Abstract
In an attempt to identify genes involved in glutathione (GSH) transport, a human mammary gland cDNA library was screened for clones capable of complementing a defect in GSH uptake in yeast cells that lack Hgt1p, the primary yeast GSH uptake transporter. Five genes capable of rescuing growth on sulfur-deficient GSH-containing medium were identified: prostate transmembrane protein, androgen induced 1 (PMEPA1); lysosomal-associated protein transmembrane 4 alpha (LAPTM4α); solute carrier family 25, member 1 (SLC25A1); lipopolysaccharide-induced TNF factor (LITAF); and cysteine/tyrosine-rich-1 (CYYR1). All of these genes encode small integral membrane proteins of unknown function, although none appear to encode prototypical GSH transporters. Nevertheless, they all increased both intracellular glutathione levels and [3H]GSH uptake rates. [3H]GSH uptake was uniformly inhibited by high concentrations of unlabeled GSH, GSSG, and ophthalmic acid. Interestingly, each protein is predicted to contain Pro-Pro-x-Tyr (PY) motifs, which are thought to be important for regulating protein cell surface expression. Uptake of the endocytotic markers lucifer yellow and FM4-64 was also enhanced by each of the five genes. Mutations of the PY motifs in LITAF largely abolished all of its effects. In summary, although the results do not reveal novel GSH transporters, they identify five PY-containing human gene products that may influence plasma membrane transport activity.
Key Words: PY-motif, Yeast, Membrane, Transport, Endocytosis, Glutathione
Introduction
Glutathione (GSH) is a major intracellular thiol, and is present in virtually all eukaryotic and most prokaryotic cells at concentrations ranging from 0.5 to 10 mM. Maintenance of GSH levels is required for many critical cell processes, and disturbances in GSH homeostasis have been implicated in the etiology and/or progression of a number of human diseases, including cancer, diseases of aging, and cardiovascular, inflammatory, immune, liver and neurodegenerative diseases [1]. Intracellular and extracellular GSH concentrations are normally tightly regulated to maintain normal physiological functions, and the two major mechanisms involved in this regulation are GSH biosynthesis and GSH efflux from cells [2]. Despite the increasing evidence that some of the multidrug resistance-associated proteins (MRP/ABCC) mediate GSH export, it is unknown whether the MRPs are the sole or even the major GSH exporters in human cells. Given the important functions of GSH in multiple cellular processes, it is possible that additional GSH transporters are present in the human genome, but have not yet been identified at molecular level [3, 4].
With the aim of identifying additional GSH transporters, we screened a human mammary gland cDNA library using an expression cloning strategy in yeast. Unlike mammalian cells, S. cerevisiae are able to take up GSH from the extracellular medium, and this uptake is mediated primarily by a high affinity GSH uptake transporter, Hgt1p [5]. In addition to Hgt1p, a low-affinity GSH uptake mechanism is present in yeast cells; however, the gene or genes responsible for this alternate GSH uptake process have not yet been identified [5, 6]. Orthologues of HGT1 are not present in higher eukaryotic organisms. The deletion of the HGT1 gene in S. cerevisiae leads to a marked impairment in the ability to grow in media in which GSH is the only sulfur source [5]. The present study took advantage of the growth deficiency of the hgt1-Δ strain to search for human genes that can facilitate or mediate GSH accumulation by these cells and therefore rescue the growth deficiency. Because the present screen tested for genes that lead to enhanced GSH uptake in yeast, it is unable to identify unidirectional GSH exporters, but should theoretically be able to identify bidirectional GSH transporters (e.g., secondary active transporters) or unidirectional GSH uptake transporters.
Five genes capable off complementing the HGT1 deletion were recovered, providing clues into the potential functions of human PMEPA1 (prostate transmembrane protein, androgen induced 1), LAPTM4α (lysosomal-associated protein transmembrane 4 alpha), SLC25A1 (solute carrier family 25, member 1), LITAF (lipopolysaccharide-induced TNF factor), and CYYR1 (cysteine/tyrosine-rich1). Because each of the encoded proteins is predicted to have PY motifs, which are thought to serve as binding sites for the ubiquitin ligases yeast Rsp5 (reverses Spt-phenotype 5) or human NEDD4 (neural precursor cell expressed, developmentally down-regulated 4), these genes may play a role in modulating protein cell surface expression.
Materials and Methods
Materials
Culture media components were obtained from Clontech Laboratories, Inc. (Mountain View, CA), Difco (Sparks, MD), Invitrogen (Carlsbad, CA) and Sigma-Aldrich (St. Louis, MO). [3H]Glutathione was purchased from Perkin Elmer (Boston, MA). Acivicin, glycine, glutamic acid, glutathione reductase, GSH, GSSG, Latrunculin A, lucifer yellow, lyticase, 0.1% poly-lysine, and yeast protease inhibitor cocktail (P8215) were obtained from Sigma (St. Louis, MO). N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl) hexatrienyl)pyridinium dibromide (FM4-64) was obtained from Molecular Probes (Carlsbad, CA). Oligonucleotides were purchased from IDT (Coralville, IA). Ophthalmic acid was purchased from Bachem Bioscience Inc. (Philadelphia, PA). Zymolyase 100T was purchased from Seikagaku Corp. (Tokyo, Japan). The anti-FLAG M2 monoclonal antibody (F3165) and anti-mouse IgG peroxidase conjugate antibody (A9044) were purchased from Sigma-Aldrich (St. Louis, MO); anti-Pep12p (A-21273) and anti-Dpm1p (A-6429) were obtained from Molecular Probe (Eugene, OR); anti-Pma1p (sc-57978) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). All chemicals used were of the highest analytical grade possible.
Yeast strains and culture condition
The hgt1-Δ yeast strain (ABC822: MATa ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801 ade2-101 hgt1Δ::LEU2) and its parental yeast strain (ABC154: MATa ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801 ade2-101)) were kindly provided by Dr. A.K. Bachhawat (Institute of Microbial Technology, Chandigarh, India). In the mutant strain, the HGT1 gene was replaced with the LEU2 gene [5]. Rich YPAD medium, (1% yeast extract, 2% Bacto peptone, 2% glucose, 0.002% adenine) was used for routine yeast culture. Yeast transformants were selectively grown on minimal selective medium (0.17% yeast nitrogen base without amino acids, 0.5% ammonium sulfate, 2% glucose, supplemented with the required amino acids and bases). Sulfur-deficient medium was prepared based on minimal selective medium with the modification that all sulfur-containing reagents were substituted with equal amounts of the corresponding chloride salt (i.e., CuCl2, MgCl2, ZnCl2, MnCl2, and NH4Cl) and free adenine base [5]. For solid media, 2% agar was added.
Yeast transformation and selection
The human mammary gland library built in the low copy yeast expression vector pMETtPGK3 was kindly provided by Dr. S.G Oliver, University of Manchester, Manchester UK. This cDNA library contains approximately 330,000 independent colonies, and protein expression is driven by the MET3 promoter [7]. The hgt1-Δ cells were transformed with plasmid DNA from the human mammary gland cDNA library or with the empty vector using the lithium acetate method [8]. Transformants were selected directly on solid sulfur-deficient medium supplemented with 200 μM or 500 μM GSH. Colonies exhibiting robust growth in comparison to the control colonies bearing the empty vector were chosen for subsequential selection in liquid sulfur-deficient medium supplemented with either 500 μM or 750 μM GSH. Growth was monitored over time by measuring the optical density expressed as OD600.
DNA preparation, sequencing and analysis
Plasmid DNA was extracted from individual positive yeast colonies and was used as the template to amplify the cDNA inserts with Promega Pfx DNA polymerase (Madison, WI). Primers used in PCR reactions were designed based on the sequences of MET3 promoter (CTC TCT GTC GTA ACA GTT GT) and of PGK1 terminator (GGC AAT TCC TTA CCT TCC AA). PCR products were purified using the Qiagen PCR purification kit (Valencia, CA) and sequenced by the Mount Desert Island Biological Laboratory DNA Sequencing Facility (Salisbury Cove, ME) with the MET3 primer. A representative of each class of cDNA insert, as determined by the alignment analysis using the Vector NTI program (Frederick, MD), was amplified by E.coli and transformed back into yeast. Once it was confirmed that the retransformed cells were able to rescue growth in liquid sulfur-deficient medium supplemented with either 500 or 750 μM GSH as the sole sulfur source repeatedly, nucleotide sequence comparisons were performed using the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST) network service against the NCBI databases. For each identified cDNA, protein topology was analyzed based on the NCBI protein reference sequence. Transmembrane domains were predicted using the HMMTOP program (Prediction of transmembrane helices and topology of proteins, http://www.enzim.hu/hmmtop/index.html), and PY motifs were predicted by ELM (The Eukaryotic Linear Motif resource for Functional Sites in protein, http://elm.edu.org/).
