponding CSPs. Pairs of residues were considered to be at the interface if their Cα atoms were within 8 Å of one other. A model was considered to satisfy a specific PRE restraint if the two residues had a Cα-Cα distance within 15 Å; otherwise the model was penalized by the addition of a weighted bounded constraint function to its score. A more detailed description of the protocol is included in the supplemental Methods.
RESULTS
We have probed the noncovalent interaction between Ube2g2 and both ubiquitin and covalently linked diubiquitin species by means of NMR CSP experiments (37). In these experiments one of the interacting protein components is isotopically labeled to enable its exclusive observation by NMR spectroscopy, and then the unlabeled cognate protein is titrated into the solution. A binding interaction between the two proteins will produce changes in the observed chemical shifts for the isotopically labeled protein. Although these experiments provide a sensitive magnetic footprint of the binding interface, details of the interaction remain ambiguous. We have, therefore, supplemented these data with site-specific data obtained from PRE experiments and then employed a novel Rosetta-based docking algorithm to produce models of the complexes.
Ube2g2 Interacts with Both Ubiquitin and Diubiquitin Species in a Very Similar Manner
The interaction between 15N-labeled Ube2g2 and ubiquitin was monitored by means of two-dimensional 1H,15N HSQC spectroscopy. Upon the addition of ubiquitin, several resonances in the HSQC spectrum of Ube2g2 shifted in position, consistent with fast exchange on the NMR timescale between bound and unbound species (Fig. 1). The Ube2g2 amide chemical shifts most strongly perturbed by ubiquitin binding correspond to residues 23–26, 29–36, 40–41, 43, 50–54, 57–58, 74, and 163–164 (Fig. 2A). Mapping of these most strongly perturbed residues onto the structure of Ube2g2 shows that the shifts are localized and occur on the opposite side of the active site in the region of the β-sheet and the C terminus (Fig. 2B). This is in general agreement with previous reports of ubiquitin binding to E2 proteins (19, 20). Similar titration experiments were then carried out with the addition of Lys-48- and Lys-63-linked diubiquitin species, respectively, into solutions of 15N-labeled Ube2g2. Both diubiquitin species (hereafter referred to as Lys-63-diUb and Lys-48-diUb) bind to Ube2g2, causing chemical shift changes that were very similar to those observed in the ubiquitin titration experiment (Fig. 2, C and D). A close inspection of HSQC peak trajectories observed for all three titrations (Ub, Lys-48-diUb, Lys-63-diUb) finds only very minor differences, suggesting a very similar if not identical mode of interaction.
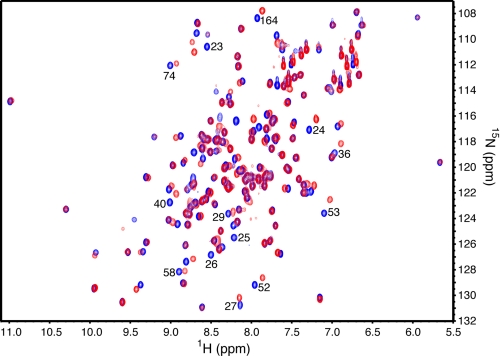
FIGURE 1.
Overlay of two-dimensional [1H, 15N] HSQC spectra of 15N-labeled Ube2g2 (blue) and 50 μm15N-labeled Ube2g2 in the presence of 540 μm ubiquitin (red).
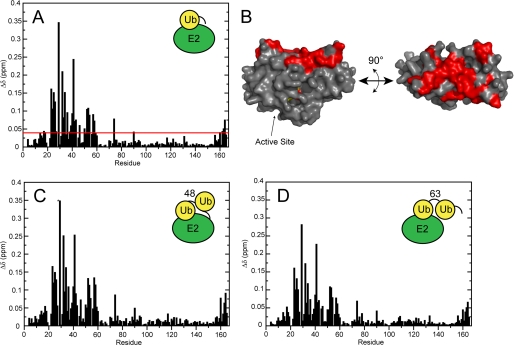
FIGURE 2.
Chemical shift perturbations observed upon the addition of different ubiquitin species to a solution of 15N-labeled Ube2g2. A, measured Ube2g2 chemical shift perturbations upon the addition of ubiquitin (50 μm Ube2g2, 500 μm Ub) is shown. B, a surface map of Ube2g2 residues affected by ubiquitin binding is shown. Residues with composite amide chemical shift perturbations >0.04 ppm are colored in red on the NMR solution state structure (PDB ID 2KLY). C, measured Ube2g2 chemical shift perturbations upon the addition of Lys-48-linked diubiquitin (60 μm Ube2g2, 500 μm Lys-48-diUb) is shown. D, measured Ube2g2 chemical shift perturbations upon the addition of Lys-63-linked diubiquitin (60 μm Ube2g2, 400 μm Lys-63-diUb) are shown.
Chemical shift perturbation experiments were also carried out in which unlabeled Ube2g2 is titrated into a solution of 15N-labeled ubiquitin and monitored by two-dimensional [1H,15N] HSQC spectroscopy. Consistent with observations for the inverse experiment, spectral changes for 15N-labeled ubiquitin upon addition of Ube2g2 are in the fast exchange regime (Fig. S1). The chemical shift perturbations observed for 15N-labeled ubiquitin upon Ube2g2 binding are shown in Fig. 3A. The most strongly shifted ubiquitin residues include 7, 13, 14, 32, 42–49, and 66–75 and form a largely contiguous patch on the surface of ubiquitin (Fig. 3B). This binding region approximately spans the area between residue Lys-48 and the flexible C-terminal tail and corresponds to the typical binding interface of ubiquitin often referred to as the hydrophobic patch.
