Abstract
Delineating ancestral gene relations among a large set of sequenced eukaryotic genomes allowed us to rigorously examine links between evolutionary and functional traits. We classified 86% of over 1.36 million protein-coding genes from 40 vertebrates, 23 arthropods, and 32 fungi into orthologous groups and linked over 90% of them to Gene Ontology or InterPro annotations. Quantifying properties of ortholog phyletic retention, copy-number variation, and sequence conservation, we examined correlations with gene essentiality and functional traits. More than half of vertebrate, arthropod, and fungal orthologs are universally present across each lineage. These universal orthologs are preferentially distributed in groups with almost all single-copy or all multicopy genes, and sequence evolution of the predominantly single-copy orthologous groups is markedly more constrained. Essential genes from representative model organisms, Mus musculus, Drosophila melanogaster, and Saccharomyces cerevisiae, are significantly enriched in universal orthologs within each lineage, and essential-gene-containing groups consistently exhibit greater sequence conservation than those without. This study of eukaryotic gene repertoire evolution identifies shared fundamental principles and highlights lineage-specific features, it also confirms that essential genes are highly retained and conclusively supports the “knockout-rate prediction” of stronger constraints on essential gene sequence evolution. However, the distinction between sequence conservation of single- versus multicopy orthologs is quantitatively more prominent than between orthologous groups with and without essential genes. The previously underappreciated difference in the tolerance of gene duplications and contrasting evolutionary modes of “single-copy control” versus “multicopy license” may reflect a major evolutionary mechanism that allows extended exploration of gene sequence space.
Keywords: orthologs, essential genes, molecular evolution, vertebrates, arthropods, fungi
Introduction
Proteins constitute the major cellular machinery and are inherited as genes encoded in genomic DNA, where the continual evolutionary processes of gene duplications, losses, and sequence mutations alter their repertoire, abundance, and sequence identity. Taking advantage of the availability of the genetic blueprints of numerous eukaryotic species, we set out to explore the trends of protein-coding gene repertoire evolution across the most sampled lineages in a large-scale and consistent manner. With a total of 95 selected species spanning several hundreds of millions of years of evolution (Hedges and Kumar 2009), the vertebrate, arthropod, and fungal lineages offer unprecedented opportunities to comprehensively catalog gene genealogies and relate these to the increasingly detailed characterizations of eukaryotic gene function.
Comparative sequence analysis allows the identification of ancestral relations among genes, that is, homology. With reference to a specific species radiation, homologous relations define orthologs, that is, genes that arose by vertical decent from a single gene of the last common ancestor (Fitch 1970; Koonin 2005). Gene duplications in descendent lineages, referred to as inparalogs, are thus also co-orthologs and comprise an orthologous group descended from a gene of the last common ancestor. Delineation of orthologs across a given lineage therefore defines the core set of genes of the last common ancestor. The conservation of protein sequence identities among orthologous group members is indicative of the strength of selection inferred by the rate of gene sequence divergence. Selection intensity on gene retention may be deduced from phyletic distributions of member genes; their presence or absence in the considered species. Constraints on gene duplicability may be manifested in gene copy-number variations in independently evolving lineages. Detailing ancestral gene relations therefore enables quantification of properties of ortholog sequence divergence, phyletic retention, and copy-number variation. Although orthologous relations are not defined by gene function, identifying “equivalent” genes in modern species nevertheless provides a working hypothesis of similar functionality, especially for single-copy orthologs. As such, confirmed and putative functional annotations of orthologous group members—including gene ontologies, protein domains, and gene essentiality—define putative biological features characterizing the group as a whole. The question can therefore be formulated to link such functional characteristics with quantifiable evolutionary properties of orthologs in sequenced genomes.
Essential genes are operationally defined in molecular genetics by gene knockouts that result in (conditional) lethality or infertility and are thus described as strongly contributing to organismal fitness. A naïve expectation for such indispensable genes in a given species would be the indispensability of their equivalents in other species. Under this assumption, studies in bacteria identified broader phyletic distributions of essential genes (Jordan et al. 2002; Gerdes et al. 2003), and indeed phyletic retention levels proved to be the most predictive feature of essentiality of bacterial and yeast genes (Gustafson et al. 2006). Similarly, gene silencing by RNA-interference (RNAi) in nematodes identified a greater proportion of mutants among targeted genes with orthologs in other eukaryotes (Castillo-Davis and Hartl 2003). Furthermore, quantifying propensity for gene loss among clusters of orthologous groups in seven distantly related eukaryotes revealed enrichment of yeast essential genes among clusters with no losses (Krylov et al. 2003).
A second expectation, as formulated by Wilson et al. (1977) and known as the “knockout-rate prediction,” anticipates more stringent constraints on the sequence evolution of essential genes. Although this gained some support from studies in bacteria (Jordan et al. 2002), confounding factors such as covariation of evolutionary rates with levels of gene expression, as well as somewhat inconsistent observations from several studies in eukaryotes, have yielded inconclusive results. Substitution rates between mouse and rat orthologs suggested that essential genes were slower evolving, but no difference was observed after controlling for fast-evolving immunity genes (Hurst and Smith 1999), whereas greater sampling of mouse gene knockouts did identify an impact of gene essentiality on the rate of protein sequence evolution (Liao et al. 2006). In Saccharomyces cerevisiae, gene evolutionary rates were negatively correlated with adverse effects of knockouts on fitness in parallel growth assays but comparing those required for maximal growth with dispensable genes failed to identify any significant difference (Hirsh and Fraser 2001). Saccharomyces cerevisiae–Candida albicans comparisons found that evolutionary rates did correlate with dispensability, however, this was only true of duplicated genes (those with within-species homologs) but not singletons (unique genes, without within-species homologs) (Yang et al. 2003). For both duplicates and singletons, Caenorhabditis elegans RNAi data suggested that amino acid replacement levels were indeed lower among essential genes (Castillo-Davis and Hartl 2003). Ignoring duplications by selecting only one ortholog from each of four compared eukaryotic species to estimate evolutionary rates, these RNAi data provided evidence that sequence evolution of indispensable proteins is constrained by selection (Luz and Vingron 2006). Dispensability was also correlated with evolutionary rates from comparisons of S. cerevisiae with more closely related fungi, but rate differences between genes with lethal and nonlethal effects were most pronounced only when comparing closer relatives (Zhang and He 2005). Thus, unequivocal support for the knockout-rate prediction has remained elusive.