Ndfip2 (Nedd4 family interacting protein 2) cDNA clone was obtained from the Mouse MGC and IMAGE Clones collection of Thermo Scientific (CAT# 8360772, Huntsville, AL). The insert was excised from pCR4-TOPO vector using EcoRI, and was subsequently subcloned into the pESC-URA vector. The pESC-Ndfip2 clone was confirmed by nucleotide sequencing. Both wild type yeast strain and the hgt1-Ä strain were transformed with pESC-URA vector and with pESC-Ndfip2 clone using the lithium acetate method [8]. [3H]GSH uptake was measured for 30 min as descried below.
Total glutathione quantification
For the determination of total glutathione, collected yeast cells were washed with sterile distilled H2O, lysed with 5% perchloric acid containing 1 mM EDTA, and vortexed vigorously in the presence of acid-washed glass beads. The supernatant was analyzed for intracellular glutathione using a microtiter plate assay containing 5,5'-dithio-bis(2-nitrobenzoic acid) and glutathione reductase [9]. Protein was analyzed using the Bio-Rad DC Protein Assay Kit (Hercules, CA) and the total glutathione concentration was normalized per mg protein.
[3H]GSH uptake assay
Transformed yeast cells were grown in minimal selective medium until an OD600 of ∼0.5 was reached. Collected cells were washed with sterile water and grown for another 11-12 h in minimal sulfur-deficient medium in the absence of GSH. As described by Bourbouloux et al. [5], [3H]GSH uptake was measured in transport buffer (20 mM MES/KOH, 5 mM CaCl2, 2.5 mM MgCl2, 2% glucose, pH 5.5) supplemented with 500 μM acivicin (an inhibitor of gamma-glutamyltranspeptidase). Twenty million yeast cells were incubated with 500 ìM [3H]GSH (final specific activity of 24.67 MBq mmol-1) at 30°C for the indicated time, and cell associated radioactivity was measured in scintillation fluid (OPTI_FLUOR, Perkin Elmer, Shelton, CA) after correction for background and quenching (LS 6500, Beckman Coulter, Fullerton, CA).
Endocytotic maker uptake assay
Lucifer yellow uptake experiments were modified from Dulic et al. [10]. Yeast cells were cultured as described in the GSH uptake assay. Fifty million yeast cells were incubated with 4 mg/ml of lucifer yellow in the sulfur-deficient medium at 0°C, 200 rpm for 2 h. Cell-associated lucifer yellow was quantified using a Spectramax GeminiXs (Molecular Devices, Sunnyvale, CA), at an excitation at 426 nm and emission at 550 nm. Uptake was determined as the amount of cell-associated lucifer yellow per mg protein after 2 h incubation.
The FM4-64 uptake protocol was adapted from Emans et al. [11]. Yeast strains were cultured as described in GSH uptake assay. Fifty yeast cells were incubated with 40 μM of FM4-64 in minimal selective medium for 2 h at 30°C or on ice. Cell-associated fluorescence was quantified using a Spectramax GeminiXs (Molecular Devices, Sunnyvale, CA) with excitation at 515 nm and emission at 650 nm. Uptake was determined as the fluorescence per mg protein after 2 h incubation.
Latrunculin A treatments
To test the inhibitory effect of latrunculin A on [3H]GSH uptake and FM4-64 accumulation, yeast cells were pre-incubated with 20 μM latrunculin A which was dissolved in DMSO, or with DMSO (final concentration is 1%) as vehicle control for 1 h at 30°C, and then the assays were carried out as described above.
LITAF PY mutagenesis and Western blotting
The two PPSY encoding domains in LITAF gene (denoted here as PY1 and PY2) were changed to PPSA using a QuikChange(®) II XL Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). All four constructs (i.e., LITAF, LITAF-PY1Δ, LITAF-PY2Δ, LITAF-PY1&2Δ) were subsequently subcloned into pESC-URA vector to generate FLAG tagged sequences. Wild type yeast cells were then transformed with the empty pESC-URA vector or with each of the four LITAF constructs. Wild type yeast cells bearing the empty pESC-URA vector or each of these four LITAF constructs were grown to mid-log phase, and proteins were isolated by vigorously vortexing with the same volume of glass beads in Cellytic Y Yeast Cell Lysis/Extraction Reagent (Sigma-Aldrich, St. Louis, MO) freshly supplemented with protease inhibitor cocktail (Sigma) and 10 mM dithiothreotol. Whole cell lysates (40 μg) were separated on 4–15% (w/v) SDS-PAGE gels (Bio-Rad). The separated polypeptides were electrotransferred on to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) for 120 min at 95 V using a wet-transfer apparatus (Bio-Rad). The membranes were blocked overnight in 5% (w/v) dried milk in TBST (20 mM Tris, 140 mM NaCl, 0.05% Tween-20, pH 7.4) at 4°C. The blots were probed with anti-FLAG at 1:4000 or with anti-Actin at 1:500 for 2 h at room temperature, and then with respective secondary antibodies at 1:3000 for 1 h at room temperature. Antibody binding was detected using Perkin-Elmer enhanced-chemiluminescence technique (Waltham, Massachusetts).
Subcellular fractionation
Subcellular fractions were isolated essentially as described by Rieder et al. [12]. Yeast cells transformed with LITAF-FLAG were grown in minimal selective medium containing raffinose as carbon source to an early log phase, and 2% galactose was added for 6 h. Collected cells were converted to spheroplasts with Zymolyase 100T (25 μg/OD600). Spheroplasts were then lysed in HEPES/KAc buffer (20 mM HEPES/KOH pH 6.8, 50 mM potassium acetate, 200 mM sorbitol, 1 mM EDTA, and freshly added 1 mM phenylmethyl sulphonyl fluoride, 1 mM dithiothreitol and 1X yeast protease inhibitor cocktail) using a Dounce homogenizer (15 strokes with a tight-fitting pestle). The lysate was cleared by centrifuging at 300 × g for 5 min. A portion of the resulting supernatant was used as the S300 fraction, and the rest was centrifuged at 13,000 × g for 10 min. The resulting pellet (P13000) was stored on ice. A portion of this supernatant was saved as the S13000 fraction, and the rest was centrifuged at 100,000 × g for 60 min to generate S100000 and P100000 fractions. Proteins were precipitated from each fraction using 10% TCA and 20 μg of each fraction were separated on BioRad 12% SDS-PAGE gels. Membranes were probed with primary antibodies (mouse anti-FLAG monoclonal antibody 1:4000, mouse anti-Pma1p antibody 1:200, mouse anti-Pep12p antibody 1:4000, and mouse anti-Dpm1p antibody 1:2000) overnight at 4°C, and then with secondary antibody for 1 h at room temperature. Antibody binding was visualized using Perkin Elmer chemiluminescence reagents.
Indirect immunofluorescence microscopy
The cellular localization of LITAF was determined essentially as described by Pringle et al. [13]. After being induced by galactose for 6 h, yeast cells were fixed with 4% formaldehyde and converted to spheroplasts with lyticase. Spheroplast suspension was applied to the multiple-well poly-lysine-coated microscope slide (PolySciences, Warrington, PA). After being blotted with anti-FLAG antibody for overnight at 4°C, the spheroplasts were stained with Rhodamine Red-X-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA) for 2 h at room temperature. The samples were viewed using an FV1000 Olympus Laser Scanning Confocal Microscope (Olympus America, Melville, NY) at 60 × magnification. Cell morphology was viewed by differential interference contrast (DIC, Nomarski), and cell nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Images were processed using FV1000 software (Olympus America, Melville, NY) and Adobe Photoshop (Adobe Systems, San Jose, CA).
