SUMMARY
Diffuse large B cell lymphoma (DLBCL) is a heterogeneous disease composed of at least two distinct subtypes: germinal centre B cell like (GCB) and activated B cell like (ABC) DLBCL. These phenotypic subtypes segregate with largely unique genetic lesions, suggesting the involvement of different pathogenetic mechanisms. In this report, we show that the BLIMP1/PRDM1 gene is inactivated by multiple mechanisms, including homozygous deletions, truncating or missense mutations, and transcriptional repression by constitutively active BCL6, in ~53% of ABC-DLBCL. In vivo, conditional deletion of Blimp1 in mouse B cells promotes the development of lymphoproliferative disorders recapitulating critical features of the human ABC-DLBCL. These results demonstrate that BLIMP1 is a bona fide tumor suppressor gene whose loss contributes to lymphomagenesis by blocking plasma cell differentiation.
INTRODUCTION
Diffuse large B cell lymphoma (DLBCL) represents the most common type of non-Hodgkin’s lymphoma in the adult, accounting for ~40% of all diagnoses (Abramson and Shipp, 2005). Based on gene expression profile analysis, distinct DLBCL subtypes have been identified whose transcriptional programs resemble that of normal B cells at various stages of differentiation (Alizadeh et al., 2000; Shaffer et al., 2002b). These include the germinal center B cell-like (GCB) DLBCL, presumably derived from a transformed germinal centre (GC) centroblast, and the activated B cell-like (ABC) DLBCL, whose cell of origin is less clear but may be related to a plasmablastic B cell. A third group of DLBCL is represented by primary mediastinal large B cell lymphoma, postulated to arise from thymic B cells (Rosenwald et al., 2003; Savage et al., 2003). A separate classification, also based on gene expression profiling, identified three discrete subsets defined by the expression of genes involved in oxidative phosphorylation (OXP), B cell receptor/proliferation (BCR), and tumor microenvironment/host inflammatory response (HR) (Monti et al., 2005).
The sub-classification of DLBCL suggests that this disease may in fact comprise several distinct entities utilizing different pathogenetic mechanisms. This notion is supported by the observation that multiple genetic lesions of plausible pathogenetic significance segregate with different subtypes of DLBCL (Lenz et al., 2008b). With a focus on the ABC/GCB-based classification, it is known that translocations of BCL2 (Huang et al., 2002), mutations within the BCL6 autoregulatory domain (Iqbal et al., 2007; Pasqualucci et al., 2003), and mutations of EZH2 (Morin et al., 2010) are associated with the GCB subtype, whereas BCL6 translocations (Iqbal et al., 2007; Ye et al., 1993), amplifications of the BCL2 locus on 18q24 (Iqbal et al., 2004) and mutations within the NF-κB (CARD11, TNFAIP3/A20) (Compagno et al., 2009; Lenz et al., 2008a) and B cell receptor signaling (CD79B) (Davis et al., 2010) pathways segregate with the ABC subtype.
Additionally, inactivating mutations of PRDM1/BLIMP1 have been found exclusively in the ABC subtype (~24% of cases) (Pasqualucci et al., 2006; Tam et al., 2006), although the precise mechanism by which these lesions contribute to lymphoma development has not yet been fully elucidated. BLIMP1 encodes a transcriptional repressor that is essential for the terminal differentiation of all B cells into plasma cells, as demonstrated by the fact that B cell conditional knockout mice fail to produce plasma cells and serum immunoglobulins (Shapiro-Shelef et al., 2003). BLIMP1 is thought to promote terminal differentiation in part by repressing genes important in B cell receptor signaling and cellular proliferation (Lin et al., 1997; Shaffer et al., 2002a). Our initial study also reported rare missense mutations of the BLIMP1 gene, but their functional consequences were not addressed. Furthermore, the majority of ABC-DLBCL studied (~77%) did not express the BLIMP1 protein despite the presence of IRF4, a transcriptional repressor which is known to be invariably co-expressed with BLIMP1 in normal GC B cells and in all plasma cells (Angelin-Duclos et al., 2000), suggesting that mechanisms alternative to mutations may be contributing to the lack of protein expression in ABC-DLBCL. Finally, in vivo evidence establishing a direct link between BLIMP1 inactivation and lymphomagenesis has yet to be reported.
