Summary
Human DNA polymerase-ι (Polι) incorporates correct nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines. In fact, the fidelity opposite template T is so poor that Polι inserts an incorrect dGTP approximately 10 times better than it inserts the correct dATP. We determine here how a template T/U is accommodated in the Polι active site and why a G is incorporated more efficiently than an A. We show that in the absence of incoming dATP or dGTP (binary complex), template T/U exists in both syn and anti conformations, but in the presence of dATP or dGTP (ternary complexes), template T/U is predominantly in the anti conformation. We also show that dATP and dGTP insert differently opposite template T/U, and that the basis of selection of dGTP over dATP is a hydrogen bond between the N2 amino group of dGTP and Gln59 of Polι.
Introduction
The survival of organisms depends critically on the ability to faithfully replicate DNA. As such, virtually all DNA polymerases replicate DNA by incorporating the nucleotide that forms the correct Watson-Crick base pair with the template base. In addition, the catalytic efficiencies with which any given polymerase forms the four possible correct base pairs are approximately the same, reflecting the near geometric identities of Watson-Crick, A.T, T.A, G.C, and C.G base pairs. This basic tenet holds for high fidelity replicative DNA polymerases and is also generally valid for the lower fidelity Y family DNA polymerases that possess the ability to replicate through DNA lesions (Prakash et al., 2005). Human DNA polymerase ι (Polι), a member of the Y family of DNA polymerases, is a notable exception to these rules. Polι incorporates nucleotides opposite template purines with a much higher efficiency and fidelity than opposite template pyrimidines. Specifically, Polι exhibits the highest efficiency and fidelity opposite template A, followed by template G, and then templates C and T. In fact, the fidelity opposite template T is so poor that Polι inserts an incorrect dGTP approximately 10 times better than it inserts the correct dATP (Haracska et al., 2001; Johnson et al., 2000; Tissier et al., 2000; Washington et al., 2004; Zhang et al., 2000).
The ternary crystal structures of Polι bound to template purines and the correct incoming dNTP have yielded major insights into the action mechanism of this polymerase. Specifically, in the structure of Polι bound to template A and an incoming dTTP, the templating A adopts a syn conformation and forms a Hoogsteen base pair with the incoming T, which remains in the anti conformation (Nair et al., 2006b; Nair et al., 2004). Similarly, in the structure of Polι paired with a template G and an incoming dCTP, a G.C+ Hoogsteen base pair is formed in the polymerase active site (Nair et al., 2005; Nair et al., 2006b). Hoogsteen base pairing provides a basis for the much higher efficiency and fidelity of nucleotide incorporation opposite template purines than opposite pyrimidines because only the templates A and G have a Hoogsteen edge by which they can establish two hydrogen bonds with the correct incoming pyrimidine nucleotide. The ability to form Hoogsteen base pairs allows Polι to promote replication through adducts such as 1, N6-ethenodeoxyadenosine that disrupt the Watson-Crick (W-C) edge but not the Hoogsteen edge of the templating purine (Nair et al., 2006a).
Compared to the purine templates, Polι is highly inefficient at incorporating the correct nucleotide A opposite template T; moreover, it incorporates a G opposite template T with an ~ 10-fold better catalytic efficiency than an A (Haracska et al., 2001; Johnson et al., 2000; Tissier et al., 2000; Washington et al., 2004; Zhang et al., 2000). However, even the incorporation of G opposite template T occurs almost 100-fold less efficiently than, for example, the incorporation of T opposite template A. We determine here how a template T is accommodated in the Polι active site and why G is incorporated more efficiently than an A. We show that in the absence of dATP or dGTP (binary complex), template T exists in both syn and anti conformations, but in the presence of dATP or dGTP (ternary complexes), template T is predominantly in the anti conformation. We also show that dATP and dGTP insert differently opposite template T, and that the basis of selection of dGTP over dATP is a hydrogen bond between the N2 amino group of dGTP and Gln59 of Polι.