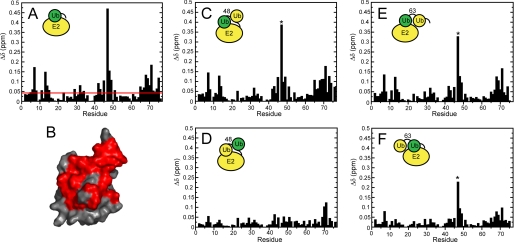
FIGURE 3.
Chemical shift perturbations observed upon the addition of Ube2g2 to different 15N-labeled ubiquitin species. A, measured Ub chemical shift perturbations upon the addition of Ube2g2 (50 μm Ub, 250 μm Ube2g2) is shown. B, a surface map of Ub residues affected by Ube2g2 binding is shown. Residues with composite amide chemical shift perturbations of >0.04 ppm are colored in red on the solution state structure (PDB ID 1D3Z). C, measured chemical shift perturbations for the distal ubiquitin subunit of Lys-48-linked diubiquitin upon the addition of Ube2g2 (70 μm Lys-48-diUb, 250 μm Ube2g2) is shown. D, measured chemical shift perturbations for the proximal ubiquitin subunit of Lys-48-linked diubiquitin upon the addition of Ube2g2 (70 μm Lys-48-diUb, 250 μm Ube2g2) is shown. E, measured chemical shift perturbations for the distal ubiquitin subunit of Lys-63-linked diubiquitin upon the addition of Ube2g2 (70 μm Lys-63-diUb, 250 μm Ube2g2) is shown. F, measured chemical shift perturbations for the proximal ubiquitin subunit of Lys-63-linked diubiquitin upon the addition of Ube2g2 (90 μm Lys-48-diUb, 250 μm Ube2g2) is shown. An asterisk indicates that the reported CSP was extrapolated from earlier titration points due to the broadening out and disappearance of the associated resonance. 15N-Labeled protein components are colored green, and unlabeled proteins are colored yellow in the inset schematic.
Chemical shift perturbation experiments in which 15N-labeled Lys-48-diUb was observed upon the addition of Ube2g2 were carried out utilizing selective 15N isotopic labeling of individual diubiquitin subunits (22, 38). The CSPs observed when the distal subunit (i.e. the ubiquitin subunit with a free Lys-48 residue) is 15N-labeled are summarized in Fig. 3C with the most affected residues including 7, 13, 45–49, 65, and 68–76. Comparison with the CSPs observed for ubiquitin shows a striking similarity (compare Figs. 3, A and C). However, when the proximal subunit (i.e. the ubiquitin subunit with a free C terminus) is 15N-labeled, the magnitude of the observed CSPs upon Ube2g2 addition is strongly diminished (Fig. 3D) relative to observations for both the distal subunit and ubiquitin alone. This difference is clearly apparent from a direct comparison of two-dimensional [1H,15N] HSQC spectra for selectively 15N-labeled distal and proximal subunits of diUb upon the addition of Ube2g2 (Fig. 4). The most strongly perturbed residues for the proximal subunit are localized to the C terminus (residues 69–71) and the amide resonance corresponding to the Lys-48 isopeptide linkage, which undergoes the largest shift (Δδ = 0.234 ppm). Furthermore, the backbone amide resonances corresponding to residues Lys-48 and Gly-47 are barely shifted at all in the case of the proximal subunit of Lys-48-diUb, in contrast with the strong CSPs observed for those two residues in all of the other 15N-labeled ubiquitin CSP experiments. These observations suggest that the presence of a covalently attached distal ubiquitin subunit may sterically interfere with the binding of Ube2g2 to the proximal subunit of a Lys-48-diUb molecule, resulting in preferential binding to the distal subunit.
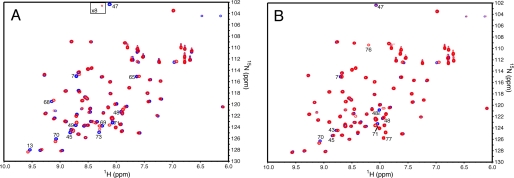
FIGURE 4.
Overlay of two-dimensional [1H,15N] HSQC spectra of selectively 15N-labeled Lys-48-linked diubiquitin species in the presence and absence of Ube2g2. A, overlaid spectra of 15N-distal-Lys-48-diUb (blue) and 90 μm 15N-distal-Lys-48-diUb in the presence of 50 μm Ube2g2 (red) is shown. The area within the inset box has been contoured at a level that is 8× lower to show the resonance corresponding to residue Gly-47 when Ube2g2 is present. B, overlaid spectra of 15N-proximal-Lys-48-diUb (blue) and 100 μm 15N-proximal-Lys-48-diUb in the presence of 50 μm Ube2g2 (red) is shown. The most strongly shifted resonance (labeled 48ϵ) corresponds to the (Lys-48)Nϵ-(Gly-76)CO isopeptide bond linking the two ubiquitin molecules.
