Abstract
DNA polymerase η (Polη) is unique among eukaryotic polymerases in its proficient ability for error-free replication through UV induced cyclobutane pyrimidine dimers, and inactivation of Polη in humans causes the variant form of xeroderma pigmentosum (XPV). We present the crystal structures of yeast Polη in ternary complex with a cis-syn thymine-thymine (T-T) dimer and with undamaged DNA. The structures reveal that the ability of Polη to efficiently replicate through the UV-induced lesion derives from a simple and yet elegant mechanism, wherein the two Ts of the T-T dimer are accommodated in an active site cleft that is much more open than in other polymerases. We also show by structural, biochemical and genetic analysis that the two Ts are maintained in a stable configuration in the active site via interactions with Gln55, Arg73, and Met74. Together, these features define the basis for Polη's action on UV damaged DNA that is crucial in suppressing the mutagenic and carcinogenic consequences of sun exposure, reducing thereby the incidence of skin cancers in humans.
Mutations in DNA polymerase η (Pol) are responsible for the variant form of xeroderma pigmentosum (XP-V)1, 2. XP-V patients are sensitive to UV radiation and they suffer from a high incidence of sunlight induced skin cancers3–6. The high incidence of skin cancers in these patients is due to the absence of functional Polη which has the remarkable ability to replicate through UV induced cyclobutane pyrimidine dimers (CPDs) such as a cis-syn thymine-thymine (T-T) dimer in an error-free way7. Unlike classical DNA polymerases, or any of the other three eukaryotic Y family polymerases - Polκ, Polι and Rev1 (Supplementary Figure 1) that become stalled at the UV induced T-T dimer, Polη can efficiently and accurately replicate past this DNA lesion8. The specificity of Polη poses two questions. First, what is the structural basis of Polη's ability to promote error-free replication through a cis-syn T-T dimer? Second, does the enzyme use the same mechanism for inserting a nucleotide opposite the thymine dimer as opposite an undamaged base? To address these longstanding questions we present here the structures of the catalytic core of yeast Polη (residues 1–513) in ternary complex with an incoming dATP and a template-primer presenting the 3'T of the T-T dimer in the active site, and of yeast Polη in ternary complex with an incoming dATP and undamaged DNA.
Crystallization and structure determination
Compared to other eukaryotic Y family polymerases, crystallization of Polη with DNA has proved very difficult because the crystals obtained have always been of the same form as the previously reported apoenzyme crystals (apo)9, and they reveal a Polη structure that is very similar to the apo form and which contains either no DNA, or DNA that is disordered and/or only partially bound (Supplementary Figure 2). In these “apo-like” crystals, there are multiple protein contacts between the “palm” domain and ”polymerase associated domain” (PAD) of one Polη molecule and palm/PAD of another molecule in the asymmetric unit, which cause dimer formation within the crystals (Supplementary Figure 3a). Amongst these, Lys140 and Ser144 of one molecule, which lie at the base of the palm domain, make hydrogen bonds or van der Waals contacts with residues Tyr199, Phe194, and Asn167 on the palm of a symmetry related molecule (Supplementary Figure 3b). We surmised that these contacts were inhibitory to DNA binding by Polη in the crystal, and to break the apo crystal contacts, we mutated Lys140 to alanine and Ser144 to tryptophan. Using the Polη K140A S144W protein, we were successful in obtaining Polη:DNA cocrystals that diffracted to high resolution, belonged to a different space group, and possessed unit cell dimensions that were different from the apoenzyme (Supplementary Table 1). To ensure that the crystal contact mutations did not alter the biological function or the biochemical activity of Polη we verified that the rad30 K140A, S144W mutant gene fully restores the UV resistance and reduces the frequency of UV induced mutations of the rad30Δ strain, identical to that of the wild type gene (Supplementary Figure 4). Also, the K140A S144W protein resembles the wild type protein in DNA synthetic activity on both undamaged and T-T dimer-containing DNAs (Supplementary Figure 5).