Results
Structure determination
We crystallized Polι with “double-ended” 18-nt template-primers designed to present template T in the active sites of two symmetrically bound Polι molecules (see Methods). We have used a similar strategy previously to crystallize Polι with undamaged and damaged DNAs (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2009). Cocrystals in the absence (binary) and presence of incoming dATP and dGTP (ternary) were obtained under identical conditions (from PEG solutions), belonging to space group P6522 with cell dimensions of a = 98 Å, b = 98 Å, c = 203Å and α=β=90°, γ=120°. The PolιT, PolιT.dATP and PolιT.dGTP structures were solved by molecular replacement (MR) using the structure of the PolιG.dCTP complex (with dCTP omitted) as a search model (Nair et al., 2005). However, in all three structures, electron densities for the template T were not as well-defined as for the template purines in previous Polι structures (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2004). To help fix the conformation(s) of the thymines, we substituted 5-bromouracil (BrU) for thymine in the template-primers and cocrystallized them with Polι in the absence and presence of incoming dATP and dGTP. We solved the PolιU, PolιU.dATP and PolιU.dGTP structures by MR and the resulting electron density maps were very similar to those obtained for the PolιT, PolιT.dATP and PolιT.dGTP complexes. However, the anomalous signal of the Br atoms permitted the calculation of anomalous difference Fourier maps, which pinpointed the positions of the Br atoms and showed that BrU (and T, by analogy) exists in both syn and anti conformations in the binary PolιU complex, but switches primarily to the anti conformation in the PolιU.dATP and PolιU.dGTP ternary complexes. The PolιU, PolιU.dATP and PolιU.dGTP complexes were selected for further refinement to 2.3Å, 2.85Å, and 2.2Å resolutions (Table 1), respectively, with the BrUs in alternate anti:syn conformations in 0.5:0.5, 0.9:0.1, and 0.7:0.3 ratios, respectively. The refined PolιU binary complex (Rfree of 28.0%; Rcrys of 25.2%) contains Polι residues 26-370, 379-394 and 404-414 (the first 25 and the last 6 residues are disordered, and residues 371-378 and 395-403 comprising two loops in the PAD have poor density), DNA nucleotides 3-11 (two of the four unpaired template nucleotides at the 5’ end are disordered), and 237 water molecules. The refined PolιU.dATP ternary complex (Rfree of 28.2%; Rcrys of 24.1 %) contains Polι residues 25-370, 379-394 and 404-414, DNA nucleotides 3-11, incoming dATP, 1 Mg2+ ion, and 156 water molecules. The refined PolιU.dGTP ternary complex (Rfree of 27.0%; Rcrys of 25.4%) contains Polι residues 25-370, 379-394 and 404-414, DNA nucleotides 3-11, incoming dATP, 2 Mg2+ ions, and 272 water molecules. All three structures have good stereochemistry, with ~86% of the residues in the most favored regions of the Ramachandran plot and <0.3% in the disallowed regions for PolιU. and PolιU.dGTP, and ~ 83% in the most favored region and < 0.6% in the disallowed region for PolιU.dATP.
Table 1.
Data Collection and Refinement Statistics
| Data Collection | PolιU | PolιU.dATP | PolιU.dGTP |
|---|---|---|---|
| Resolution (Å) | 2.3 | 2.85 | 2.2 |
| No. of Measured | 371962 | 198630 | 429934 |
| No. of Unique | 26617 | 14231 | 30091 |
| Data Coverage (%) | 99.7(97.2) | 100(100) | 99.9(100.0) |
| Rmerge (%)a, b | 6.8(36.1) | 9.4(38.0) | 8.6 (43.3) |
| I/σ | 44.0(6.0) | 31.3 (6.1) | 31.4 (5.9) |
| Refinement Statistics | |||
| Resolution Range | 50-2.3 | 50-2.85 | 50-2.2 |
| Reflections | 25488 | 13690 | 28795 |
| Rcryst (%)c | 25.2 | 24.1 | 25.4 |
| Rfree (%)d | 28.0 | 28.2 | 27.0 |
| Non-hydrogen atoms | |||
| Protein | 2872 | 2877 | 2877 |
| DNA | 320 | 320 | 320 |
| Incoming NTP | 30 | 31 | |
| Mg2+ ion | 1 | 2 | |
| Water | 237 | 156 | 272 |
| B factors (Å2) | |||
| Protein | 52.9 | 52.4 | 51.5 |
| DNA | 54.1 | 44.1 | 50.1 |
| Incoming NTP | 76.2 | 50.9 | |
| Mg2+ ion | 57.2 | 61.4 | |
| Water | 42.1 | 38.9 | 45.4 |
| Rms deviations | |||
| Bonds (Å) | 0.006 | 0.007 | 0.006 |
| Angles (°) | 1.3 | 1.3 | 1.2 |
| Ramachandran Plot Quality | |||
| Most favored (%) | 86.4 | 83.8 | 84.6 |
| Generously allowed | 13.3 | 15.6 | 15.1 |
| Disallowed (%) | 0.3 | 0.6 | 0.3 |
Values for outermost shells are given in parentheses
Rmerge =Σ|I-<I>|/ΣI, where I is the integrated intensity of a given reflection.
Rcryst = Σ∥Fobserved| − |Fcalculated∥/Σ|Fobserved|
For Rfree calculations ~ 8% of data excluded from refinement was used for the PolιU, PolιU.dATP, and PolιU.dGTP complexes.
Overall arrangement
In all three complexes, a Polι molecule binds to each end of the double-ended template primer (Fig. 1). The two molecules are related by a crystallographic two-fold axis and thus make identical contacts with the template-primer. Polι has the familiar right-handed architecture with palm (residues 25-37, 99-224), fingers (38-98), and thumb (225-288) domains, and the PAD (polymerase associated domain; residues 298-414) unique to Y-family polymerases (Fig.1). The palm forms the floor of the DNA binding cavity and carries the active site residues (Asp34, Asp126 and Glu127), while the fingers domain lies over the templating base (and the incoming nucleotide in the ternary complexes). The thumb and the PAD are connected by a long linker that spans the width of the DNA. The thumb skims the minor groove surface on one side the template-primer, while the PAD docks in the major groove on the other side (Fig. 1). The interface between the two Polι molecules is relatively small, with ~700 Ả2 of surface area buried between the thumb domain of one polymerase and the PAD of the other. The majority of Polι-DNA interactions are mediated by the PAD, wherein the main chain amides on “outer” β-strands of the PAD β-sheet make a series of hydrogen bonds with the template and primer strands.
Figure 1. Polι binary and ternary complexes.
