Abstract
Retinoids are promising agents for the treatment/prevention of breast carcinoma. We examined the role of microRNAs in mediating the effects of all-trans-retinoic acid (ATRA), which suppresses the proliferation of estrogen receptor-positive (ERα+) breast carcinoma cells, such as MCF-7, but not estrogen receptor-negative cells, such as MDA-MB-231. We found that pro-oncogenic miR-21 is selectively induced by ATRA in ERα+ cells. Induction of miR-21 counteracts the anti-proliferative action of ATRA but has the potentially beneficial effect of reducing cell motility. In ERα+ cells, retinoid-dependent induction of miR-21 is due to increased transcription of the MIR21 gene via ligand-dependent activation of the nuclear retinoid receptor, RARα. RARα is part of the transcription complex present in the 5′-flanking region of the MIR21 gene. The receptor binds to two functional retinoic acid-responsive elements mapping upstream of the transcription initiation site. Silencing of miR-21 enhances ATRA-dependent growth inhibition and senescence while reverting suppression of cell motility afforded by the retinoid. Up-regulation of miR-21 results in retinoid-dependent inhibition of the established target, maspin. Knockdown and overexpression of maspin in MCF-7 cells indicates that the protein is involved in ATRA-induced growth inhibition and contributes to the ATRA-dependent anti-motility responses. Integration between whole genome analysis of genes differentially regulated by ATRA in MCF-7 and MDA-MB-231 cells, prediction of miR-21 regulated genes, and functional studies led to the identification of three novel direct miR-21 targets: the pro-inflammatory cytokine IL1B, the adhesion molecule ICAM-1 and PLAT, the tissue-type plasminogen activator. Evidence for ICAM-1 involvement in retinoid-dependent inhibition of MCF-7 cell motility is provided.
Keywords: Cancer Therapy, Gene Regulation, MicroRNA, Nuclear Receptors, Retinoid
Introduction
All-trans-retinoic acid (ATRA)2 and derivatives (retinoids) are promising agents in the treatment/chemoprevention of hematologic and other malignancies (1, 2), including breast carcinoma (3). Breast cancer is an heterogeneous group of tumors with variable response to therapeutic agents, including retinoids. Generally, breast carcinoma cells expressing estrogen receptor type α (ERα+) are sensitive to, whereas the ERα-negative (ERα−) counterparts are refractory to, the anti-proliferative activity of retinoids (1).
MicroRNAs (miRNAs) are short RNAs controlling the stability of target mRNAs or their translation into protein products (4). They influence cell homeostasis and response to drugs (5, 6), modulating the activity of numerous target transcripts simultaneously, via binding to the 3′-untranslated region. Little is known about the effects of retinoids on miRNAs in breast carcinoma and/or other neoplasias (7).
MCF-7 breast carcinoma cells are ERα+, whereas the MDA-MB-231 counterparts are ERα− (8) (9). MCF-7 are sensitive, whereas MDA-MB-231 cells are refractory to the transcriptional and proliferative effects of E2. The pair of cell lines is a model (10–16) for the association between ERα positivity and response to the anti-proliferative effects of retinoids.
We used predominantly the MCF-7 and MDA-MB-231 cell lines to study the effects of ATRA and derivatives on miRNA expression. miR-21 was the only miRNA whose expression was perturbed by the retinoid. Retinoid-dependent induction of the miRNA was observed in MCF-7 and other ERα+ cell lines. The consequences of miR-21 induction were evaluated in terms of retinoid-dependent functional responses and gene expression.
EXPERIMENTAL PROCEDURES
Cell Lines and Chemicals
All of the cell lines were from the American Type Culture Collection. Breast cancer cells were grown in F12 medium (Invitrogen) containing 5% charcoal-stripped calf serum (Lonza, Walkersville, MD) with 0.01 μm E2. ATRA and E2 were from Sigma. AM580 and CD437 have been described (17, 18).
Single-cell Motility
Single-cell motility assays were performed on BSA-coated substrate (19) using the Imaging Station CellR̂ (Olympus, Segrate, Italy) and the software Image J (Rasband W, National Institutes of Health, Bethesda, MD).
Microarrays
miRNA microarrays were generated by spotting 1,450 miRNAs, (Exiqon miRNA probe set v8.1) in quadruplicate onto Corning epoxide-coated slides. Samples from TRIzol-extracted RNA (20 μg) were enriched for microRNA using the flash PAGE fractionator system (Ambion, Austin, TX) and subsequently labeled for hybridization using the mirVana miRNA labeling kit (Ambion). Three competitive hybridization experiments were performed in duplicate, using microRNA fractions pooled from three independent cell cultures (20). Arrays were scanned using a GenePix 4000B Scanner driven by GenePix Pro 4.0 (Molecular Devices). All of the analyses were performed with the statistical programming and graphics environment R. Differentially expressed miRNAs were identified using the empirical Bayes approach, which ranks genes on a combination of magnitude and consistency of differential expression (20, 21). Gene expression microarray (G4112F; Agilent, Palo Alto, CA) experiments were performed as detailed (22). miRNA and gene expression microarray results were deposited in the GEO database (GSE18693).
Cell Growth and Senescence
Cell growth was evaluated using MTT (23), and senescence was determined with a β-galactosidase kit (Cell Signaling Technology, Beverly, MA) or with the EpiQuick global trimethyl histone H3-K9 quantification kit (EPIGENTEK, Brooklyn, NY).
ChIP, Oligonucleotides, Plasmid Constructs, and Transfections
ChIP assays (24) were performed with anti-RARα (sc-551x), anti-ERα (sc-542x), and anti-CYP1A1 (sc-20772) irrelevant antibodies (Santa Cruz Biotechnology). A list of the oligonucleotides used in the study are described in supplemental Table S1. In vitro mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). The RARE-tk-Luc, RARα, and RARγ expression plasmids were described (24, 25). The anti-miR-21 (AM10206), pre-miR-21 (PM10206) oligonucleotides, and Silencer Select siRNAs for ICAM-1 (s7087) and for maspin (s10466) were from Ambion Inc. FlexiTube siRNAs for PLAT (SI00018746 and SI02779903) were obtained from Qiagen. The green fluorescent protein plasmid (pEGFP-N1) was from Clontech. The cDNAs coding for ICAM-1, maspin, and PLAT were amplified by RT-PCR from MCF-7 RNA and subcloned first in pCR2.1 vector (TA cloning kit; Invitrogen) and subsequently reamplified by PCR and subcloned in pcDNA3 (Invitrogen) gene (supplemental Table S1). To obtain the luciferase reporter constructs driven by the MIR21 promoter, a 1.5-kb fragment of the 5′-flanking region was amplified by PCR and was inserted in the corresponding firefly luciferase reporter plasmid both in the sense (miR-21 S) and antisense (miR-21 AS) orientations as follows. The fragment was first subcloned in the plasmid pCR2.1 (AT cloning kit; Invitrogen) and subsequently digested either by XhoI and HindIII (for the sense orientation) or by XhoI and KpnI (for the anti sense orientation). These fragments were inserted in the plasmid pGL-3 Basic Vector (Promega Italia, Milan, Italy) to obtain the plasmid DNA constructs in the appropriate orientations. The cells were transfected with 50 nm of pre-miR-21, anti-miR-21, or control oligonucleotides using the siPORT NeoFX reagent (Ambion). siRNAs (5 nm of Silencer Select siRNA or 30 nm of FlexiTube siRNA) were transfected with siPORT NeoFX reagent, whereas cDNA constructs were introduced using FuGENE HD transfection reagent (Promega). Transactivation experiments with the miR-21 promoter constructs and 3′-UTR-luciferase assays were performed using MCF-7 and 293T cells (5, 24), respectively.
Real Time PCR
Mature miR-21 miRNA was measured using TaqMan assays (Applied Biosystems). The primary transcript, pri-miR-21, and potential miR-21 target transcripts were determined by PCR with SYBR green or custom-designed TaqMan assays (24). For the detection of pre-miR21, miRNAs were fractionated on a polyacrylamide gel, and Northern blot analysis was performed with a 212-bp probe (nucleotides 2,385–2,596 of AY699265). A DNA fragment complementary to RUN-6 (106 nucleotides) was amplified from MCF-7 RNA by RT-PCR using the oligonucleotide primers listed on supplemental Table S1 and subsequently subcloned in pCR2.1 vector (TA cloning kit; Invitrogen). The RUN-6 insert was reamplfied by PCR using T7 and M13 reverse primers, 32P-labeled, and used as a probe to show the equal amounts of RNA were loaded on the gel. The following TaqMan assays (Applied Biosystems) were used for the quantitative determination of the following mRNAs: PLAT, Hs00938315_m1; PTX3, Hs00173615_m1; ICAM-1, Hs99999152_m1; IL1B, Hs01555413_m1; CCR1, Hs00174298_m1; TNFAIP3, Hs01574287_m1; SERPINB5 (Maspin), Hs00985283_m1; and ACTB (β-actin endogenous control), 433762F.
Western Blot, ELISA, and FACS Analyses
Western blot experiments were performed on total cellular extracts according to routine procedures (24) using the following antibodies from Cell Signaling Inc. (Danvers, MA): anti-maspin (catalog number 9117). Anti-β-actin antibody (catalog number sc-8432) were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-CCR1 antibodies were from Abcam (Cambridge, UK; catalog number ab1681), and anti-PLAT antibodies were from Affinity Biologicals Inc. (Ancaster, Canada). IL1B and PLAT proteins were measured in cell extracts using commercially available ELISA assays (IL1B Quantikine, catalog number DLB50; R & D Systems, Minneapolis, MN; PLAT, ZYMUTEST tPA antigen; Hyphen BioMed, Neuville sur Oise, France, catalog number sRK011A). FACS analysis of ICAM-1 was performed with the anti ICAM-1 (Invitrogen; catalog number 07-5403) following co-transfection of anti-miR-21 or anti-NC oligonucleotides and pEGFP-N1 (containing the enhanced GFP gene). After staining with secondary antibodies conjugated with phycoerythrin, dual color FACS analysis was performed, and the mean associated red fluorescence caused by ICAM-1 surface binding was measured on the GFP+ fraction of cells, using a FACSCalibur instrument (Becton Dickinson, Milano, Italy).