Overall structure
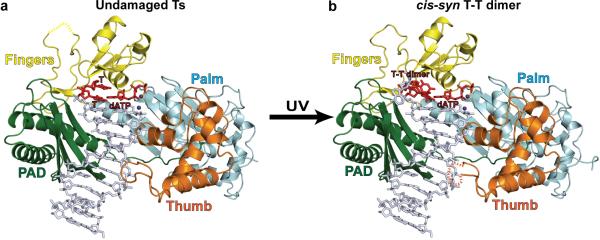
In both the structures with undamaged or damaged DNAs, Polη embraces the template-primer with its palm (residues 1–30 and 130–286), fingers (residues 34–127), and thumb (residues 289–378) as well as the polymerase associated domain (PAD; residues 395–509) unique to Y-family polymerases (Fig.1). The palm carries the active site residues, Asp30, Asp155 and Glu156, which catalyze the nucleotidyl transfer reaction10 (Fig. 2a). In both structures, incoming dATP is bound with its triphosphate moiety interlaced between the fingers and palm domain, making identical hydrogen bonds with Tyr64 and Arg67 from the fingers domain and Lys279 from the palm domain, and its sugar packed against the aromatic ring of Phe35 (Fig. 2a). The catalytic residues Asp30, Asp155 and Glu156 are arrayed between the dATP triphosphate moiety and the primer terminus, and two Mg2+ ions – analogous to metals “A” and “B” in replicative DNA polymerases11–13 – complete the Polη active site (Fig. 2a). Thus, Polη is well poised for dATP insertion in both structures: opposite the 3'T of the UV-induced cis-syn T-T dimer as well as opposite undamaged T (Figs. 1 & 2a). The putative 3'oxygen (at the primer terminus) is located ~3.3Å from the dATP α-phosphate and is aligned more or less linearly with respect to the Pα-O3' bond (angle of about 155°) in each structure.
Figure 1.
Polη-DNA-dATP ternary complexes. a, Structure of Polη with undamaged DNA. The palm, fingers, thumb domains and the PAD are shown in cyan, yellow, orange and green, respectively. The DNA is in gray, and the putative Mg2+ ions are in dark blue. The templating 3'T (and the 5'T) and incoming dATP are in red. b, Structure of Polη with a cis-syn T-T dimer in DNA. The cis-syn T-T dimer and incoming dATP are shown in red. Yellow and orange dashed lines depict unstructured loops in the fingers and thumb domains, respectively.
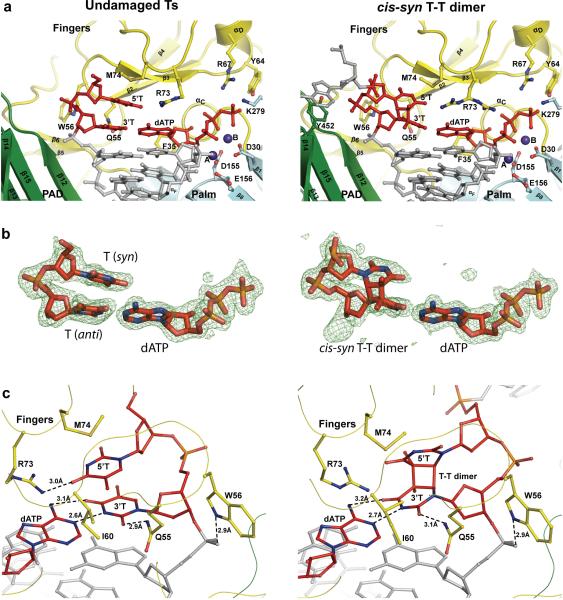
Figure 2.
Close-up views of the active site regions. a, The active site regions with undamaged (left) and T-T dimer-containing (right) DNAs. The palm and fingers domains and the PAD are shown in cyan, yellow, and green, respectively. The DNA is colored gray, and the putative Mg2+ ions (A and B) are dark blue. The undamaged templating T (and the 5'T) (left), the cis-syn T-T dimer (3'T and 5'T), and incoming dATP are in red. Highlighted and labeled are the catalytic residues (D30, D155, and E156), residues that interact with the triphosphate moiety of incoming dATP (Y64, R67, and K279), residues that interact with templating T and the 5'T (Q55, W56, M74, and R73), and F35, which stacks against the dATP sugar. R73 is shown in two orientations in the T-T dimer structure. Y452 is also shown (right), which stacks against the base of the nucleotide 5' to the T-T dimer. The residues are colored to match the domain they belong to. b, Simulated annealing Fo-Fc omit maps (contoured at 3.0σ) of undamaged templating T (and the 5'T) and incoming dATP (left), and cis-syn T-T dimer and incoming dATP (right). c, Hydrogen bonding and van der Waals interactions between residues on the fingers domain (Q55, W56, I60, R73 and M74) and undamaged Ts (left) and cis-syn T-T dimer (right).