In similar fashion to the Ube2g2·Lys-48-diUb experiments, Ube2g2 was titrated into Lys-63-diUb with selective 15N-labeling of either the distal or proximal subunits (21). Upon binding of Ube2g2, both the distal (Fig. 3E) and proximal (Fig. 3F) subunits demonstrate CSP profiles that are quite similar to that observed for the interaction of Ube2g2 with ubiquitin (Fig. 3A). The resonance corresponding to the Lys-63 isopeptide linkage exhibits only a very small chemical shift perturbation (Δδ = 0.019). Although there does appear to be some preference for Ube2g2 binding to the distal subunit relative to the proximal subunit, taken together these observations are indicative of independent binding to each subunit.
Determination of Binding Affinities
For the described CSP experiments, the measured Δδs as a function of added titrant were fit to a simple two state binding model,
 |
in which P and T denote the NMR observe protein and titrant protein, respectively. This approach assumes a 1:1 binding stoichiometry, which is supported here by the absence of any serious spectral broadening that would necessarily result if higher order complexes (with a molecular mass of at least 50 kDa) were being formed. The combined CSP data (e.g. corresponding to both Ube2g2 and Ub observe experiments) for the 15–20 resonances with the largest observed chemical shift changes were then globally fit to a single dissociation constant, Kd, and the residue specific changes in chemical shifts, Δδmax(i), between bound (PT) and free (P) states.
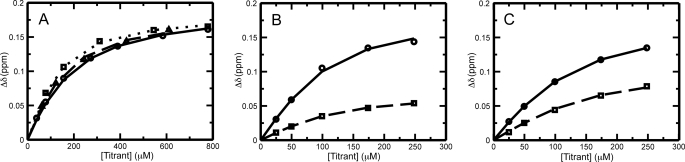
Initial fitting attempts exhibited substantial systematic differences in results between different or even replicate datasets that were subsequently identified as arising from the uncertainty in protein concentrations. This was an especially pronounced problem for ubiquitin, which has a propensity for glass adsorption (39) as well as a low extinction coefficient at 280 nm. Furthermore, erroneous A280 readings can be obtained as a result of additives leaching out from polypropylene plasticware (40, 41). This problem was overcome by allowing the concentration of the ubiquitin stock solution to float as an independent fitting parameter for each distinct titration experiment. This allowed the measured Δδ data to be fit with precisions in the vicinity of 1 ppb. Representative fits to the data as a function of added titrant protein are shown in Fig. 5. Using a two-state binding model, the best fit dissociation constants between Ube2g2 and ubiquitin, Lys-63-diUb, and Lys-48-diUb are estimated to be 90 ± 14, 63 ± 10, and 47 ± 5 μm, respectively (Tables 1 and S1). The reported errors were estimated based on the observed S.D. for 500 independent fits performed on random selections of 75% of the residues with double counting allowed.
FIGURE 5.
Representative fits to the chemical shift perturbation data. A, fit to CSP data for residue Gly-23 of 15N-labeled Ube2g2 upon the addition of ubiquitin (open circles and solid line), Lys-48-linked diubiquitin (open triangles and dashed line), and Lys-63-linked diubiquitin (open squares and dotted line) is shown. B, fit to CSP data for residue Thr-7 of the distal subunit (open circles and solid line) and the proximal subunit (open squares and dashed line) of Lys-48-linked diubiquitin upon the addition of Ube2g2 is shown. C, fit to CSP data for residue Thr-7 of the distal subunit (open circles and solid line) and the proximal subunit (open squares and dashed line) of Lys-63-linked diubiquitin upon the addition of Ube2g2 is shown.
TABLE 1.
Ube2g2 binding affinities for different ubiquitin species
| Observed species | Kda | K1, K2b |
|---|---|---|
| μm | μm | |
| Ubiquitin | 90 ± 14 | |
| Lys-63-diUb | 63 ± 10 | Distal, 103 ± 16 |
| Proximal, 162 ± 26 | ||
| Lys-48-diUb | 47 ± 5 | Distal, 65 ± 7 |
| Proximal, 168 ± 17 |
a Fit to a two-state binding model.
b Fit to competitive a three-state model.
Although all of the CSP data can be very well fit and a Kd estimated from a two-state model, our CSP results for diubiquitin suggest that binding to Ube2g2 may be dominated by interactions with individual Ub subunits. As such, we have also considered a three-state binding model in which Ube2g2 interacts independently with each individual ubiquitin subunit,
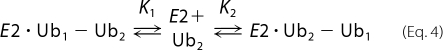 |
Assuming fully competitive binding (which implies 1:1 stoichiometry), the apparent affinity Kapp resulting from a fit to a two-state model will be related to the actual affinities K1 and K2 according to
 |
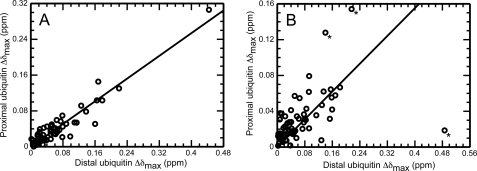
For the special case when the nature of the binding interaction of Ube2g2 with each subunit is identical, the ratio of observed CSPs for each subunit will correspond inversely to the ratio of affinities. This is illustrated in Fig. 6, with the estimated maximum CSPs (Δδmax) plotted, respectively, for residues in the distal and proximal subunits of both Lys-48-diUb and Lys-63-diUb. In the case of Lys-63-diUb, the CSPs between the two subunits exhibit a good correlation (Fig. 6A), supporting the interpretation that each subunit binds Ube2g2 in an independent but similar manner. A best fit line to the points has a slope of 0.63, leading to estimated dissociation constants of 103 ± 16 and 162 ± 26 μm for the interaction of Ube2g2 with the distal and proximal subunits of Lys-63-diUb, respectively (Table 1). Notably the affinity of Ube2g2 for the distal subunit of Lys-63-diUb is very close to that estimated for ubiquitin. On the other hand, the correlation between the CSPs observed for the distal and proximal subunits of Lys-48-diUb is much weaker (Fig. 6B). This suggests that Ube2g2 interacts differently with the two ubiquitin subunits, rendering the applicability of the three-state binding model questionable. Nevertheless, a best fit line to the points excluding prominent outliers yields a slope of 0.39, resulting in an approximate 3-fold preference for the distal subunit using this three-state model (Table 1). This is consistent with the observed differences in CSPs between subunits (Fig. 4) as well as the strikingly narrower lines observed for the proximal subunit relative to the distal subunit at high Ube2g2 concentrations, indicating greater mobility as would occur if Ube2g2 bound primarily to the distal subunit. Regardless of which model is a better representative (two or three state), it is clear from the primary data alone that the distal subunit of the Lys-48-diUb molecule dominates the interaction with Ube2g2, with more limited contacts made between Ube2g2 and the proximal subunit of Lys-48-diUb.