Data analysis
Data are given as mean ± SE. Mean values were considered to be significantly different when P<0.05 by use of a two-way ANOVA or a one-way ANOVA followed by Bonferroni's multiple comparison test. Software used for statistical analysis was GraphPad Prism (version 5 for windows; GraphPad Software, San Diego CA).
Results
PMEPA1, LAPTM4α, SLC25A1, LITAF, and CYYR1 rescue the impaired growth of hgt1-Δ in medium in which GSH is the only sulfur source
hgt1-Δ cells were transformed with a human mammary cDNA library which contains about 300,000 individual clones, and the transformants were subjected to selection on sulfur-deficient medium supplemented with 200 or 500 μM GSH. Approximately 300 colonies grew faster on this medium than colonies from cells transformed with the empty pMETtPGK3 vector, giving rise to colonies that were at least two times larger than the cells containing only vector. The growth of these larger colonies was subsequently assayed quantitatively in liquid sulfur-deficient medium supplemented with either 500 or 750 μM GSH. About one-half of these colonies showed increased growth. Plasmid DNAs isolated from these positive colonies were used as the template to amplify the cDNA inserts, which were subsequently partially sequenced. Plasmid DNA of each clone was amplified in E. coli and transformed back into the hgt1-Δ cells. The growth of retransformed cells was again monitored in liquid sulfur-deficient medium supplemented with either 500 or 750 μM GSH as the sole sulfur source. This screening process resulted in the identification of five genes that showed a strong ability to increase cell growth of hgt1-Δ cells. Each of these five genes was fully sequenced to reveal five relatively small membrane proteins (Fig. 1): PMEPA1, LAPTM4α, SLC25A1, LITAF, and CYYR1. Interestingly, multiple colonies contained LAPTM4α and LITAF. The possible cellular localizations, biological functions, and associations with human diseases of all five genes are summarized in Table 1. With the exception of SLC25A1, none of the encoded proteins are predicted to be a transporter. An examination of their predicted molecular structures using the HMMTOP and ELM programs revealed that CYYR1, LITAF and PMEPA1 are expected to be single transmembrane domain proteins. LAPTM4α and SLC25A1 are predicted to have 4 and 5 transmembrane domains, respectively. Remarkably, all five genes are predicted to encode proteins containing PY motifs (xPPxY, P = Pro, Y = Tyr, x = any amino acid; Fig. 1).
Fig. 1.
Predicted amino acid sequences and protein topologies of the five human genes identified in the screening. Transmembrane domains (underlined sequence) are predicted by the HMMTOP program and PY motifs (sequence in bold italic) are predicted by ELM program.
Table 1.
Summary of subcellular localization, possible cellular functions and predicted associations with human diseases. Charcot-Marie-Tooth disease type 1C (CMT1C); Extramammary Paget disease (EMPD); Crohn's Disease (CD); Ulcerative colitis (UC); DiGeorge syndrome (DGS); Velo-cardio-facial syndrome (VCFS); conotruncal anomaly face syndrome (CTAF).
| NCBI accession # | Protein name | Size (aa) | Subcellular localization | Human diseases association | Putative cellular functions | References |
|---|---|---|---|---|---|---|
| NP_43186.1 | CYYR1 | 154 | Membrane (predicted) | Neuroendocrine tumors | Unknown | [32, 33] |
| NP_055528.1 | LAPTM4α | 233 | Intracytoplasmic membrane (predicted) | Unknown | Nucleosides and its derivatives transport (by similarity) | [31, 46, 47] |
| NP_004853.2 | LITAF | 161 | Intracytoplasmic membrane | CMT-1C; EMPD; CD/UC; B-cell lymphomas | Lysosomal sorting; tumor suppression;trans-cription regulation | [16, 26, 39, 40, 41, 42, 43, 43, 48, 49] |
| NP_064567.2 | PMEPA1 | 287 | Golgi-associated | Cancers | Cell growth inhibition | [24, 35, 36, 44, 50] |
| NP_005 975.1 | SLC25A1 | 311 | Mitochondria membranes (predicted) | DGS; VCFS; CTAF | Citrate-H+/malate exchange (by similarity) | [51, 52] |
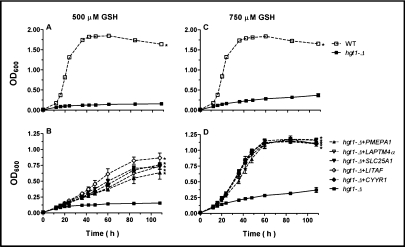
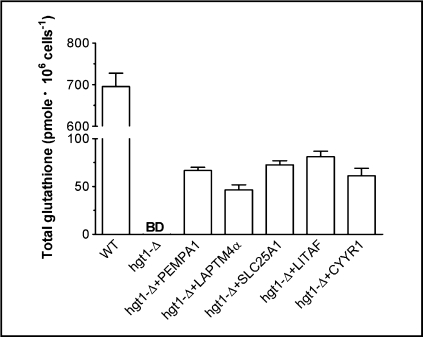
As illustrated in Fig. 2, these five human genes were able to rescue the impaired growth of hgt1-Δ cells in sulfur-deficient medium supplemented with 500 μM GSH (Fig. 2B) or 750 μM GSH (Fig. 2D), restoring cell growth to nearly the same level as that of wild type cells (Figs. 2A, 2C). To confirm that the growth recovery was due to an increase in intracellular glutathione concentration, hgt1-Δ transformants and wild type cells transformed with empty vector were cultured in sulfur-deficient medium supplemented with 750 μM GSH. After 48 h, the total intracellular glutathione concentration in wild type cells was about 700 pmol/106 cells (∼10 mM), which represents the normal physiological glutathione concentration in yeast cells, whereas glutathione concentration in hgt1-Δ cells was below the detection limit of the current method (25 pmole) (Fig. 3). Glutathione levels in hgt1-Δ cells expressing each of the five genes were about 10% of those of wild type cells (∼1 mM) (Fig. 3). Nevertheless, these modest increases in intracellular glutathione concentration may be sufficient to restore their growth (Figs. 2B, 2D).
Fig. 2.
Expression of five human genes rescues the growth impairment of hgt1-Δ cells in sulfur-deficient medium in the presence of 500 μM or 750 μM GSH as the sole sulfur source. Cells were grown overnight at 30°C in minimal selective medium. Subsequently, they were washed and inoculated at a cell density (expressed as OD600) of 0.01 in sulfur-deficient medium supplemented with either 500 or 750 μM GSH. Cell growth was monitored over a period of 108 h. Values are given as means ± SEM, N=3. * Significantly different from hgt1-Δ cells transformed with the empty vector pMETtPGK3 (p<0.05).
Fig. 3.
Expression of five human genes increases the intracellular total glutathione concentration in hgt1-Δ cells. Cells were grown overnight at 30°C in minimal selective medium. Subsequently they were washed and innoculated at a cell density (expressed as OD600) of 0.01 in sulfur-deficient medium supplemented with 750 μM GSH at 30°C for 48 h. Collected cells were lysed and intracellular total glutathione content was measured. Values are given as means ± SEM, N=3. BD = below detection limit.
The expression of these five human genes in hgt1-Δ cells leads to enhanced [3H]GSH uptake, which can be inhibited by high concentrations of GSH, GSSG and ophthalmic acid
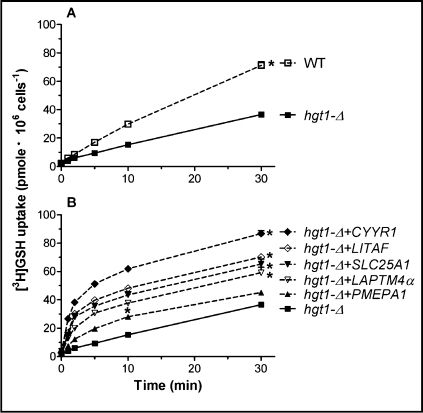
To determine whether the higher intracellular glutathione levels seen in hgt1-Δ cells transformed with the five human genes were due to increased GSH uptake, [3H]GSH transport activities of the wild type and hgt1-Δ transformants were measured after being cultured overnight in sulfur-deficient medium. Fig. 4A demonstrates that while hgt1-Δ cells are impaired in [3H]GSH uptake compared to the WT cells, these cells are able to accumulate some GSH by other mechanisms. hgt1-Δ cells transformed with LAPTM4α, SLC25A1, LITAF or CYYR1 all showed increased uptake of [3H]GSH (Fig. 4B). The uptake of [3H]GSH in hgt1-Δ cells transformed with PMEPA1 was also increased, however to a smaller extent (Fig. 4B). Note that the initial [3H]GSH uptake rate (1 min uptake) in hgt1-Δ cells transformed with LAPTM4α, SLC25A1, LITAF or CYYR1 was even higher than that in wild type cells (Fig. 5).