The thumb and the PAD straddle the duplex portion of the template-primer, connected by a long linker that cradles one side of the DNA (Fig. 1). The thumb skims the DNA minor groove surface, while the PAD is anchored in the major groove on the opposite side. The duplex portion of the template primer has a B-DNA-like conformation in both structures, with average helical twist and rise values of ~34° and 3.2Å, respectively, and the phosphates of most nucleotides are in the standard BI configuration.
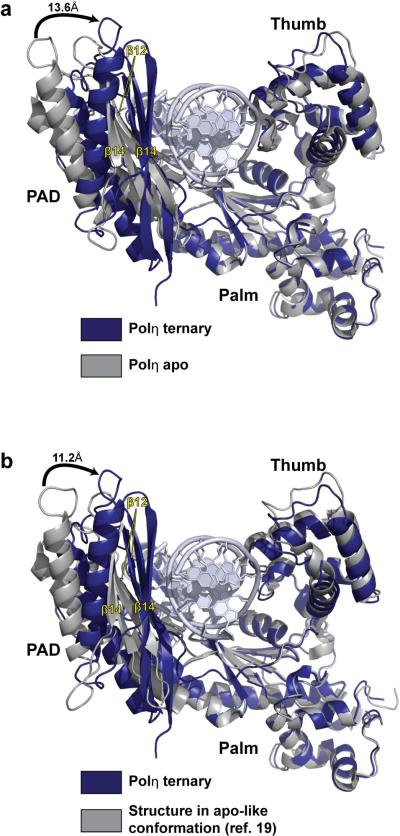
Strikingly, in both structures, the PAD moves by as much as 13.6Å from its position in the apo Polη structure9, making multiple contacts within the DNA major groove (Figs. 3a & Supplementary Figure 6). Amongst these contacts is a series of hydrogen bonds between the main chain amides of the outer β-strands (β12 and β14) of the PAD and the template and primer strands; including, for example, hydrogen bonds between the main chain amides of Ser394, Met396, Asn398, Asn400, and Ser458 and the phosphates of nucleotides T4, T3, T2, T1, and P7 (where T and P refer to template and primer strands, respectively, and the subscripts refer to the number of base pairs from the templating base position), respectively (Supplementary Figure 6). These PAD:DNA hydrogen bonds are conserved in almost all Y-family polymerase ternary structures14–18, but some or all are absent in the structures of yeast Polη with cisplatin adducts19. This is likely because the reported cocrystals (3.1–3.3Ǻ resolution) belong to the same space group and have similar unit cell dimensions as the apo Polη crystals9, and thus the PAD in these structures is constrained to a position similar to that in the apo Polη structure (Fig. 3b). The inability of the PAD to “close” around the DNA in these crystals, due to crystal contacts (Supplementary Figure 7), may explain the observation of more than one DNA orientation in these structures19. The thumb also rotates towards the DNA, but to a much lesser extent than the PAD (Fig. 3A). Together, the palm, fingers, and thumb domains, and the PAD bury ~2500Å2 of solvent accessible surface area at the Polη-DNA-dNTP interface.
Figure 3.
Conformational changes in Polη ternary complex. a, Conformational change in Polη upon complex formation. Upon DNA and dNTP binding, the PAD swings by as much as ~13.6A towards the major groove of the DNA. The thumb also rotates towards the DNA but to a much lesser extent than the PAD. b, Comparison between our Polη ternary complex and that reported by Alt et al between Polη and a cisplatin adduct19. In the Alt et al. structure, Polη has an apo-like conformation (cf Figure 3a), wherein the PAD occupies a position similar to that in the apo Polη structure
An open active site cleft
The Polη active site cleft is uniquely adapted to accommodate a cis-syn T-T dimer (Fig. 2a). The Polη fingers domain lies further away from the templating base than in any other DNA polymerase, allowing for the two Ts of the T-T dimer to be accommodated unhindered within the open active site cleft (Figs. 1 & 2). The 3'T of the T-T dimer makes Watson-Crick (WC) hydrogen bonds with incoming dATP (Fig. 2b). The nascent base pair is highly propeller twisted (−25.1°) and appears to be stabilized in part by a hydrogen bond between O2 of 3'T and Nε2 of Gln55 from the fingers domain (Figs. 2a, b & c). The 5'T of the T-T dimer inclines towards the fingers domain and makes van der Waals contacts with Met74 and Arg73 from the loop between helices αC and αD on the fingers domain (Figs. 2a & c). The guanidinium group of Arg73 is relatively mobile and assumes two orientations, one interacting with the 5'T and the other with incoming dATP. Notably, the path of the T-T dimer through the active site is guided (in part) by the nucleotide 5' to the T-T dimer, which splays out of the active site and its base stacks above Tyr452 from the PAD (Fig. 2a).