In the present study, we investigated the full spectrum of BLIMP1 lesions by comprehensively characterizing a large panel of DLBCL for the presence of mutations, copy number alterations and expression of the BLIMP1 protein. We analyzed the functional consequences of the BLIMP1 missense mutations and explored additional epigenetic mechanisms to inactivate BLIMP1 in ABC-DLBCL. Finally, we assessed the contribution of BLIMP1 inactivation to the pathogenesis of ABC-DLBCL in vivo.
RESULTS
Inactivation of BLIMP1 by truncating mutations and biallelic gene deletions in DLBCL
To investigate the full complement of genetic lesions affecting BLIMP1 in DLBCL, we characterized 158 DLBCL samples (139 primary biopsies and 19 cell lines) representative of the major phenotypic subtypes for the presence of mutations and copy number changes affecting the BLIMP1 gene. The study panel included 51 ABC, 61 GCB and 10 unclassified DLBCL, as determined by gene expression profile analysis. The remaining 36 cases were classified by immunohistochemistry into GC (n=12) and non-GC type (n=24) (Hans et al., 2004).
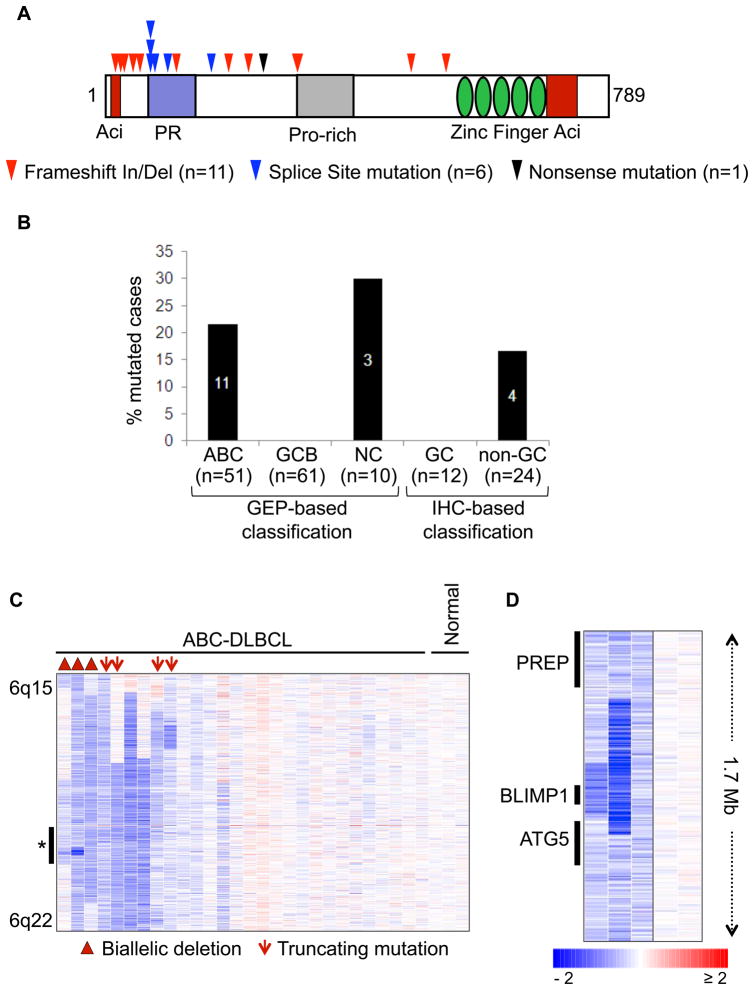
Sequencing analysis of the BLIMP1 coding exons identified 18 truncating mutations in 16 biopsies and 2 cell lines, including frameshift insertions and deletions (n=11), splice site mutations (n=6) and one nonsense mutation. Most of the changes clustered towards the N-terminal portion of the BLIMP1 protein and were predicted by sequence analysis to inactivate protein function by removing critical functional domains, including the PR, proline-rich and DNA binding Zinc finger domains (Figure 1A). Consistent with initial reports (Pasqualucci et al., 2006; Tam et al., 2006), all of these mutations were found in cases displaying an ABC/NC (n=14/51, 27%) or non-GC (n=4/24, 17%) phenotype, indicating that genetic inactivation of BLIMP1 is specific for this DLBCL subtype (Figure 1B). Analysis of 6q21 deletions, examined by a combination of copy number analyses, revealed that the majority of the mutated cases (n=16/18, 89%) had lost expression of the wild-type allele due to deletion, epigenetic silencing, or uniparental disomy of the mutated allele, demonstrating biallelic inactivation of the gene (Table S1 and Figure S1). Moreover, 3 ABC-DLBCL cases were found to harbor biallelic gene deletions of 21 Mb, 742 Kb and 271 Kb, with the latter only encompassing BLIMP1, but not the two proximal genes ATG5 and PREP (Figure 1C). This finding provides direct evidence that BLIMP1 is a critical target gene in the 6q21 deleted region commonly observed in DLBCL (Gaidano et al., 1992).