RESULTS
miR-21 Is the Only miRNA Induced by ATRA in MCF-7 Cells
We determined the differential profiles of miRNA expression in MCF-7 and MDA-MB-231 cells cultured with or without ATRA (Fig. 1A). During logarithmic growth, divergent miRNA profiles were evident in the two cell lines, with MDA-MB-231 cells having high expression of miR-142–3p, miR-18a, and miR-135b and low expression of miR-342, as observed in ERα− and basal breast tumors (26). Relative to MCF-7, MDA-MB-231 cells showed low expression of three miRNA clusters (miR-200c/miR-141; miR-200a/miR-200b/miR-429; and miR-182/miR-96/miR-183) down-regulated in cancer stem cells (27).
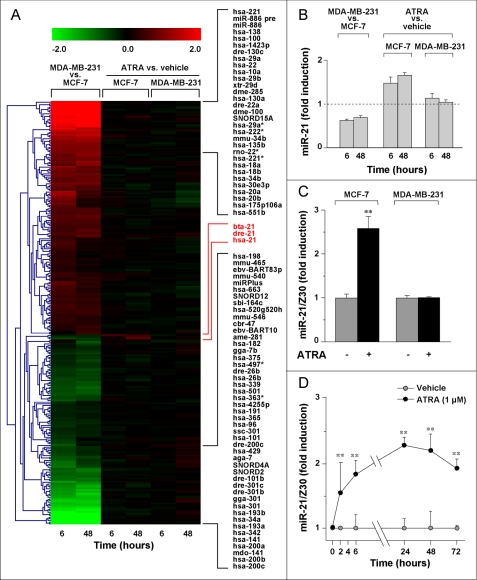
FIGURE 1.
MicroRNA profiles in MCF-7 and MDA-MB-231 cells. A, heat map of microRNAs showing significant differences (adjusted p < 0.05) in at least one of the comparisons indicated. Relevant microRNAs are listed on the right. B, relative levels of miR-21 (means ± S.D. of two independent microarray experiments performed in triplicate). C, RT-PCR validation of microarray results (mean ± S.D.; three independent cultures). The cells were treated with ATRA (1 μm) for 6 h. The data are normalized for the content of the Z30 microRNA endogenous standard. The data are representative of three independent experiments. **, significantly higher than the corresponding control value (p < 0.01, Student's t test). D, time course of miR-21 induction. The results were obtained as in C and are representative of two independent experiments. **, significantly higher than the corresponding control value (p < 0.01, Student's t test).
Although ATRA did not affect the miRNA profile of MDA-MB-231 cells, in the MCF-7 counterparts, the retinoid caused significant increases of a single miRNA, miR-21 (Fig. 1, A and B). The result was confirmed by RT-PCR (Fig. 1C). Induction of miR-21 was detectable after 2 h and leveled off at 24 h (Fig. 1D).
ATRA Induces Transcription of the MIR21 Gene via Retinoic Acid-responsive Elements
To determine the mechanism of miR-21 induction in MCF-7 cells, we measured the levels of the primary transcript (pri-miR-21) and the 77-nucleotide hairpin pre-miRNA (Fig. 2A). Both pri-miR-21 and pre-miR-21 were induced by ATRA, suggesting increased transcription of the MIR21 gene. Similar effects were not observed in MDA-MB-231 cells, in which the basal levels of pri-miR-21 and pre-miR-21 were left unaffected by the retinoid (data not shown).
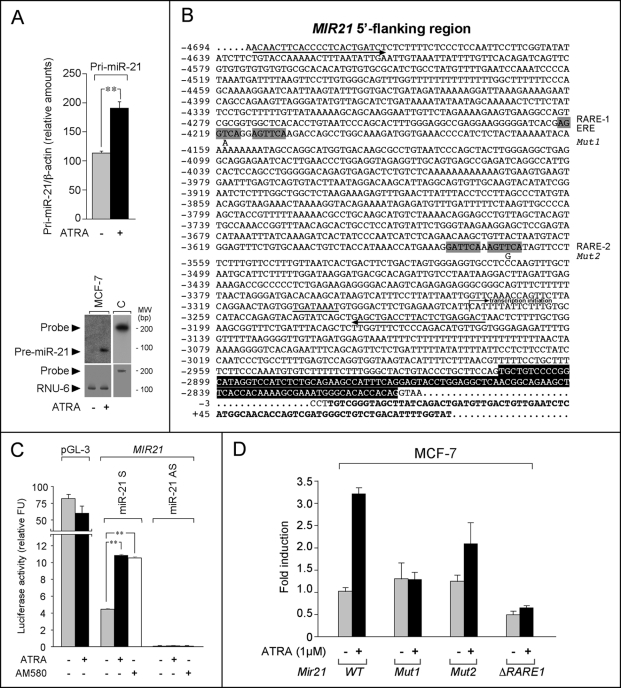
FIGURE 2.
Transcriptional regulation of MIR21 by retinoids in ERα+ cell lines via RARα. A, MCF-7 cells were treated with ATRA (1 μm) or vehicle (Me2SO) for 48 h. Upper panel, RT-PCR analysis of pri-miR-21, using a primer pair corresponding to sequences placed outside the miR-21 containing stem-loop region of the primary transcript. Two sets of primers were selected from the published sequence (AY699265) and used for the amplification of pri-miR-21. Because the results obtained with the two pairs of primers were superimposable, only the results obtained with the first pair are shown. Each value is the mean ± S.D. of three separate cultures. Lower panel, Northern blot analysis of pre-miR-21 (radiolabeled probe encompassing the stem-loop region). MW, molecular weight. The blot was rehybridized with a U6 snRNA probe (RNU-6) to confirm that similar amounts of low molecular weight RNA were loaded in each lane. The results are representative of two independent experiments. B, nucleotide sequence of the 5′-flanking region of the MIR21 gene. The two predicted RAREs (retinoic acid responsive elements, RARE-1 and RARE-2) and the estrogen responsive element (ERE) overlapping with RARE-1 are indicated by gray boxes. The point mutations (Mut1 and Mut2) introduced in RARE-1 and RARE-2 are indicated. The putative TATA box is underlined with a solid line. The putative transcription initiation site is indicated. The sequence boxed in black represents exon 11 of the TMEM-49 coding gene, which overlaps the MIR21 gene. The sequence in bold type corresponds to the miR-21 stem-loop. Residue +1 corresponds to the first nucleotide of the miR-21 stem-loop sequence. The two arrows below the sequence pointing in opposite directions indicate the position of the two oligonucleotides used for the amplification of the 5′-flanking region of the miR-21. C, MCF-7 cells were transfected with firefly luciferase reporter constructs driven by the 5′-flanking region of human MIR21 in the sense or antisense orientation. A plasmid containing a CMV-based enhancer/promoter was used as a negative control for the effect of ATRA (pGL-3). The cells were treated with ATRA (1 μm) or AM580 (0.1 μm) for 24 h. The results are expressed in arbitrary units following normalization with Renilla luciferase (mean ± S.D., two replicate transfections). D, functional analysis of the putative RARE-1 and RARE-2 sequences. Constructs containing the firefly luciferase reporter gene driven by the wild type MIR21 promoter (WT) or the same promoter with point mutations in RARE-1 and RARE-2 (Mut1 and Mut2, see text) or deletion of RARE-1 sequences (ΔRARE1) were transfected in MCF-7 cells, before treatment with vehicle or ATRA (1 μm) for 24 h.
Two predicted retinoid-regulated elements (RARE-1 and RARE-2) are present in the 5′-flanking region of MIR21 (Fig. 2B) (28, 29). To define the functional activity of the predicted RARE-1 and RARE-2 sequences, the MIR21 5′-flanking region was inserted upstream of a luciferase reporter. In MCF-7 cells, the reporter was induced by ATRA only when the promoter was in the sense orientation (Fig. 2C). Transactivation of MIR21 was specific, because ATRA did not affect the activity of a control reporter (p-GL3). In MDA-MB-231 cells, the basal level of MIR21 promoter activity was left unaltered by ATRA (data not shown). Point mutations were introduced in critical residues of the RAR-binding consensus repeats ((A/G)G(G/T)TCA) of RARE-1 (Mut1: AGGACA, nucleotide −4,218) and RARE-2 (Mut2: AGTGCA, nucleotide −3,570) (Fig. 2B). Mut1 abrogated transcriptional activation of MIR21 by ATRA, although a minor reduction was also observed with Mut2 (Fig. 2D). The role of RARE-1 in ligand-dependent transactivation of MIR21 transcription was confirmed by deletion of both RAR-binding consensus repeats (ΔRARE1). All of this indicates that both RARE-1 and RARE-2 control the transcriptional activity of MIR21 in a retinoid-dependent manner.