Polη adopts a nearly identical configuration in the T-T dimer and undamaged TT structures, superimposing with an rmsd of 0.24Å (for 506 Cαs) (Fig. 1). The palm, fingers and thumb domains and the PAD are positioned almost identically with respect to the template-primer and incoming nucleotide. The two undamaged Ts are also accommodated in much the same way as a cis-syn T-T dimer, except that the two bases are coplanar and the 5'T lies deeper within the open active site cleft (Fig. 2). An unexpected difference between the two complexes is that the undamaged 5'T is in a syn conformation, primarily as the result of favorable van der Waals contacts between its C5 methyl group and Ile60 from the fingers domain (Fig. 2c). Correspondingly, the undamaged 5'T makes somewhat different contacts with Met74 and Arg73, wherein Met74 lies more directly over the base and Arg73 makes a hydrogen bond with the O4 atom (Figs. 2a & c). However, despite these differences in interactions with Arg73 and Met74, the mode of dATP binding and the active site geometry are indistinguishable in the two ternary complexes (Fig. 2a), and provide a basis for the ability of Polη to replicate UV-damaged and undamaged DNAs with the same efficiency and fidelity.
Biochemical and genetic analysis
To evaluate the contributions of the Gln55 (Q55), Arg73 (R73), and Met74 (M74) residues to Polη's ability to replicate through the T-T dimer, we examined the effects of the Q55A, R73A, and M74A mutations on Polη's DNA synthetic activity on undamaged and damaged DNAs. As shown in Supplementary Figure 8, while the R73A and M74A mutations have no effect on synthetic activity on undamaged or T-T dimer-containing DNA, the R73A M74A double mutation and the Q55A mutation displayed some loss in DNA synthetic activity on both DNAs, whereas the DNA synthetic activity of the Q55A R73A mutant protein was reduced very considerably on both the undamaged and damaged DNAs. As determined from steady state kinetic analyses while the combination of the R73A M74A mutations had no effect on the efficiency (kcat/Km) of nucleotide incorporation on undamaged DNA, the incorporation efficiency opposite the 3'T of the dimer was reduced ~ 8-fold (Supplementary Table 2). These results are in keeping with our observation that R73 and M74 interact somewhat differently with the undamaged and damaged DNA templates.
Hydrogen bonding of Q55 with the 3'T is important for Polη activity, as the Q55A mutation results in an ~ 15 fold loss in dATP incorporation opposite both the undamaged and damaged 3'T (Supplementary Table 2). Although the R73A mutation has no effect on dATP incorporation, the Q55A R73A double mutation confers an ~ 50-fold reduction in dATP incorporation opposite both the undamaged and damaged 3'T (Supplementary Table 2). We infer from these biochemical observations that Q55 and R73 play an important role in maintaining the 3'T and 5'T template residues in the Polη active site, regardless of whether the DNA is damaged or undamaged.
To examine the biological consequences of the Q55A, R73A, and M74A mutations in yeast cells, we examined their effects on UV sensitivity and UV mutability. Importantly, the Q55A R73A double mutation imparts a high degree of UV sensitivity and confers an increase in UV mutability (Supplementary Figure 9). The Q55A and the R73A M74A mutations also cause increased UV sensitivity and UV hypermutability; however, the biological effects of these mutations are less severe than those of the Q55A R73A double mutation. The R73A and M74A single mutations also affect Polη function in vivo, but to a lesser degree than the other mutations (Supplementary Figure 9). We conclude from the biochemical and biological data presented here that the Q55, R73, and M74 residues play an important role in maintaining both the residues of a pyrimidine dimer in a stable configuration in the Polη active site.