Figure 1. Inactivation of BLIMP1 by truncating mutations and biallelic deletions in ABC-DLBCL.
(A) Distribution of truncating mutations along the BLIMP1 protein, with known functional domains annotated. Aci=acidic domain; PR=PR domain; Pro-rich=proline-rich domain (see also Table S1). (B) Percentage of cases with truncating BLIMP1 mutations in various DLBCL subtypes classified by gene expression profiling (GEP) or by immunohistochemistry (IHC)(see text). (C) dChip SNP inferred copy number heatmap of the 6q15-q22.1 region in ABC-DLBCL cases and three normal DNA controls. Cases harboring homozygous deletions or truncating mutations are denoted by symbols (see also Figure S1). The region indicated by an asterisk is shown at higher magnification in panel (D) for the three homozygously deleted cases (and two normal DNAs), with the approximate position of the BLIMP1, ATG5 and PREP genes on the left.
Missense mutations affect BLIMP1 protein stability and its trans-repression activity
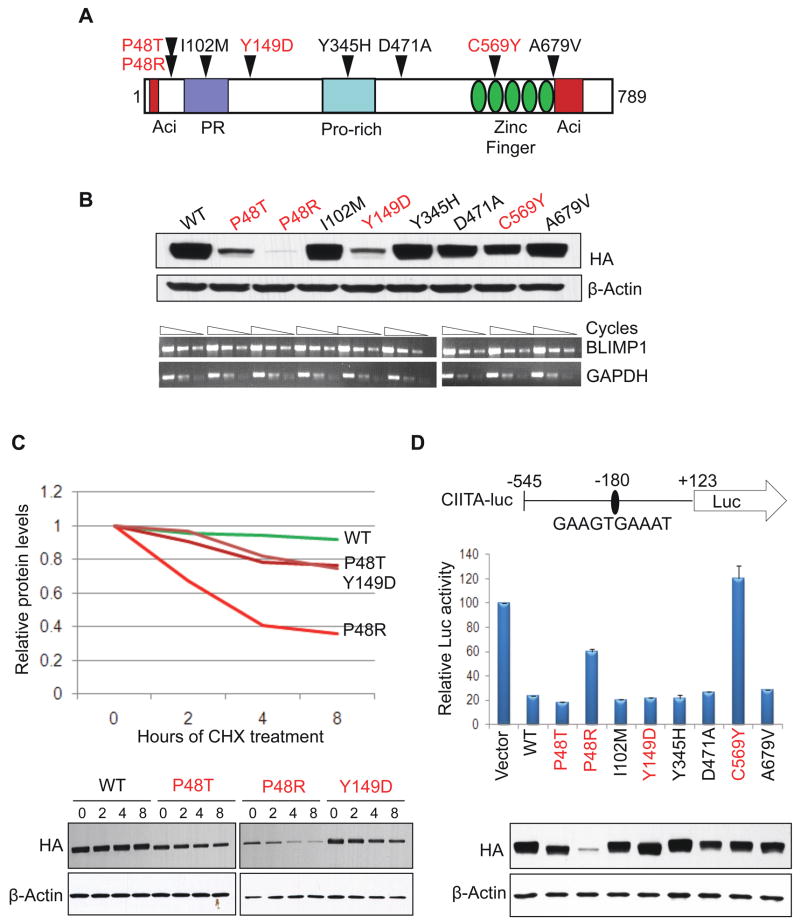
In 7 additional cases (5 ABC/non-GC and 2 GCB/GC DLBCL), we uncovered missense mutations affecting various residues along the entire BLIMP1 protein (Figure 2A), and associated with loss of the second allele in three cases (Table S2). Sequencing analysis of matched normal DNA in one sample with available material demonstrated the somatic origin of the mutation (Figure S1). The remaining variants were not found in public SNP databases (see methods), with one exception (Y345H), which was therefore excluded from the calculation of the mutation frequency.