RARα Is a Primary Determinant of Retinoid Sensitivity in MCF-7 Cells and Is the Retinoid Receptor Responsible for miR-21 Induction by ATRA
In basal conditions (Fig. 3A), MCF-7 and MDA-MB-231 cells express RARα, RARγ, RXRα, and RXRβ mRNAs. The levels of these transcripts reflect the relative amounts of the corresponding proteins, as demonstrated for RARα, RARγ, and RXRα (Fig. 3B). The major quantitative difference in the complement of receptors is observed for RARα, which is much more abundant in MCF-7 than in MDA-MB-231 cells (30). This is consistent with the observation that ERα+ breast carcinomas generally express higher levels of RARα mRNA.3
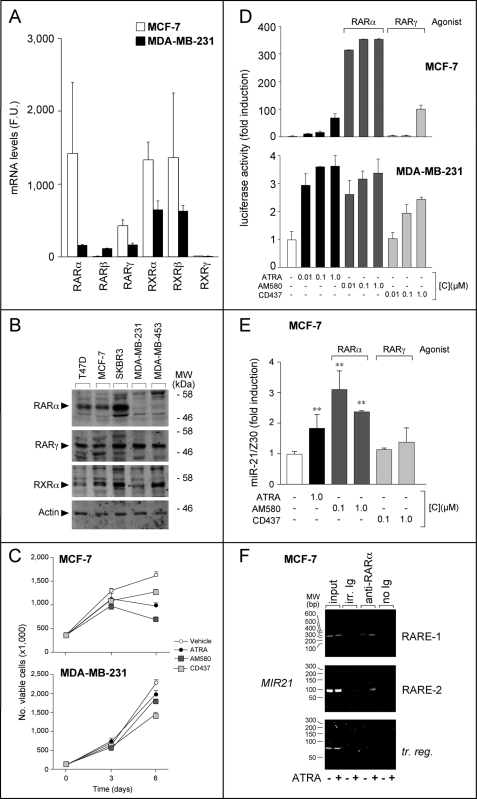
FIGURE 3.
Significance of RARα for miR-21 induction by ATRA and retinoid sensitivity in MCF-7. A, the panel shows the levels of the transcripts encoding RAR and RXR nuclear retinoic acid receptors in MCF-7 and MDA-MB-231 cells. The results were obtained from the whole genome gene expression microarray data set and represent the means ± S.D. of six separate slides. The data are expressed in absolute fluorescent units, and the values determined in MCF-7 and MDA-MB-231 are directly comparable. B, Western blot analyses of the indicated RAR and RXR proteins are shown in the panel. The results were obtained using extracts of the indicated breast carcinoma cell lines. The total amount of protein present in each lane is similar, as indicated by the β-actin signal obtained. C, the growth curves of MCF-7 and MDA-MB-231 cells incubated in the presence of vehicle (Me2SO), ATRA (1 μm), AM580 (0.1 μm), or CD437 (0.1 μm) for the indicated amounts of time are shown. Each value is the mean ± S.D. of three replicate cultures. D, MCF-7 and MDA-MB-231 cells were transfected with the retinoid-dependent reporter construct, RARE-tk-Luc, and treated for 24 h with the indicated concentrations of ATRA, AM580, or CD437 (mean ± S.D., two replicate transfections). All of the results are representative of at least two independent experiments. E, RT-PCR determination of miR-21 in MCF-7 cells treated with the indicated concentrations of ATRA, AM580, or CD437, for 48 h (mean ± S.D., three independent cultures). **, significantly higher than control (Student's t test, p < 0.01). F, ChIP assays were performed on MCF-7 cells E2-depleted for 5 days and subsequently treated with vehicle or ATRA in the presence of E2 for 2 h, using anti-RARα antibodies and relative negative controls. irr. Ig, irrelevant antibodies of the same Ig type as the anti-RARα counterparts. For the amplification of the transcribed region (tr. reg.) of MIR21, we used the same pair of amplimers described for Fig. 2A.
RARα is the primary determinant of the sensitivity to the anti-proliferative and transcriptional effects of ATRA in MCF-7 cells, as indicated by the results obtained with the RARα-selective agonist, AM580; the RARγ-specific agonist, CD437; and the pan-RAR agonist, ATRA (17) (2, 18). In fact, AM580 (0.1 μm) inhibited MCF-7 growth more efficiently than ATRA (1 μm) (Fig. 3C) and was more powerful in activating the retinoid reporter, RARE-tk-Luc (Fig. 3D). At receptor-selective concentrations (0.01 or 0.1 μm) (31), CD437 exerted modest anti-proliferative and transcriptional effects. As expected, in MDA-MB-231 cells, AM580 and CD437 were devoid of growth inhibitory activity and caused an activation of RARE-tk-Luc 2 orders of magnitude lower relative to MCF-7 cells.
We evaluated the relative importance of RARα and RARγ for miR-21 induction in MCF-7 cells, comparing the effects of AM580, CD437, and ATRA. Induction of miR-21 in MCF-7 cells was observed with the RARα ligand, AM580, but not with the RARγ agonist, CD437 (Fig. 3E). AM580 was also capable of transactivating the luciferase reporter construct driven by the MIR21 promoter (Fig. 2C). All of this indicates that transcriptional activation of MIR21 is mediated predominantly by RARα. The contention is supported by the fact that RARα binds to the endogenous MIR21 promoter in MCF-7 cells, as demonstrated by ChIP experiments (Fig. 3F). In MCF-7 cells treated with ATRA, RARα-specific signals were determined with amplimers encompassing the RARE-1- and RARE-2-containing regions, whereas no binding to a transcribed region of MIR21 was detected. This demonstrates that RARα is recruited to the promoter of MIR21 upon ligand activation. The results of ChIP analysis complement the functional data on the promoter and support the concept that MIR21 is a direct RARα-target gene.
Induction of miR-21 by ATRA Is Observed Only in ERα+ Cells
To investigate the determinants of cell-specific miR-21 induction, we selected three other breast carcinoma cell lines with different sensitivity to ATRA and distinct patterns of ERα/RAR/RXR expression (Fig. 3B). ERα+ T47D cells were sensitive to the anti-proliferative action of ATRA and responded to the retinoid with miR-21 induction (Fig. 4A). In contrast, expression of the miRNA was unaltered in ERα− SKBR3 and MDA-MB-453 cells, showing sensitivity and resistance to ATRA, respectively. Our results suggest a correlation between ERα positivity and ATRA-dependent miR-21 up-regulation. The data obtained with SKBR3 cells support the concept that sensitivity to retinoids and high levels of RARα in the context of ERα negativity are insufficient for miR-21 induction.
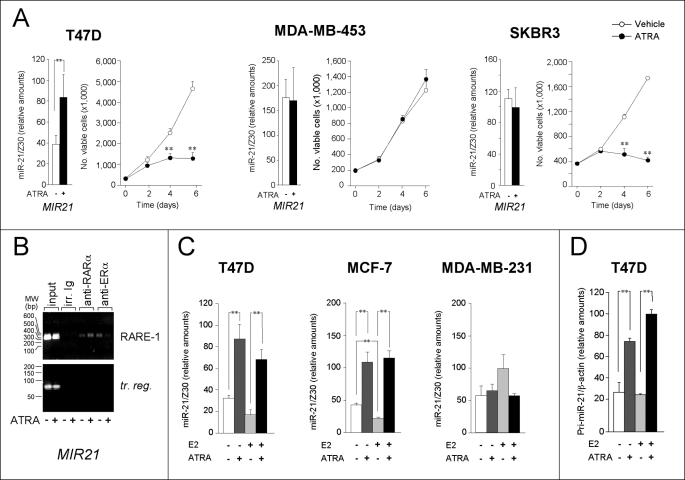
FIGURE 4.
miR-21 induction in ERα− and ERα+ cell lines. A, in the bar graphs, the indicated cell lines were treated with ATRA (1 μm) for 48 h. The microRNA fraction was extracted and subjected to the determination of mature miR-21 by quantitative RT-PCR. The data are normalized for the content of the Z30 microRNA endogenous standard and are the means ± S.D. of three independent cultures. **, significantly higher than the corresponding control (Student's t test, p < 0.01). The data are representative of at least two independent experiments. In the line graphs, ERα+ T47D as well as ERα− SKBR3 and MDA-MB-453 cells were treated with ATRA (1 μm) for the indicated amount of time. The growth curves of the three cell lines are shown. Each value is the mean ± S.D. of three replicate cultures. **, significantly lower than the corresponding vehicle-treated value (Student's t test, p < 0.01). B, ChIP assays were performed on MCF-7 cells treated with vehicle or ATRA as in Fig. 3F, using anti-RARα and anti-ERα antibodies as well as relative negative controls. irr. Ig, irrelevant antibodies of the same Ig type as the anti-ERα counterparts. For the amplification of the transcribed region (tr. reg.) of MIR21, we used the same pair of amplimers described in Fig. 2A. The results are representative of two independent experiments. C, the indicated cell lines were depleted of E2 by culturing in F12 medium supplemented with charcoal-stripped serum for 5 days. The cells (125,000/ml) were replated and treated with vehicle, E2 (0.01 μm), ATRA (1 μm), or the combination of the two agents for 48 h. Mature miR-21 was determined as above. The data are the means ± S.D. of three independent cultures. The results shown are representative of two independent experiments. **, significantly different (Student's t test, p < 0.01). D, T47D cells were depleted of E2 and treated as in C. Total RNA was extracted and subjected to quantitative RT-PCR for the determination of pri-miR-21. The data are the means ± S.D. of three independent cultures. The results shown are representative of two independent experiments. **, significantly higher (Student's t test, p < 0.01).
Selective induction of miR-21 by ATRA in ERα+ cells may be controlled by multiple factors, including antagonistic (32) or cooperative (33) interactions between estrogen and retinoid receptors (34, 35). Interestingly, the MIR21 promoter contains an estrogen-regulated element, overlapping to RARE-1 (Fig. 2B) (36). For all these reasons, the effect of ATRA treatment on ERα binding to the miR-21 promoter was evaluated by ChIP analysis in MCF-7 cells (Fig. 4B). In the absence of ATRA, ERα bound to the promoter region containing the putative estrogen-regulated element site. Treatment of cells with ATRA resulted in release of ERα from the promoter and concomitant recruitment of RARα to the same regulatory region. This is in line with similar antagonistic effects reported for other ERα- and RAR-dependent genes (32).