Gene essentiality indicates a critical contribution to organismal fitness, but it does not necessarily describe any specific biological roles. Detailed functional characterizations are thus required to further explore the evolutionary traits of genes linked to particular cellular processes, facilitated by state of the art functional annotations provided by the Gene Ontology (GO) (GO-Consortium 2010) and InterPro (Hunter et al. 2009) resources. The most rigorously curated and, thus, the most accurate and comprehensive gene functional annotations are represented by the GO’s founding model organisms: mouse (Mus musculus), fly (Drosophila melanogaster), and yeast (S. cerevisiae). Detailed mouse, fly, and yeast GO annotations therefore, respectively, facilitate inferred putative functionality of orthologs across the vertebrate, arthropod, and fungal lineages. Matches to InterPro signatures of protein domains, the majority of which are well annotated, provide further hints describing the likely biological roles of genes with recognizable sequence signatures. Together, GO and InterPro resources offer large-scale functional characterization describing confirmed and putative biological features for the majority of eukaryotic protein-coding genes.
A number of previous studies have investigated the connections between evolutionary traits and functional properties on the premise that gene functionality impacts on the strength of negative selection. Although several studies have focused on relating gene essentiality and rates of sequence evolution as summarized above, additional examined characteristics included gene expression levels, propensity for gene loss, gene compactness, or placements within protein interaction or gene regulatory networks, for example, gene sequence evolutionary rates were negatively correlated with expression level and positively correlated with propensity for gene loss (Krylov et al. 2003). Beyond correlating pairs of specific traits, composite variables derived from multivariate statistical approaches may reveal intercorrelations among traits and highlight emergent gene properties. Using these approaches, Wolf et al. (2006) interpreted the principal component among seven such characteristics as reflecting a gene’s “importance” or “status”, with strong positive contributions from expression level, number of paralogs, essentiality and protein interactions, large negative contributions from evolutionary rate, and propensity for gene loss (Wolf et al. 2006). Concomitant analysis of several gene characteristics applying multivariate approaches can therefore be useful to provide perspectives on the principal factors influencing the evolution of genes and gene repertoires.
In this study, applying a consistent methodology to the delineation of orthologous gene relations through comparative analysis of 40 vertebrates, 23 arthropods, and 32 fungi allowed us to identify the core sets of genes descended from the last common ancestor in each lineage. By quantifying evolutionary properties of gene retention, sequence divergence, and duplicability, we were able to examine correlations between traits and links with gene functional characteristics. We focused on testing specific hypotheses using intuitively interpretable pairwise relations among gene characteristics in three major eukaryotic lineages, supported by principal component analysis to examine the intercorrelations among these traits. With essential genes from representative model organisms, we aimed to explore the expectation of broad phyletic distributions of essential genes across dozens of eukaryotic species from three distinct lineages. Furthermore, utilizing consistent measures of sequence evolution, we set out to test the knockout-rate prediction across the fungal, arthropod, and vertebrate lineages. Employing functional attributes of gene essentiality with comprehensive gene ontologies and protein domain signatures, we explored relations between gene functionality and evolutionary traits reflecting strengths of selection on gene retention, sequence divergence, and duplicability.
Materials and Methods
Data Sources
Gene Sets
The complete predicted protein-coding gene sets of 95 eukaryotic species were retrieved from publically available genomic resources. These included 40 vertebrates from Ensembl (Release 55, July 2009), 23 arthropods from AphidBase, BeeBase, BeetleBase, FlyBase (FB), SilkDB, VectorBase and wFleaBase (current releases in July 2009), and 32 fungi from UniProt (Release 15.0, March 2009) (supplementary table 1, Supplementary Material online). Preprocessing of the gene sets selected the longest protein-coding transcript of any gene annotated with multiple transcripts resulting in a nonredundant set of 1,363,300 protein-coding genes for subsequent orthology delineation analysis.
Gene Annotations
Gene essentiality data for mouse (M. musculus), fly (D. melanogaster), and yeast (S. cerevisiae) were retrieved from the Database of Essential Genes (DEG 5.4) (Zhang and Lin 2009). Alternative gene essentiality data were retrieved by querying: 1) Mouse Genome Informatics (MGI) (Bult et al. 2010) resources for pre/peri/postnatal lethality phenotypes, 2) FB (Tweedie et al. 2009) for “phenotypic class: lethal”, and 3) Saccharomyces Genome Database (SGD) (Engel et al. 2010) for “systematic deletion phenotype: inviable.” InterPro and GO annotations describing putative gene functional attributes were retrieved from InterPro and UniProt (UniProt-Consortium 2010) resources, respectively. GO term parent–child relationships as well as InterPro to GO mappings were retrieved from GO.
Orthologous Group Classification
Orthology Delineation
The classification of protein-coding genes into orthologous groups was based on a clustering procedure of all-against-all Smith–Waterman protein sequence comparisons using PARALIGN (Saebø et al. 2005) as implemented in the OrthoDB methodology (Waterhouse et al. 2010). The clustering procedure starts with the identification of all best reciprocal hits with an e value cutoff of 1 × 10−6, followed by their triangulation with an e value cutoff of 1 × 10−3, requiring all member sequences to overlap; the clusters are further expanded to include more closely related within-species inparalogs. This procedure has been scrutinized as part of several genome projects (Richards et al. 2008; Elsik et al. 2009; Kirkness et al. 2010; Werren et al. 2010), and extensive manual examination of orthologous groups (Waterhouse et al. 2007; Wyder et al. 2007; Lemay et al. 2009; Matsui et al. 2009) has confirmed their biological relevance and acceptable accuracy. Because orthology is defined relative to the last common ancestor of the species being considered, thereby determining the hierarchical nature of orthologous classifications, the procedure built orthologous groups at each radiation along the three phylogenies of 40 vertebrates, 23 arthropods, and 32 fungi.
Orthology Type
Orthologous groups exhibit different phyletic distributions of their member genes, allowing them to be classified into different types according to their gene copy-numbers across each of the three phylogenies. Universal groups, separately defined for each lineage, were required to have gene members in more than 90% of the species (missing from no more than two arthropods or no more than three vertebrates or fungi), thereby accounting for possible artifacts from incomplete genome sequencing and/or annotation. The remaining orthologous groups were deemed nonuniversal. Universal orthologous groups were further partitioned into single-copy groups, with only one gene member in more than half of the species, and the remaining multicopy groups, with more than one gene in each of at least half of the species. This binary distinction between universal and nonuniversal orthologous groups as well as between single-copy and multicopy orthologous groups was chosen for simplicity and consistency over alternative (stricter and possibly more intuitive) categorizations presented in the Supplementary Material online that support the same conclusions (supplementary figs. 1–3, Supplementary Material online).
