Abstract
The centromeric DNA of all eukaryotes is assembled upon a specialized nucleosome containing a histone H3 variant known as CenH3. Despite the importance and conserved nature of this protein, the characteristics of the centromeric nucleosome are still poorly understood. In particular, the stoichiometry and DNA-binding properties of the CenH3 nucleosome have been the subject of some debate. We have characterized the budding yeast centromeric nucleosome by biochemical and biophysical methods and show that it forms a stable octamer containing two copies of the Cse4 protein and wraps DNA in a left-handed supercoil, similar to the canonical H3 nucleosome. The DNA-binding properties of the recombinant nucleosome are identical to those observed in vivo demonstrating that the octameric structure is physiologically relevant.
Keywords: Centromeres, DNA-Protein Interaction, DNA Topology, Histones, Nucleosome, Protein-DNA Interaction, CENP-A, CenH3
Introduction
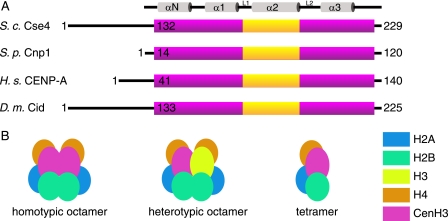
The centromere is the chromosomal locus responsible for attachment to the mitotic spindle via the kinetochores, and it is essential for correct chromosome segregation in all eukaryotes (1). It has been recognized for some time that centromeric chromatin has somewhat different properties to bulk chromatin; in particular, centromeric nucleosomes are distinguished by containing a histone H3 variant, known as CENP-A in humans (2), Cse4 in budding yeast (3), Cnp1 in fission yeast (4), and CID in fruit flies (5). For simplicity, we shall refer to these generically as CenH3 nucleosomes. This histone H3 variant includes a conserved histone fold domain (HFD)2 preceded by an N-terminal extension that is highly divergent in both size and amino acid composition (Fig. 1A). The HFD also contains a CENP-A targeting domain that appears to be responsible for the special mechanical properties of the centromere-specific nucleosome (6), and it is sufficient to impart centromere targeting and functionality in hybrid H3-CENP-A targeting domain nucleosomes (7). Although there are now numerous crystal structures of histone H3-containing nucleosomes, there are currently no crystal structures of an intact CenH3 nucleosome, and it is unclear exactly how these specific modifications contribute to the assembly and overall structure of the chromatin in which they are embedded.
FIGURE 1.
Centromeric nucleosome structure. A, schematic diagram of four CenH3 proteins from S. cerevisiae (S.c.), Schizosaccharomyces pombe (S.p.), Homo sapiens (H.s.), and Drosophila melanogaster (D.m.). The purple bar shows the extent of the histone fold domain, and the yellow box shows the position of the CENP-A targeting domain motif. The secondary structure elements of the histone fold are indicated at the top of the figure. B, possible stoichiometries of centromeric nucleosomes showing the two possible octameric complexes and a tetramer.
Recently, it has been proposed that CenH3 nucleosomes might be tetrameric instead of octameric (8, 9) and wrap DNA in the reverse direction to canonical nucleosomes, introducing positive supercoiling (10), suggestions that have proved somewhat controversial (11, 12). It is also unclear whether a centromeric nucleosome octamer might contain two copies of CenH3 (homotypic) or one each of CenH3 and H3 proteins (heterotypic), as shown in Fig. 1B. In addition, the non-histone protein Scm3 has also been shown to bind Cse4 and H4 (13) and may be involved in recruitment of the centromeric nucleosome. Despite original suggestions (13), it does not appear that it remains part of the core nucleosome once at the active centromere (14), although it may remain associated with it. In vivo studies of centromeric nucleosome composition, for example by immunoprecipitation analyses, may be complicated by the chromatin preparation procedure or co-purification of other non-CenH3 components bound to chromatin. Here, we present the results of a rigorous biophysical analysis of the budding yeast centromeric nucleosome, using purified recombinant proteins to try and clarify some of these issues.
EXPERIMENTAL PROCEDURES
Cloning
For protein expression, the plasmids pET28, pET22, pETDuet, and pCDFDuet (Novagen) were manipulated by standard procedures to yield derivatives containing histone genes, which were obtained by PCR.