In a further set of experiments, we evaluated whether induction of miR-21 by ATRA was influenced by E2. MCF-7, T47D, and MDA-MB-231 cells were estrogen-starved and subsequently exposed to ATRA, E2, or E2 + ATRA (Fig. 4C). miR-21 levels in MDA-MB-231 were unaffected by E2 and/or the retinoid, whereas the estrogen inhibited miR-21 expression in MCF-7 (36) and T47D cells. In these conditions, ATRA-dependent increases in miR-21 were evident both in the presence and absence of E2. As observed in MCF-7 cells, induction of miR-21 in T47D cells involved ATRA-dependent and E2-independent up-regulation of pri-miR-21 (Fig. 4D). Thus, ligand-activated ERα does not exert significant effects on miR-21 induction by ATRA, suggesting that undefined permissive mechanisms underlay retinoid-dependent up-regulation of miR-21 in ERα+ cells.
miR-21 Modulates Growth, Senescence, and Motility in MCF-7 Cells
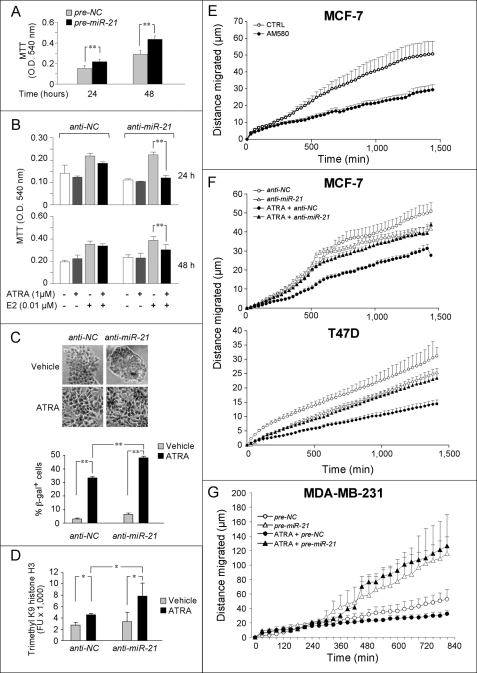
Induction of miR-21 by an anti-proliferative agent like ATRA in ERα+ breast carcinoma cells was unexpected, because the miRNA is endowed with oncogenic properties (37–41). To study the role of miR-21 induction in some of the cellular responses to ATRA, we overexpressed or silenced miR-21 by transient transfection of validated oligonucleotides (pre-miR-21 and anti-miR-21). Pre-miR-21 stimulated MCF-7 cell proliferation at the time points considered (Fig. 5A). In complementary studies, anti-miR-21 did not alter the proliferative action of E2 (Fig. 5B). ATRA exerted only minimal effects on the growth of control oligonucleotide (anti-NC)-treated and E2-stimulated MCF-7 cells at these early time points. In contrast, significant growth inhibition by ATRA was observed upon miR-21 silencing, indicating sensitization of MCF-7 cells. These results suggest that miR21 induction is part of negative feedback loops counteracting the anti-proliferative activity of ATRA.
FIGURE 5.
Effects of overexpression or silencing of miR-21 on growth/senescence of MCF-7 cell and random motility of MCF-7, T47D, and MDA-MB-231 cells. A, E2-depleted MCF-7 cells were transfected with pre-miR-21 or the negative control pre-NC. Cell growth was evaluated 24 or 48 h later by the MTT assay (mean ± S.D., three independent cultures). B, E2-depleted MCF-7 cells were transfected with anti-miR-21 or the negative control, anti-NC. After 24 h from the transfection, the cells were treated with ATRA and/or E2 for further 24 or 48 h and proliferation evaluated as in A. C, E2-depleted MCF-7 cells were transfected with anti-miR-21 or anti-NC. After transfection, the cells were treated with vehicle or ATRA (1 μm) for a further 4 days in the presence of E2 (0.01 μm) and stained for the senescence marker β-galactosidase. The pictures show representative light micrographs, and the bar graph illustrates the percentage of β-galactosidase-positive cells. More than 500 cells/field, 4 fields/experimental point, were counted (mean ± S.D., three independent cultures). For A–C, **, p < 0.01 (Student's t test). The results are representative of three independent experiments. D, E2-depleted MCF-7 cells were transfected with anti-miR-21 or the negative control, anti-NC. Twenty-four hours after transfection, the cells were treated with ATRA (1 μm) or vehicle in the presence of E2 for another 48 h. Nuclear extracts were evaluated for the amounts of trimethyl K9 histone H3, a marker of senescence-associated changes in chromatin structure, using a specific ELISA assay. The results are the means ± S.D. of three independent cultures. *, p < 0.05 (Student's t test). E, MCF-7 cells were treated with vehicle or AM580 (0.1 μm) for 48 h. Single cell random motility was monitored by time lapse microscopy for the indicated amount of time. Each value is the mean ± S.E. of at least 16 cells/experimental point. CTRL, control. F, E2-depleted MCF-7 or T47D cells (5 days) were transfected with anti-miR-21 or anti-NC along with pEGFP-N1 (containing the enhanced GFP gene) before treatment with ATRA (1 μm) or vehicle in the presence of E2 (0.01 μm). The displacement of individually tracked GFP-positive cells (mean ± S.E., at least 10 GFP-positive cells/experimental point) was measured by time lapse microscopy, starting 48 h after transfection. The results are representative of three independent experiments. All of the points of the curves corresponding to the anti-NC and ATRA + anti-NC treatments are significantly different (Student's t test, p < 0.01). G, MDA-MB-231 cells were transfected with pre-miR-21 or pre-NC along with pEGFP-N1 before treatment with ATRA (1 μm) or vehicle. Random motility was measured as in E and F. The results are representative of three independent experiments. All of the points of the curves corresponding to the pre-miR-21 and pre-NC treatments are significantly different regardless of the presence/absence of ATRA (Student's t test, p < 0.01).
Senescence is a modality by which cells stop dividing and is the final destiny of ATRA-treated MCF-7 cells (13). The effect of miR-21 silencing on ATRA-induced senescence was evaluated, using two molecular markers: β-galactosidase and trimethyl K9 histone H3 (Fig. 5, C and D). Control-transfected cells became positive for β-galactosidase after 4 days of treatment with ATRA. The percentage of senescent cells was significantly augmented in ATRA- and anti-miR-21-treated cultures. Similar effects were observed with trimethyl K9 histone H3, a marker of senescence-associated changes in chromatin structure (42). Thus, miR-21 induction counteracts not only the anti-proliferative but also the cell aging responses to retinoids.
miR-21 is proposed to promote cancer cell dissemination (43), whereas retinoids exert opposite effects (44). Given this dichotomy, we evaluated the action of ATRA on random cell motility, one component of the complex process of invasion, using time lapse microscopy (Fig. 5, E–G). This assay was selected because MCF-7 and T47D cells, unlike MDA-MB-231 cells, are characterized by very low motility in Matrigel (45), the semi-solid matrix commonly used in three-dimensional invasion assays. ATRA (1 μm) inhibited the ability of both MCF-7 and T47D cell lines to move in a nondirectional manner while exerting no appreciable effect on MDA-MB-231 cells (data not shown). AM580 (0.1 μm) suppressed random motility of MCF-7 cells, indicating involvement of RARα (Fig. 5E). The significance of miR-21 for ATRA-dependent decrease in motility was examined by inhibiting the miRNA in MCF-7 and T47D cells. In the absence of ATRA, transfection of anti-miR-21 in both MCF-7 and T47D cells did not cause significant alterations in cell motility (Fig. 5F). In contrast, retinoid-dependent inhibition of MCF-7 and T47D motility was reproducibly and significantly attenuated by transfection of anti-miR21. On the other hand and as expected on the basis of available reports (43), transfection of control or ATRA-treated MDA-MB-231 cells with pre-miR-21 always resulted in augmentation of motility (Fig. 5G). Our findings demonstrate that the ERα−, MDA-MB-231 cells, and the ERα+, MCF-7, or T47D cells have different responses to modulation of miR-21 levels in terms of motility. In addition, they indicate that miR-21 induction mediates at least part of the reduction in cell motility afforded by ATRA in ERα+ cells.
miR-21 Regulates the Established Target, Maspin, in MCF-7 Cells
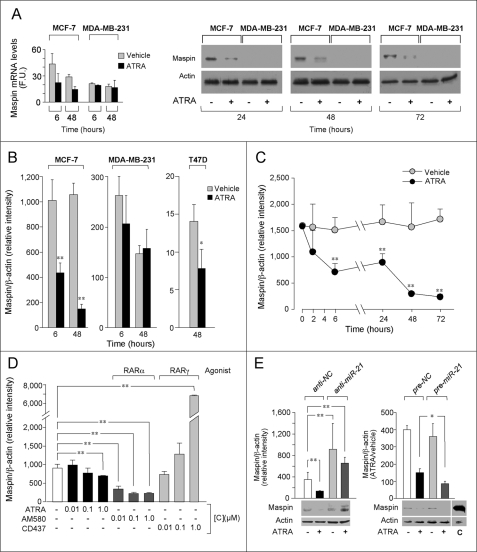
To identify functionally relevant genes regulated by ATRA-dependent miR-21 induction, first, we focused on differential expression of established miR-21 targets (46) in MCF-7 and MDA-MB-231 cells, defining the expression of the mRNAs encoding SERPINB5 (maspin) (47, 48), PTEN (49), PDCD4 (37), HNRP4 (50), Spry2 (51), NFIB (29), TPM1 (39), and RECK (52) by microarray analysis (supplemental Table S2). Six of the eight target mRNAs considered were left unaffected by ATRA treatment in both cell lines. In MCF-7 cells, the retinoid caused up-regulation of PDCD4 mRNA and protein (48 and 72 h) (data not shown). The only miR-21 target mRNA down-regulated by ATRA in MCF-7 cells was maspin (Fig. 6A). Reduction of maspin mRNA by the retinoid was accompanied by a decrease of the protein (24–72 h). Subsequent RT-PCR experiments showed ATRA-dependent decrease of maspin not only in MCF-7 but also in T47D cells and confirmed the lack of modulation by the retinoid in MDA-MB-231 cells (Fig. 6B). In MCF-7 cells, down-regulation of maspin mRNA was evident at 6 h and maximal at 48 h (Fig. 6C). Consistent with the relative ability to induce miR-21, AM580 was more effective than ATRA in reducing maspin mRNA levels (Fig. 6D), which were not decreased by CD437. The lower levels of maspin protein observed in MDA-MB-231 cells (53) were not affected by ATRA treatment (data not shown).