Comparison to Polκ
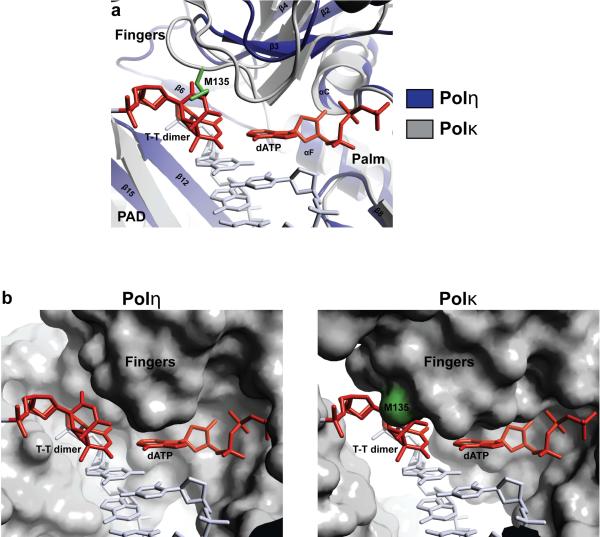
In contrast to Polη, the fingers domain in most DNA polymerases impinges on the templating base, so that only the templating base is held in the active site and the rest of the 5' unpaired template strand is directed out of the active site cleft20. And, since the two Ts of a cis-syn T-T dimer are covalently linked by the cyclobutane ring and cannot be separated, the active site clefts of these polymerases are not equipped to handle a T-T dimer. This is true for classical polymerases as well as other eukaryotic Y family polymerases15–18, 20. Fig. 4 shows the structure of Polκ18 superimposed onto that of Polη (via their palm domains). The Polκ fingers domain sterically overlaps with the 5'T of the T-T dimer, whereas the Polη fingers domain is displaced “upwards” and towards the minor groove side of the 5'T (by >5 Å), allowing both Ts of the T-T dimer to bind unhindered within the active site cleft (Fig. 4). The Polη fingers domain appears to be propped up in part by a short β-strand (β6; residues 123–127) that lies between the core of the fingers domain and the PAD (Figs. 2a, 4, & Supplementary Figure 10). Extensive van der Waals interactions between Val126 (β6) and Gln55/Trp56 (fingers) appear to draw the core of the fingers domains away from the templating base (Supplementary Figure 10).
Figure 4.
Comparison between Polη and Polκ. a, Polκ (gray) superimposed on the Polη ternary complex via the palm domain. In Polκ, the fingers domain is in close proximity to the cis-syn T-T dimer (red) and it collides with the 5'T of the T-T dimer. b, Left, molecular surface of Polη in the UV-damaged ternary complex. Both Ts of the T-T dimer fit unhindered within the active site cleft. Right, molecular surface of Polκ when superimposed on the Polη ternary complex via the palm domain. The 5'T of the T-T dimer collides with M135 and other residues of the Polκ fingers domain.
The fingers and palm domains are pre-oriented for dNTP binding and catalysis
Pre-steady state kinetic studies have suggested that Polη undergoes a rate-limiting conformational change step before the phosphoryl transfer of the incoming nucleotide to the primer terminus21. From the structures presented here, this rate-limiting step does not appear to correspond to a conformational change in the fingers domain, which occupies almost the same position with respect to the palm (and the active site residues) as in the apo-enzyme structure (Figs. 2a and Supplementary Figure 11). The most significant change in the fingers domain on DNA binding is the reorientation of Arg73 and Met74 side chains (and a portion of the loop carrying them), which swing towards the 5'T in both the undamaged and the damaged complexes (Supplementary Figure 11). On the other hand, the conformations of Tyr64 and Arg67 side chains that make hydrogen bonds with the triphosphate moiety of incoming dATP are almost the same as in the apo Polη structure; and, amongst the active site residues (Asp30, Asp155 and Glu156), Asp30 undergoes a change in rotamer in order to bind the catalytic metals (Supplementary Figure 11). Together, the structures reinforce the notion that the fingers and palm domains in Y-family polymerases are pre-oriented for dNTP binding and catalysis9, 18, 22, 23, and that most of the change in these domains corresponds to the local reorientation of some side chains.
We show here that the ability of Polη to efficiently replicate through a T-T dimer derives from a simple and yet elegant mechanism, wherein the two Ts of the T-T dimer are accommodated unhindered in an open active cleft. The “openness” of the active site cleft is the critical feature which distinguishes Polη from the classical polymerases, as well as other Y-family polymerases; and we show that the T-T dimer is stabilized in the active site by interactions with Gln55, Arg73, and Met74. Together, these structural features and interactions define the basis for Polη's action on UV damaged DNA that is crucial in suppressing the mutagenic and carcinogenic consequences of sun exposure in humans.