Average Percent Identity
The average amino acid percent identity among the members of each orthologous group provides a measure of the level of overall group protein sequence conservation. The average percent identity for each orthologous group was calculated as the mean of all between-species pairwise percent identities of member proteins. Excluding within-species pairwise identities effectively calculates a measure of group sequence conservation across all the member species that is independent of gene copy-number. For the universal orthologous groups defined in each lineage, which by definition all have broad phyletic distributions, this provides an absolute measure of group sequence conservation.
Related Groups
The all-against-all Smith–Waterman protein sequence comparisons also enabled identification of homologous relations among orthologous groups. Comparing two orthologous groups, the average alignment score of all between-group gene comparisons would provide a basis for calculating a corresponding e value, indicative of the number of matches with a score at least as good that would be expected to occur by chance. The number of orthologous groups in the database as well as the scoring system used (substitution matrix and gap penalties) would have to be taken into account to calculate such an e value. Because empirical calculations of averaged scores and the corresponding log-scaled averaged e values from the all-against-all gene comparisons show a correlation that matches almost exactly to the lambda value of the scoring system used by PARALIGN (supplementary fig. 4, Supplementary Material online), averaged between-group gene-to-gene e values were used as an approximation of the e values describing homologous relations among orthologous groups, considering homology support at e value cutoffs of 1 × 10−3 and 1 × 10−10.
Essential Orthologous Groups
Essential genes from model organisms were mapped to orthologous groups in vertebrates, arthropods, and fungi. Mapping essential genes from the DEG resource to orthologous groups in each lineage identified 1,673 mouse orthologous groups (2,054 mouse genes), 324 fly orthologous groups (334 fly genes), and 1,074 yeast orthologous groups (1,110 yeast genes). Employing alternative species-specific resources identified 1,560 mouse orthologous groups (1,954 MGI genes), 2,066 fly orthologous groups (2,234 FB genes), and 1,074 yeast orthologous groups (1,109 SGD genes). Analyses of DEG-defined essential genes in mouse and yeast and FB-defined fly genes are presented in the main text, whereas alternative sets supporting the same conclusions are described in the Supplementary Material online.
Results and Discussion
Core Gene Sets of Vertebrates, Arthropods, and Fungi
The Majority of Genes Exhibit Traceable Orthology and Functional Annotations
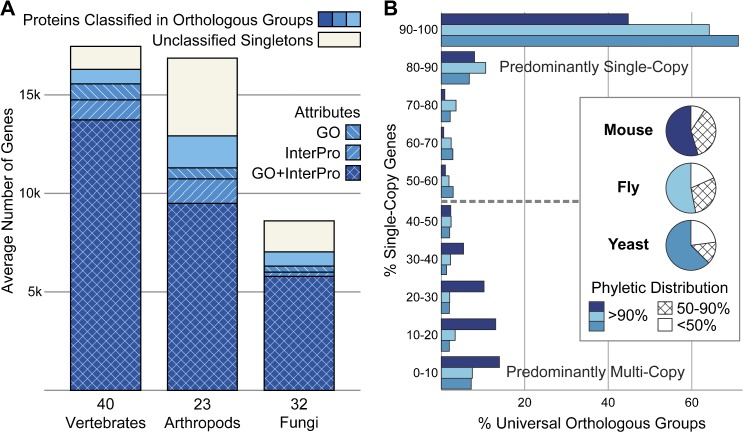
Our orthology delineation procedure established ancestral gene relations among 95 eukaryotic species at each level of the three major lineages. Of the total of 1,363,300 protein-coding genes, 86% were classified into 16,031, 18,937, and 13,535 orthologous groups at the levels of the last common ancestor of vertebrates, arthropods, and fungi, respectively (fig. 1A). Assuming that genes descended from a common ancestor are likely to share general functionality enabled tentative extrapolation of functional attributes ascribed to one or more members to the group as a whole. Accordingly, orthologous group descriptions were summarized from associated InterPro and GO annotations of individual member genes such that 92% of the almost 1.18 million orthologous group member genes were classified in orthologous groups described by either InterPro domains or GO terms and 81% by both attributes. The larger classified proportion of vertebrate genes (93.4%) compared with arthropods (76.5%) and fungi (81.6%) likely reflects the higher levels of evolutionary divergence among the species sampled along the arthropod and fungal phylogenies, which may limit the detection of distant homology. Estimated divergence times of major eukaryotic lineages (Hedges and Kumar 2009) suggest that compared with about 450 My of vertebrate evolution, the arthropods have diverged over some 700 My and the fungi probably span at least a billion years. In addition, rates of vertebrate protein sequence evolution are significantly slower compared with arthropods (Wyder et al. 2007) and fungi (Dujon 2006). The smaller proportion of classified genes in arthropods may also be influenced by the more variable annotation approaches and resulting total gene counts (supplementary fig. 5 and table 1, Supplementary Material online).
FIG. 1.—
Orthology classification across 40 vertebrates, 23 arthropods, and 32 fungi. (A) The average numbers of classified and annotated proteins per proteome are shown for each lineage. Eighty-six percent of a total of 1.36 million genes were classified into orthologous groups, and 92% of classified genes were assigned to orthologous groups that can be described by either GO or InterPro attributes or both. (B) Copy-number distributions of universal orthologous groups in mouse (Mus musculus), fly (Drosophila melanogaster), and yeast (Saccharomyces cerevisiae). More than half of orthologous groups are universal in each lineage and are either almost all single copy or all multicopy. Orthologous groups with members in more than 90% of the vertebrate, arthropod, or fungal species define the sets of universal orthologous groups in each lineage and constitute more than half of mouse, fly, and yeast orthologous groups (inset pie charts). The proportion of predominantly multicopy universal orthologous groups is notably larger in mouse compared with fly or yeast.
The comprehensive catalog of hierarchical orthologous groups of protein-coding genes in these three eukaryotic lineages with associated GO and InterPro attributes is freely accessible through the OrthoDB resource, http://cegg.unige.ch/orthodb (Waterhouse et al. 2010).