FIGURE 6.
Maspin is controlled by ATRA-dependent miR-21 induction in retinoid-responsive cells. A, effect of ATRA on the mRNA and protein levels of maspin in MCF-7 and MDA-MB-231 cells. Left panel, MCF-7 or MDA-MB-231 was treated for 6 or 48 h with vehicle or ATRA (1 μm) prior to RNA extraction and microarray analysis. The levels of maspin mRNA were determined from the whole genome gene expression microarray data set. The results are expressed in absolute fluorescence units and are the means ± S.D. of three biological replicates. Right panel, MCF-7 and MDA-MB-231 cells were treated for the indicated amount of time with vehicle or ATRA (1 μm). The same amounts of total cellular extracts (a pool of three independent cultures) were separated on SDS-PAGE and subjected to Western blot analysis with antibodies recognizing maspin (molecular mass, 40 kDa). β-Actin was used to demonstrate that identical amounts of MCF-7 and MDA-MB-231 proteins were added to each lane of the gel. B–D, the levels of maspin transcripts were determined by RT-PCR using a TaqMan assay. The results are the means ± S.D., three independent cultures. B, MCF-7, MDA-MB-231, or T47D cells were treated with 1 μm ATRA for the indicated amount of time. C, MCF-7 cells treated with 1 μm ATRA (time course). D, MCF-7 cells were treated with the indicated compounds. E, MCF-7 cells were transfected with anti-miR-21, pre-miR-21, or the negative controls, anti-NC and pre-NC, before treatment with ATRA for 72 h. The graphs indicate the results obtained by quantitative RT-PCR analysis of the maspin mRNA. The Western blots show the results obtained on the maspin protein (40 kDa). The same cell extracts were subjected to Western blot analysis with anti-maspin and anti-β-actin antibodies. Lane C shows that the maspin signal detected in MDA-MB231 cells is shown as a control to indicate that different levels of the protein are expressed in the two cell lines. The blot with β-actin (43 kDa) indicates that the same amounts of protein were loaded in each lane. All of the data are representative of at least two independent experiments. *, p < 0.05; **, p < 0.01 (Student's t test).
To gain direct evidence for miR-21 involvement in retinoid-dependent control of maspin, MCF-7 cells were transfected with anti-miR-21 and pre-miR-21 (Fig. 6E). Anti-miR-21 increased the basal levels and prevented retinoid-dependent down-regulation of maspin mRNA. Similar effects were observed at the protein level. In contrast, pre-miR-21 enhanced the down-regulation of maspin mRNA and protein afforded by ATRA. Altogether these results demonstrate that ATRA-triggered induction of miR-21 in ERα+ cells controls the amounts of maspin protein primarily acting on the corresponding transcript.
IL1B, ICAM-1, and PLAT Are New and Direct miR-21 Targets
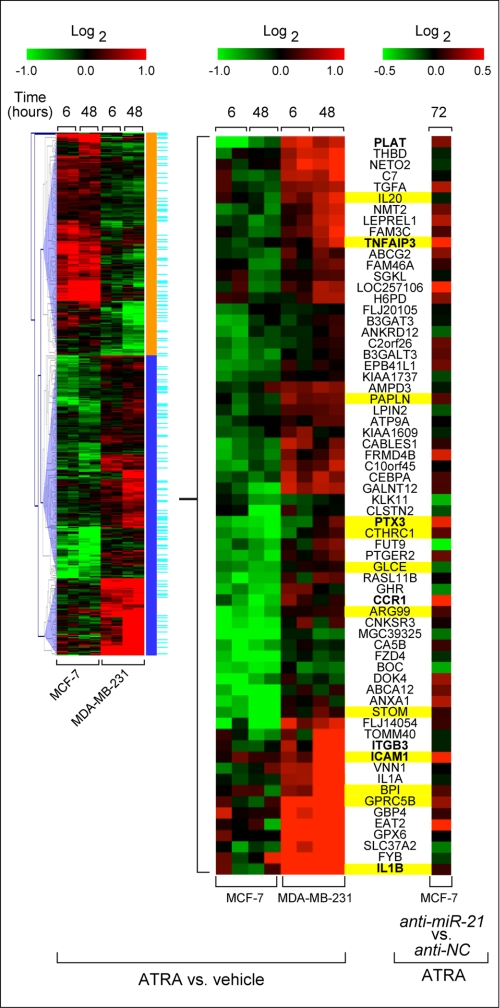
We exploited our cellular model to identify novel transcripts negatively controlled by ATRA via miR-21, using gene expression microarrays in combination with a bio-informatic approach. Given the large pool of miR-21 target genes predicted by the commonly available algorithms, we progressively restricted their number. Initially, ATRA-induced changes in the transcriptomes of MCF-7 and MDA-MB-231 cells were determined. Because miR-21 was induced by ATRA in MCF-7 cells but not in MDA-MB231 cells, we focused on genes whose retinoid-dependent regulation was significantly different (p < 0.01) in the two cellular contexts (Fig. 7, left panel, and supplemental Table S2). Of the 481 genes identified, 276 were either down-regulated in MCF-7 but not in MDA-MB-231 or up-regulated by the retinoid in MDA-MB-231 but not in MCF-7 cells (blue cluster). This is the regulation pattern expected for direct targets of miR-21. Subsequently, we focused on the 66 genes of the blue cluster (17%) predicted to be potential miR-21 targets by at least one of the algorithms considered and for which functional information is available. Interestingly, genes involved in inflammation and immunity (Fig. 7, middle panel, yellow marks) are enriched in this last group.
FIGURE 7.
Identification of putative new miR-21 target genes. Gene expression profiling of MCF-7 and MDA-MB-231 cells treated with vehicle or 1 μm ATRA (Log2 ratio of ATRA versus vehicle). Left panel, heat map of genes whose retinoid-dependent regulation is significantly different in the two cell lines (p < 0.01) and classification in patterns consistent with miR-21 regulation (blue cluster) or not (orange cluster). Genes predicted to be target of miR-21 by Miranda, PITA, or TargetScan are indicated in light blue. Middle panel, partial blow-out of the blue cluster containing the 66 predicted and annotated targets of miR-21. Right panel, heat map of the same transcripts derived from MCF-7 cells treated with 1 μm ATRA (72 h) after transfection with anti-miR-21 or anti-NC (Log2 ratio of ATRA+anti-miR-21 versus ATRA+anti-NC). Genes belonging to inflammatory/immune responses/leukocyte migration pathways are marked in yellow.
To examine the role of miR-21 in the regulation of these genes, MCF-7 cells were transfected with anti-miR-21 or anti-NC and treated with ATRA for 72 h before analysis of global gene expression. The two transcriptomes were compared, and the data obtained for the 66 predicted miR-21 target genes were extracted (Fig. 7, right panel). We expressed the results as the Log2 ratio of the anti-miR-21/anti-NC signal, anticipating that this value would be positive for direct miR-21 targets. Using a threshold value of 0.2, the prediction was fulfilled for 15 (22%) of the genes.
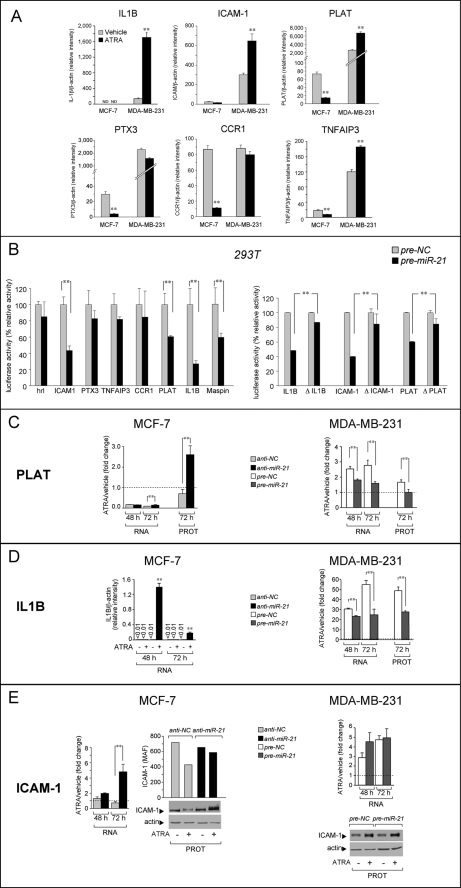
Among the 15 genes up-regulated by anti-miR-21 transfection, we selected six candidates (IL1B, ICAM-1, PLAT, PTX3, CCR1, and TNFAIP3) of biological interest for further validation (see supplemental Table S3 for the predicted seed sequences of the corresponding 3′-UTRs). IL1B, PTX3, and TNFAIP3 were chosen for their role in mediating inflammatory responses, which are involved in tumor growth and invasion, given the enrichment of this type of genes in the blue cluster. ICAM-1, PLAT, and CCR1 were considered, because they are involved in cancer cell dissemination, a process potentially affected by miR-21 via regulation of cell motility (Fig. 5). Differential regulation of these target mRNAs by ATRA in MCF-7 and MDA-MB-231 cells was validated by RT-PCR, at the RNA level (Fig. 8A), following treatment with vehicle or ATRA. IL1B, ICAM-1, and PLAT mRNAs were induced by ATRA in MDA-MB-231. In MCF-7 cells, IL1B and ICAM-1 mRNAs were left unaffected, whereas PLAT was down-regulated by the retinoid. This is consistent with ATRA-dependent elevation of miR-21 in MCF-7 cells limiting the induction, otherwise observed in MDA-MB-231 cells, and/or reducing expression of the transcripts. PTX3 and CCR1 mRNAs were not induced by ATRA in MDA-MB-231 cells but were down-regulated in MCF-7 cells, which is also consistent with targeting by miR-21. With the exception of TNFAIP3, which was not significantly induced by ATRA in MDA-MB-231 cells, the mRNA regulation pattern was generally confirmed at the protein level (48 or 72 h) (data not shown).