Fig. 4.
[3H]GSH uptake is enhanced in hgt1-Δ cells transformed with the five human genes. After growth at 30°C in minimal selective medium to a cell density (expressed as OD600) of approximately 0.5, cells were washed and grown for another 11–12 h at 30°C in the sulfur-deficient medium in the absence of GSH. For the uptake experiments, cells were incubated with 500 μM 3[H]GSH in MES buffer. Values are given as means ± SEM, N=3-7 individual experiments. * Significantly different from hgt1Δ cells transformed with the empty pMETtPGK3 vector (p<0.05).
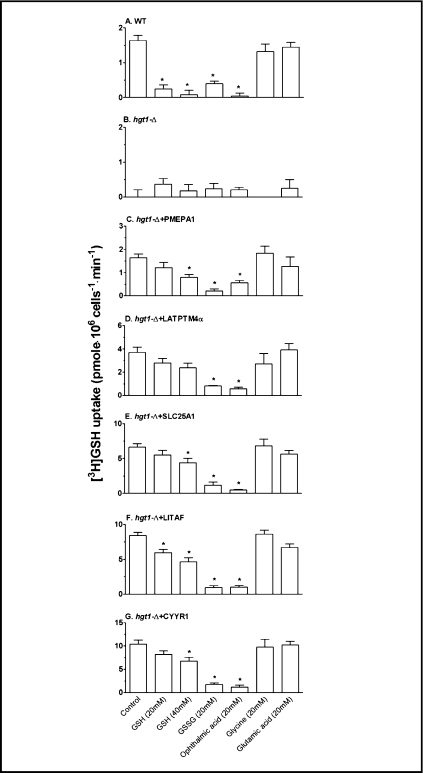
Fig. 5.
The initial [3H]GSH uptake rate of hgt1-Δ cells transformed with the five human genes is inhibited by GSH, GSSG and ophthalmic acid. After growth at 30°C in minimal selective medium to a cell density (expressed as OD600) of approximately 0.5, cells were washed and grown for another 11-12 h at 30°C in sulfur-deficient medium in the absence of GSH. The initial uptake rates of 500 μM 3[H]GSH solution (control) were determined in the presence of various compounds at a concentration of 20 mM each. GSH was tested as an inhibitor at both 20 and 40 mM. Values are given as means ± SEM, N=4-8 separate experiments. * Significantly different from control (p<0.05).
To gain insight into the mechanism for the enhanced [3H]GSH uptake by these transformants, initial uptake rates were measured at 1 min in the presence and absence of various unlabeled potential competitive inhibitors. As shown in Fig. 5A, addition of 20 mM GSH, GSSG, or ophthalmic acid (L-γ-glutamyl-L-α-aminobutyryl-glycine, a GSH analog) resulted in the inhibition of the initial uptake rate of [3H]GSH in the wild type cells transformed with the empty pMETtPGK3 vector. However, the amino acids glutamic acid and glycine had no effect (Fig. 5A), consistent with the known substrate specificity of Hgt1p (5). In contrast, no inhibitory effect of any of these compounds was observed in the hgt1-Δ cells transformed with the empty pMETtPGK3 vector (Fig. 5B). Addition of these compounds to hgt1-Δ cells transformed with PMEPA1 (Fig. 5C), LAPTM4α (Fig. 5D), SCL25A1 (Fig. 5E), LITAF (Fig. 5F) and CYYR1 (Fig. 5G) resulted in a uniform pattern of inhibition. These results suggest that the enhanced GSH uptake in the hgt1-Δ cells transformed with these five human genes is occurring by a common transport mechanism that is inhibitable by the compounds GSH, GSSG, and ophthalmic acid that also inhibit Hgt1p. However, because the inhibition was evident only at high mM concentrations of these compounds, the relative affinity for GSH uptake by this as yet undefined transport mechanism appears to be relatively low.
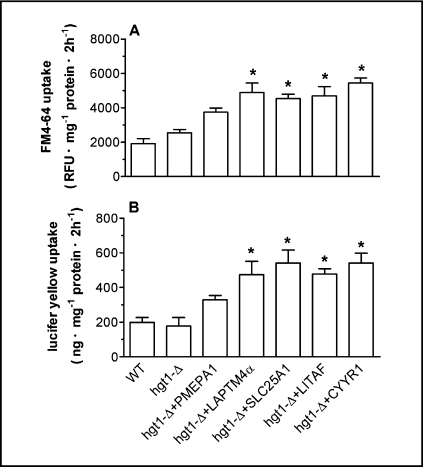
hgt1-Δ cells transformed with the five human genes show enhanced uptake of endocytotic markers
As illustrated in Fig. 1, all five identified proteins are predicted to contain PY motifs, which have been implicated to play a role in protein-protein interactions involving binding to WW domains in target proteins, including NEDD4 and its yeast homolog Rsp5. NEDD4/Rsp5p E3 ligases mediate ubiquitin-dependent endocytosis, which is a key component of the regulation of plasma membrane protein abundance [14]. To examine whether these five human genes affect endocytotic processes in yeast, the uptake of lucifer yellow (a commonly used marker for fluid phase endocytosis) and FM4-64 (a marker for membrane labeling and membrane turnover) was measured in wild type and hgt1-Δ transformants after overnight culture in sulfur-deficient medium. hgt1-Δ cells transformed with LAPTM4α, SLC25A1, LITAF or CYYR1 showed a significant increase in FM4-64 uptake (Fig. 6A) and in lucifer yellow uptake (Fig. 6B) when compared to the wild type cells or to hgt1-Δ cells, consistent with the hypothesis that these genes are stimulating membrane turnover in hgt1-Δ cells. PMEPA1 expressing cells also exhibited slightly increased accumulation of these two endocytotic markers, although the difference was not statistically significant (Fig. 6).
Fig. 6.
Lucifer yellow uptake and FM4-64 uptake are increased in hgt1-Δ cells transformed with the five human genes. After being cultured at 30°C in minimal selective medium to a cell density (expressed as OD600) of 0.5, cells were washed and grown for another 11–12 h at 30°C in sulfur-deficient medium in the absence of GSH. To start the uptake experiment, cells were incubated with 4 mg/ml lucifer yellow or 40 μM FM4-64 at 30°C for 2 h, shaking at a speed of 200 rpm for Lucifer yellow or 75 rpm for FM4-64. Values are given as means ± SEM, N=4-7. *Significantly different from hgt1-Δ cells transformed with the empty pMETtPGK3 vector (p<0.05).
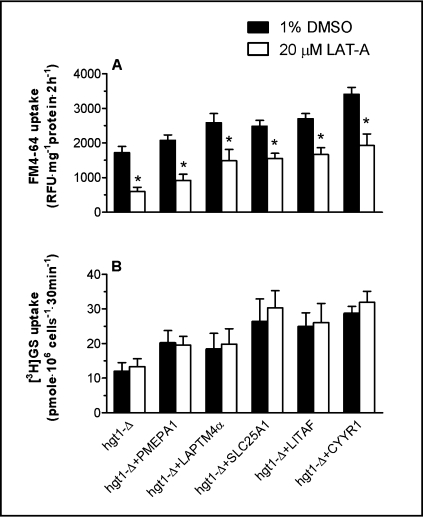
The enhanced [3H]GSH uptake in hgt1-Δ cells transformed with PMEPA1, LAPTM4α, SLC25A1, LITAF or CYYR1 is not inhibited by latrunculin A
To test for possible connections between the enhanced GSH uptake and the increased endocytotic marker uptake, latrunculin A (LAT-A) treatment was applied to hgt1-Δ transformants prior to measuring the uptake of FM4-64 and of [3H]GSH in these cells. LAT-A disrupts the actin cytoskeleton by sequestering actin monomers, and therefore has been widely used as an endocytotic inhibitor in yeasts [15]. hgt1-Δ transformants were pre-treated with 20 μM LAT-A or with 1% DMSO (vehicle control) for 60 minutes. As shown in Fig. 7A, LAT-A pretreatment resulted in decreased FM4-64 uptake in each tested strain, suggesting that endocytosis in these cells was inhibited by LAT-A treatment. In agreement with the results of figure 6A, cells expressing each of the five genes had higher rates of FM4-64 uptake than that of hgt1-Δ cells; however, LAT-A did not appear to be effective at inhibiting the FM4-64 uptake that was stimulated by each of the five genes (Fig. 7A). Note for example, that whereas LAT-A decreased FM4-64 uptake by about 65% in the control hgt1-Δ cells, it only decreased FM4-64 uptake by 38% in cells expressing LITAF (Fig. 7A).