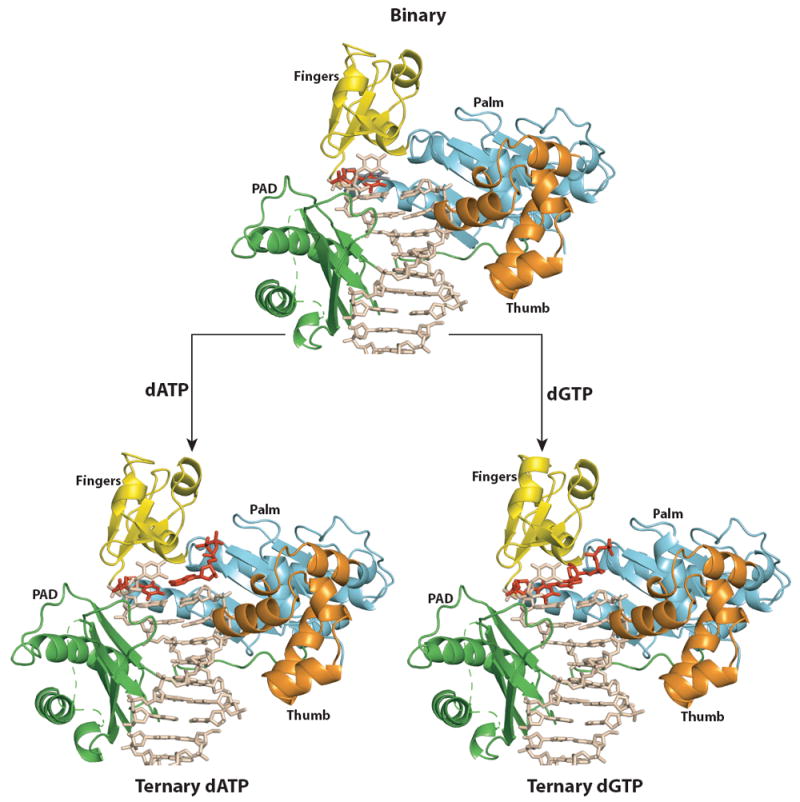
Overall structures of PolιU (top), PolιU.dATP (bottom left) and PolιU.dGTP (bottom right). Palm, fingers, thumb, and PAD domains are shown in cyan, yellow, orange, and green respectively. DNA is shown in tan; the incoming nucleotide and the template U in anti conformation are shown in red. The syn conformation of templating U in the binary structure is shown in gray and has been omitted from the ternary structures for clarity.
The PolιU binary complex
The palm, fingers and thumb domains and the PAD occupy identical positions with respect to the template-primer as in previous Polι structures (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2004), and the PolιU.dATP and PolιU.dGTP ternary complexes described below. Thus, there is no open or closed form of Polι analogous to replicative DNA polymerases, wherein the fingers domain rotates by as much as ~40° in response to dNTP binding (Doublie et al., 1999; Rothwell and Waksman, 2005). The major difference between PolιU and the PolιA and PolιG binary complexes determined previously is in the DNA (Nair et al., 2006b). In the PolιA and PolιG binary complexes, the purines are in the anti conformation, which flip to the syn conformation in reaction to dNTP binding (Nair et al., 2006b). Once in the syn conformation, the Hoogsteen edge of A and G is available for hydrogen bonding with the W-C edge of incoming dTTP and dCTP, respectively. In contrast, in the PolιU binary complex, template U adopts a mixture of anti and syn conformations (Fig. 2). In response to dATP or dGTP binding (described below), occupancy for the anti conformation increases while that for the syn conformation decreases. Unlike purines, pyrimidines lack a Hoogsteen edge. Thus, it is only in the anti conformation that the polar atoms on T/U (N3, O2, and O4) are available for hydrogen bonding with dATP or dGTP in the Polι active site.
Figure 2. Active site in the PolιU binary complex.
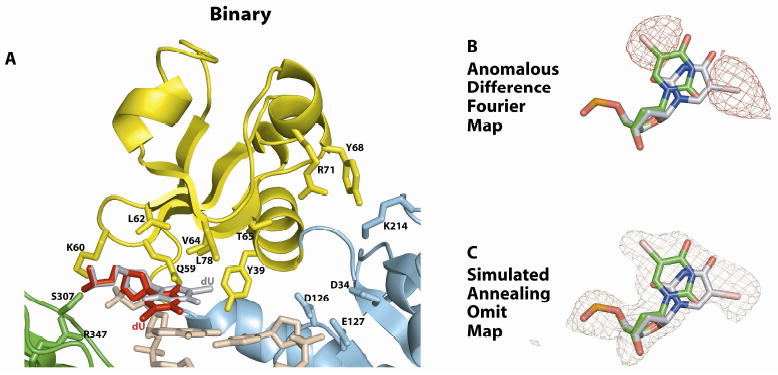
(A) Close-up view of the of PolιU active site region. The catalytic residues (D34, D126, and E127), the residues apposed to the template U (Q59, K60, L62, V64, L78, S307, and R347) and some other residues near the active site (Y39, T65, Y68, R71, and K214) are highlighted and labeled. The anti conformation of template is shown in red and the syn conformation in gray. (B) Anomalous difference Fourier map (contoured at 3σ) showing the two positions of the C5 Br atom of the templating base. Based on the map the templating base was modeled in both anti and syn conformations. (C) Simulated annealing Fo-Fc omit map (contoured at 3σ) with the atoms of the templating base of PolιU omitted.