Protein Purification
Histone proteins were co-expressed in Escherichia coli strain BL21(DE3) RIL. Cells were grown to A600 = 0.6 and induced with 0.3 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. Cells were sonicated in buffer containing 0.5 m NaCl, 20 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 10 mm β-mercaptoethanol, and complete protease inhibitor (Roche Applied Science). Supernatant was loaded onto a 5-ml HiTrap heparin column (GE Healthcare) and eluted with a gradient of 0.5–2 m NaCl. Fractions containing histone complex were further purified by size exclusion chromatography in buffer containing 2 m NaCl, 20 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, 10 mm β-mercaptoethanol. Samples were analyzed by SDS-PAGE. Reconstitution of nucleosomes from individually purified histones was carried out as described previously (15).
DNA Preparation
DNA binding, MNase assays, and EM studies used salmon sperm DNA (Sigma). Topoisomerase assays used a derivative of pET28 containing the centromere sequence from Saccharomyces cerevisiae chromosome 3 cloned between EcoRV and XhoI sites. Variations of the α-satellite core sequence (16) were used for AUC and obtained by self-priming DNA synthesis (17). The sequence of interest was purified by ion exchange (Mono Q, GE Healthcare) in 50 mm KCl, 50 mm phosphate buffer, pH 6.8, with a gradient to 1 m KCl.
Preparation of Nucleosome-DNA Complex
DNA and histone complex were added at equimolar ratios in either 2 m NaCl (MNase and EM studies) or 2 m KCl (AUC), 10 mm Tris-HCl, pH 7.5. Serial dilutions were carried out to reduce the salt concentration to 1, 0.85, 0.67, 0.2, and 0.1 m, using 50 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 1 mm CaCl2, 1 mm EDTA, 1 mm DTT. For EM studies, the buffer consisted of 50 mm Tris-HCl, pH 7.5.
Analytical Ultracentrifugation
Histone complexes were analyzed in a buffer containing 2 m NaCl, 20 mm Tris-HCl, pH 8.0, 0.1 mm EDTA. Reconstituted DNA and histones were purified by DEAE ion exchange chromatography in buffer containing 0.25 m KCl, 10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT with a linear gradient to 0.6 m KCl. Samples containing nucleosomes reconstituted with DNA were diluted 5-fold in buffer containing 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT. All samples were concentrated to >0.1 mg/ml before loading into cells with identical buffer as a reference. Samples were run for 15 h at 28,000 rpm (nucleosomes) or 18,000 rpm (nucleosome: DNA) at 16 °C. Data were analyzed using SEDFIT (18), using a continuous c(s) distribution.
Micrococcal Nuclease Assay
Reconstituted DNA and histones were subjected to digestion with micrococcal nuclease at 2 units/ml for various time points (0, 15, and 30 s and 1 and 5 min). The reaction was quenched by addition of EDTA, and samples were purified using Qiagen PCR purification kit before analysis by agarose gel electrophoresis. The length of the DNA bound to mono- or di-nucleosomes was determined by gel excision of bands, blunt end ligation into pBR322 cut with EcoRV, and sequencing of the region of interest.
Negative Staining Electron Microscopy
Reconstituted nucleosomes were deposited on glow discharged ultra-thin carbon-coated copper grids and stained with 0.5% (w/v) uranyl acetate. Grids were visualized using a Tecnai Twin microscope operated at 120 kV. Images were analyzed using ImageJ software (National Institutes of Health).
DNA Supercoiling Assay
Plasmid DNA in a 2 m NaCl buffer was added to increasing concentrations of nucleosomes (H3 or Cse4 variants). The salt concentration was then reduced as described in the methods for the preparation of the complex. The plasmid was relaxed by Vaccinia topoisomerase I (kindly provided by I. Grainge) incubation for 1 h at 37 °C in dilution buffer. DNA was de-proteinized by addition of SDS and proteinase K, followed by purification with the Qiagen PCR purification kit. Samples were analyzed by TAE-agarose gel electrophoresis in the absence of EtBr at 100 V for 5 h, and bands were visualized by post-run staining with EtBr and UV illumination. Gels were run in the absence and presence of 1 μg/ml chloroquine.