FIGURE 8.
Validation of selected novel miR-21 targets. A, MCF-7 or MDA-MB-231 cells were treated with ATRA (1 μm) for 48 h. Total RNA was extracted and used for the determination of the indicated transcripts by quantitative RT-PCR. The data are normalized for the content of the β-actin control RNA and are the means ± S.D. of three independent cultures. The data are representative of three independent experiments. B, 293T cells were co-transfected with pre-NC or pre-miR-21 and plasmids containing the Renilla luciferase reporter upstream of the indicated cDNA 3′-UTR. Forty-eight hours later, luciferase activity was determined. The results were normalized for the transfection efficiency using firefly luciferase. Each value is the mean ± S.D. of three replicates and is representative of three independent experiments. Deletion mutants of the miR-21 seed sequences present in the 3′-UTR of the indicated cDNAs were generated. Wild type Renilla luciferase-based constructs (hrl) or deletion mutants thereof were co-transfected with pre-miR-21 or pre-NC oligonucleotide in the presence of the normalizing firefly luciferase plasmid (pGL-3) in 293T cells. After 48 h, the luciferase activities were measured in cell extracts. The data are the means ± S.D. of three independent cultures and are representative of two independent experiments. **, significantly different (Student's t test, p < 0.01). C–E, effect of anti-miR-21 in MCF-7 and pre-miR-21 in MDA-MB-231 cells on the ATRA-dependent regulation of PLAT, IL1B, and ICAM-1 mRNAs and proteins. The cells were transfected with the indicated oligonucleotides, and the extracted RNA was subjected to RT-PCR. The results were normalized for the expression of the β-actin mRNA, are the means ± S.D. of triplicate cell cultures, and are representative of three independent experiments. **, significantly different (p < 0.01). Total protein extracts were prepared from cells treated as above and subjected to ELISA (PLAT and IL1B) Western blot or FACS (ICAM-1) analysis. PROT, protein.
To define whether any of the six genes considered is a direct miR-21 target, we cloned the 3′-UTR of the selected transcripts downstream of a luciferase reporter and evaluated the effect of miR-21 in 293T cells, which contain low levels of the miRNA (54) (Fig. 8B, left panel). miR-21 inhibited the expression of the ICAM-1, PLAT, and IL1B constructs, indicating that they are direct targets. Pre-miR-21 inhibition was reversed by deletion of the predicted “seed” sequence in the 3′-UTR (Fig. 8B, right panel), confirming that miR-21 targets these sites.
To verify that miR-21 induced by ATRA is relevant for the regulation of the three identified targets: PLAT, IL1B, and ICAM-1, we transfected MCF7 cells with anti-miR-21 (Fig. 8, C–E, left panels). At the mRNA level, transfection of anti-miR-21 attenuated ATRA-dependent down-regulation of PLAT (72 h). Furthermore, in cells treated with ATRA, challenge of MCF-7 cells with anti-miR-21 resulted in the appearance of detectable amounts of IL1B (48 and 72 h) and caused a switch of ICAM-1 down-regulation into up-regulation (72 h). At the protein level, treatment of cells with anti-miR-21 for 72 h increased the amounts of PLAT and total (Western blot) or membrane-associated (FACS) ICAM-1 proteins in an ATRA-dependent fashion, consistent with what was observed on the corresponding transcripts. The amounts of IL1B protein were always below the limit of detection in these cells, even in the presence of anti-miR-21.
The relevance of miR-21 induction for the control of IL1B and PLAT is supported by mirror results obtained in MDA-MB-231 cells exposed to pre-miR-21 and ATRA (Fig. 8, C–E, right panels). Forced expression of pre-miR-21 reduced ATRA-dependent induction of PLAT and IL1B mRNAs and proteins. By converse, the mRNA or protein expression pattern of ICAM-1 observed in ATRA-treated MDA-MB-231 cells was not affected by pre-miR-21 transfection. These last results suggest that direct regulation of ICAM-1 by miR-21 in this cell context is superseded by other regulatory circuits activated by the retinoid.
Relevance of Maspin, ICAM-1, and PLAT for MCF-7 Growth and/or Motility
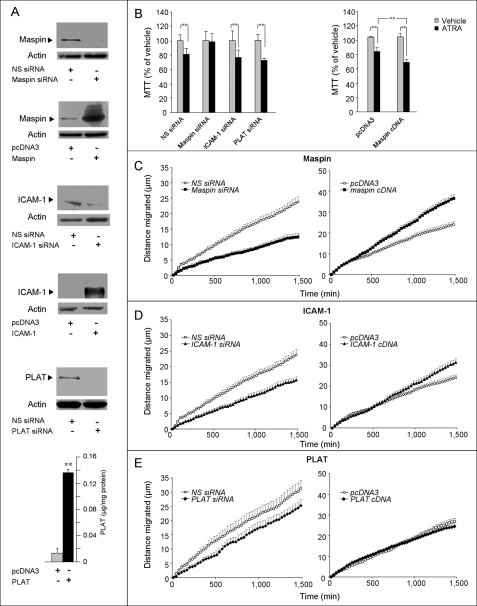
The roles of maspin and the two newly identified miR-21 targets, ICAM-1 and PLAT, in the growth and/or motility of MCF-7 cells were evaluated, after silencing and overexpression of the three genes by transient transfection of validated siRNAs and cDNAs (Fig. 9A). Silencing of ICAM-1 and PLAT did not affect the growth of MCF-7 cells in basal conditions or after treatment with ATRA (Fig. 9B, left panel). In contrast, down-regulation of maspin prevented growth inhibition by the retinoid. In line with a role for maspin in the process, overexpression of the protein enhanced retinoid-dependent MCF-7 cell growth inhibition (Fig. 9B, right panel). This is consistent with the onco-suppressive role of maspin (55) and indicates that down-regulation of the protein by ATRA may be part of the negative feedback loop set in motion by miR-21 induction on the retinoid-dependent anti-proliferative action (Fig. 5, A and B).
FIGURE 9.
Effect of maspin, ICAM-1, and PLAT silencing and/or overexpression on MCF-7 growth and motility in the presence and absence of ATRA. A, MCF-7 cells were transfected with the indicated siRNAs or cDNAs. Seventy-two hours after transfection, protein extracts were subjected to Western blot analysis. The same amounts of protein extracts (derived from a pool of three independent cultures) were separated on PAGE and detected by antibodies against maspin (40 kDa), ICAM-1 (90 kDa), and PLAT (68 kDa). The last panel illustrates the levels of PLAT measured with a specific ELISA assay. NS siRNA, irrelevant oligonucleotide provided by the same commercial source of the validated siRNAs that serves as a negative control in the experiments. B, left panel, MCF-7 cells were transfected with validated siRNAs targeting maspin, ICAM-1, and PLAT or the appropriate negative control NS siRNA. Cell growth was evaluated 48 h after the addition of vehicle (Me2SO) or ATRA (1 μm) by the MTT assay (mean ± S.D., six independent cultures). **, p < 0.01 (Student's t test). The results are representative of two independent experiments. Right panel, MCF-7 cells were transfected with maspin cDNA or the corresponding void vector (pcDNA3). Forty-eight hours later, cell growth was monitored by the MTT assay. **, significantly lower relative to all the other experimental groups (p < 0.01). C–E, MCF-7 cells were transfected with maspin (C), ICAM-1 (D), PLAT (E), or control siRNA (left panels) or the corresponding cDNAs (right panels) along with pEGFP-N1 (containing the enhanced GFP gene) in the presence of E2 (0.01 μm). The displacement of individually tracked GFP-positive cells (mean ± S.E., at least 15 GFP-positive cells/experimental point) was measured by time lapse microscopy, starting 48 h after transfection. The results are representative of two independent experiments. Starting from 500 min, all of the points of the curves corresponding to the ICAM-1 siRNA or maspin siRNA treatments are significantly lower than the corresponding NC siRNA points (Student's t test, p < 0.05). Starting from 500 and 1,000 min, respectively, all the points of the curves corresponding to the maspin and ICAM-1 cDNAs are significantly higher than the corresponding vector points (Student's t test, p < 0.05) in untreated MCF-7 cells. The results are representative of two independent experiments.