The PolιU.dATP ternary complex
In the PolιA.dTTP and PolιG.dCTP ternary complexes, incoming dNTP effectively pushes the templates A and G into the syn conformation for if they were to remain in anti conformation they would clash with the incoming nucleotide (Nair et al., 2005; Nair et al., 2006b; Nair et al., 2004). By contrast, in the PolιU.dATP ternary complex (and the PolιU.dGTP complex – described below), it is the syn rather than the anti conformation of the template base that clashes with the incoming nucleotide, wherein the C5 Br (or CH3 in the case of T) sterically overlaps with the W-C edge of incoming dNTP (Fig. 3). The small fraction of template T in the syn conformation in the ternary complex crystals likely reflects a fraction of complexes without incoming dNTP in the binary state. In the description below, we refer mainly to the anti conformation.
Figure 3. Comparison of the Polι active site in PolιU.dATP and hPolιU.dGTP ternary complexes.
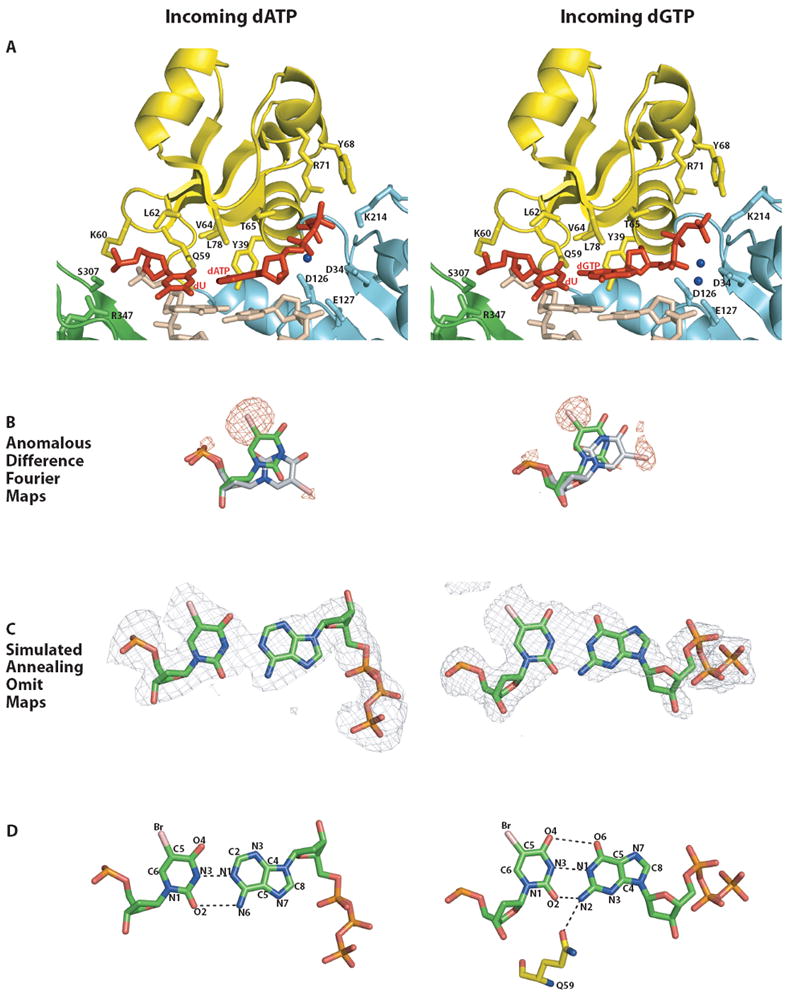
(A) Close-up views of the active site regions in PolιU.dATP (left) and PolιU.dGTP (right). Coloring scheme is same as for Figure 1. Putative magnesium ions are shown as blue spheres. The catalytic residues (D34, D126, and E127), residues apposed close to the template base (Q59, K60, L62, V64, L78, S307, and R347), and those near the incoming nucleotide (Y39, T65, Y68, R71, and K214) are highlighted and labeled. The anti conformation of templating U and the incoming nucleotide are shown in red. The minor syn conformation of the templating base in both the structures has been omitted for clarity. (B) Anomalous difference Fourier maps (contoured at 3σ) showing the position of the C5 Br atoms in the templating base of PolιU.dATP (left) and PolιU.dGTP (right). Note that the density corresponding to the position of the bromine atoms in the anti conformation of the template is stronger than that for the syn conformation in both ternary complexes. (C) Simulated annealing Fo-Fc omit maps (contoured at 3σ) around the templating base and the incoming nucleotide in the PolιU.dATP (left) and PolιU.dGTP (right) structures. The template U and incoming nucleotide are in trans in the PolιU.dATP complex (left), and in cis in the PolιU.dGTP complex (right). (D) U.dATP (left) versus U.dGTP (right) base pairing in the active site of Polι ternary complexes. trans orientation coupled with anti conformation of U and dATP results in a non-canonical Watson-Crick pair in the active site of PolιU.dATP (left). In PolιU.dGTP (right), incoming dGTP inserts in cis, and its N2 amino group forms a hydrogen bond with Gln59 emanating from the fingers domain.