More Than Half of Orthologous Groups Are Universal across Each Lineage and Are Almost All Single Copy or All Multicopy
Defining universal orthologous groups within each lineage as having gene members in more than 90% of the sampled species classified over half of mouse, fly, or yeast groups as universal across the vertebrate, arthropod, or fungal lineages, respectively (fig. 1B). These orthologs that exhibit broad phyletic distributions are likely to be under stringent selection for gene retention. Nevertheless, even seemingly indispensable universal single-copy orthologs can be lost (Wyder et al. 2007) or missed by whole-genome sequencing or annotation. Partitioning these fractions of universal orthologous groups according to their member gene copy-numbers revealed that the majority of groups are either almost all single copy or all multicopy (fig. 1B). This distribution suggests that strong gene dosage constraints likely preserve the single-copy status of the majority of universal orthologs, especially in arthropods and fungi, allowing only some exceptional cases of “copy-number runaways.” At the other end of the spectrum, relaxation of copy-number restrictions appears to lead to multiple independent duplications in the majority of the descendant lineages, most prominently among vertebrates, effectively issuing a “multicopy license” across the entire lineage.
This trend of “single-copy control” versus multicopy license, observed in each of the three lineages, suggests two major modes through which gene function influences eukaryotic gene repertoire evolution. Investigations correlating gene evolutionary and functional traits should therefore consider appropriate controls to account for this dichotomy of gene duplicability.
Sequence Conservation of Universal Orthologous Groups Is Markedly Higher among Single-Copy Than Multicopy Orthologs
Single- and multicopy orthologous groups considered universal across the vertebrate, arthropod, or fungal lineages originate from the last common ancestor of each considered species phylogeny. Despite being of the same ages, single-copy groups exhibit significantly higher average protein sequence identities compared with multicopy groups (fig. 2, supplementary fig. 1, Supplementary Material online). Although the vertebrate lineage exhibits many more accumulated duplicates than in arthropods or fungi (fig. 1B), the striking contrast between the sequence conservation of single-copy genes versus the divergence of multicopy genes is consistently observed in each of the three lineages.
FIG. 2.—
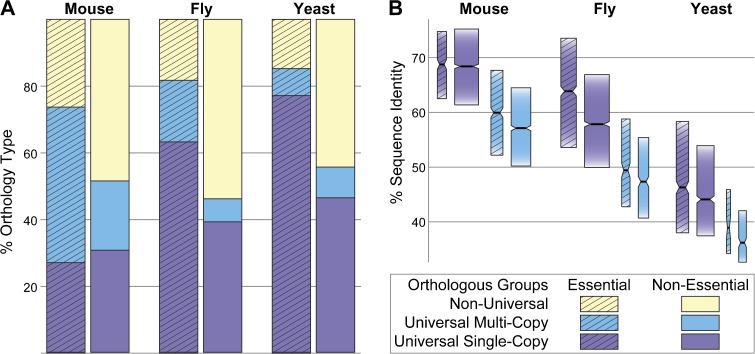
Phyletic profiles and sequence conservation levels of essential compared with nonessential genes in mouse (Mus musculus), fly (Drosophila melanogaster), or yeast (Saccharomyces cerevisiae). Orthologous groups with essential genes are enriched in universal orthologs and show constrained sequence evolution. (A) The majority of mouse (74%), fly (82%), and yeast (85%) essential gene-containing orthologous groups are universal—they belong to orthologous groups with members in more than 90% of the vertebrate, arthropod, or fungal species, whereas only about 50% of the remaining, nonessential, orthologous groups are universal. Orthologous groups with essential genes are therefore significantly more likely to be universal than nonessential groups (Fisher’s Exact Tests P < 1 × 10−65). Distinguishing between predominantly single-copy and predominantly multicopy universal orthologous groups reveals that most universal essential groups are single copy in fly and yeast. (B) Among both single- and multicopy universal orthologous groups, those with essential genes display greater sequence conservation, measured as the mean of interspecies protein sequence identities among orthologous group members, than those without. Notched boxes show medians of orthologous group percent identities with the limits of the upper and lower quartiles, and box widths are proportional to the number of orthologous groups in each category. For significance tests, see supplementary table 2 (Supplementary Material online).
This is indicative of a substantially greater impact on organismal fitness of mutations in genes evolving under single-copy control. Mutations may be better buffered in the case of multicopy genes, which may be maintained after duplication principally due to gene dosage constraints rather than because of functional innovations (Aury et al. 2006). Nevertheless, the subset of eukaryotic genes with a multicopy license may be allowed to explore protein sequence space, which may in turn facilitate functional divergence through fine-tuning or novelties under models of subfunctionalization or neofunctionalization (Lynch and Conery 2000; Hahn 2009).
The Ancestral Repertoire of Distinct Protein-Coding Sequences Has Grown from Fungi to Arthropods to Vertebrates
Detectable homology between orthologous groups (see Materials and Methods) is indicative of the common sequence ancestry of their full-length member genes (ancient paralogs) or of individual sequence regions (protein domains). At an e value cutoff of 1 × 10−3, about half of universal orthologous groups in mouse (41.2%), fly (59.5%), and yeast (43.9%) are homologous to at least one other universal orthologous group of their respective lineages (supplementary fig. 6, Supplementary Material online). Counting sets of these homologous universal groups and summing them with groups without homologs provides a lower estimate of the total numbers of distinct protein-coding sequences in the genomes of the last common ancestors of vertebrates, arthropods, or fungi. At an e value cutoff of 1 × 10−3, 4,869 unique gene sequences are estimated for the vertebrate ancestor, which is more than 1.5 times that of arthropods (3,145) and more than double that of fungi (2,212).
These consistent insights into the ancestral gene contents of three major eukaryotic lineages support the ideas of an expanding gene universe and perceived organismal complexity.
Evolutionary Traits of Essential Genes
The delineation of orthologous groups across the phylogenies of vertebrate, arthropod, and fungal species with sequenced genomes describes the core gene sets of three different eukaryotic lineages. This provided the context from which to examine the evolutionary characteristics of the subset of experimentally defined essential genes through concomitant analysis of gene dispensability with evolutionary traits of orthologous group phyletic distributions and sequence diversities. Employing viability data from model organisms (see Materials and Methods) identified 14.2% of mouse, 19.6% of fly, and 23.7% of yeast orthologous groups that contained an essential gene (essential groups). The mapping of these essential genes to orthologous groups within each of the vertebrate, arthropod, and fungal lineages allowed us to explore the broader evolutionary context beyond a single model organism species, and thus derive predictive associations of essentiality.