Fig. 7.
Latrunculin A treatment decreases the basal uptake rate of FM4-64, but has no effect on [3H]GSH uptake. Cells were grown at 30°C in minimal selective medium until a cell density reaches 0.5 OD600. Subsequently, cells were washed and grown for another 11–12 h at 30°C in sulfur-deficient medium in the absence of GSH. After pre-incubation with 20 μM latrunculin A at 30°C for 1 h, FM4-64 uptake or [3H]GSH uptake of yeast cells was carried out as described earlier. Values are given as means ± SEM, N=5-6. * Significantly different from cells treated with 1% DMSO (p<0.05).
LAT-A had no effect on either the basal [3H]GSH uptake or the enhanced [3H]GSH uptake observed in cells transformed with the five genes (Fig. 7B). These results suggest that neither the increased GSH uptake, nor the increased uptake of general endocyttotic markers in hgt1-Δ cells transformed with PMEPA1, LAPTM4α, SLC25A1, LITAF or CYYR1 is directly due to an increase in conventional endocytotic processes. Although the mechanism of the enhanced [3H]GSH uptake is not known, the present results suggest that it is due to increased activity or membrane delivery of the as yet unidentified endogenous GSH uptake mechanism.
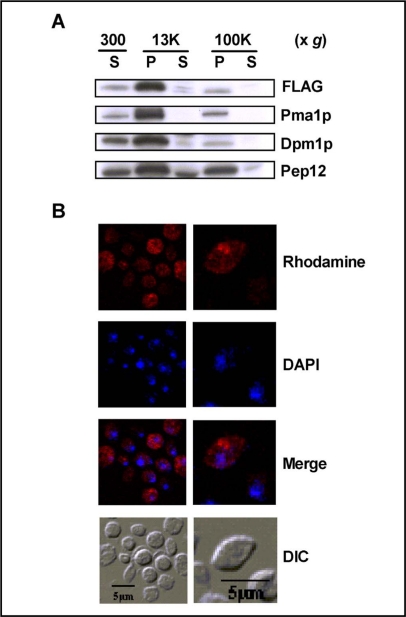
Subcellular localization of LITAF
To further characterize the potential mechanism by which these proteins stimulate GSH uptake, additional studies examined the intracellular localization of LITAF in yeast cells. When expressed in yeast, FLAG tagged LITAF was recognized as a protein of about 25 kDa, which is similar to the size of LITAF detected in mammalian cells [16]. Upon subcellular fractionation, LITAF was mainly present in the P13000 fraction, but was also detected in the P100000 fraction, albeit at much lower levels (Fig. 8A). Dpm1p (Dol-P-Man Synthase, an endoplasmic reticulum membrane marker protein) and Pma1p (Plasma Membrane H+-ATPase, a plasma membrane marker protein) also showed a predominant distribution in the P13000 fraction with low levels in the P100000 fraction, whereas Pep12p (carboxyPEPtidase Y-deficient, an endosome/Golgi membrane marker protein) was detected in the P13000 and P100000 fractions at roughly comparable levels (Fig. 8A). Essentially no LITAF was detected in the S100000 fraction, suggesting that LITAF is membrane protein. The distribution of LITAF between P13000 and P100000 indicates that LITAF is likely associated with large organelles such as the endoplasmic reticulum and the plasma membrane.
Fig. 8.
LITAF is associated with membrane-enriched fractions. The expression of LITAF-FLAG in wild type cells (ABC154) was induced by culturing the cells in galactose containing medium for 6 h. Subcellular fractions isolated by differential centrifugation were analyzed by SDS-PAGE gel. LITAF was blotted with FLAG antibody, whereas anti-Pma1p antibody, anti-Dpm1p and anti-Pep12p antibody were used to detected plasma membrane marker protein, ER marker protein and endosome/Golgi marker protein, respectively (8A). Wild type yeast cells expressing LITAF were fixed with formaldehyde, converted to spheroplasts with lyticase, and stained with Rhodamine X-Red. Nuclei were visualized by counterstaining with DAPI. Cell morphology is shown in DIC.
Intracellular localization of LITAF was also examined using indirect immunostaining and confocal fluorescence microscopy (Fig. 8B). The results demonstrate that LITAF was expressed diffusely throughout the cell, and was found around the periphery of the cell. Taken together, the results in Fig. 8 indicate that LITAF is a membrane protein, and is associated with the membrane of intracellular organelles and with the plasma membrane (Fig. 8B).
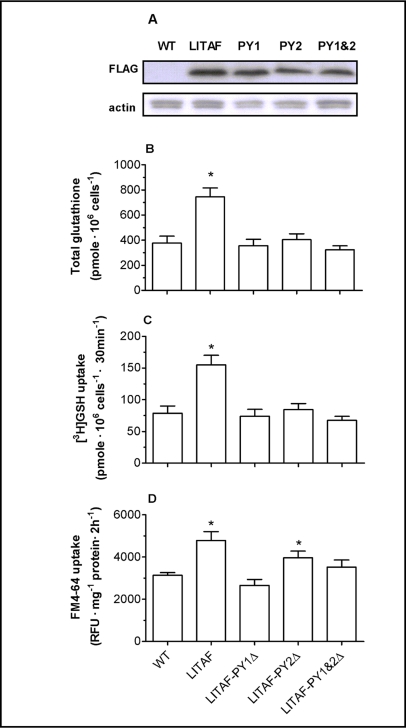
Mutation of the two predicted PY motifs of LITAF largely abolishes the ability of this protein to increase intracellular glutathione levels, and to stimulate [3H]GSH and FM4-64 uptake
To examine the functional significance of the PY motifs in these genes, LITAF was selected for mutational analysis. The two predicted PY motifs of LITAF were mutated from PPSY to PPSA, and wild type yeast cells were transformed with both wild type LITAF and three LITAF mutants. Mutation of these two PY motifs had only minimal effects on the expression level of LITAF protein (Fig. 9A); however, they essentially abolished the ability of LITAF to increase intracellular glutathione levels, and to stimulate [3H]GSH and FM4-64 uptake (Fig. 9B-D). As illustrated in figure 9, wild type cells transformed with LITAF had higher intracellular glutathione levels, and enhanced [3H]GSH and FM4-64 uptake, compared with untransformed cells, consistent with results obtained from hgt1-Δ cells transformed with LITAF (Figs. 3, 4 and 6) suggesting that the functions of LITAF are not specifically to complement the HGT1 deficiency, but are more likely to regulate membrane turnover by an common mechanism. Intracellular glutathione levels, [3H]GSH uptake rates and FM4-64 uptake rates of wild type cells transformed with LITAF mutants were generally comparable to those of control cells (wild type cells transformed with empty vector pESC-URA), indicating that these effects of LITAF in yeast depend on its PY motifs. The subcellular localization of these mutants in yeasts was also examined, however no differences were detected using indirect immunofluorescence (data not shown).
Fig. 9.