Figure 2. Missense mutations affect BLIMP1 protein stability and trans-repression activity.
(A) Distribution of missense mutations along the BLIMP1 protein; in red, mutants that showed an effect in any of the assays performed in (B–D)(see also Table S2). (B) Western blot (top) and semi-quantitative RT-PCR (bottom) analysis of exogenous BLIMP1 expression in 293T cells transfected with equimolar amounts of vectors expressing HA-tagged wild-type or mutant BLIMP1 alleles. (C) Analysis of exogenous BLIMP1 protein expression in 293T cells transfected with the indicated mutant alleles and treated with cycloheximide for 2, 4 or 8 hours. Data were quantitated by densitometric analysis, normalized to β-actin levels, and graphed relative to time zero (top). The western blot analysis is shown on the bottom panel. (D) Trans-repression activity of wild-type and mutant BLIMP1 proteins in 293T cells co-transfected with a luciferase reporter construct driven by the human CIITA promoter (region −545 to +123, encompassing a consensus BLIMP1 binding site at position −180)(top). Luciferase activities are represented as percent change relative to the basal activity of the reporter (set to 100), after normalization to Renilla luciferase activity (mean ± SD, as obtained from three independent experiments). In the bottom panel, western blot analysis using anti-HA antibodies monitors for the corresponding exogenous BLIMP1 expression levels; note that, for the three unstable mutants, higher amounts of plasmid DNA were transfected to achieve comparable levels. Nevertheless, expression of the P48R mutant protein remained significantly lower than wild-type, presumably due to its marked instability (see also Figure S2).
In order to test the functional significance of these mutations, we first assessed the basal expression levels of the corresponding alleles upon transient transfection in 293T cells. As shown in Figure 2B, three ABC/non-GC DLBCL mutants (P48T, P48R and Y149D) were expressed at significantly lower protein levels compared to wild-type BLIMP1, despite similar mRNA levels, suggesting that these mutations may destabilize the BLIMP1 protein. Treatment with cycloheximide documented that the proteins encoded by these three mutants had a significantly decreased half-life, with the P48R polypeptide being most unstable (Figure 2C). Accordingly, the Polyphen algorithm (Sunyaev et al., 2001), used to make in silico predictions of the potential functional consequences of missense mutations, determined that these three mutations may affect the BLIMP1 protein structure and/or folding (data not shown). Furthermore, the SUDHL2 ABC-DLBCL cell line, which harbors the P48R mutation, expresses significantly reduced BLIMP1 protein levels despite elevated amounts of mRNA (comparable to the multiple myeloma cell line U266) (Figure S2), consistent with protein instability.