Essential Genes Are Enriched in Universal Orthologs
Comparing proportions of universal and essential groups in vertebrates, arthropods, and fungi revealed that essential groups are significantly more likely to be universal than nonessential groups (Fisher’s Exact Tests P < 1 × 10−65; fig. 2A, supplementary fig. 2 and table 2, Supplementary Material online). The preferentially universal phyletic distributions of essential gene-containing groups in each of the examined eukaryotic lineages are consistent with observed broad phyletic distributions of essential genes in bacteria, yeasts, and nematodes (Jordan et al. 2002; Castillo-Davis and Hartl 2003; Gerdes et al. 2003; Krylov et al. 2003; Gustafson et al. 2006). This correlation is also supported by principal component analysis (supplementary fig. 7 and table 3, Supplementary Material online), which, in agreement with observed opposing contributions of essentiality and propensity for loss to a gene’s importance (Wolf et al. 2006), highlighted major coordinated contributions of gene essentiality and phyletic retention levels. Furthermore, comparing the three lineages reveals that universal vertebrate, arthropod, or fungal groups make up a similar majority of essential groups in mouse (74%), fly (82%), and yeast (85%), respectively, which suggests that at least about three quarters of experimentally identified essential genes in individual organisms are likely to have orthologs within each lineage. Nevertheless, a minor fraction of essential groups exhibit nonuniversal phyletic distributions, suggesting that these essential genes may be crucial to biological processes specific to the selected model organisms and their closer relatives.
Our large-scale study, therefore, ultimately confirms the propensity of essential genes to belong to universal orthologous groups in fungi and confidently extends the same trend to the arthropod and vertebrate lineages.
The Majority of Universal Essential Groups Are Single Copy in Yeast and Fly
Distinguishing between single- and multicopy universal orthologous groups reveals marked differences among the three lineages with respect to essentiality. Yeast essential groups are enriched in universal single-copy but not multicopy groups, in fly they are mostly single copy but there is also a greater proportion of multicopy groups, whereas in mouse, only multicopy groups are enriched (Fisher’s Exact Tests P < 1 × 10−50; fig. 2A, supplementary fig. 2 and table 2, Supplementary Material online). This is reflected in the total proportions in each lineage, where mouse exhibits a much larger proportion of universal multicopy groups (44.9%) compared with fly (17.4%) and yeast (14.4%) (fig. 1B). Principal component analysis also suggests that the relaxation of copy-number constraints is the most distinguishing feature of the vertebrates (supplementary fig. 7 and table 3, Supplementary Material online). Thus, even among highly constrained essential genes, maintained duplications appear prevalent along the vertebrate lineage, whereas the majority of arthropod and fungal universal groups are single copy.
This prompts speculation that the negative effects of gene expression level imbalances are diminished in vertebrates, thereby relaxing single-copy controls and allowing multicopy gene buffering of mutations that lead to extended exploration of protein sequence space. Such supposedly redundant gene copies may provide a buffer against gene inactivation, however, compensation of essential functions by paralogs should not be assumed, as duplicates in mouse do not necessarily confer functional redundancy (Liao and Zhang 2007). In fact, retention of gene duplicates may be facilitated by coordinated reduction of expression levels, rendering both copies necessary to fulfill the biological role of the ancestral gene whereas creating redundancy at the level of their molecular function (Qian et al. 2010). However, these differences in gene duplicability may also reflect relative strengths of purifying selection in the three lineages imposed by factors such as effective population sizes. The greater permissiveness of slightly deleterious events in vertebrates would allow the accumulation of redundant gene copies that would be purged under more intensive selection pressures (Lynch and Conery 2000).
Sequence Evolution of Essential Genes Is More Constrained
Among both single- and multicopy universal orthologous groups, those with essential genes exhibit greater sequence conservation than those without (fig. 2B, supplementary fig. 2 and table 2, Supplementary Material online). This difference is clear when orthologous groups are partitioned into universal single- versus multicopy groups to control for the effects of age and copy-number constraints on sequence evolution. Universal groups are made up of descendants of ancestral genes retained across each lineage and thus represent orthologous groups of a common age at least as old as the last common ancestor. The remaining nonuniversal groups may be either ancient groups that have experienced multiple gene losses, or younger clade-specific groups, which display variable ranges of sequence identities (supplementary fig. 3, Supplementary Material online). Subsequent partitioning of the age-controlled universal orthologous groups into single- and multicopy groups (as shown in fig. 1B) then distinguishes between the effects of copy-number constraints and the effects of essentiality on sequence evolution.
The lower level of amino acid substitutions among essential groups is indicative of stronger purifying selection throughout the evolution of vertebrates, arthropods, and fungi and supports the knockout-rate prediction of slower sequence evolution of essential genes. However, this quantitative distinction between genes with known essential functions and those without is substantially less prominent than the distinction between single-copy constrained genes and those with a multicopy license. Thus, through a consistent, whole-genome scale analysis, we show higher constraints on the sequence evolution of essential genes across vertebrates, arthropods, and fungi.
Essential Groups in Animals Are More Likely to Have Homologs
If essential gene duplications resulted in severely imbalanced functionality of critical biological processes then essential groups might be expected to be limited to appearing only once in each lineage as unique orthologous groups. Instead, essential groups appear more likely to have relatives, suggesting that essential gene ancestors have more frequently given rise to novel related orthologous groups in vertebrates and arthropods (table 1, supplementary table 4, Supplementary Material online). Alternatively, ancient genes with frequently maintained duplicates have gained essential functions. Although 54% of universal mouse essential groups have universal group relatives, only 38% of universal nonessential groups are related to other universal orthologous groups at the 1 × 10−3 cutoff. The difference is similar for fly groups, where 67% of universal essential versus 56% of universal nonessential groups have relatives. However, in yeast, the small 3% difference is not statistically significant (46% essential vs. 43% nonessential).
Table 1.