RESULTS
Preparation and Mass of Nucleosome Particles
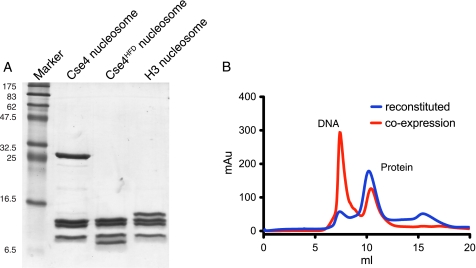
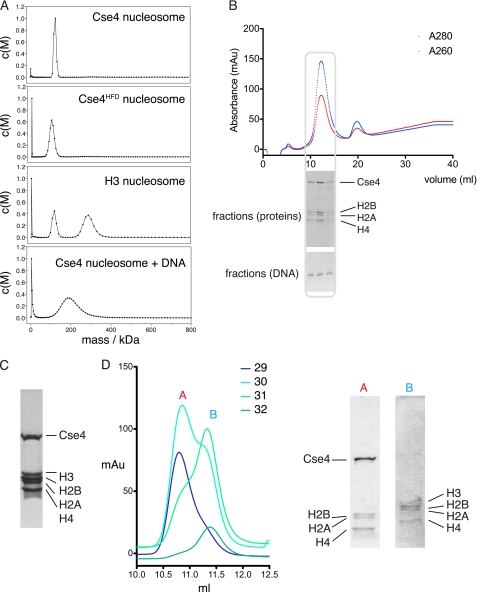
To avoid any possible artifacts resulting from traditional reconstitution procedures involving refolding and dialysis of individually produced histones from denaturants, we co-expressed all four budding yeast histone proteins in E. coli and purified intact, untagged nucleosomes by conventional means (Fig. 2A). These nucleosomes had an identical size exclusion profile to those prepared by reconstitution from separate refolded histones (Fig. 2B). Although a previous study has suggested that the Cse4 nucleosomes are octameric both in vivo and in vitro (14), the molecular masses of the nucleosomes were not accurately determined. We have used AUC to determine hydrodynamic properties of the intact nucleosomes. We initially analyzed wild-type Cse4 nucleosomes, a variant truncated Cse4 molecule encompassing just the histone fold domain (Cse4HFD, residues 129–229), and canonical histone H3-containing nucleosomes. The experimental data (Fig. 3A and Table 1) from sedimentation velocity experiments clearly show that all three types of nucleosome have molecular masses consistent with an octameric stoichiometry containing two copies each of histones H2A, H2B, Cse4/H3, and H4. Moreover, there is no evidence for any tetrameric subcomplex in any of the samples that might result from breakdown of an unstable octamer. To test the possibility that the oligomeric state of the nucleosome may change upon DNA binding, we reconstituted our Cse4 nucleosomes with a 125-bp fragment of DNA, based upon the nuclease protection characteristics (see below). The particles so formed are homogeneous both in protein and DNA composition (Fig. 3B). We determined the molecular mass of the nucleosome-DNA complex by AUC and found it to again be consistent with a single octamer wrapped by one copy of the DNA sequence (Fig. 3A and Table 1).
FIGURE 2.
Expression of recombinant histone complexes. A, SDS-polyacrylamide gel of purified nucleosomes containing the Cse4, Cse4HFD, or H3 protein. B, size exclusion profiles for Cse4-containing nucleosomes prepared by simultaneous co-expression of all subunits (red trace) and reconstitution from individual re-folded histones (blue trace). The first peak in the red trace at ∼7 ml arises from excess DNA contamination.
FIGURE 3.
Mass and stoichiometry of centromeric nucleosomes. A, sedimentation velocity AUC traces of purified histone complexes and Cse4 nucleosomes reconstituted with 125-bp DNA sequence. The second peak in the H3 complex trace probably arises from association of two nucleosomes. B, purification of Cse4 nucleosome with defined length DNA as used for AUC experiments. A gel filtration trace of the complex is shown with SDS-PAGE and agarose gel analysis of the fractions containing the nucleosome. C, SDS-PAGE gel of co-expression of all five histone proteins, including Cse4. Roughly equal levels of expression are seen for each protein. D, high resolution size exclusion chromatography traces of fractions from expression and purification of nucleosomes with simultaneous expression of both Cse4 and H3 variant complexes. Fractions 29–32 from the initial gel filtration peak were re-run individually on a high resolution column and analyzed by SDS-PAGE. The elution profile demonstrates that the peak splits into two populations of homotypic octamer, and no heterotypic species (which one would expect to see at an intermediate elution volume) appears. The gel shows mutual exclusion of Cse4-containing and H3-containing complexes.
TABLE 1.