We then assessed the relative trans-repression activity of the mutant proteins by measuring their ability to down-regulate the expression of a luciferase reporter gene driven by a portion of the CIITA gene promoter, a known BLIMP1 direct target (Piskurich et al., 2000). Compared to wild-type, two mutants (P48R, C569Y) failed to efficiently repress the reporter (Figure 2D). In the P48R mutant, this effect can be attributed to the observed protein instability and consequently reduced expression levels; however, the C569Y mutant had lost its activity despite comparable protein amounts, suggesting that this mutation may directly impair the BLIMP1 trans-repression function. Indeed, this mutation substitutes a critical cysteine within the second zinc finger of the BLIMP1 DNA binding domain, and abrogated DNA binding to the endogenous CIITA gene promoter, as assessed in B cells by chromatin immunoprecipitation assays (Figure S2). Overall, these results indicate that a subset of BLIMP1 missense mutants, specifically those associated with ABC/non-GC DLBCL, impair the stability and/or function of the BLIMP1 protein.
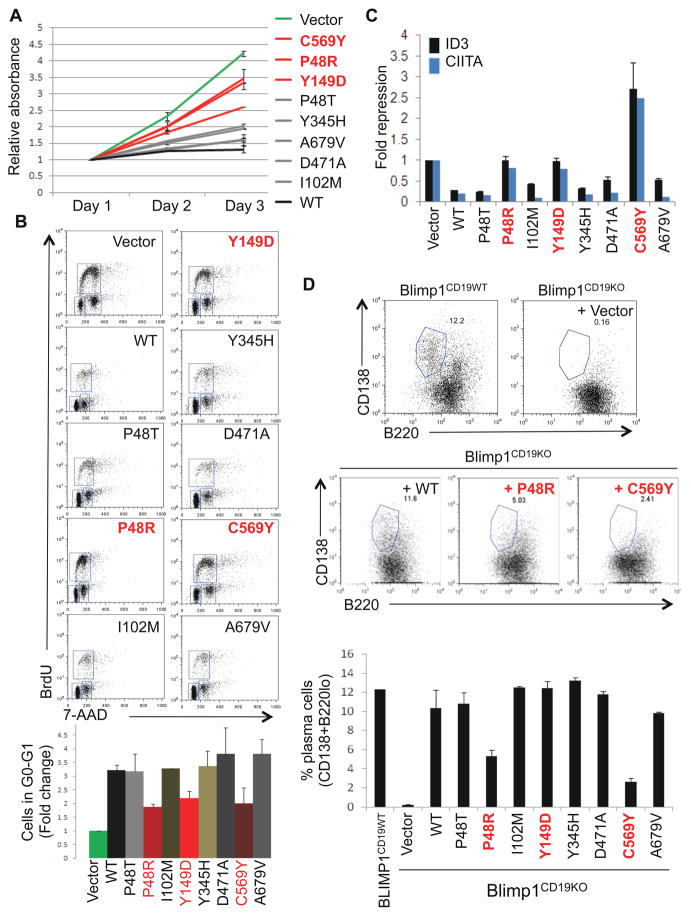
To further investigate the consequences of these mutations in B cells, we over-expressed the mutant alleles in the GCB-DLBCL cell line BJAB via lentiviral-mediated transduction. Compared to empty vector-transduced cells, those infected with wild-type BLIMP1 failed to proliferate (Figure 3A), consistent with the previously reported ability of BLIMP1 to repress multiple genes involved in cell cycle progression (Lin et al., 1997; Shaffer et al., 2002a). On the contrary, cells infected with vectors expressing either unstable (P48R, Y149D) or trans-repression defective (C569Y) missense mutants proliferated at rates similar to those of control cells. Flow cytometric analysis of the cell cycle profile revealed a 3-fold increase in the G0/G1 population of cells expressing wild-type BLIMP1, but not in cells transduced with the above three mutants (Figure 3B), further documenting their loss of function.
Figure 3. BLIMP1 missense mutations impair its ability to induce cell cycle arrest and promote plasma cell differentiation.