FIGURE 6.
Correlation between chemical shift perturbations for the distal and proximal subunits of diubiquitin upon Ube2g2 binding. The open circles correlate the best fit Δδmax values obtained for residues of the distal and proximal ubiquitin subunits of Lys-63-linked diubiquitin (A) and Lys-48-linked diubiquitin (B). Shown also are the best fit lines with zero intercept and with corresponding slopes of 0.63 (in A) and 0.39 (in B). Those points indicated by an asterisk were excluded from the fit.
Paramagnetic Relaxation Enhancement Experiments
PRE experiments were carried out to obtain site-specific intermolecular distance restraints that could be used together with the CSP data to build a model (42). Our general approach was to covalently link a paramagnetic nitroxide containing species (MTSL) to ubiquitin (or Lys-48-diUb) at a specifically introduced cysteine residue and then titrate this spin-labeled species into a solution of 15N-labeled Ube2g2. Any Ube2g2 residues that are in proximity to the MTSL tag will experience a greatly enhanced relaxation rate manifesting as a reduction in corresponding resonance intensity in the two-dimensional 1H,15N HSQC spectrum. Once a sufficient amount of the spin-labeled titrant protein has been added, the spin label is then typically reduced with ascorbate, and a reference spectrum is acquired (32). As ascorbate reacts with Ube2g2, dithionite was used here to remove the tag entirely. It is important to test for binding of the spin label itself when performing these experiments. To this end, a compound similar to MTSL but lacking a free thiol (l-2-amino-3-[thiomethyl-1-(1-oxyl-2,2,5,5-tetramethyl-3 -pyrrolin-3-yl)]propanoic acid dihydrochloride) was used to test for binding to Ube2g2. The addition of this free spin label to a concentration of 150 μm did not perceptibly alter chemical shifts or resonance intensities of Ube2g2, suggesting that no native interaction exists between the spin label and Ube2g2.
Spin-labeled Ubiquitin
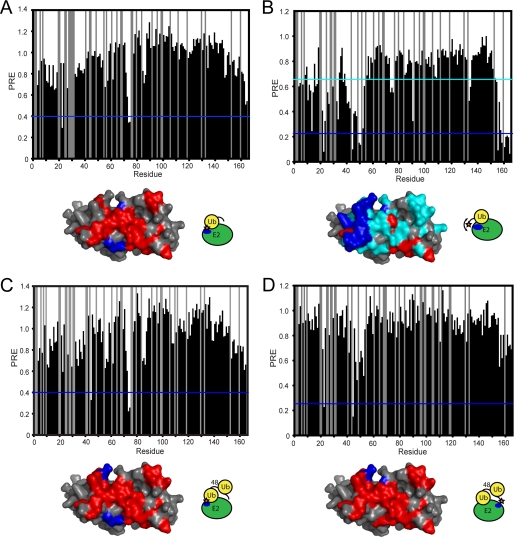
An MTSL spin label was attached to ubiquitin in two different positions followed by titration into a solution of 15N-labeled Ube2g2. In the first experiment, the MTSL spin label was attached to a K48C ubiquitin mutant to report on the position of the side chain of residue 48 in the Ube2g2·Ub complex. The results are shown in Fig. 7A, with those residues exhibiting a very strong PRE colored in dark blue (residues Gly-23, Thr-74, and Cys-75). The Ube2g2 residues that exhibited significant CSPs are colored in red. As anticipated, the strongest PREs are highly localized and are at the edge of the binding interface. Unexpectedly, however, strong PREs were observed in two distinct regions centered on residues Thr-74/Cys-75 and Gly-23. Furthermore, no strong PREs are observed between the sites, suggesting that these two regions reflect well ordered and distinct bound states. It may be possible that the K48C-MTSL side chain exists in two different conformations, but this appears improbable given that the K48C-MTSL side chain, which has a Lys-48-Cα to nitroxide distance of up to ∼8 Å, would be required to span the 18.3 Å distance between the amides of residues 23 and 75. Alternatively, the PRE data may reflect the existence of two different bound state conformations in which ubiquitin binds to Ube2g2 such that its Lys-48 side chain is positioned in proximity to Thr-74/Cys-75 of Ube2g2 in one conformation and toward Gly-23 of Ube2g2 in the other.