METHODS SUMMARY
For structure determinations, we overexpressed the catalytic core of yeast Polη (residues 1–513) containing the K140A and S144W mutations in E.coli. For biochemical analyses, Polη proteins were expressed in and purified from yeast. The DNA substrates for cocrystallization consisted of an 11-nt primer harboring a dideoxycytosine at its 3' end (5'-GTCCTCCCCTCdd-3') annealed to either an undamaged 16-nt template (5'-TAATTGAGGGGAGGAC-3') or a 16-nt template bearing a cis-syn T-T dimer (5'-TAATTGAGGGGAGGAC-3'). The ternary complex crystals with the T-T dimer (with incoming dATP) belong to space group C2221 with unit cell dimensions of a=88.1Å, b=227.0Å, and c=85.9Å. The ternary complex crystals with undamaged DNA similarly belong to space group C2221, with unit cell dimensions of a=88.8Å, b=227.9Å, and c=86.0Å. Both cocrystals contain one protein-DNA complex in the asymmetric unit. Importantly, these cocrystals are different from the “apo-like” crystals reported previously for the Polη apoenzyme9 (P41212; a = b= 105Å, c=292Å) and for structures of Polη with cisplatin adducts19 (P41212; a = b= 104Å, c=293Å). They also diffract to high resolution, namely 1.76Å resolution for the T-T dimer ternary complex and 2.0Å for the undamaged DNA ternary complex (Supplementary Table 1). The structures were solved by molecular replacement methods using the apo-Polη structure as a search model. The refined structures have excellent stereochemistry, with 97.7% of residues in the undamaged complex and 98.4% of residues in the T-T dimer complex in the most favored regions of the Ramachandran plot. For kinetic analyses, full length wild-type Polη and the mutants were assayed for nucleotide incorporation on a 75-nt template containing a cis-syn T-T dimer or its undamaged equivalent annealed to a 33-nt (running start) or a 44-nt (standing start) primer. For genetic complementation studies, yeast strain YR30.2 (MATahis3Δ-100, leu2–3,112, trp1Δ, ura3–52 rad30Δ) was transformed with plasmids carrying the wild type or mutant versions of the RAD30 gene (Supplementary Table 3).
METHODS
Protein and DNA preparation
The catalytic core (residues 1–513) of wild-type S. cerevisiae Polη was expressed as an N-terminal GST-fusion protein in E. coli from plasmid pBJ875 as previously described9. Mutations K140A and S144W were generated in plasmid pBJ875 using the QuickChange site-directed mutagenesis kit (Stratagene) to yield plasmid pSL414. The GST-Polη1–513 K140A S144W fusion was expressed in E. coli and purified by affinity chromatography using a glutathione-Sepharose column. The GST tag was removed on the column by incubation with PreScission protease (Amersham), and the eluted Polη1–513 K140A S144W was further purified by ion exchange (Q Sepharose) before being concentrated and used for cocrystallization. The 11-nt DNA primer oligonucleotide was synthesized with a 2',3'-dideoxycytosine at its 3'-end (5'-GTCCTCCCCTCdd-3'). This primer was annealed to either the undamaged 16-nt template strand (5'-TAATTGAGGGGAGGAC-3'), or the 16-nt template strand containing a cis-syn T-T dimer (5'-TAATTGAGGGGAGGAC-3'). The primer and undamaged template strands were first purified by ion exchange chromatography, while the T-T dimer containing template was first purified by reverse phase chromatography, prior to annealing.
Plasmids for biochemical and genetic analysis
The Polη Q55A, R73A, M74A mutations and the indicated combinations were made by mutagenic PCR using oligonucletides containing the appropriate codon changes. PCR fragments were cloned into the full length RAD30 open reading frame and the mutations were verified by sequencing. Mutant fragments were then used to replace the wildtype regions in either pR30.175 for protein expression, or pBJ1587 (3.3 kB hinDIII/PstI fragment) for genetic complementation assays. pBJ1587 harbors a 3.3 kBp HindIII/PstI fragment containing the wildtype RAD30 gene in the single copy vector Ycplac11124. Plasmids used for expression and complementation assays of mutants are listed in Supplementary Table 3.