Nucleosome masses
Predicted and experimental molecular masses obtained by analytical ultracentrifugation of nucleosome variants assuming either tetrameric or octameric compositions.
| Complex | Cse4 | H3 | Cse4HFD | Cse4 + DNA |
|---|---|---|---|---|
| kDa | kDa | kDa | kDa | |
| Predicted mass, tetramer | 66.4 | 55.0 | 51.5 | 143.5 |
| Predicted mass, octamer | 132.9 | 109.9 | 102.9 | 210.0 |
| Experimental mass | 121.0 | 114.0 | 103.0 | 206.0 |
Homotypic Versus Heterotypic Octamer
It is possible that the centromeric nucleosome might contain one copy of Cse4 and one copy of histone H3 as part of a so-called heterotypic octamer. To test this possibility, we expressed histones H2A, H2B, H3, H4, and Cse4 simultaneously and purified the resultant complexes. All five proteins appeared to be produced in roughly equal proportions (Fig. 3C), ensuring that availability of a given histone was not a limiting factor in complex formation. Gel filtration analysis shows a single peak containing both H3-containing and Cse4-containing variant complexes, but we were unable to discriminate between them. Fractions taken from this peak were then re-run on a high resolution gel filtration column and split into two populations, one consisting of H3 homotypic octamers and the other of Cse4 homotypic octamers (Fig. 3D). We see no evidence of heterotypic octamer formation, although we cannot preclude that they might form with very low yield.
DNA Protection Characteristics
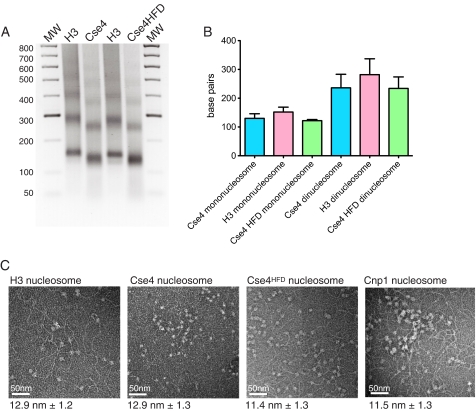
We next examined the DNA-binding properties of the nucleosomes using MNase protection assays. In this assay, normal histone H3 nucleosomes protect a region of ∼150 bp. Digestion of DNA bound to Cse4 and Cse4HFD nucleosomes showed a characteristic ladder of bands with a repeat size slightly less than seen with H3 nucleosomes (Fig. 4A). Sequencing these bands showed the repeat size to be ∼125 bp (Fig. 4B). Analysis of the protected sections of DNA showed a random nucleotide composition, indicating that the centromeric nucleosomes have no intrinsic sequence bias and suggesting that other factors are required to direct assembly at a specific locus, for example the Scm3 protein (19). It is unclear what the structural basis for the reduced size of MNase-protected DNA might be. It could be argued that the circumference of the Cse4 nucleosome is less than that of an H3 nucleosome due to a more compact octamer; indeed, it has been shown that a tighter structure is adopted CENP-A nucleosomes (6, 20). Alternatively, the entrance and exit sites of the DNA wrap might be more loosely bound by the nucleosome, resulting in increased MNase accessibility. To try and resolve these possibilities, we visualized DNA-bound nucleosomes by negative-stain electron microscopy. We analyzed H3, Cse4, Cse4HFD complexes as well as the fission yeast Cnp1 nucleosome bound to mixed sequence DNA (Fig. 4C). In all cases the dimensions of the core nucleosome were similar to the canonical H3 complex, with an observed diameter of ∼12 nm. It therefore seems unlikely that the smaller protection size results from a significantly smaller core particle alone, but rather that the interactions between the nucleosome and DNA at the end of the superhelical gyres are weakened, allowing increased access for the nuclease to the DNA.
FIGURE 4.
DNA binding of histone complexes. A, agarose gel electrophoresis of MNase-digested DNA bound to histone complexes containing Cse4, Cse4HFD, or H3 variants. B, quantification of lengths of DNA protected from MNase digests on both mono- and di-nucleosomes as described in A. C, electron microscopy of nucleosomes (S. cerevisiae H3, Cse4, and Cse4HFD variants and S. pombe Cnp1 variant) bound to salmon sperm DNA. Average diameters of the nucleosomes are shown below each image (n = 100). The image scale bar, 50 nm. MW, molecular weight.