FIGURE 7.
Paramagnetic relaxation enhancement observed for 15N-labeled Ube2g2 upon the addition of different spin labeled ubiquitin species. A, the addition of K48C-MTSL-tagged ubiquitin to 15N-labeled Ube2g2 is shown. B, the addition of G75CΔ-MTSL tagged ubiquitin to 15N-labeled Ube2g2 is shown. C, the addition of distal-K48C-MTSL tagged Lys-48-diUb to 15N-labeled Ube2g2 is shown. D, the addition of proximal-G75CΔ-MTSL-tagged Lys-48-diUb to 15N-labeled Ube2g2 is shown. Residues that exhibit a PRE less than the level indicated by the horizontal dark blue and cyan bars are colored correspondingly on the surface map of Ube2g2 (PDB ID 2KLY, model 6). Residues that have a composite chemical shift perturbation of 0.04 ppm or greater are colored red unless they are also affected by PREs, in which case they remain colored in blue or cyan.
In the second experiment, the MTSL tag was attached to a G75CΔ mutant of ubiquitin and then titrated into a solution of 15N-labeled Ube2g2. As can be seen in Fig. 7B, a broad swath of residues in Ube2g2, including 22, 45, 50, 51, 52, 156, 159, and 161–165, exhibit very strong PRE effects (in dark blue). Furthermore, there is a large group of residues that shows significant but more moderate PRE effects (Fig. 7B, cyan). The large surface area of Ube2g2 exhibiting strong PRE effects is indicative of a C-terminal tail of ubiquitin that remains dynamic upon binding to Ube2g2. The approximate division of the Ube2g2 ubiquitin binding surface into separate regions of strong (Fig. 7B, dark blue) and moderate (Fig. 7B, cyan) PREs supports the possible existence of two different binding conformations that have different orientations with respect to the surface of Ube2g2.
Spin-labeled Lys-48-linked Diubiquitin
PRE experiments were also conducted using a spin-labeled Lys-48-diUb molecule in analogy to the ubiquitin experiments described above. In the first experiment, the MTSL tag was attached to a K48C ubiquitin mutant that was placed in the distal position of Lys-48-diUb. Upon the addition to a solution of 15N-labeled Ube2g2, strong PREs are observed for residues Asp-45, Thr-74, and Cys-75 (Fig. 7C). These PRE observations are strikingly similar to what was observed in the MTSL-K48C-ubiquitin PRE experiments (compare Fig. 7A), including the apparent requirement that ubiquitin either binds to Ube2g2 with two different orientations or that the K48C-MTSL side chain assumes two very different conformations.
A second set of PRE measurements were made with an MTSL tag attached to a G75CΔ ubiquitin mutant placed in the proximal position of a Lys-48-diUb molecule. Upon the addition to 15N-labeled Ube2g2, strong PRE effects are observed for a distinct region encompassing residues Gly-23 and Asp-45 (Fig. 7D, dark blue). It is apparent that in the case of Lys-48-diUb binding to Ube2g2, the free C-terminal tail of the proximal Ub subunit can assume a well ordered conformation. This is consistent with the observation of significant CSPs corresponding to the C-terminal region of the proximal subunit of Lys-48-diUb. Yet, if one looks closely at the PREs in Fig. 7D, there is a group of modest PREs (residues 21–22, 45–54, and 155–165) that appear to coincide with the strongest PREs observed in the MTSL-G75Δ-ubiquitin experiments. This suggests that there may also be a direct, albeit weakly populated interaction between the proximal subunit of Lys-48-diUb and Ube2g2.
NMR Restraint-driven Docking Using Rosetta
A novel docking protocol was developed using the Rosetta biomolecular structure prediction suite (33) to predict the interactions between Ube2g2 and ubiquitin as well as Lys-48-diUb. This poses a challenge due to the 90 μm Ube2g2·Ub binding affinity, which is weaker than most protein-protein complexes successfully predicted using standard RosettaDock (34). Nevertheless, RosettaDock can take fruitful advantage of cases in which biochemical information (43, 44) or multiple/flexible backbone conformations can be incorporated (35, 36, 44). We have addressed the challenge posed here by low binding affinity by utilizing NMR CSP and PRE measurements to guide sampling toward physically realistic solutions and by allowing for the known flexibility of the ubiquitin C-terminal tail. In this case, successful prediction is indicated by convergence to a tightly clustered model which has both low Rosetta scores and which best satisfies the experimental restraints.
Docking of Ube2g2 and Ubiquitin
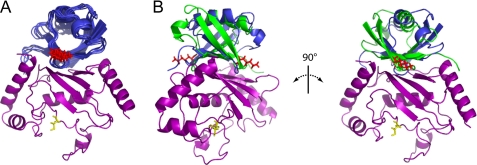
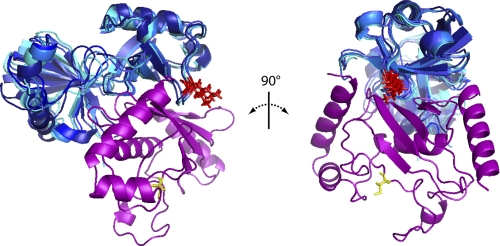
Docking of ubiquitin and Ube2g2 utilized the CSP measurements from both the Ube2g2- and Ub-observed titration experiments as well as PRE restraints using both K48C- and G75CΔ-tagged ubiquitin molecules. As discussed in the previous section, the measured PREs were not consistent with a single docked conformation. As such, the PRE restraints were divided into two sets and utilized in two different conformational searches, respectively. For the primary docking simulation, so named because of its apparent similarity to the UbcH5c·Ub solution structure (20), 1000 models were built of which 6 of the 10 with the lowest scores formed a tight cluster. This cluster is shown in Fig. 8A with the corresponding statistics presented in Table 2. The member of this cluster with the lowest energy was chosen as representative and is referred to as the primary Ube2g2·Ub model. The primary model agrees very closely with the UbcH5c·Ub complex (PDB ID 2FUH) (20), with only a slight displacement (Cα r.m.s.d. = 4.4 Å) of the ubiquitin molecules when the E2 proteins are superimposed.