Essential and Nonessential Orthologous Groups with and without Related Groups in Mouse (Mus musculus), Fly (Drosophila melanogaster), or Yeast (Saccharomyces cerevisiae)
| Mouse |
Fly |
Yeast |
||||||||
| Cutoff | ES | NE | ft | ES | NE | ft | ES | NE | ft | |
| 1 × 10−3 | Relatives | 667 | 1,978 | 1.2 × 10−24 | 1,123 | 2,207 | 9.0 × 10−13 | 420 | 828 | 7.6 × 10−2 |
| No relatives | 565 | 3,214 | 565 | 1,701 | 495 | 1,099 | ||||
| 1 × 10−10 | Relatives | 192 | 376 | 5.7 × 10−12 | 653 | 1,187 | 9.8 × 10−3 | 173 | 321 | 7.8 × 10−2 |
| No relatives | 1,040 | 4,816 | 1,035 | 2,721 | 742 | 1,606 | ||||
Note.—Universal essential orthologous (ES) groups and nonessential (NE) groups are compared with universal orthologous groups with and without related groups at e value cutoffs of 1 × 10−3 and 1 × 10−10. Essential groups in animals are more likely to have relatives (P values of Fisher’s Exact Tests [ft] for enrichment). See Materials and Methods for definitions of universal, essential, and related orthologous groups. For results with alternative essentiality data sets, see supplementary table 4 (Supplementary Material online).
This trend is consistent with studies that compared evolutionary rates of duplicated genes (with within-species homologs) with singletons (unique, without within-species homologs) and showed that conserved genes with constrained sequence evolution were more likely to give rise to maintained duplicates (Davis and Petrov 2004; Jordan et al. 2004). Thus, functionally important genes or their constituent domains appear to have been utilized more frequently throughout the evolution of eukaryotes.
Functional Perspective
The Majority of Biological Processes Are Enriched for Essential Genes in Animals
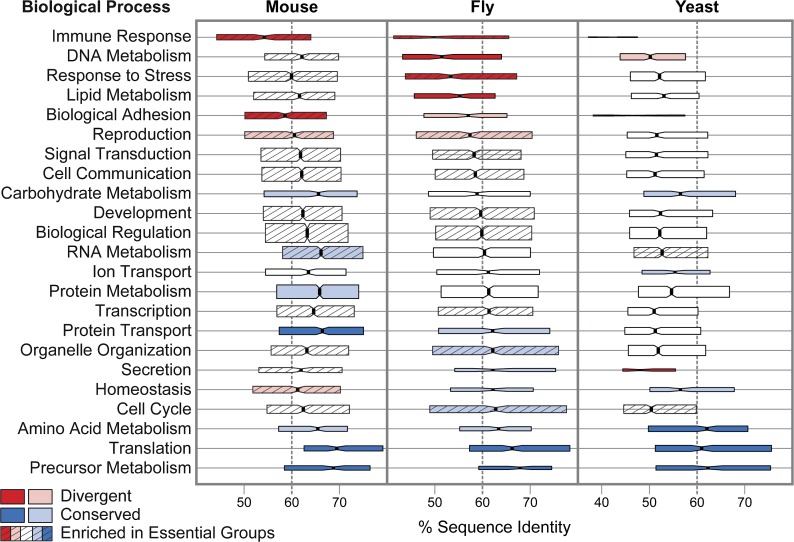
Identifying proportions of essential gene-containing orthologous groups in GO functional categories revealed significant enrichment of essential genes among principal biological processes in mouse and fly (fig. 3, supplementary table 5, Supplementary Material online). Member gene GO and InterPro attributes facilitated the assignment of principal GO terms (e.g., generic GO Slim terms) to universal mouse, fly, and yeast orthologous groups using GO child to parent relations and InterPro to GO mappings (GO-Consortium 2010). Of the 23 biological processes, 16 and 10 are enriched in essential genes in mouse and fly, respectively, whereas only 2 show significant enrichment in yeast. Examining all categories defined by the generic GO Slim subsets of biological processes, molecular functions, and cellular components confirms this difference among processes and functions whereas components instead exhibit similar proportions of enriched terms in the three lineages (supplementary table 5 and fig. 8, Supplementary Material online).
FIG. 3.—
Orthologous group sequence conservation of principal GO biological processes in mouse (Mus musculus), fly (Drosophila melanogaster), and yeast (Saccharomyces cerevisiae). Processes are ordered by median orthologous group sequence identities of fly categories with the most divergent in red and the most conserved in blue (more than one standard deviation greater than—dark blue—or less than—dark red—the mean identity). Several mouse and fly biological processes are enriched in essential gene-containing orthologous groups (striped shading) in contrast to only two processes in yeast. Notched boxes show medians of orthologous group percent identities with the limits of the upper and lower quartiles, and box heights are proportional to the number of orthologous groups in each category. For GO identifiers, median identities, orthologous group counts, and Fisher’s Exact Tests for enrichment of essential groups, see supplementary table 5 (Supplementary Material online).
Although variations in annotation strategies may contribute to some observed differences, the general paucity of categories enriched in essential groups in fungi compared with animals appears to suggest that fungal processes and functions are more robust to gene knockouts. Robustness is a key attribute of complex systems that facilitates evolvability and may itself be selected (Kitano 2004). It is therefore tempting to speculate that stronger effective selection throughout the evolution of fungi may confer greater robustness on fungal processes and functions.
Divergence Rates Vary among Functional Categories and Lineages
Orthologous groups of genes involved in the processes of translation and precursor metabolism (generation of precursor metabolites and energy) show the highest levels of sequence conservation in vertebrates, arthropods, and fungi (fig. 3). At the other end of the scale, immune responses and adhesion processes are shared divergent categories and reproductive processes are relatively divergent in both vertebrates and arthropods, whereas divergence of DNA metabolism is common to arthropods and fungi. Structural molecule and translation factor activities are commonly conserved molecular functions, whereas the most divergent include receptor, signal transducer, and nuclease activities, as well as carbohydrate and chromatin binding (supplementary table 5 and fig. 8, Supplementary Material online). Among cellular components, the ribosome appears highly conserved, whereas the most divergent are extracellular elements in vertebrates and arthropods and components of the nucleus in arthropods and fungi. These results are in agreement with pairwise studies such as fly–mosquito (Zdobnov et al. 2002) or human–chicken (Chicken-Genome-Consortium 2004) comparisons that identified structural molecules and those involved in protein transport as the most conserved and signal transducers and immune-related genes as the most divergent. Similarly, multispecies comparisons from human to yeast have associated fast-evolving protein families with regulatory roles and responses to stimuli such as immune challenges, which contrast slow-evolving families involved in transport, protein synthesis, or primary metabolism (Lopez-Bigas et al. 2008).
Orthologous group average sequence identities, therefore, clearly distinguish between fast- and slowly evolving functional categories, highlighting both common and distinct evolutionary pressures on functional subsets of genes in the three eukaryotic lineages.