PY motif mutations largely abolish the effect of LITAF on intracellular GSH levels, and on [3H]GSH and FM4-64 uptake. PPSY motifs in LITAF were changed to PPSA by site-directed mutagenesis. Wild type strain ABC154 was transformed with empty vector, LITAF, or LITAF with PY mutations and whole cell lysate of each transformant was subjected to SDS-PAGE gel. The blot was probed with anti-FLAG antibody for LITAF expression, and with anti-actin for loading control (9A). Values are given as means ± SEM, N=3-6. * Significantly different intracellular glutathione concentration (9B), [3H]GSH uptake (9C) and FM4-64 uptake (9D) when compared to wild type cells transformed with the empty pESC-URA vector (p<0.05).
Expression of mouse Ndfip2 (N4WBP5) has no effect on [3H]GSH uptake in either the wild type or the hgt1-Δ yeast strain
To examine whether another PY-containing protein has similar effects on [3H]GSH uptake in yeast cells, mouse Ndfip2 (Nedd4 family interacting protein 2) was expressed in both the wild type yeast strain and the hgt1-Ä strain. [3H]GSH uptake was measured for 30 min in yeast cells transformed with the pESC-URA vector or Ndfip2 after being grown overnight in a sulfur-deficient medium that contained galactose. However, the expression of Ndfip2 had no effect on [3H]GSH uptake in either strain (data not shown), suggesting that the transport activity of the endogenous yeast low affinity GSH uptake mechanism is not modulated by Ndfip2.
Discussion
Bourbouloux et al. [5] previously demonstrated that Hgt1p is quantitatively the major GSH uptake transporter in S. cerevisiae. Without this transporter, the cells have minimal ability to take up GSH from the culture medium and show marked growth retardation when GSH is the only sulfur source. However, hgt1-Δ cells do retain some ability to take up GSH at a slow rate (e.g., see Fig. 4A), and to grow slowly using GSH as a sulfur source (Fig. 2). Thus, yeast cells have an alternate mechanism for taking up GSH, although this alternative mechanism has not been identified at the molecular level and has not been characterized biochemically.
The present study identifies five human genes, PMEPA1, LAPTM4α, SLC25A1, LITAF, and CYYR1 that can functionally complement the growth defect of hgt1-Δ cells cultured in a medium in which GSH is the only sulfur source. Each of these genes encodes a protein containing PY motifs that are thought to be important for regulating protein cell surface expression via interacting with the ubiquitin ligases NEDD4/Rsp5p [14, 17]. The first PY motif was identified through a functional cDNA library screen for putative ligands of the WW domain of the Yes-associated protein [18]. The presence of these motifs allows the PY motif-containing proteins to bind to WW domains in target proteins, including the E3 ubiquitin ligase NEDD4 [19]. NEDD4 protein belongs to the Hect-domain family of E3 ubiquitin-protein ligase. A major function of such proteins is to down-regulate the cell surface expression of membrane proteins via ubiquitin-dependent endocytosis, followed by degradation in both proteasomes and lysosomes [14, 20]. Mutations in the PY motifs of the epithelial Na+ channel disrupt its interaction with NEDD4, which leads to constitutively increased channel activity and hypertension in the Liddle syndrome [21]. Rsp5p is the unique yeast homologue of NEDD4. The direct interaction between Rsp5p and its target protein through PY-WW binding is essential for multiple-vesicular body targeting and endosomal sorting of several yeast membrane proteins [22, 23] Indeed, some studies have also indicated possible associations of NEDD4 with PMEPA1 [24], LAPTM4α [25], and LITAF [26]. Regulation of plasma membrane protein turnover by this mechanism may either occur directly, by binding of Rsp5p/NEDD4 to specific membrane proteins that contain PY motifs, or indirectly, by binding of Rsp5p/NEDD4 to PY-containing adapter molecules that then target the ubiquitin ligase activity to specific plasma membrane proteins [17, 27].
The present results are consistent with a model in which these five human genes enhance GSH uptake by increasing the activity of an undefined yeast low affinity GSH uptake mechanism [5, 6]. As illustrated in figure 5, the enhanced [3H]GSH uptake in cells expressing the five human genes was uniformly inhibited by high concentrations of unlabeled GSH, GSSG, and ophthalmic acid, suggesting that the effects of these genes on [3H]GSH uptake involves a common low affinity uptake pathway. Although the mechanism by which these genes enhance the activity of this alternate mode of GSH uptake is unknown, they may do so by increasing production, intracellular targeting, or the stability of components of the uptake machinery. One possibility is that expression of the PY-containing genes titrates the Rsp5p-associated degradation pathway away from some endogenous yeast transporter, thereby enhancing its activity. The enhanced turnover of general endocytotic markers detected in cells expressing the PY-containing genes might then reflect increased turnover of membrane proteins via an alternative latrunculin-A-insensitive pathway.
By analogy, the PY-containing protein Ndfip2 appears to increase cell surface expression of the amiloride-sensitive epithelial sodium channel (ENaC) by interfering with the Nedd4-mediated regulation of ENaC [28]. Ndfip2 contains three transmembrane domains and two amino terminal PPxY motifs, which physically interact with the WW domains of Nedd4 family proteins. However, this stimulatory effect of Ndfip2 on ENaC activity appears to be relatively selective, as Ndfip1, a highly related protein that also contains PY motifs, has no effect on ENaC activity [28]. In addition, Ndfip2 did not increase the transport activity of CFTR or Kir1. 1a, indicating selectivity for the target proteins. In the present study, expression of Ndfip2 in yeast cells did not produce stimulatory effects in GSH uptake, but this negative result does not necessarily exclude the proposed model, as there are many possible reasons for the lack of effect, including differential specificity, sensitivity, and affinity for possible binding sites in yeast proteins.
Expression of the five genes also led to greater accumulation of the endocytotic markers lucifer yellow and FM4-64 in yeast. Whereas the basal accumulation of FM4-64 was inhibited by LAT-A, neither the higher rates of FM4-64 uptake nor the enhanced GSH uptake observed in cells expressing the five human genes was affected by LAT-A, indicating that the enhanced GSH uptake associated with expression of the human genes is not mediated by a LAT-A-sensitive endocytotic mechanism. Upon subcellular fractionation, LITAF was found in the P13000 fraction which primarily contains large organelles and the plasma membrane. Indirect immunostaining of LITAF resulted in a staining pattern characteristic of localization to the plasma membrane and intracellular membranes. Mutation of the two PY motifs of LITAF resulted in loss of the observed effects, indicating that this motif plays an essential role in the effects of LITAF expression in yeast.
To date, little is known about the biological functions of the five genes identified in the present study. SLC25A1 is mapped to chromosomal region 22q11, and micro-deletions in this region are associated with DiGeorge syndrome, velo-cardio-facial syndrome and conotruncal anomaly face syndrome [29]. Although it is not known whether SLC25A1 plays a role in GSH homeostasis, some members of SLC25A family may contribute to mitochondrial GSH uptake. In particular, the dicarboxylate (SLC25A10) and 2-oxoglutarate (SLC25A11) carriers of the inner mitochondrial membrane are believed to catalyze uptake of GSH into the matrix through an exchange mechanism [30]. The three members of the lysosomal-associated protein transmembrane (LAPTM) family, LAPTM4α, LAPTM4β and LAPTM5 are thought to function as transporters or channels, although their precise function is unknown. When expressed in a drug-sensitive strain of S. cerevisiae, mouse LAPTM4α regulates intracellular compartmentalization of amphipathic solutes and confers cellular resistance or hypersensitivity to a range of drugs [31]. Both CYYR1 and PMEPA1 have been implicated in human diseases, although their functions are also unknown. CYYR1 is expressed in a broad range of human tissues, and the central cysteine-tyrosine-rich domain is strongly conserved from lower vertebrates to humans [32]. Alternative splicing and expression of the CYYR1 transcript has been reported in human neuroendocrine tumors [33]. PMEPA1 was first described as an androgen-induced gene. It is widely expressed in normal and solid malignant tissues, with the highest expression in prostate tissue [34]. The expression of PMEPA1 is increased in primary and metastasized colon tumors and several other solid tumors [35, 36], whereas it is decreased in prostate tumor tissues [34]. In addition, PMEPA1 is also induced by tumor growth factor β [36]. LITAF is present in most human tissues and its expression can be induced by multiple pathways, including TNFα- and p53-activated pathways [16, 37, 38]. Elevated mRNA and protein expression of LITAF has been found in intestinal tissues from patients with Crohn's disease and ulcerative colitis [39, 40]. In addition, mutations of LITAF cause Charcot-Marie-Tooth disease type 1C (CMT1C), an autosomal dominant demyelinating peripheral neuropathy [41]. Although the etiology of CMT1C is not understood yet, a role of LITAF in modulating protein trafficking and turnover has been proposed to be involved [26, 42].