DNA Supercoiling by Nucleosome
We also determined the manner in which DNA is wrapped by the Cse4 complex. Studies on reconstituted Drosophila CID-containing chromatin suggest that the centromeric nucleosome wraps DNA in a “reverse” right-handed gyre, introducing positive supercoils (10). This was supported by an in vivo minichromosome assay in yeast, which also indicated this unusual DNA configuration. We repeated the in vitro supercoiling assays using our reconstituted Cse4 nucleosomes. The binding of both Cse4 and H3 nucleosomes to DNA introduced supercoils into a plasmid template as expected (Fig. 5A), with no requirement for a histone chaperone in assembly. By running the assay in the presence of chloroquine (Fig. 5B), we see that the topoisomers migrate more slowly in the gel, due to the compensatory positive writhe introduced by the drug. This shows that the nucleosomes induce negative supercoiling, and this is seen to be identical for both H3- and Cse4-containing complexes. A similar result has recently been observed using the human CENP-A nucleosome (20) suggesting that negative supercoiling is a conserved property of the centromeric nucleosome.
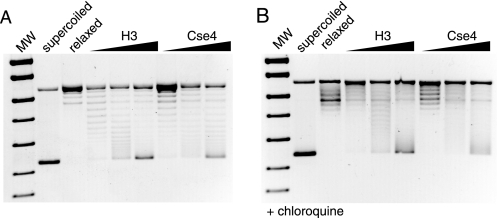
FIGURE 5.
Nucleosome-induced DNA topology assays. A, effect of nucleosome binding on DNA topology. Plasmid DNA was bound to increasing concentrations of Cse4 or H3 nucleosomes, before topoisomerase relaxation. The de-proteinized plasmids were analyzed by agarose gel electrophoresis. Controls show fully supercoiled and topoisomerase-relaxed plasmid. Increasing degrees of supercoiling are observed upon nucleosome addition. B, same experiment repeated in the presence of 1 μg/ml chloroquine shows retarded migration of the topoisomers, indicating relaxation of the negative supercoiling. MW, molecular weight.
DISCUSSION
In this study, we have demonstrated that the budding yeast centromeric nucleosome forms a stable octamer, containing two copies of the Cse4 protein and that this structure is maintained upon DNA binding. At no stage in our work have we seen any evidence for a species corresponding to a tetrameric hemisome, and the Cse4-H3 mixing experiments strongly argue that a symmetrical CenH3-CenH3 (or H3-H3) interaction is strongly favored over a CenH3-H3 heterodimer.
While this manuscript was in preparation, the structure of the core nucleosomal CenH3/H4 tetramer was described, using a truncated version of the human CENP-A protein (20). The conclusions reached by analysis of the crystal structure are essentially the same as those presented in this study, which together show that both a subnucleosomal structure and an intact DNA-bound nucleosome contain a (CenH3-H4)2 tetramer that acts as a core for the full octameric complex.
This study, together with previously published results (14, 20), unequivocally demonstrates that the centromeric nucleosome is octameric in vitro. We believe that this is also true in vivo for several reasons. The symmetric octamer is the favored and most stable form of the complex, and this must be true regardless of the pathway by which it was formed. The presence in vivo of an intrinsically unstable tetrameric hemisome could only occur if it were stabilized by another non-histone factor. This would have to bind tightly in stoichiometric quantities, be an essential gene product, as well as being highly conserved throughout evolution. No such factor has yet been found.
A second reason to think that the reconstituted nucleosomes are equivalent to those found at the centromere is provided by analysis of the DNA-binding properties. Whereas the Cse4 octamer is similar in size to canonical H3 nucleosomes, the site size of DNA protected from MNase digestion is somewhat reduced and less well defined. This altered protection relative to canonical nucleosomes has previously been seen in multiple in vivo and in vitro studies (8, 21, 22) and has been interpreted as evidence for a tetrameric nucleosome (8). Our results clearly show that this smaller site size is compatible with octameric nucleosomes, and it is not necessary to invoke alternative assembly models. Crucially, this demonstrates that the DNA-binding properties of the recombinant nucleosomes reflect the situation in vivo and argues against the possibility that experiments on reconstituted nucleosomes are artifactual.