(A) Proliferative capacity of BJAB B cells transduced with lentiviral vectors expressing the indicated BLIMP1 proteins along with GFP, as assessed by an MTT assay on sorted GFP+ cells. Shown are representative data from one of two independent experiments performed in duplicate (mean ± SD) (see also Figure S3). (B) Left, representative flow cytometric analysis of BJAB B cells transduced with the indicated vectors and stained for incorporated BrdU and 7-amino-actinomycin D (7-AAD). Region gates define cells residing in G0-G1, S and G2-M phases of the cell cycle. The G0-G1 population was quantitated relative to empty vector-transduced cells (set at 1), and the mean ± SD from two independent experiments is shown below. (C) Expression of the BLIMP1 targets ID3 and CIITA in sorted GFP+ BJAB B cells, as determined by quantitative real-time RT-PCR (n=3; mean ± SD). Levels were normalized to both BLIMP1 and GAPDH, and are shown as fold changes relative to vector-transduced cells (set as 1). (D) Representative flow cytometric analysis of CD138 and B220 staining in Blimp1CD19KO splenic B cells, reconstituted with the indicated vectors and stimulated to undergo plasma cell differentiation by LPS treatment for 3 days, as compared to wild-type (Blimp1CD19WT) B cells. The percentage of cells in the gated (plasma cell) population is shown. Data from two independent experiments are quantitated in the bottom panel (mean ± SD). Mutants that showed an effect in any of the assays (A-D) are indicated in red.
Consistent with results obtained in the previous assays, the P48R, Y149D and C569Y mutants were also unable to repress the expression of endogenous ID3 and CIITA, two known direct target genes of BLIMP1 (Shaffer et al., 2002a) (Figure 3C). Interestingly, when compared to cells transduced with wild-type BLIMP1 or other functionally active mutants, cells expressing these three proteins showed consistently higher levels of GFP, which is co-regulated with BLIMP1 via the bicistronic lentiviral cassette, suggesting counter-selection against high, toxic levels of wild-type BLIMP1 but not against the functionally deficient mutants (Figure S3).
Finally, we assessed the ability of the various missense mutants to promote plasma cell differentiation, the primary physiological function of BLIMP1 in B cells. To address this question, we utilized Blimp1 conditional-knockout mouse B cells (Blimp1CD19KO), which are unable to undergo plasma cell differentiation after stimulation with lipopolysaccharide but can be rescued by retroviral-mediated delivery of wild-type BLIMP1 (Shapiro-Shelef et al., 2003). In this system, approximately 10% of the cells reconstituted with wild-type BLIMP1 efficiently differentiated into CD138+B220lo plasma cells, analogous to control cells isolated from wild-type animals (12%)(Figure 3D). Conversely, plasma cell differentiation was significantly reduced in cells reconstituted with the P48R and C569Y mutants (5% and 2% respectively). The Y149D mutant, which was defective in previous assays, appeared to retain its activity in this system, likely due to the fact that sufficiently high protein levels were achieved via retroviral over-expression (data not shown).
In summary, data from a variety of assays in both 293T and B cells demonstrate that four of the seven somatic missense mutants analyzed displayed functional defects, ranging in their severity from a complete loss of multiple functions (P48R and C569Y) to more subtle effects (P48T and Y149D). Notably, the defective mutants were all associated with ABC/non-GC DLBCL cases.