FIGURE 8.
Rosetta models for the Ube2g2·Ub complexes. A, shown is the primary RosettaDock ensemble of ubiquitin docked to Ube2g2 (ubiquitin (blue); Ube2g2 (violet)). Shown is the six-member ensemble of tightly clustered solutions (within 2 Å of one another) found among the 10 lowest energy solutions. B, overlay of the two best RosettaDock-generated ubiquitin binding models corresponding to the primary (blue) and alternate (green) ubiquitin conformations with Ube2g2 are colored violet. Residues Cys-89 of Ube2g2 and Lys-48 of ubiquitin are rendered in sticks and colored yellow and red, respectively. This figure was generated using the program PyMOL.
TABLE 2.
RosettaDock statistics
| Model | Clustera | CSPs satisfied | PRE violationsb | Interface energyc |
|---|---|---|---|---|
| Ube2g2·Ub (primary) | 6 | 42/59 (71%) | Gly-23(E2)-Gly-75(Ub) (16.1 Å)d | −3.3 |
| Ube2g2·Ub (alternate) | 7 | 42/59 (71%) | Thr-4(E2)-Gly-75(Ub) (15.6 Å)e | −5.7 |
| Ube2g2·Lys-48-diUb | 4f | 52/74 (70%) | None | −9.3 |
a Number of models clustered within 2 Å r.m.s.d. among the 10 lowest scoring solutions.
b PRE violations greater than 15 Å.
c The intermolecular component of the total energy of the complex expressed in Rosetta energy units, where negative indicates stronger binding.
d The PREs utilized for the primary solution were: Ub-K48C-MTSL ⇆ Ube2g2-(74, 75); Ub-G75CD-MTSL ⇆ Ube2g2-(23, 45, 50–52, 156, 159, 161–165).
e The PREs utilized for the alternate solution were: Ub-K48C-MTSL ⇆ Ube2g2-23; Ub-G75CΔ-MTSL ⇆ Ube2g2-(4, 5, 11–15, 17, 18, 26, 27, 30, 33, 34, 38).
f The proximal subunit clustered within 5 Å r.m.s.d.
A second simulation was then carried out using the alternate set of PRE restraints. As summarized in Table 2, this simulation also resulted in a tight cluster of solutions with a satisfactory agreement with the CSP and PRE restraints and a negative Rosetta interface energy. Both the primary and alternate models satisfied ∼70% of the CSP restraints and violated a single PRE restraint involving the G75CΔ-MTSL tag. This violation was not considered significant given the known flexibility of the C terminus of ubiquitin. Strikingly, the primary and alternate conformations are related by nearly 180° as shown in Fig. 8B. Apparently, ubiquitin presents a hydrophobic rod-like surface that can form an acceptable interaction interface with the binding groove of Ube2g2 in either of two opposite orientations.
Docking of Lys-48-diubiquitin and Ube2g2
Due to the close similarity of the primary Ube2g2·Ub model to the solution structure of the UbcH5c·Ub complex, the Ube2g2·Lys-48-diUb docking simulation was carried out utilizing the subset of the PRE restraints, which were consistent with positioning of the distal subunit of diubiquitin in the primary conformation. One thousand models were built and subsequently sorted using the all-atom Rosetta score. A tight cluster of seven models was found among the 10 lowest-scoring structures which exhibited a maximum of a 2 Å intramodel Cα r.m.s.d. between the distal ubiquitin subunits. Among that cluster, four models exhibited a 5 Å intramodel Cα r.m.s.d. for the proximal ubiquitin subunit (Fig. 9). The member of this ensemble with the lowest energy was chosen to be representative with the corresponding statistics summarized in Table 2. The Ube2g2·Lys-48-diUb model satisfies 52 of 74 observed CSPs (70%), has a negative interface energy (−9.3 Rosetta energy units), and satisfies all four PRE restraints. After superposition of the corresponding E2 proteins, the positioning of the distal ubiquitin subunit of Lys-48-diUb deviated from the positioning of ubiquitin in the UbcH5c·Ub complex (PDB ID 2FUH) (20) and the Ube2g2·Ub RosettaDock model by a Cα r.m.s.d. of 5.3 and 4.1 Å, respectively.
FIGURE 9.
RosettaDock ensemble of Lys-48-linked diubiquitin (blue shades) bound to Ube2g2 (violet). Shown is the ensemble of four low-energy solutions which clustered to within 2 Å for the distal subunit and 5 Å for proximal ubiquitin subunits. Residues Cys-89 of Ube2g2 and Lys-48 of ubiquitin are rendered in sticks and colored yellow and red, respectively. This figure was generated using the program PyMOL.