Nucleotide incorporation assays and kinetic analysis
The full length wild-type and mutants of GST tagged yeast Polη were expressed in yeast, and purified by glutathione-Sepharose affinity chromatography as described above. All proteins were treated with PreScission protease to remove the GST tag. Proteins were quantified by Bradford assay and Coomassie stained SDS-PAGE analysis. The standard DNA polymerase assay (5 μl) contained 25 mM Tris-HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 10% glycerol and were carried out at 30° C. The DNA substrate consisted of a 75mer oligonucleotide template containing a cis-syn T-T dimer or its undamaged equivalent annealed to a 33nt (running start) or a 44 nt (standing start) primer. For running start assays, 25 μM of each dNTP were used and reactions were carried out for 10 min. For standing start kinetic assays, reactions contained only dATP at concentrations varying from 0.05 to 1000 μM, and were carried out from 2.5 to 10 minutes. Reaction products were separated on 10% TBE-PAGE gels containing 8M urea, and visualized by a phophorimager. Kinetic parameters were determined by plotting the rate of product formation versus dATP concentration and fit to the Michealis-Menten equation as described25.
Genetic analysis
For genetic complementation studies, yeast strain YR30.2 (MATa his3Δ-100, leu2–3,112, trp1Δ, ura3–52 rad30Δ) was transformed with plasmids carrying the wild type or mutant versions of the RAD30 gene (Supplementary Table 3). Cells were grown to midlogarithmic phase in synthetic complete media lacking leucine (SC-leu) for maintenance of the plasmids, washed, resuspended to a density of 2 × 108 cells per ml, and sonicated to disperse clumps. Appropriate dilutions of cells were spread onto the surface of SC-leu for viability determinations and on SC-leu-arg+can for mutagenesis determinations, after which, plates were UV irradiated followed by incubation at 30°C in the dark.
Cocrystallization
The undamaged and the T-T dimer ternary complexes were prepared by incubating Polη1–513 K140A S144W with the respective primer-template in a molar ratio of 1:2 along with 10mM dATP and 10mM MgCl2. Cocrystals of the undamaged ternary complex were obtained from solutions containing 24% PEG 4000 (w/v), 0.2 M Li2SO4, and 0.1 M TRIS.HCL buffer (pH 8.5). Cocrystals of the T-T dimer ternary complex were obtained from solutions containing 33% PEG 4000 (w/v), 0.2 M Li2SO4, and 0.1 M TRIS.HCL buffer (pH 8.5). For data collection, the cocrystals were cryoprotected by soaking them in mother liquor solutions plus increasing amounts of glycerol (0 to 15%, in 5% increments), and then flash frozen in liquid nitrogen. Cocrystals of the undamaged complex belong to the space group C2221, with unit cell dimensions of a=88.8Å, b=227.9Å, and c=86.0Å. Cocrystals of the T-T dimer complex belong to space group C2221 with unit cell dimensions of a=88.1Å, b=227.0Å, and c=85.9Å. Both cocrystals contain one protein-DNA complex in the asymmetric unit. Importantly, these cocrystals are different from the “apo-like” crystals reported previously for the Polη apoenzyme9 (P41212; a = b= 105Å, c=292Å) and for structures of Polη with cisplatin adducts19 (P41212; a = b= 104Å, c=293Å).