Epigenetic inactivation of BLIMP1 in ABC-DLBCL cases carrying BCL6 translocations
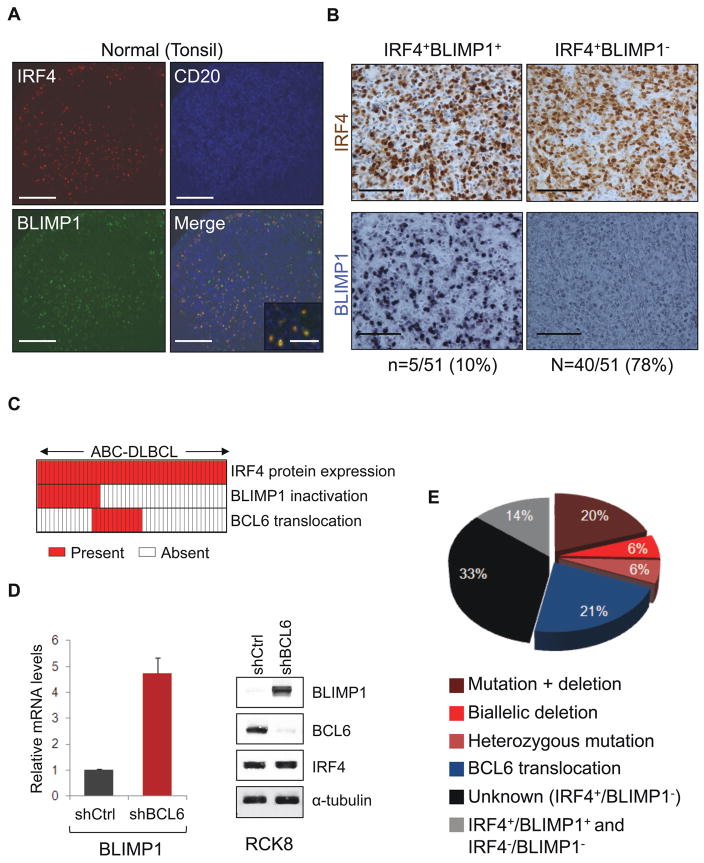
Taking together cases with functionally-deficient missense mutants and cases with truncating mutations and biallelic deletions, our data demonstrate that BLIMP1 is genetically inactivated in approximately one-third of ABC-DLBCL (n=16/51). Nonetheless, additional mechanisms appeared to be contributing to BLIMP1 inactivation in this tumor subtype, because an even larger fraction of cases (n=27/51, 53%) lacked BLIMP1 protein expression despite the absence of BLIMP1 structural alterations and the presence of IRF4, a protein invariably co-expressed with BLIMP1 in a subset of normal human GC B cells (Angelin-Duclos et al., 2000) and involved in the terminal differentiation pathway leading to BLIMP1 activation (Klein et al., 2006; Saito et al., 2007) (Figure 4A,B). Given that the BCL6 transcriptional repressor can directly repress BLIMP1 expression (Tunyaplin et al., 2004) and is deregulated by chromosomal translocations preferentially in ABC-DLBCL (Ye et al., 1995), we assessed the relationship between structural alterations of BCL6 and BLIMP1. Notably, ~26% (n=13/51) of ABC-DLBCL cases contained translocations of BCL6 that were, with two exceptions, mutually exclusive with BLIMP1 structural alterations (Figure 4C). Cases with BCL6 translocations also had significantly lower BLIMP1 mRNA levels compared to cases that lacked translocations and expressed the BLIMP1 protein (Figure S4). Furthermore, shRNA-mediated knockdown of BCL6 expression in the BCL6-translocated RCK8 and OCI-Ly8 ABC/non-GC-DLBCL cell lines promoted up-regulation of BLIMP1 mRNA and protein (Figure 4D and data not shown). Overall, these data support the hypothesis that deregulated BCL6 expression via chromosomal translocations is responsible for suppressing BLIMP1 expression in a fraction of IRF4+ ABC-DLBCLs. Thus, over half (53%, n=27/51) of these tumors have inactivated BLIMP1 through predominantly mutually exclusive structural alterations of BLIMP1 and BCL6 (Figure 4E).
Figure 4. BCL6 translocations and BLIMP1 inactivation are mutually exclusive in ABC-DLBCL.
(A) Immunofluoresence analysis of BLIMP1 (green), IRF4 (red) and CD20 (blue) expression in normal GC cells of a human tonsil (scale bar, 325μm; inset, 150μm). (B) IRF4 (brown) and BLIMP1 (blue) immunostaining in representative ABC-DLBCL cases displaying a normal expression pattern (IRF4+BLIMP1+)(left panels) or specific lack of BLIMP1 expression (IRF4+BLIMP1−)(right panels)(scale bar: 125 μm). The percentage of cases in each group is provided below. Six additional cases (12%) were negative for expression of both proteins (not shown; see also Table S1 and S2 for a detailed characterization of individual cases). (C) Distribution of BLIMP1 and BCL6 structural alterations in IRF4+ ABC-DLBCL. Columns represent individual patients, with color-codes indicating the presence or absence of the corresponding feature. (D) BLIMP1 mRNA (left) and protein (right) levels in the BCL6-translocated RCK8 cell line, transduced with lentiviral vectors expressing a control shRNA (shCtrl) or a BCL6-specific shRNA (shBCL6). BLIMP1 mRNA levels were determined by quantitative real-time RT-PCR and are shown as fold change relative to shCtrl-transduced cells, after normalization for GAPDH (n=3; mean ±SD). Western blot analysis of BCL6 controls for efficient BCL6 knockdown (see also Figure S4). (E) Overall frequency of BLIMP1 and BCL6 structural alterations in ABC-DLBCL. The unknown category denotes cases that lack BLIMP1 protein expression, in the absence of BLIMP1 or BCL6 structural alterations. Two cases carrying both BCL6 translocations and biallelic BLIMP1 inactivation were included into the “BLIMP1 mutation + deletion” category.