DISCUSSION
Ube2g2 binds to the hydrophobic patch of ubiquitin with an interface composed of residues in its β-sheet and C-terminal helix, which is consistent with reported observations for other E2 apoenzymes including HsUbc2b (19) and UbcH5c (20). Our PRE experiments suggest that ubiquitin must interact with Ube2g2 such that the MTSL-K48C-linked tag assumes two different positions relative to the Ube2g2 surface. Based on the 18 Å distance between the two regions with strong PREs (corresponding respectively to residues Gly-23 and Thr-74/Cys-75), it appears quite unlikely that these data can be explained by two very different K48C-MTSL side-chain conformations. Indeed, when the RosettaDock algorithm modeled the Ube2g2·Ub interaction based on PRE and CSP restraints, it found two tight solution clusters for ubiquitin binding that are related by a 180° rotation. The possibility that ubiquitin binds in two orientations appears to be plausible as the interaction is hydrophobic in nature and involves the packing of a convex rod-like surface (of ubiquitin) into the concave groove on the surface of Ube2g2. Although one might assign populations to these two states based on the observed PRE intensities, we expect large uncertainties associated with such an estimate given the very simple PRE experiments that were employed in this study. Furthermore, considering the strong nonlinear (r−6) dependence of the PRE on the distance between the nuclear and paramagnetic centers, it remains conceivable that the alternate binding conformation corresponds to an encounter complex or a weakly populated state (45). We have presumed that the so-called primary conformation is the relevant one based on its close agreement with the UbcH5c·Ub complex (Cα r.m.s.d. = 4.4 Å). The conservation of this ubiquitin binding site across different E2 proteins implies that the site plays an important biological role that is not well understood at present.
The investigation of Ube2g2 binding to both Lys-63-diUb and Lys-48-diUb resulted in a slightly higher affinity for Lys-48-diUb. However, we do not consider this difference to be significant as it could simply arise due to deviations from the simple binding model employed or due to differences in the side-chain of residue 48 (i.e. Lys-48 versus Arg-48 for Lys-63-diUb and Lys-48-diUb, respectively). Rather, the results from this study are consistent with the possibility that Ube2g2 could just as well utilize polyubiquitin chains with non-Lys-48 linkages as a substrate.
The most significant result was the observation of differences in Ube2g2 binding affinity for individual ubiquitin subunits of diubiquitin. In the case of Lys-63-diUb, Ube2g2 binds to each subunit as if it were an independent ubiquitin molecule but with some preference for binding to the distal relative to the proximal subunit. These results are generally consistent with reports proposing that subunits of Lys-63-linked diubiquitin bind with indistinguishable dissociation constants to ubiquitin-associated domains (46, 47).
In contrast, the interaction of Lys-48-diUb with Ube2g2 is dominated by the distal subunit. Indeed, the results reported for ubiquitin and the distal subunit of Lys-48-diUb are difficult to distinguish, including the observation of two distinct regions with strong PREs. There is some evidence for direct binding of the proximal subunit of Lys-48-diUb to Ube2g2but it must be a considerably weaker interaction. The CSP data as a whole support a model in which the proximal subunit of Lys-48-diUb interacts with Ube2g2 differently than ubiquitin alone. The much broader range of proximal subunit positions in the Ube2g2·Lys-48-diUb Rosetta model is reflective of the NMR evidence for a proximal subunit that is dynamically sampling different conformations.
Ube2g2 prefers to bind to the distal subunit of both Lys-48-diUb and Lys-63-diUb. This preference is much more striking in the case of Lys-48-diUb and can be attributed to steric hindrance of proximal subunit binding due to the presence of the Lys-48-(proximal)-Gly-76 (distal) isopeptide linkage. By extension of this reasoning it can be expected that the terminal subunit of a Lys-48-linked polyubiquitin chain will be the most accessible for binding interactions with other proteins. If so, then this potentially constitutes a general mechanism whereby the free Lys-48 side chain of a growing polyubiquitin chain can be positioned for reaction with a second donor ubiquitin molecule. We propose that access to the terminal subunit of a polyubiquitin chain is largely unimpeded due to the mobility of ubiquitin C-terminal tail, whereas access is restricted to the other subunits in a manner that depends on the linkage type and the size of the binding partner. Under this model a smaller ubiquitin-binding protein would exhibit less preference for terminal subunit binding, whereas a larger protein would exhibit more preference. In the case of Ube2g2 (18.6 kDa), the strong preference observed for binding to the distal subunit of Lys-48-linked diUb manifests as a milder preference for distal subunit binding in Lys-63-linked diUb because Lys-63 is more distant from the binding site.
Supplementary Material
Acknowledgments
We acknowledge the Johns Hopkins University Biomolecular NMR Facility and thank Prof. Juliette Lecomte for many insightful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants T32GM008403 (to W. E. B.), GM075310–04S1 (to J. R. T.), and GM078221 (to J. J. G.). This work was also supported by UCB SA (to A. S., J. J. G., and J. R. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Fig. S1, and Table S1.
- CSP
- chemical shift perturbation
- MTSL
- S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl) methyl methanesulfonothioate
- Lys-48-diUb
- Lys-48-linked diubiquitin
- Lys-63-diUb
- Lys-63-linked diubiquitin
- PRE
- paramagnetic relaxation enhancement
- r.m.s.d.