In summary, the present study identifies five human genes that rescue growth of a HGT1-deficient yeast strain, and provides clues into the potential function of these genes. Currently, the functions of the five genes are unknown, although four of them have been implicated in human diseases [35, 39, 41, 43, 44, 45]. A better understanding of their physiological functions may be expected to clarify the etiology and pathogenesis of these diseases.
Abbreviations
CYYR1 (cysteine/tyrosine-rich1); FM4-64 (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl)hexatrienyl) pyridinium dibromide); GSH (reduced glutathione); LAPTM4α (lysosomal-associated protein transmembrane 4 alpha); LITAF (lipopolysaccharide-induced TNF factor); Ndfip2 (Nedd4 family interacting protein 2); NEDD4 (neural precursor cell expressed, developmentally down-regulated 4); PY (xPPxY, P = Pro, Y = Tyr, x = any amino acid); RSP5 (reverses Spt-phenotype); SLC25A1 (solute carrier family 25, member 1); PMEPA1 (prostate transmembrane protein, androgen induced 1).
Acknowledgements
This work was supported in part by National Institutes of Health (NIH) Grants DK048823 and DK067214, and National Institute of Environmental Health Sciences Center Grant ES01247 and Training Grant ES07026. We thank Dr. Anand K. Bachhawat (Institute of Microbial Technology, Chandirgarh, India) for providing the hgt1-Δ S. cerevisiae strain, Dr. Stephen G. Oliver (University of Manchester, Manchester, UK) for providing the human mammary gland cDNA library, Dr. David A. Pearce (University of Rochester, Rochester, New York) for sharing anti-Dpm1p antibody, Dr. Zhifeng Cui for his advice in this project, and David McMillan for technical assistance.
References
- 1.Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol Chem. 2009;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 3.Ballatori N, Hammond CL, Cunningham JB, Krance SM, Marchan R. Molecular mechanisms of reduced glutathione transport: Role of the MRP/CFTR/ABCC and OATP/SLC21A families of membrane proteins. Toxicol Appl Pharmacol. 2005;204:238–255. doi: 10.1016/j.taap.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Ballatori N, Krance SM, Marchan R, Hammond CL. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol Aspects Med. 2008;30:13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourbouloux A, Shahi P, Chakladar A, Delrot S, Bachhawat AK. Hgt1p, a high affinity glutathione transporter from the yeast saccharomyces cerevisiae. J Biol Chem. 2000;275:13259–13265. doi: 10.1074/jbc.275.18.13259. [DOI] [PubMed] [Google Scholar]
- 6.Miyake T, Hazu T, Yoshida S, Kanayama M, Tomochika K, Shinoda S, Ono B. Glutathione transport systems of the budding yeast saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1998;62:1858–1864. doi: 10.1271/bbb.62.1858. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Osborn M, Gitsham P, Yen K, Miller, Oliver SG. Using yeast to place human genes in functional categories. Gene. 2003;303:121–129. doi: 10.1016/s0378-1119(02)01142-3. [DOI] [PubMed] [Google Scholar]
- 8.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 9.Baker MA, Cerniglia GJ, Zaman A. Microtiter plate assay for the measurement of glutathione and glutathione disulfide in large numbers of biological samples. Anal Biochem. 1990;190:360–365. doi: 10.1016/0003-2697(90)90208-q. [DOI] [PubMed] [Google Scholar]
- 10.Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H. Yeast endocytosis assays. Methods Enzymol. 1991;194:697–710. doi: 10.1016/0076-6879(91)94051-d. [DOI] [PubMed] [Google Scholar]
- 11.Emans N, Zimmermann S, Fischer R. Uptake of a fluorescent marker in plant cells is sensitive to brefeldin A and wortmannin. Plant Cell. 2002;14:71–86. doi: 10.1105/tpc.010339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rieder SE, Emr SD. Overview of subcellular fractionation procedures for the yeast saccharomyces cerevisiae. Curr Protoc Cell Biol. 2001;Chapter 3 doi: 10.1002/0471143030.cb0307s07. Unit 3.7. [DOI] [PubMed] [Google Scholar]
- 13.Pringle, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 14.Rotin D, Staub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: Role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J Membr Biol. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- 15.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moriwaki Y, Begum NA, Kobayashi M, Matsumoto M, Toyoshima K, Seya T. Mycobacterium bovis bacillus calmette-guerin and its cell wall complex induce a novel lysosomal membrane protein, SIMPLE, that bridges the missing link between lipopolysaccharide and p53-inducible gene, LITAF(PIG7), and estrogen-inducible gene, EET-1. J Biol Chem. 2001;276:23065–23076. doi: 10.1074/jbc.M011660200. [DOI] [PubMed] [Google Scholar]
- 17.Staub O, Abriel H, Plant P, Ishikawa T, Kanelis V, Saleki R, Horisberger JD, Schild L, Rotin D. Regulation of the epithelial na+ channel by Nedd4 and ubiquitination. Kidney Int. 2000;57:809–815. doi: 10.1046/j.1523-1755.2000.00919.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen HI, Sudol M. The WW domain of yes-associated protein binds a proline-rich ligand that differs from the consensus established for src homology 3-binding modules. Proc Natl Acad Sci U S A. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial na+ channel deleted in liddle's syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey KF, Kumar S. Nedd4-like proteins: An emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- 21.Abriel H, Loffing J, Rebhun JF, Pratt JH, Schild L, Horisberger JD, Rotin D, Staub O. Defective regulation of the epithelial na+ channel by Nedd4 in liddle's syndrome. J Clin Invest. 1999;103:667–673. doi: 10.1172/JCI5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–1296. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharya S, Zoladek T, Haines DS. WW domains 2 and 3 of Rsp5p play overlapping roles in binding to the LPKY motif of Spt23p and Mga2p. Int J Biochem Cell Biol. 2008;40:147–157. doi: 10.1016/j.biocel.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu LL, Shi Y, Petrovics G, Sun C, Makarem M, Zhang W, Sesterhenn IA, McLeod DG, Sun L, Moul JW, Srivastava S. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res. 2003;63:4299–4304. [PubMed] [Google Scholar]
- 25.Pak Y, Glowacka WK, Bruce MC, Pham N, Rotin D. Transport of LAPTM5 to lysosomes requires association with the ubiquitin ligase Nedd4, but not LAPTM5 ubiquitination. J Cell Biol. 2006;175:631–645. doi: 10.1083/jcb.200603001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirk AJ, Anderson SK, Hashemi SH, Chance PF, Bennett CL. SIMPLE interacts with NEDD4 and TSG101: Evidence for a role in lysosomal sorting and implications for charcot-marie-tooth disease. J Neurosci Res. 2005;82:43–50. doi: 10.1002/jnr.20628. [DOI] [PubMed] [Google Scholar]
- 27.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Konstas AA, Shearwin-Whyatt LM, Fotia AB, Degger B, Riccardi D, Cook DI, Korbmacher C, Kumar S. Regulation of the epithelial sodium channel by N4WBP5A, a novel Nedd4/Nedd4-2-interacting protein. J Biol Chem. 2002;277(33):29406–29416. doi: 10.1074/jbc.M203018200. [DOI] [PubMed] [Google Scholar]