Structure determination and refinement
X-ray data on cryocooled T-T dimer and undamaged DNA cocrystals were measured at the Advanced Photon Source (beamline 24ID) and the Brookhaven National Laboratory (beamline X6A), extending to 1.76Å and 2.0Å resolutions, respectively. The data were indexed and integrated using DENZO and scaled using SCALEPACK26. These data were used to obtain molecular replacement (MR) solutions using the apo-Polη structure (without the PAD) as a search model. Specifically, the program PHASER27 gave a unique MR solution for each data set, which was then used to place the PAD (using PHASER). The solutions were refined and the electron density maps calculated with the refined models showed clear densities for the respective primer-templates, incoming nucleotide, and metal ions. Both models were subjected to iterative rounds of building with COOT28 and refinement with REFMAC29. During the later rounds of refinement with REFMAC, protein residues 1–104, 105–149, 150–306, 307–388, and 389–512 and each individual DNA strand were treated as TLS groups, as identified using the TLSMD server (http://skuld.bmsc.washington.edu/~tlsmd/). TLS refinement lowered the Rfree of the T-T dimer complex to 18.7% with an Rcryst of 16.3%, and the Rfree of the undamaged complex to 22.7% with an Rcryst of 19.4%. The final refined model of the T-T dimer complex includes residues −2 to 356 and 360 to 511 (residues −2 to 0 correspond to the linker region between the GST tag and Polη, and residues 357 to 359 of the thumb domain could not be modeled due to poor electron density), nucleotides 1–11 of the primer strand, nucleotides 3–16 of the template strand, one dATP molecule, two magnesium ions, two sulfate ions, and 709 water molecules. The final refined model of the undamaged complex includes amino residues 1 to 112 and 115 to 512 (residues 113 and 114 of the fingers domain could not be modeled due to poor electron density), nucleotides 1–11 of the primer strand, nucleotides 4–16 of the template strand, one dATP molecule, two magnesium ions, two sulfate ions, and 627 water molecules. Both structures have excellent stereochemistry, with 97.7% of residues in the undamaged complex and 98.4% of residues in the T-T dimer complex in the most favored regions of the Ramachandran plot, according to MolProbity30. Simulated annealing omit maps were generated with CNS31, and all structural figures were generated with PyMOL32.
Supplementary Material
Acknowledgements
We thank the staff at Brookhaven National Laboratory (beamlines X6A and X29) and the Advanced Photon Source (24ID) for facilitating X-ray data collection. We thank D.T. Nair, S. Lone, R. Vasquez-Del Carpio, and M. Swan for discussions. This work was supported by NIH grants to A.K.A, S.P. and L.P.
Footnotes
Author Information. Reprints and permissions information is available at www.nature/com/reprints. Atomic coordinates and structure factors files have been deposited in the Protein Data Bank under accession codes 3MFH and 3MFI. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–5. doi: 10.1126/science.285.5425.263. see comments. [DOI] [PubMed] [Google Scholar]
- 2.Masutani C, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–4. doi: 10.1038/21447. see comments. [DOI] [PubMed] [Google Scholar]
- 3.Broughton BC, et al. Molecular analysis of mutations in DNA polymerase eta in xeroderma pigmentosum-variant patients. Proc Natl Acad Sci U S A. 2002;99:815–820. doi: 10.1073/pnas.022473899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratchev A, Strein P, Utikal J, Sergij G. Molecular genetics of Xeroderma pigmentosum variant. Exp Dermatol. 2003;12:529–36. doi: 10.1034/j.1600-0625.2003.00124.x. [DOI] [PubMed] [Google Scholar]
- 5.Inui H, et al. Xeroderma pigmentosum-variant patients from America, Europe, and Asia. J Invest Dermatol. 2008;128:2055–68. doi: 10.1038/jid.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanioka M, et al. Molecular analysis of DNA polymerase eta gene in Japanese patients diagnosed as xeroderma pigmentosum variant type. J Invest Dermatol. 2007;127:1745–51. doi: 10.1038/sj.jid.5700759. [DOI] [PubMed] [Google Scholar]
- 7.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Poleta. Science. 1999;283:1001–4. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 8.Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 9.Trincao J, et al. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–26. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 10.Kondratick CM, Washington MT, Prakash S, Prakash L. Acidic residues critical for the activity and biological function of yeast DNA polymerase eta. Mol Cell Biol. 2001;21:2018–25. doi: 10.1128/MCB.21.6.2018-2025.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–8. doi: 10.1038/34593. see comments. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. Embo J. 1998;17:7514–25. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J Biol Chem. 1999;274:17395–8. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 14.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 15.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–80. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 16.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair. Structure (Camb) 2005;13:1569–77. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–22. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 18.Lone S, et al. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–14. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Alt A, et al. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–70. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 20.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–40. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 21.Washington MT, Prakash L, Prakash S. Yeast DNA polymerase eta utilizes an induced-fit mechanism of nucleotide incorporation. Cell. 2001;107:917–27. doi: 10.1016/s0092-8674(01)00613-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol Cell. 2001;8:427–37. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 23.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. Embo J. 2005;24:2957–67. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–34. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 25.Johnson RE, Prakash L, Prakash S. Yeast and human translesion DNA synthesis polymerases: expression, purification, and biochemical characterization. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–21. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 30.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunger AT, et al. Crystallography & NMR system: A software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Delano WL. Delano Scientific LLC; San Carlos, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.