There are probably multiple reasons for the reduced DNA protection exhibited by CenH3 nucleosomes. A more compact structure is probably partly responsible, but we feel the main cause is weaker protein-DNA interactions. Crystal structures of intact, DNA-bound nucleosomes have shown that the interaction closest to the entrance-exit points of the DNA is formed with the N-terminal section of histone H3 (16, 23), which is highly diverged in all known CenH3 orthologs. Weaker interactions here would allow increased access to nucleases and explain the species-specific differences seen in MNase protection assays, as well as explain the more diffuse appearance of the protected bands. It is unclear what the physiological relevance of this might be, but it may well involve the as-yet unknown role of the extended CenH3 N-terminal tails.
Our results and those of others (20) show that the octameric CenH3 nucleosome is only capable of wrapping DNA in a conventional left-handed manner, with induction of negative supercoils. It is possible that that previous results from the in vivo minichromosome assay indicating Cse4-induced positive supercoiling in yeast (10) could be explained by transcriptional artifacts or nonspecific binding of Ndc10 to the centromeric DNA complicating the analysis. This work, together with previously published observations on a range of CenH3 orthologs, strongly suggests that the octamer is the physiologically relevant form. Further work will be required to understand how the CenH3-specific histone contributes to the overall structure of the centromeric nucleosome and what effect this has on the mechanical properties of the centromere.
Acknowledgments
We thank T. Toda for helpful comments on the manuscript, D. J. Scott for advice on AUC analysis, and I. Grainge for assistance with topology assays. We also wish to thank the LRI EM unit for access to their microscopes.
This work was supported by Cancer Research UK.
- HFD
- histone fold domain
- MNase
- micrococcal nuclease
- AUC
- analytical ultracentrifugation.
REFERENCES
- 1. Allshire R. C., Karpen G. H. (2008) Nat. Rev. Genet. 9, 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Earnshaw W. C., Rothfield N. (1985) Chromosoma 91, 313–321 [DOI] [PubMed] [Google Scholar]
- 3. Meluh P. B., Yang P., Glowczewski L., Koshland D., Smith M. M. (1998) Cell 94, 607–613 [DOI] [PubMed] [Google Scholar]
- 4. Takahashi K., Chen E. S., Yanagida M. (2000) Science 288, 2215–2219 [DOI] [PubMed] [Google Scholar]
- 5. Henikoff S., Ahmad K., Platero J. S., van Steensel B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black B. E., Foltz D. R., Chakravarthy S., Luger K., Woods V. L., Jr., Cleveland D. W. (2004) Nature 430, 578–582 [DOI] [PubMed] [Google Scholar]
- 7. Black B. E., Jansen L. E., Maddox P. S., Foltz D. R., Desai A. B., Shah J. V., Cleveland D. W. (2007) Mol. Cell 25, 309–322 [DOI] [PubMed] [Google Scholar]
- 8. Dalal Y., Wang H., Lindsay S., Henikoff S. (2007) PLoS Biol. 5, e218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furuyama T., Dalal Y., Henikoff S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6172–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Furuyama T., Henikoff S. (2009) Cell 138, 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dechassa M. L., D'Arcy S., Luger K. (2009) Cell 138, 22–24 [DOI] [PubMed] [Google Scholar]
- 12. Lavelle C., Recouvreux P., Wong H., Bancaud A., Viovy J. L., Prunell A., Victor J. M. (2009) Cell 139, 1216–1218 [DOI] [PubMed] [Google Scholar]
- 13. Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C. (2007) Cell 129, 1153–1164 [DOI] [PubMed] [Google Scholar]
- 14. Camahort R., Shivaraju M., Mattingly M., Li B., Nakanishi S., Zhu D., Shilatifard A., Workman J. L., Gerton J. L. (2009) Mol. Cell 35, 794–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dyer P. N., Edayathumangalam R. S., White C. L., Bao Y., Chakravarthy S., Muthurajan U. M., Luger K. (2004) Methods Enzymol. 375, 23–44 [DOI] [PubMed] [Google Scholar]
- 16. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 17. Muecke M., Samuels M., Davey M., Jeruzalmi D. (2008) Structure 16, 837–841 [DOI] [PubMed] [Google Scholar]
- 18. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Visnapuu M. L., Greene E. C. (2009) Nat. Struct. Mol. Biol. 16, 1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekulic N., Bassett E. A., Rogers D. J., Black B. E. (2010) Nature 467, 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Polizzi C., Clarke L. (1991) J. Cell Biol. 112, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yoda K., Ando S., Morishita S., Houmura K., Hashimoto K., Takeyasu K., Okazaki T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7266–7271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White C. L., Suto R. K., Luger K. (2001) EMBO J. 20, 5207–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]