To account for the poor efficiency of Polι in inserting A opposite template T, we had considered a model in which the template T/U is evicted from the active site and dATP is inserted opposite an essentially “abasic” site. This is clearly not the case. The structure reveals unexpected trans U:A base pairing (Figs, 3A and B) (Leontis et al., 2002). By comparison to a standard Watson-Crick (WC) T/U:A base pair, where the glycosidic bonds of the two nucleosides are in cis, the U and A in our structure are in trans or antiparallel to one another. The C1’-C1’ distance is 11.2Å, as compared to ~10.5Å in a standard W-C base pair. Also, because the glycosidic angles of both U and A are anti, they couple with the trans orientation of the glycosidic bonds to produce local parallel-stranded DNA at the nascent base-pair (Leontis et al., 2002). In a standard cis T/U:A base pair, the N1 and N6 atoms of A form hydrogen bonds with the N3 and O4 atoms of T/U, respectively. In a trans T/U:A base pair, we can imagine A inverting around the N1(A)…N3(T) hydrogen bonds to generate a potential new N6(A)…O2(T/U) hydrogen bond in place of the N6(A)…4(T/U) hydrogen bond (Fig. 3). The N1(A)…N3(U) and N6(A)…O2(U) distances in our structure are 2.71Å and 3.52Å, respectively.
In all previous Polι ternary complexes, the incoming dNTP sugar packs against the aromatic ring of Tyr39 and a hydrogen bond is also made between its 3’OH and the main chain amide of the tyrosine (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2009; Nair et al., 2004). In the present structure, as the result of dATP inverting to the trans conformation, the sugar is removed from the aromatic ring of Tyr39 and is close to surface of the major groove. Although, the dATP triphosphate moiety lies between the palm and fingers domains as in previous Polι structures it weaves a different path (Fig. 3A). The α- and β-phosphates, for example, are displaced by >4Å from their positions in previous Polι structures, and as a consequence some interactions are lost while others are maintained. For example, Thr65 and Tyr68 on the fingers domain are unable to make hydrogen bonds with the β- and γ- phosphates, respectively, but Arg71 from the fingers domain and Lys214 from the palm domain are still able to reach and make bonds with the β- and γ- phosphates (Fig. 3A). Also, because of the displacement of the α-phosphate the active geometry is incompatible with catalysis, with the putative 3’OH at the primer terminus located >6Å from the α-phosphate of incoming dNTP.
The PolιU.dGTP ternary complex
In contrast to dATP, dGTP inserts in cis opposite template U (Fig. 3). The structure suggests distorted U:G base pairing in which the U is either ionized or exists as an enol tautomer (Lawley and Brookes, 1962; Sowers et al., 1987; Topal and Fresco, 1976; Watson and Crick, 1953). That is, the U and G bases lie opposite one another in distorted W-C geometry rather than the more common wobble geometry (Fig. 3). Although T/U occurs predominantly as a keto tautomer, it can also exist as a rare enol tautomer and in an ionized form that can pair with G via W-C geometry (Lawley and Brookes, 1962; Sowers et al., 1987; Topal and Fresco, 1976; Watson and Crick, 1953). In the structure, the N3 and O2 atoms of U make putative hydrogen bonds with the N1 atom and N2 amino group of G, respectively. The U and G bases are staggered and propeller twisted with respect to each other, and the hydrogen bond between O2(U) and N2(G) is relatively short (~2.45Å), which may be indicative of an ionized form of U (rather than the enol tautomer). Interestingly, BrU (used in our structural analysis) has a greater tendency than T/U to pair with G and this property has been ascribed to the increased propensity of BrU to form the enol tautomer or the ionized form (Danilov et al., 2009; Lawley and Brookes, 1962; Sowers et al., 1987). However, in crystal and NMR structures of DNAs containing BrUs (Brown et al., 1986; Sowers et al., 1989), BrU pairs with G in much the same way as unmodified U/T, namely via wobble geometry. Intriguingly, NMR spectra at high pH (9.8) (Sowers et al., 1989), suggests an ionized BrU;G W-C base pair with a geometry similar to what we observe here in the PolιU.dGTP complex (crystallized at a pH of 6.5).
The incoming dGTP triphosphate moiety interleaves between the palm and fingers domains, and makes hydrogen bonds with Thr65, Tyr68, and Arg71 from the fingers domain, and Lys214 from the palm domain. The triphosphate moiety and the catalytic residues Asp34, Asp126 and Glu127 coordinate two putative Mg2+ ions in the active site. The geometry of the active site is better poised for catalysis than in the PolιU.dATP structure, with the putative 3’OH at the primer terminus located ~3.3Ả from the dGTP α-phosphate and aligned more or less linearly with respect to the scissile Pα O3’ bond.