- root mean square deviation
- Ub
- ubiquitin
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 2. Haas A. L., Rose I. A. (1982) J. Biol. Chem. 257, 10329–10337 [PubMed] [Google Scholar]
- 3. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 4. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kostova Z., Wolf D. H. (2003) EMBO J. 22, 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 7. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai B., Ye Y., Rapoport T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246–255 [DOI] [PubMed] [Google Scholar]
- 10. Tanaka K., Suzuki T., Hattori N., Mizuno Y. (2004) Biochim. Biophys. Acta 1695, 235–247 [DOI] [PubMed] [Google Scholar]
- 11. Kim I., Xu W., Reed J. C. (2008) Nat. Rev. Drug Discov. 7, 1013–1030 [DOI] [PubMed] [Google Scholar]
- 12. Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., Weissman A. M. (2007) Nat. Med. 13, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 13. Tsutsumi S., Yanagawa T., Shimura T., Kuwano H., Raz A. (2004) Clin. Cancer Res. 10, 7775–7784 [DOI] [PubMed] [Google Scholar]
- 14. Li W., Tu D., Brunger A. T., Ye Y. (2007) Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 15. Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li W., Tu D., Li L., Wollert T., Ghirlando R., Brunger A. T., Ye Y. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raasi S., Varadan R., Fushman D., Pickart C. M. (2005) Nat. Struct. Mol. Biol. 12, 708–714 [DOI] [PubMed] [Google Scholar]
- 19. Miura T., Klaus W., Gsell B., Miyamoto C., Senn H. (1999) J. Mol. Biol. 290, 213–228 [DOI] [PubMed] [Google Scholar]
- 20. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 21. Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., Fushman D. (2004) J. Biol. Chem. 279, 7055–7063 [DOI] [PubMed] [Google Scholar]
- 22. Varadan R., Walker O., Pickart C., Fushman D. (2002) J. Mol. Biol. 324, 637–647 [DOI] [PubMed] [Google Scholar]
- 23. Ju T., Bocik W., Majumdar A., Tolman J. R. (2010) Proteins Struct. Funct. Genet. 78, 1291–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beal R., Deveraux Q., Xia G., Rechsteiner M., Pickart C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marley J., Lu M., Bracken C. (2001) J. Biomol. NMR 20, 71–75 [DOI] [PubMed] [Google Scholar]
- 26. Piotrowski J., Beal R., Hoffman L., Wilkinson K. D., Cohen R. E., Pickart C. M. (1997) J. Biol. Chem. 272, 23712–23721 [DOI] [PubMed] [Google Scholar]
- 27. Hofmann R. M., Pickart C. M. (2001) J. Biol. Chem. 276, 27936–27943 [DOI] [PubMed] [Google Scholar]
- 28. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29. Goddard T. D., Kneller D. G. (2004) SPARKY 3, University of California, San Francisco [Google Scholar]
- 30. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 31. Gill S. C., von Hippel P. H. (1989) Anal. Biochem. 182, 319–326 [DOI] [PubMed] [Google Scholar]
- 32. Battiste J. L., Wagner G. (2000) Biochemistry 39, 5355–5365 [DOI] [PubMed] [Google Scholar]
- 33. Das R., Baker D. (2008) Annu. Rev. Biochem. 77, 363–382 [DOI] [PubMed] [Google Scholar]
- 34. Gray J. J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C. A., Baker D. (2003) J. Mol. Biol. 331, 281–299 [DOI] [PubMed] [Google Scholar]
- 35. Chaudhury S., Gray J. J. (2008) J. Mol. Biol. 381, 1068–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sircar A., Gray J. J. (2010) PLoS Comput. Biol. 6, e1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zuiderweg E. R. P. (2002) Biochemistry 41, 1–7 [DOI] [PubMed] [Google Scholar]
- 38. Varadan R., Assfalg M., Raasi S., Pickart C., Fushman D. (2005) Mol. Cell 18, 687–698 [DOI] [PubMed] [Google Scholar]
- 39. Raasi S., Pickart C. M. (2005) Methods Mol. Biol. 301, 47–55 [DOI] [PubMed] [Google Scholar]
- 40. Lewis L. K., Robson M., Vecherkina Y., Ji C., Beall G. W. (2010) Biotechniques 48, 297–302 [DOI] [PubMed] [Google Scholar]
- 41. McDonald G. R., Hudson A. L., Dunn S. M., You H., Baker G. B., Whittal R. M., Martin J. W., Jha A., Edmondson D. E., Holt A. (2008) Science 322, 917. [DOI] [PubMed] [Google Scholar]
- 42. Kalbitzer H., Kremer W., Schumann F., Spörner M., Gronwald W. (2007) Protein Rev. 5, 189–229 [Google Scholar]
- 43. Chaudhury S., Sircar A., Sivasubramanian A., Berrondo M., Gray J. J. (2007) Proteins Struct. Funct. Genet. 69, 793–800 [DOI] [PubMed] [Google Scholar]
- 44. Sircar A., Chaudhury S., Kilambi K. P., Berrondo M., Gray J. J. (2010) Proteins Struct. Funct. Genet. 78, 3115–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Iwahara J., Clore G. M. (2006) Nature 440, 1227–1230 [DOI] [PubMed] [Google Scholar]
- 46. Sims J. J., Cohen R. E. (2009) Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sims J. J., Haririnia A., Dickinson B. C., Fushman D., Cohen R. E. (2009) Nat. Struct. Mol. Biol. 16, 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.