Distinct Domain Proliferations Characterize Vertebrate, Arthropod, and Fungal Proteomes
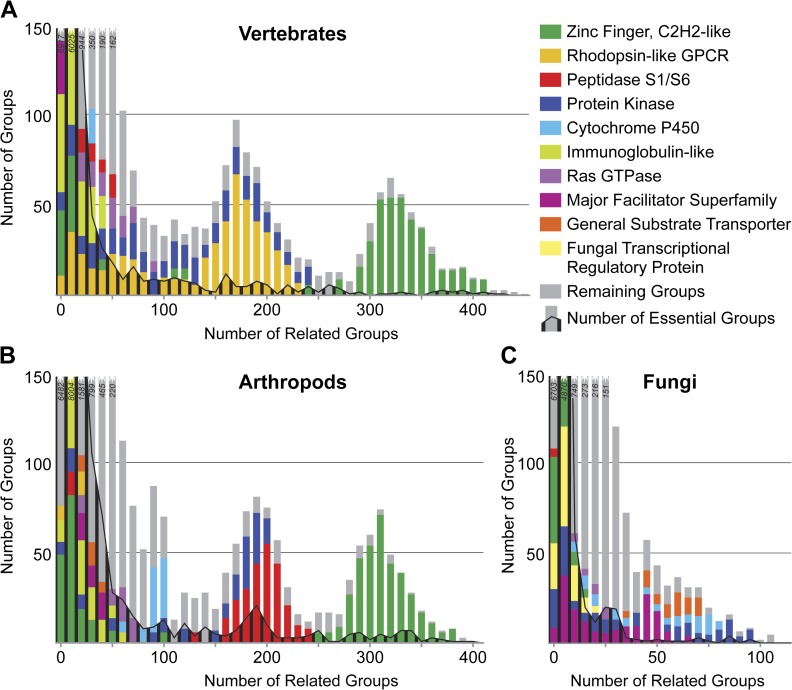
Shared sequence homology among orthologous groups may derive from expanded repertoires of functionally beneficial protein domains in each lineage. Many orthologous groups are related (share common sequence ancestry) to only a few or a few tens of other groups but examining the protein domain signatures of orthologous groups with the most numerous related groups reveals the proliferation of a few functional domains that has created distinguishing superfamilies in each lineage (fig. 4). At the e value cutoff of 1 × 10−3 (see Materials and Methods), 9.0% of vertebrate and 7.3% of arthropod orthologous groups have more than 50 related groups, whereas in fungi, only 5.4% of orthologous groups have more than 20 related groups. Even essential genes may be found among these orthologous groups with the highest numbers of relatives (fig. 4), in agreement with the observation that functionally important genes or domains have been frequently reutilized throughout eukaryotic evolution (table 1).
FIG. 4.—
Independent proliferation of a few functional domains has created distinguishing protein superfamilies in vertebrates (A), arthropods (B), and fungi (C). The majority of the orthologous groups with identifiable homology to other groups are related to only a few other groups, however, those with numerous relatives are often characterized by specific protein domains. The superfamily of zinc finger C2H2-like proteins is common to vertebrates and arthropods, whereas the expansion of vertebrate rhodopsin-like GPCRs contrasts that of the arthropod peptidases. The fungi are characterized by families of transporters of the major facilitator superfamily and general substrate transporters. Cytochrome P450s are prominent in arthropods and fungi and the protein kinases feature in all three lineages. Orthologous groups with essential genes may be found among those with some of the highest numbers of relatives. Related groups are defined by the average pairwise Smith–Waterman e value between all the members of each group in each lineage with a cutoff of 1 × 10−3.
The highly abundant zinc finger proteins feature in both vertebrates and arthropods, whereas the proliferation of rhodopsin-like G protein–coupled receptors (GPCRs) in vertebrates contrasts the expanded superfamily of arthropod peptidases. Cytochrome P450s are prominent in arthropods and fungi, whereas families of transporters characterize the fungi, and the protein kinases are prominent in all three lineages. These domains characterize some of the most abundant eukaryotic protein-coding genes, whereas other domains highlight lineage-specific biology such as vertebrate Kruppel-associated boxes involved in transcriptional repression and major histocompatibility complex proteins of the acquired immune system (supplementary table 6, Supplementary Material online). Arthropod-specific domains include pheromone/odorant-binding proteins and oxygen-carrying hemocyanins, whereas subfamilies of glycoside hydrolases and cellulose-binding domains characterize fungal-specific biology.
Zinc finger motifs form part of DNA-binding domains of many transcription factors and mediate protein–protein interactions (Brayer and Segal 2008). The evolution of zinc finger proteins in the two animal lineages have followed distinct paths, with expansions of the AD-type in arthropods that are not mirrored in vertebrates, and proliferation in vertebrates of zinc finger proteins with the acquired vertebrate-specific Kruppel-associated boxes (Copley 2008). The proliferation of such proteins with regulatory and interaction-mediating functions may have facilitated evolution of organismal complexity in higher eukaryotes. GPCRs are signal-transducer transmembrane proteins that include hormone, olfactory, neurotransmitter, and light receptors (Fredriksson et al. 2005). Vertebrates exhibit extensive GPCR family expansions, most notably of rhodopsin-like GPCRs, which include vertebrate olfactory and chemokine receptors that are absent from arthropods. In contrast, arthropod chemoreceptors are not related to rhodopsin-like GPCRs but operate instead as ligand-gated ion channels that form a large repertoire of gustatory receptors across Arthropoda and olfactory receptors in terrestrial insects (Sato et al. 2008; Peñalva-Arana et al. 2009). The usually secreted S1/S6 peptidases employ histidine–aspartate–serine catalysis to cleave target proteins in diverse processes including fertilization, development, intestinal digestion, blood coagulation, apoptosis, and immunity (Di Cera 2009). Arthropod diversity is reflected in the variety of food sources upon which they rely, especially as larvae or nymphs, and the abundance of arthropod proteases may be driven by such digestive requirements. These expansions may also reflect key roles of proteases in characteristic arthropod processes of molting and metamorphosis or the immune defense of the open circulatory system. Cytochrome P450s are important for the synthesis and breakdown of hormones as well as in detoxification processes critical for the clearance of many potentially harmful xenobiotics. Their prominent diversity in arthropods is almost certainly linked to the incredible variety of environmental challenges, particularly with respect to the coevolution of arthropods and plants (Feyereisen 2006). The numerous cytochrome P450s in fungi contribute to their ability to occupy a large variety of ecological niches that often require specialized secondary metabolism (Kelly et al. 2009), and their numerous transporters are critical for exchanging solutes with their environments.