The C1’-C1’ distance across the U:G base pair is ~9.6Å, as compared to ~10.5Å in a standard W-C base pair. In previous Polι structures with undamaged and damaged DNAs, the C1’-C1’ distance across the nascent pairs ranges from ~8.5 to ~9.1Å, wherein the template sugar is entrenched in a cavity lined by residues from the fingers domain, including the side chain of Leu62 and the aliphatic portions of Gln59 and Lys60 (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2004). On the other side of the active site, the dNTP sugar is fixed by packing against the aromatic ring of Tyr39, as well as by a hydrogen bond between its 3’OH and the main chain amide of the tyrosine. The resulting constriction in C1’-C1’ distance across the sugars is conducive to Hoogsteen base pairing opposite template purines (Nair et al., 2005; Nair et al., 2006b; Nair et al., 2004). In the present structure, a change in the Leu62 rotamer (from χ2 of 169° in PolιG.dCTP to 116° PolιU.dGTP) allows the template U sugar to shift by ~1Å and still fit within the hydrophobic cavity delineated by Leu62, Gln59, Lys60. The most revealing feature of our structure is a hydrogen bond between the N2 amino group of incoming dGTP and OE1 of Gln59 (2.9Å) on the fingers domain (Figs. 3A and D). This direct hydrogen bond between G and the polymerase appears to be the key stabilizing interaction favoring the insertion of G over A opposite template T in the Polι active site. That is, even if incoming dATP was to assume a cis configuration with respect to template T/U it would be unable to make this hydrogen bond because it lacks an N2 amino group.
To evaluate the significance of the observed hydrogen bond between Gln59 and the N2 amino group of incoming dGTP, we compared the catalytic efficiencies (kcat/Km) of dGTP vs. dITP incorporation using steady-state kinetic analyses (Johnson et al., 2000; Nair et al., 2006a). Since dITP lacks the N2 amino group, any reduction in the efficiency of dITP incorporation would result from the contribution of hydrogen bonding between the N2 amino group of dGTP and OE1 of Gln59 to dGTP incorporation. As shown in Table 2, compared to dGTP, dITP incorporation is reduced by 10-fold opposite template T. We infer from this observation that hydrogen bonding of Gln59 with the N2 amino group of dGTP contributes positively to G incorporation opposite template T and could account for the preferential incorporation of G over an A opposite template T.
Table 2.
Catalytic efficiency of dGTP vs. dITP incorporation opposite template T by Polι
| Incoming nucleotide | kcat (min-1) | Km (μM) | kcat/ Km | Efficiency relative to dGTP insertion |
|---|---|---|---|---|
| dGTP | 2.97±0.07 | 29.8 ±1.74 | 0.10 | - |
| dITP | 1.36±0.03 | 144.3±13.0 | 0.01 | 0.10 (10x↓) |
The data represent an average of three experiments
Discussion
Compared to the purine templates, Polι is highly inefficient at incorporating the correct nucleotide A opposite template T. In fact, the fidelity opposite template T is so poor that Polι inserts an incorrect G roughly 10 times better than it inserts the correct A (Haracska et al., 2001; Johnson et al., 2000; Tissier et al., 2000; Washington et al., 2004; Zhang et al., 2000). However, even the incorporation of G opposite template T occurs almost 100-fold less efficiently than, for example, the incorporation of T opposite template A. Perhaps reflective of this inefficiency opposite template T, the electron density for template U/T (and incoming nucleotides in the ternary complexes) is not as well-defined as in Polι structures with template purines (Nair et al., 2005; Nair et al., 2006b; Nair et al., 2004). Nonetheless, via the use of bromouracils we have gained some insights into how a template U/T may be accommodated in the Polι active site and why a G is incorporated more efficiently than an A.
In the PolιU binary complex, template U/T exists in both syn and anti conformations. In contrast, in the PolιA and PolιG binary complexes the purine is exclusively in the anti conformation (Nair et al., 2006b); which then flips over to the syn conformation in the presence of incoming dATP or dGTP (in ternary complexes). The existence of template U/T in syn conformation is somewhat surprising since pyrimidines are considered less likely than purines to adopt the syn conformation due to potential steric overlap between the O2 and the sugar atoms (Haschemeyer and Rich, 1967; Saenger, 1984). However, any such steric repulsion may be counteracted in the Polι active site cleft by favorable van der Waals contacts between the C5 substituent and Leu78 emanating from the fingers domain. Interestingly, dGTP and dATP insert differently opposite template U/T, namely dGTP in cis and dATP in trans. However, the incorporation of A or G onto the primer terminus, although highly inefficient, can only occur in the cis conformation where the putative 3’OH at the primer terminus is <4Ả from the dNTP α-phosphate. We postulate that dATP and dGTP alternate between cis and trans conformations opposite template T, and that dGTP is (on balance) favored in the cis conformation by a hydrogen bond between its N2 amino group and Gln59 of Polι. In all, the structures offer a plausible basis for the inefficiency of nucleotide incorporation opposite template T; wherein, dATP and dGTP interconvert between distorted W-C base-pairing in cis and catalytically incompatible base pairing in trans. We also show that a hydrogen bond between Gln59 and the N2 amino group of dGTP contributes positively to G incorporation opposite template, whereby dITP (lacking an N2 amino group) is incorporated at ~10-fold lower catalytic efficiency than dGTP and which could account for the preferential incorporation of G over an A opposite template T.