- 29.Goldmuntz E, Wang Z, Roe BA, Budarf ML. Cloning, genomic organization, and chromosomal localization of human citrate transport protein to the DiGeorge/velocardiofacial syndrome minimal critical region. Genomics. 1996;33:271–276. doi: 10.1006/geno.1996.0191. [DOI] [PubMed] [Google Scholar]
- 30.Lash LH. Mitochondrial glutathione transport: Physiological, pathological and toxicological implications. Chem Biol Interact. 2006;163:54–67. doi: 10.1016/j.cbi.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogue DL, Kerby L, Ling V. A mammalian lysosomal membrane protein confers multidrug resistance upon expression in saccharomyces cerevisiae. J Biol Chem. 1999;274:12877–12882. doi: 10.1074/jbc.274.18.12877. [DOI] [PubMed] [Google Scholar]
- 32.Vitale L, Casadei R, Canaider S, Lenzi L, Strippoli P, D'Addabbo P, Giannone S, Carinci P, Zannotti M. Cysteine and tyrosine-rich 1 (CYYR1), a novel unpredicted gene on human chromosome 21 (21q21.2), encodes a cysteine and tyrosine-rich protein and defines a new family of highly conserved vertebrate-specific genes. Gene. 2002;290:141–151. doi: 10.1016/s0378-1119(02)00550-4. [DOI] [PubMed] [Google Scholar]
- 33.Vitale L, Frabetti F, Huntsman SA, Canaider S, Casadei R, Lenzi L, Facchin F, Carinci P, Zannotti M, Coppola D, Strippoli P. Sequence, “subtle” alternative splicing and expression of the CYYR1 (cysteine/tyrosine-rich 1) mRNA in human neuroendocrine tumors. BMC Cancer. 2007;7:66. doi: 10.1186/1471-2407-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu LL, Shanmugam N, Segawa T, Sesterhenn IA, McLeod DG, Moul JW, Srivastava S. A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics. 2000;66:257–263. doi: 10.1006/geno.2000.6214. [DOI] [PubMed] [Google Scholar]
- 35.Rae FK, Hooper JD, Nicol DL, Clements JA. Characterization of a novel gene, STAG1/PMEPA1, upregulated in renal cell carcinoma and other solid tumors. Mol Carcinog. 2001;32:44–53. doi: 10.1002/mc.1063. [DOI] [PubMed] [Google Scholar]
- 36.Brunschwig EB, Wilson K, Mack D, Dawson D, Lawrence E, Willson JK, Lu S, Nosrati A, Rerko RM, Swinler S, Beard L, Lutterbaugh JD, Willis J, Platzer P, Markowitz S. PMEPA1, a transforming growth factor-beta-induced marker of terminal colonocyte differentiation whose expression is maintained in primary and metastatic colon cancer. Cancer Res. 2003;63:1568–1575. [PubMed] [Google Scholar]
- 37.Myokai F, Takashiba S, Lebo R, Amar S. A novel lipopolysaccharide-induced transcription factor regulating tumor necrosis factor alpha gene expression: Molecular cloning, sequencing, characterization, and chromosomal assignment. Proc Natl Acad Sci U S A. 1999;96:4518–4523. doi: 10.1073/pnas.96.8.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baumann B, Seufert J, Rolf O, Jakob F, Goebel S, Eulert J, Rader CP. Upregulation of LITAF mRNA expression upon exposure to TiAlV and polyethylene wear particles in THP-1 macrophages. Biomed Tech (Berl) 2007;52:200–207. doi: 10.1515/BMT.2007.037. [DOI] [PubMed] [Google Scholar]
- 39.Stucchi A, Reed K, O'Brien M, Cerda S, Andrews C, Gower A, Bushell K, Amar S, Leeman S, Becker J. A new transcription factor that regulates TNF-alpha gene expression, LITAF, is increased in intestinal tissues from patients with CD and UC. Inflamm Bowel Dis. 2006;12:581–587. doi: 10.1097/01.MIB.0000225338.14356.d5. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Bennett CL. Litaf/Simple protein is increased in intestinal tissues from patients with CD and UC, but is unlikely to function as a transcription factor. Inflamm Bowel Dis. 2007;13:120–121. doi: 10.1002/ibd.20010. [DOI] [PubMed] [Google Scholar]
- 41.Street VA, Bennett CL, Goldy JD, Shirk AJ, Kleopa KA, Tempel BL, Lipe HP, Scherer SS, Bird TD, Chance PF. Mutation of a putative protein degradation gene LITAF/SIMPLE in charcot-marie-tooth disease 1C. Neurology. 2003;60:22–26. doi: 10.1212/wnl.60.1.22. [DOI] [PubMed] [Google Scholar]
- 42.Saifi GM, Szigeti K, Wiszniewski W, Shy ME, Krajewski K, Hausmanowa-Petrusewicz I, Kochanski A, Reeser S, Mancias P, Butler I, Lupski SIMPLE mutations in charcot-marie-tooth disease and the potential role of its protein product in protein degradation. Hum Mutat. 2005;25:372–383. doi: 10.1002/humu.20153. [DOI] [PubMed] [Google Scholar]
- 43.Matsumura Y, Matsumura Y, Nishigori C, Horio T, Miyachi Y. PIG7/LITAF gene mutation and overexpression of its gene product in extramammary paget's disease. Int J Cancer. 2004;111:218–223. doi: 10.1002/ijc.20251. [DOI] [PubMed] [Google Scholar]
- 44.Giannini G, Ambrosini MI, Di Marcotullio L, Cerignoli F, Zani M, MacKay AR, Screpanti I, Frati L, Gulino A. EGF-and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol Carcinog. 2003;38:188–200. doi: 10.1002/mc.10162. [DOI] [PubMed] [Google Scholar]
- 45.Williams NM, Spurlock G, Norton N, Williams HJ, Hamshere ML, Krawczak M, Kirov G, Nikolov I, Georgieva L, Jones S, Cardno AG, O'Donovan MC, Owen MJ. Mutation screening and LD mapping in the VCFS deleted region of chromosome 22q11 in schizophrenia using a novel DNA pooling approach. Mol Psychiatry. 2002;7:1092–1100. doi: 10.1038/sj.mp.4001188. [DOI] [PubMed] [Google Scholar]
- 46.Hogue DL, Ellison MJ, Young JD, Cass CE. Identification of a novel membrane transporter associated with intracellular membranes by phenotypic complementation in the yeast saccharomyces cerevisiae. J Biol Chem. 1996;271:9801–9808. doi: 10.1074/jbc.271.16.9801. [DOI] [PubMed] [Google Scholar]
- 47.Cabrita MA, Hobman TC, Hogue DL, King KM, Cass CE. Mouse transporter protein, a membrane protein that regulates cellular multidrug resistance, is localized to lysosomes. Cancer Res. 1999;59:4890–4897. [PubMed] [Google Scholar]
- 48.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, Beltran E, Agirre X, Marugan I, Marin M, Rosenwald A, Sugimoto KJ, Wheat LM, Karran EL, Garcia JF, Sanchez L, Prosper F, Staudt LM, Pinkel D, Dyer MJ, Martinez-Climent JA. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. [DOI] [PubMed] [Google Scholar]
- 49.Bennett CL, Shirk AJ, Huynh HM, Street VA, Nelis E, Van Maldergem L, De Jonghe P, Jordanova A, Guergueltcheva V, Tournev I, Van Den Bergh P, Seeman P, Mazanec R, Prochazka T, Kremensky I, Haberlova J, Weiss MD, Timmerman V, Bird TD, Chance PF. SIMPLE mutation in demyelinating neuropathy and distribution in sciatic nerve. Ann Neurol. 2004;55:713–720. doi: 10.1002/ana.20094. [DOI] [PubMed] [Google Scholar]
- 50.Cardillo MR, Di Silverio F. Prostate-specific G protein couple receptor genes and STAG1/PMEPA1 in peripheral blood from patients with prostatic cancer. Int J Immunopathol Pharmacol. 2006;19:871–878. doi: 10.1177/039463200601900416. [DOI] [PubMed] [Google Scholar]
- 51.Stoffel M, Karayiorgou M, Espinosa R, 3rd, Beau MM. The human mitochondrial citrate transporter gene (SLC20A3) maps to chromosome band 22q11 within a region implicated in DiGeorge syndrome, velo-cardio-facial syndrome and schizophrenia. Hum Genet. 1996;98:113–115. doi: 10.1007/s004390050169. [DOI] [PubMed] [Google Scholar]
- 52.Heisterkamp N, Mulder MP, Langeveld A, ten Hoeve J, Wang Z, Roe BA, Groffen J. Localization of the human mitochondrial citrate transporter protein gene to chromosome 22Q11 in the DiGeorge syndrome critical region. Genomics. 1995;29:451–456. doi: 10.1006/geno.1995.9982. [DOI] [PubMed] [Google Scholar]