Conclusions
This study provides a consistent view on gene evolutionary traits across a large set of vertebrate, arthropod, and fungal species spanning millions of years of divergence and sheds light on several fundamental principles of eukaryotic gene repertoire evolution. Classification of protein-coding genes into orthologous groups of genes descended from their common ancestors reveals that the majority of groups exhibit broad phyletic distributions with genes almost universally present within each lineage. Most of these genes evolve under single-copy control but those with a multicopy license frequently duplicate across the entire lineage. Evolution of single-copy orthologs is also constrained at the sequence level and contrasts the elevated divergence among maintained duplicates. In addition, the evolutionary perspective on gene essentiality assessed in model organisms firmly supports the hypotheses that such genes are under stronger selection for both gene retention and gene sequence conservation in vertebrates, arthropods, and fungi.
Our methodologically consistent and large-scale approach provides evidence that most of these principles are shared among these three major eukaryotic lineages and highlights lineage-specific idiosyncrasies that should be taken into account for cross-lineage comparisons. Some of these principles, such as the stronger constraints on the sequence evolution of essential genes, were previously hypothesized or evidenced using smaller data sets, but the most quantitatively prominent distinction on the tolerance of gene copy-number variations has been largely underappreciated.
Supplementary Material
Supplementary figures 1–8 and tables 1–6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/.)
Acknowledgments
The authors would like to thank Prof Iwona Stroynowski, Dr Ivo Pedruzzi, Dr Fredrik Tegenfeldt, and all members of the Computational Evolutionary Genomics Group for useful discussions, as well as the anonymous referees for their constructive suggestions.
References
- Aury J, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Brayer K, Segal D. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50(3):111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- Bult C, Kadin J, Richardson J, Blake J, Eppig J. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis C, Hartl D. Conservation, relocation and duplication in genome evolution. Trends Genet. 2003;19(11):593–597. doi: 10.1016/j.tig.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Chicken-Genome-Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Copley R. The animal in the genome: comparative genomics and evolution. Philos Trans R Soc Lond B Biol Sci. 2008;363(1496):1453–1461. doi: 10.1098/rstb.2007.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Petrov D. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2(3):E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cera E. Serine proteases. IUBMB Life. 2009;61(5):510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22(7):375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Elsik C, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyereisen R. Evolution of insect P450. Biochem Soc Trans. 2006;34(Pt 6):1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- Fitch W. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19(2):99–113. [PubMed] [Google Scholar]
- Fredriksson R, Lagerström M, Schiöth H. Expansion of the superfamily of G-protein-coupled receptors in chordates. Ann N Y Acad Sci. 2005;1040:89–94. doi: 10.1196/annals.1327.011. [DOI] [PubMed] [Google Scholar]
- Gerdes S, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol. 2003;185(19):5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GO-Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson A, Snitkin E, Parker S, DeLisi C, Kasif S. Towards the identification of essential genes using targeted genome sequencing and comparative analysis. BMC Genomics. 2006;7:265. doi: 10.1186/1471-2164-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered. 2009;100(5):605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Kumar S. The timetree of life. Oxford: Oxford University Press; 2009. [Google Scholar]
- Hirsh A, Fraser H. Protein dispensability and rate of evolution. Nature. 2001;411(6841):1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst L, Smith N. Do essential genes evolve slowly? Curr Biol. 1999;9(14):747–750. doi: 10.1016/s0960-9822(99)80334-0. [DOI] [PubMed] [Google Scholar]
- Jordan I, Rogozin I, Wolf Y, Koonin E. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 2002;12(6):962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan I, Wolf Y, Koonin E. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Krasevec N, Mullins J, Nelson D. The CYPome (Cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S53–S61. doi: 10.1016/j.fgb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Kirkness E, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A. 2010;107(27):12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5(11):826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Koonin E. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Krylov D, Wolf Y, Rogozin I, Koonin E. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13(10):2229–2235. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay D, et al. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 2009;10(4):R43. doi: 10.1186/gb-2009-10-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B, Scott N, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23(11):2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Liao B, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23(8):378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, De S, Teichmann S. Functional protein divergence in the evolution of Homo sapiens. Genome Biol. 2008;9(2):R33. doi: 10.1186/gb-2008-9-2-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz H, Vingron M. Family specific rates of protein evolution. Bioinformatics. 2006;22(10):1166–1171. doi: 10.1093/bioinformatics/btl073. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Matsui T, Yamamoto T, Wyder S, Zdobnov E, Kadowaki T. Expression profiles of urbilaterian genes uniquely shared between honey bee and vertebrates. BMC Genomics. 2009;10:17. doi: 10.1186/1471-2164-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva-Arana D, Lynch M, Robertson H. The chemoreceptor genes of the waterflea Daphnia pulex: many Grs but no Ors. BMC Evol Biol. 2009;9:79. doi: 10.1186/1471-2148-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Liao B, Chang A, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26(10):425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Saebø P, Andersen S, Myrseth J, Laerdahl J, Rognes T. PARALIGN: rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005;33:W535–W539. doi: 10.1093/nar/gki423. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Pellegrino M, Nakagawa T, Vosshall L, Touhara K. Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature. 2008;452(7190):1002–1006. doi: 10.1038/nature06850. [DOI] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt-Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R, Zdobnov E, Tegenfeldt F, Li J, Kriventseva E. OrthoDB: the hierarchical catalog of eukaryotic orthologs in 2011. Nucl. Acids Res. doi: 10.1093/nar/gkq930. Advance Access October 23, 2010. 2010 doi: 10.1093/nar/gkq930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren J, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327(5963):343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Carlson S, White T. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Wolf Y, Carmel L, Koonin E. Unifying measures of gene function and evolution. Proc Biol Sci. 2006;273(1593):1507–1515. doi: 10.1098/rspb.2006.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyder S, Kriventseva E, Schröder R, Kadowaki T, Zdobnov E. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 2007;8(11):R242. doi: 10.1186/gb-2007-8-11-r242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gu Z, Li W. Rate of protein evolution versus fitness effect of gene deletion. Mol Biol Evol. 2003;20(5):772–774. doi: 10.1093/molbev/msg078. [DOI] [PubMed] [Google Scholar]
- Zdobnov E, et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298(5591):149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- Zhang J, He X. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 2005;22(4):1147–1155. doi: 10.1093/molbev/msi101. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37:D455–D458. doi: 10.1093/nar/gkn858. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]