As with maspin, ICAM-1 and PLAT have been involved in cancer cell invasion (55–57); we investigated their significance for MCF-7 cell random motility (Fig. 9, C–E). Silencing of maspin and ICAM-1 with specific siRNAs resulted in a significant reduction (p < 0.01) of cell motility. In contrast, silencing of PLAT was not associated with a significant effect on this parameter. Mirror experiments conducted by transfection of cDNAs corresponding to the coding sequences of maspin, ICAM-1, and PLAT support these observations. In fact, ICAM-1 and maspin overexpression resulted in increased basal cell motility. Based on these data, we propose that at least part of the anti-motility effect of ATRA is mediated by down-regulation of ICAM-1 and maspin following retinoid-dependent induction of miR-21.
DISCUSSION
We demonstrated that treatment of retinoid-sensitive ERα+ breast carcinoma cells with ATRA resulted in the induction of miR-21. Up-regulation of miR-21 was the consequence of increased transcription of the corresponding gene via selective activation of RARα. This was due to a direct effect of the ligand-activated receptor on two functional RAREs, mapping to the 5′-flanking region of MIR21.
Induction of the oncogenic miR-21 by an anti-proliferative agent like ATRA in ERα+ breast carcinoma cells was unexpected. For this reason, we deemed it important to establish whether miR-21 induction was involved in some of the cellular responses potentially underlying the therapeutic activity of ATRA. In the MCF-7 context, miR-21 counteracted the anti-proliferative and pro-senescence effects of ATRA. This is consistent with the reported role of miR-21 on the growth and progression of breast carcinoma, suggesting that induction by ATRA is part of an uncharacterized negative feedback loop similar to the one activated by E2 in ERα+ breast cancer cells (6, 58) or interferon in colon cancer cells (59). On the other hand, the results of the studies on the motility of MCF-7 and T47D cells performed with anti-miR-21 imply a role for the miRNA in mediating inhibition of this process by ATRA. The anti-motility action of miR-21 in ATRA-treated ERα+ cells is apparently against the idea that the miRNA is pro-invasive for breast cancer cells, when three-dimensional in vitro assays are used (54). This discrepancy probably reflects the absence of localized stimuli guiding the direction of the cell movement in the motility assay used in this study. Another point to be considered stems from the results obtained after calculation of persistence (ratio of the direct distance from start point to end point divided by the total track distance) in ATRA-treated MCF-7 and T47D cells transfected with anti-miR-21 or anti-NC. In these conditions, silencing of miR-21 was associated with decreased persistence in both cell lines (MCF-7: anti-miR-21 = 0.17 ± 0.01, anti-NC = 0.24 ± 0.02; mean ± S.E., n = 30; and T47D: anti-miR-21 = 0.15 ± 0.01, anti-NC = 0.27 ± 0.02; mean ± S.E., n = 30). Altogether, our data suggest that miR-21 enhances instantaneous velocity. However, this phenomenon does not translate into efficient migration because of the observed decrease in persistence. Clearly the effect of miR-21 on motility is complex and further complicated by the concomitant cellular responses activated by ATRA.
Apparently, our results provide contrasting evidence regarding the role of miR-21 induction in terms of the overall anti-cancer activity of retinoids. In fact, induction of miR-21 seems to be detrimental for the growth inhibitory effect of ATRA. As such, strategies aimed at suppressing this effect should enhance the anti-proliferative action of the retinoid. On the other hand, the observations on cell motility suggest a potentially beneficial role of miR-21 induction for the anti-metastatic activity of retinoids. However, caution should be exercised in drawing any conclusion on this point. Indeed, it must be stressed that motility is clearly only one component of the complex process of cancer cell dissemination, which is modulated by many other factors that are not considered with the random motility assay employed in this study.
Among the few validated miR-21 targets, maspin is the only one modulated by ATRA via induction of the miRNA in MCF-7 cells. In this context, it is of particular relevance that expression of two established miR-21 target genes, PTEN and PDCD4, inhibiting growth and survival of cancer cells (37, 60) was left unaltered (PTEN) or was increased (PDCD4) in ATRA-treated MCF-7 cells. This indicates that the two proteins do not play a significant role in miR-21-dependent suppression of retinoid-triggered growth inhibition. In contrast, maspin, which is also endowed with onco-suppressor properties, is one of the miR-21 targets contributing to ATRA-dependent growth inhibition of MCF-7 cells, as suggested by our silencing and overexpression experiments. In line with down-regulation of maspin by miR-21, our data support a role for the protein in mediating ATRA- and miR-21-dependent inhibition of cell motility as well. The relevance of our observations for the reported role of certain forms of maspin in suppressing the metastatic potential of cancer cells (47, 61) remains to be established.
One of the most important outcomes of our study is the identification of three novel and functionally validated miR-21 targets: ICAM-1, PLAT, and IL1B. These three genes are regulated by miR-21 primarily at the mRNA level.
ICAM-1 is an adhesion molecule, and its down-regulation by miR-21 is not involved in cell growth. In contrast, silencing of ICAM-1 resulted in reduction of MCF-7 random cell motility, whereas overexpression of the protein exerted an opposite effect. These data indicate that inhibition of ICAM-1 by ATRA-dependent induction of miR-21 mediates at least part of the anti-motility effects exerted by the miRNA in retinoid-treated cells, consistent with the described pro-motility action of the protein in breast carcinoma cells (62, 63).
PLAT codes for a fibrinolytic factor whose induction in cells lacking miR-21 up-regulation may be part of a stereotyped response to ATRA, which is anti-thrombotic (64). Inhibition of PLAT expression by miR-21, as observed in MCF-7 cells, does not seem to play a role in the anti-proliferative effects exerted by ATRA. Nevertheless, PLAT down-regulation may be therapeutically desirable, because stimulation of a thrombotic response in cancer cells is associated with tumor growth and progression in vivo (65). Although PLAT activates the motility of neural crest cells (66), involvement of the protein in the miR-21-dependent anti-motility effects observed in ATRA-treated ERα+ cells is once again ruled out by silencing and overexpression studies.
Finally, IL1B is an inflammatory protein, and our microarray data indicate that ATRA up-regulates inflammation-related genes in ERα− MDA-MB-231 cells but not in ERα+ MCF-7 cells. Based on the presence of several potential miR-21 targets in this group of genes, including IL1B, we propose that retinoid-dependent miR-21 induction inhibits, either directly or indirectly, certain aspects of the inflammatory responses otherwise activated by retinoids in breast cancer cells. Because inflammation is known to be involved in tumor growth in vivo (67), the proposed anti-inflammatory action of miR-21 may play a positive role in the overall therapeutic responses to retinoids.
In conclusion, this is the first demonstration that MIR21 is a direct retinoid target gene only in retinoid-sensitive ERα+ breast carcinoma cells. The mechanisms underlying cell context specificity are unknown, although simple anti-estrogenic effects are unlikely. The role of miR-21 and the three novel targets: ICAM-1, PLAT, and IL1B, in retinoid-induced anti-proliferative, pro-senescence, and anti-motility effects requires further study, because it has implications for the therapeutic use of these agents.
Supplementary Material
Acknowledgments
We thank Gianfranco Bazzoni and Andrew Bert for critical reading of the manuscript and for useful suggestions. We also thank Felice Deceglie and Alessandro Soave for the artwork.
This work was supported by the Associazione Italiana Ricerca contro il Cancro, Ministero della Università e della Ricerca, Fondazione “Italo Monzino,” and Fondazione “Negri-Weizmann.”

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3.
E. Garattini, unpublished observations.
- ATRA
- all-trans-retinoic acid
- ERα
- estrogen receptor type α
- miRNA
- microRNA
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ICAM-1
- intercellular adhesion molecule 1.