An interesting question is why dATP or dGTP would flip over to the trans conformation. We believe that the answer lies in the same structural feature of Polι that make it conducive to Hoogsteen base pairing opposite a template purine, namely a constricted active site that reduces the C1’-C1’ distance across the nascent base pair from ~10.5Å in most DNA polymerases to <9Å in Polι (Nair et al., 2005; Nair et al., 2006a; Nair et al., 2006b; Nair et al., 2009; Nair et al., 2004). The template purine sugar is entrenched in a cavity lined by residues from the fingers domain, including the side chain of Leu62 and the aliphatic portions of Gln59 and Lys60, while on the other side of the active site the dNTP sugar is fixed by packing against the aromatic ring of Tyr39, as well as by a hydrogen bond between its 3’OH and the main chain amide of the tyrosine. Template A or G is driven to the syn conformation by this “narrowing” of the distance across the incipient base pair in the Polι active site cleft (Nair et al., 2006b). In contrast, a pyrimidine would not free up as much “space” as a purine by rotating to the syn conformation due to its smaller size and more isotropic shape. Also, a pyrimidine in the syn conformation does not offer the same hydrogen bonding opportunities as a purine in the syn conformation, and, in the case of T the C5 methyl group can sterically overlap with incoming dATP or dGTP. It is not surprising therefore to see BrU in the anti conformation when paired with dATP or dGTP, and the “narrowness” of the Polι active site cleft appears to be managed in two ways. First, distorted W-C pairing in cis where the bases are somewhat staggered with respect to each other and the C1’-C1’ is ~9.6Å and, second, where the incoming dNTP flips over to a trans orientation and thereby effectively “removes” the sugar from the constraint of packing against aromatic ring of Tyr39. Interestingly, although trans or reverse base pairing is more common in RNA than DNA structures (Fortsch et al., 1996; Lee and Gutell, 2004; Leontis et al., 2002), it has also been observed in a G:T mismatch in the active site of Dpo4 (Trincao et al., 2004), an archaeal Y-family DNA polymerase. In all, the structures we present here complement earlier Polι structures with template purines and provide a basis by which Polι inserts (albeit, highly inefficiently) dATP and dGTP opposite T. Importantly, almost all of the biochemical properties of Polι appear to stem from a constricted active site cleft that permits the bypass of minor-groove purine adducts such as 1, N6-ethenodeoxyadenosine through Hoogsteen base-pairing (Nair et al., 2006a), while at the same time hindering the ability of Polι to insert a nucleotide opposite template pyrimidines.
Experimental Procedures
Crystallization
The GST-Polι (residues 1-420) fusion protein was expressed and purified as described previously (Johnson et al., 2000). A self-complementary 18-mer oligonucleotide was synthesized containing dideoxycytosine at its 3’ ends (5’-TCT-BrU-GGGTCCTAGGACCCdd-3’, BrU = 5-bromo-deoxyuridine). The oligonucleotide was purified by reverse phase HPLC (on a C18 column), desalted, and lyophilized. Prior to crystallization, the oligonucleotide was annealed with itself to give a “double-ended” template-primer with two replicative ends. For crystallization of the PolιU.dGTP ternary complex, Polι and DNA were mixed in the ratio of 1:1.2, followed by the addition of dGTP and MgCl2 to final concentrations of 20 mM and 10 mM respectively. For the PolιU binary complex, only protein and the corresponding DNA were mixed together. The ternary and binary complexes were crystallized from solutions containing 10-15 % PEG 5000 MME and 0.2 –0.4 M (NH4)2SO4 in 0.1 M MES buffer (pH=6.5). Crystals belong to space group P6522 with cell dimensions of a = 98Å, b = 98Å, c = 203Å and α=β=90°, γ=120°. For data collection, the crystals were cryoprotected by soaks for 5 minutes in mother liquor solutions containing 5%, 10% and 15% glycerol, respectively, and then flash frozen in liquid nitrogen.
Structure Determination and Refinement
X-ray data on cryocooled crystals were measured at Advanced Photon Source (APS, beamline 17-ID) of Argonne National Laboratory. Single wavelength anomalous diffraction data on the binary and ternary crystals were collected at a wavelength of 0.920Å corresponding to the peak of the Br K-edge absorption profile. Data sets were indexed and integrated using DENZO and reduced using SCALEPACK (Otwinowski and Minor, 1997). The positions of the bromine atoms belonging to 5-bromouracil of the templating base were determined from anomalous difference Fourier maps and helped establish the orientation of the templating base in all three structures. The PolιU, PolιU.dATP, and PolιU.dGTP structures were solved by molecular replacement (MR), using the earlier PolιG.dCTP complex as a search model (with dCTP omitted) (Nair et al., 2005). The program AmoRe (Navaza, 1994) gave a unique MR solution in each case. The first round of refinement and map calculation was carried out without the template and the 5’ unpaired nucleotides. The electron density maps showed unambiguous densities for the template BrU and the 5’ nucleotide, which were then included in the model for subsequent refinement. Iterative rounds of refinement and water picking were performed with CNS (Brunger et al., 1998) and model building with program O (Jones et al., 1991). All models have good stereochemistry, as shown by PROCHECK (Laskowski et al., 1993) with > 83 % of the residues in the most favored regions of the Ramachandran plot and ≤ 0.6 % in the disallowed regions. Figures were prepared using PyMol (Delano, 2002).