REFERENCES
- 1. Garattini E., Gianni M., Terao M. (2007) Curr. Pharm. Des. 13, 1375–1400 [DOI] [PubMed] [Google Scholar]
- 2. Garattini E., Gianni M., Terao M. (2004) Curr. Pharm. Des. 10, 433–448 [DOI] [PubMed] [Google Scholar]
- 3. Uray I. P., Brown P. H. (2006) Expert Opin. Investig. Drugs 15, 1583–1600 [DOI] [PubMed] [Google Scholar]
- 4. Brodersen P., Voinnet O. (2009) Nat. Rev. Mol. Cell Biol. 10, 141–148 [DOI] [PubMed] [Google Scholar]
- 5. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 6. Bhat-Nakshatri P., Wang G., Collins N. R., Thomson M. J., Geistlinger T. R., Carroll J. S., Brown M., Hammond S., Srour E. F., Liu Y., Nakshatri H. (2009) Nucleic Acids Res. 37, 4850–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. (2005) Cell 123, 819–831 [DOI] [PubMed] [Google Scholar]
- 8. Mandal S., Davie J. R. (2007) BMC Cancer 7, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jönsson G., Staaf J., Olsson E., Heidenblad M., Vallon-Christersson J., Osoegawa K., de Jong P., Oredsson S., Ringnér M., Höglund M., Borg A. (2007) Genes Chromosomes Cancer 46, 543–558 [DOI] [PubMed] [Google Scholar]
- 10. Patel J. B., Mehta J., Belosay A., Sabnis G., Khandelwal A., Brodie A. M., Soprano D. R., Njar V. C. (2007) Br. J. Cancer 96, 1204–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. del Rincón S. V., Guo Q., Morelli C., Shiu H. Y., Surmacz E., Miller W. H. (2004) Oncogene 23, 9269–9279 [DOI] [PubMed] [Google Scholar]
- 12. Kogai T., Schultz J. J., Johnson L. S., Huang M., Brent G. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8519–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Dokmanovic M., Stein W. D., Ardecky R. J., Roninson I. B. (2006) Cancer Res. 66, 8749–8761 [DOI] [PubMed] [Google Scholar]
- 14. Fu H., Yang G., Lu F., Wang R., Yao L., Lu Z. (2006) Biochem. Biophys. Res. Commun. 343, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 15. Lu M., Mira-y-Lopez R., Nakajo S., Nakaya K., Jing Y. (2005) Oncogene 24, 4362–4369 [DOI] [PubMed] [Google Scholar]
- 16. Zhu W. Y., Jones C. S., Amin S., Matsukuma K., Haque M., Vuligonda V., Chandraratna R. A., De Luca L. M. (1999) Cancer Res. 59, 85–90 [PubMed] [Google Scholar]
- 17. Gianní M., Li Calzi M., Terao M., Guiso G., Caccia S., Barbui T., Rambaldi A., Garattini E. (1996) Blood 87, 1520–1531 [PubMed] [Google Scholar]
- 18. Parrella E., Giannì M., Fratelli M., Barzago M. M., Raska I., Jr., Diomede L., Kurosaki M., Pisano C., Carminati P., Merlini L., Dallavalle S., Tavecchio M., Rochette-Egly C., Terao M., Garattini E. (2006) Mol. Pharmacol. 70, 909–924 [DOI] [PubMed] [Google Scholar]
- 19. Hirano M., Hashimoto S., Yonemura S., Sabe H., Aizawa S. (2008) J. Cell Biol. 182, 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powell J. A., Thomas D., Barry E. F., Kok C. H., McClure B. J., Tsykin A., To L. B., Brown A., Lewis I. D., Herbert K., Goodall G. J., Speed T. P., Asou N., Jacob B., Osato M., Haylock D. N., Nilsson S. K., D'Andrea R. J., Lopez A. F., Guthridge M. A. (2009) Blood 114, 4859–4870 [DOI] [PubMed] [Google Scholar]
- 21. Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article3 [DOI] [PubMed] [Google Scholar]
- 22. Terao M., Kurosaki M., Barzago M. M., Fratelli M., Bagnati R., Bastone A., Giudice C., Scanziani E., Mancuso A., Tiveron C., Garattini E. (2009) Mol. Cell Biol. 29, 357–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plumb J. A. (2004) Methods Mol. Med. 88, 165–169 [DOI] [PubMed] [Google Scholar]
- 24. Gianni' M., Boldetti A., Guarnaccia V., Rambaldi A., Parrella E., Raska I., Jr., Rochette-Egly C., Del Sal G., Rustighi A., Terao M., Garattini E. (2009) Cancer Res. 69, 1016–1026 [DOI] [PubMed] [Google Scholar]
- 25. Ciana P., Di Luccio G., Belcredito S., Pollio G., Vegeto E., Tatangelo L., Tiveron C., Maggi A. (2001) Mol. Endocrinol. 15, 1104–1113 [DOI] [PubMed] [Google Scholar]
- 26. Blenkiron C., Goldstein L. D., Thorne N. P., Spiteri I., Chin S. F., Dunning M. J., Barbosa-Morais N. L., Teschendorff A. E., Green A. R., Ellis I. O., Tavaré S., Caldas C., Miska E. A. (2007) Genome Biol. 8, R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., Dirbas F. M., Somlo G., Pera R. A., Lao K., Clarke M. F. (2009) Cell 138, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ribas J., Lupold S. E. (2010) Cell Cycle 9, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujita S., Ito T., Mizutani T., Minoguchi S., Yamamichi N., Sakurai K., Iba H. (2008) J. Mol. Biol. 378, 492–504 [DOI] [PubMed] [Google Scholar]
- 30. Rishi A. K., Shao Z. M., Baumann R. G., Li X. S., Sheikh M. S., Kimura S., Bashirelahi N., Fontana J. A. (1995) Cancer Res. 55, 4999–5006 [PubMed] [Google Scholar]
- 31. Mologni L., Ponzanelli I., Bresciani F., Sardiello G., Bergamaschi D., Gianní M., Reichert U., Rambaldi A., Terao M., Garattini E. (1999) Blood 93, 1045–1061 [PubMed] [Google Scholar]
- 32. Hua S., Kittler R., White K. P. (2009) Cell 137, 1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ross-Innes C. S., Stark R., Holmes K. A., Schmidt D., Spyrou C., Russell R., Massie C. E., Vowler S. L., Eldridge M., Carroll J. S. (2010) Genes Dev. 24, 171–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elgort M. G., Zou A., Marschke K. B., Allegretto E. A. (1996) Mol. Endocrinol. 10, 477–487 [DOI] [PubMed] [Google Scholar]
- 35. Mattie M. D., Benz C. C., Bowers J., Sensinger K., Wong L., Scott G. K., Fedele V., Ginzinger D., Getts R., Haqq C. (2006) Mol. Cancer 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wickramasinghe N. S., Manavalan T. T., Dougherty S. M., Riggs K. A., Li Y., Klinge C. M. (2009) Nucleic Acids Res. 37, 2584–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008) J. Biol. Chem. 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 38. Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007) Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 39. Zhu S., Si M. L., Wu H., Mo Y. Y. (2007) J. Biol. Chem. 282, 14328–14336 [DOI] [PubMed] [Google Scholar]
- 40. Bourguignon L. Y., Spevak C. C., Wong G., Xia W., Gilad E. (2009) J. Biol. Chem. 284, 26533–26546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Y., Chaerkady R., Beer M. A., Mendell J. T., Pandey A. (2009) Proteomics 9, 1374–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu C. H., van Riggelen J., Yetil A., Fan A. C., Bachireddy P., Felsher D. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13028–13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li T., Li D., Sha J., Sun P., Huang Y. (2009) Biochem. Biophys. Res. Commun. 383, 280–285 [DOI] [PubMed] [Google Scholar]
- 44. Nwankwo J. O. (2002) Anticancer Res. 22, 4129–4135 [PubMed] [Google Scholar]
- 45. Gozgit J. M., Pentecost B. T., Marconi S. A., Otis C. N., Wu C., Arcaro K. F. (2006) Mol. Cancer Res. 4, 905–913 [DOI] [PubMed] [Google Scholar]
- 46. Krichevsky A. M., Gabriely G. (2009) J. Cell Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Latha K., Zhang W., Cella N., Shi H. Y., Zhang M. (2005) Mol. Cell Biol. 25, 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zou Z., Anisowicz A., Hendrix M. J., Thor A., Neveu M., Sheng S., Rafidi K., Seftor E., Sager R. (1994) Science 263, 526–529 [DOI] [PubMed] [Google Scholar]
- 49. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007) Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Papagiannakopoulos T., Shapiro A., Kosik K. S. (2008) Cancer Res. 68, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 51. Sayed D., Rane S., Lypowy J., He M., Chen I. Y., Vashistha H., Yan L., Malhotra A., Vatner D., Abdellatif M. (2008) Mol. Biol. Cell 19, 3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oh J., Takahashi R., Kondo S., Mizoguchi A., Adachi E., Sasahara R. M., Nishimura S., Imamura Y., Kitayama H., Alexander D. B., Ide C., Horan T. P., Arakawa T., Yoshida H., Nishikawa S., Itoh Y., Seiki M., Itohara S., Takahashi C., Noda M. (2001) Cell 107, 789–800 [DOI] [PubMed] [Google Scholar]
- 53. Domann F. E., Rice J. C., Hendrix M. J., Futscher B. W. (2000) Int. J. Cancer 85, 805–810 [DOI] [PubMed] [Google Scholar]
- 54. Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y. Y. (2008) Cell Res. 18, 350–359 [DOI] [PubMed] [Google Scholar]
- 55. Oliveira A. M., Ross J. S., Fletcher J. A. (2005) Am. J. Clin. Pathol. 124, (Suppl.) S16–S28 [DOI] [PubMed] [Google Scholar]
- 56. Qin L. X., Tang Z. Y. (2004) J. Cancer Res. Clin. Oncol. 130, 497–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. DeClerck Y. A., Imren S., Montgomery A. M., Mueller B. M., Reisfeld R. A., Laug W. E. (1997) Adv. Exp. Med. Biol. 425, 89–97 [DOI] [PubMed] [Google Scholar]
- 58. Castellano L., Giamas G., Jacob J., Coombes R. C., Lucchesi W., Thiruchelvam P., Barton G., Jiao L. R., Wait R., Waxman J., Hannon G. J., Stebbing J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15732–15737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang C. H., Yue J., Fan M., Pfeffer L. M. (2010) Cancer Res. 70, 8108–8116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Knobbe C. B., Lapin V., Suzuki A., Mak T. W. (2008) Oncogene 27, 5398–5415 [DOI] [PubMed] [Google Scholar]
- 61. Joensuu K. M., Leidenius M. H., Andersson L. C., Heikkila P. S. (2009) Hum. Pathol. 40, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 62. Rosette C., Roth R. B., Oeth P., Braun A., Kammerer S., Ekblom J., Denissenko M. F. (2005) Carcinogenesis 26, 943–950 [DOI] [PubMed] [Google Scholar]
- 63. Strell C., Lang K., Niggemann B., Zaenker K. S., Entschladen F. (2010) Exp. Cell Res. 316, 138–148 [DOI] [PubMed] [Google Scholar]
- 64. Marchetti M., Vignoli A., Bani M. R., Balducci D., Barbui T., Falanga A. (2003) Haematologica 88, 895–905 [PubMed] [Google Scholar]
- 65. Rickles F. R. (2009) Thromb. Res. 123, (Suppl. 2) S16–S20 [DOI] [PubMed] [Google Scholar]
- 66. Erickson C. A., Isseroff R. R. (1989) J. Exp. Zool. 251, 123–133 [DOI] [PubMed] [Google Scholar]
- 67. Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.