Steady State Kinetic Analyses
The DNA substrate used contained a 32 nt oligonucleotide primer (5’-GTTTTCCCAG TCACGACGATGCTCCGGTAC-3’) annealed to a 52 nt template (5’-TTCGTATAATGCCTACACTTGAGTACCGGAGCATCGTCGT GACTGGGAAAAC-3’). The standard DNA polymerase reaction (5μl) contained 25 mM Tris-HCl (pH-7.5), 5 mM MgCl2, 1 mM dithiothreitol, 100 μg/ml BSA, 10% glycerol, various concentrations of dGTP or dITP, 10 nM DNA substrate, and 0.5 nM Polι. All reactions were carried out for 5 min at 37°C. Reaction products were resolved on 13% polyacrylamide gels containing 8M urea. The band intensities of substrate and products of deoxynucleotide incorporation reactions were quantitated by using a PhosphorImager and ImageQuant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, Vobs, was determined by dividing the amount of product formed by the time of reaction protein concentration used. The Vobs was then plotted as a function of deoxynucleotide concentration and the data fitted to Michaelis-Menten equation describing a hyperbola Vobs=(kcat[Enz] × [dNTP]/km+ [dNTP].
Acknowledgments
We thank the staff at the Advanced Photon Source (beamline 17-ID) for facilitating X-ray data collection. We thank Anshu Bhatnagar for contributing to kinetic studies. This work was supported by grant CA115856 from the U. S. National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brown T, Kneale G, Hunter WN, Kennard O. Structural characterisation of the bromouracil.guanine base pair mismatch in a Z-DNA fragment. Nucleic Acids Res. 1986;14:1801–1809. doi: 10.1093/nar/14.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve R, Jiang W, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Danilov VI, van Mourik T, Kurita N, Wakabayashi H, Tsukamoto T, Hovorun DM. On the mechanism of the mutagenic action of 5-bromouracil: a DFT study of uracil and 5-bromouracil in a water cluster. J Phys Chem A. 2009;113:2233–2235. doi: 10.1021/jp811007j. [DOI] [PubMed] [Google Scholar]
- Delano WL. The PyMol Molecular Graphics System. Delano Scientific LLC; San Carlos, USA: 2002. [Google Scholar]
- Doublie S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure Fold Des. 1999;7:R31–35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- Fortsch I, Fritzsche H, Birch-Hirschfeld E, Evertsz E, Klement R, Jovin TM, Zimmer C. Parallel-stranded duplex DNA containing dA.dU base pairs. Biopolymers. 1996;38:209–220. doi: 10.1002/(SICI)1097-0282(199602)38:2%3C209::AID-BIP7%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, Phillips BB, Hurwitz J, Prakash L, Prakash S. Targeting of human DNA polymerase iota to the replication machinery via interaction with PCNA. Proc Natl Acad Sci U S A. 2001;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haschemeyer AE, Rich A. Nucleoside conformations: an analysis of steric barriers to rotation about the glycosidic bond. J Mol Biol. 1967;27:369–384. doi: 10.1016/0022-2836(67)90026-5. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases iota and zeta act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- Jones AT, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;A47:110–119. [Google Scholar]
- Lawley PD, Brookes P. Ionization of DNA bases or base analogues as a possible explanation of mutagenesis, with special reference to 5-bromodeoxyuridine. J Mol Biol. 1962;4:216–219. doi: 10.1016/s0022-2836(62)80053-9. [DOI] [PubMed] [Google Scholar]
- Lee JC, Gutell RR. Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs. J Mol Biol. 2004;344:1225–1249. doi: 10.1016/j.jmb.2004.09.072. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, Westhof E. The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA Polymerase iota Incorporates dCTP Opposite Template G via a G.C+ Hoogsteen Base Pair. Structure (Camb) 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006a;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site. Structure. 2006b;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. DNA synthesis across an abasic lesion by human DNA polymerase iota. Structure. 2009 doi: 10.1016/j.str.2009.02.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- Navaza J. AMoRe: an automated package for molecular replacement. Acta Cryst. 1994;50:157–163. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L. Eukaryotic Translesion Synthesis DNA Polymerases: Specificity of Structure and Function. Annu Rev Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acid structure (New York, Springer-Verlag) 1984 [Google Scholar]
- Sowers LC, Goodman MF, Eritja R, Kaplan B, Fazakerley GV. Ionized and wobble base-pairing for bromouracil-guanine in equilibrium under physiological conditions. A nuclear magnetic resonance study on an oligonucleotide containing a bromouracil-guanine base-pair as a function of pH. J Mol Biol. 1989;205:437–447. doi: 10.1016/0022-2836(89)90353-7. [DOI] [PubMed] [Google Scholar]
- Sowers LC, Shaw BR, Veigl ML, Sedwick WD. DNA base modification: ionized base pairs and mutagenesis. Mutat Res. 1987;177:201–218. doi: 10.1016/0027-5107(87)90003-0. [DOI] [PubMed] [Google Scholar]
- Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Wolfle WT, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Dpo4 is hindered in extending a G.T mismatch by a reverse wobble. Nat Struct Mol Biol. 2004;11:457–462. doi: 10.1038/nsmb755. [DOI] [PubMed] [Google Scholar]
- Washington MT, Johnson RE, Prakash L, Prakash S. Human DNA polymerase iota utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol Cell Biol. 2004;24:936–943. doi: 10.1128/MCB.24.2.936-943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Wu X, Wang Z. Preferential incorporation of G opposite template T by the low-fidelity human DNA polymerase iota. Mol Cell Biol. 2000;20:7099–7108. doi: 10.1128/mcb.20.19.7099-7108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