Conditional deletion of Blimp1 in the mouse results in the development of B cell lymphoproliferative disorders and DLBCL
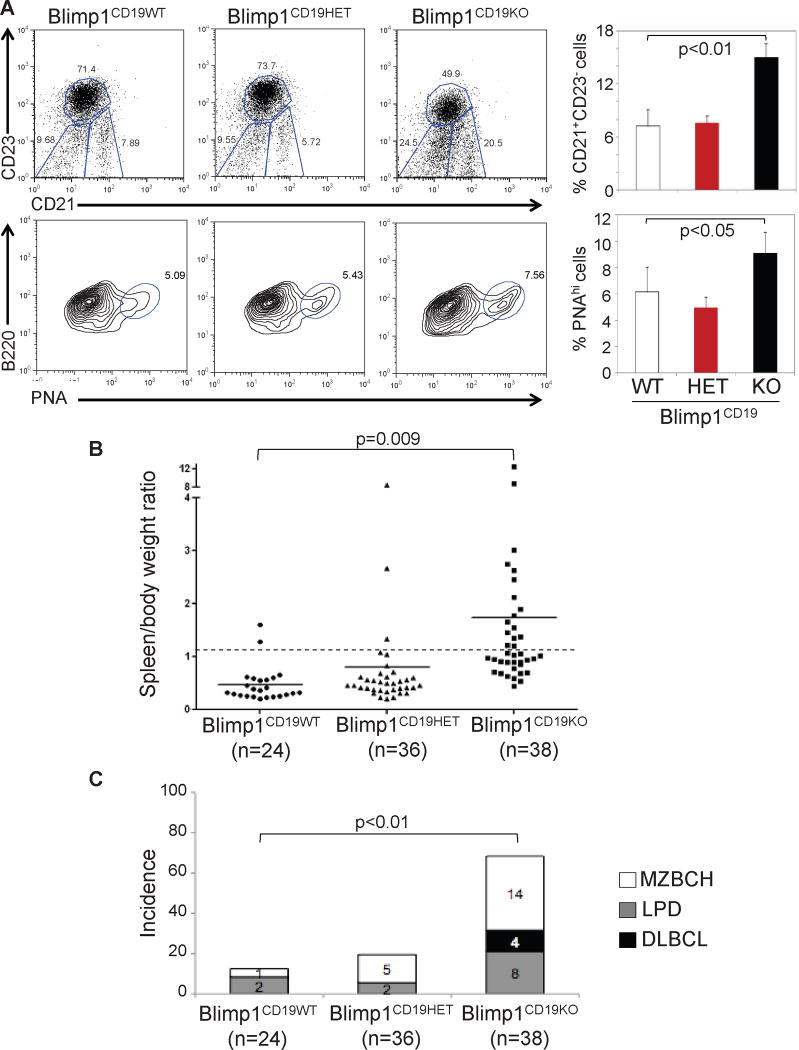
To assess the role of BLIMP1 inactivation in lymphomagenesis in vivo, we generated mice lacking Blimp1 specifically in B cells by crossing Blimp1 conditional-knockout mice (Ohinata et al., 2005) with CD19-Cre (Rickert et al., 1997) or Cγ1-Cre (Casola et al., 2006) mice, leading to deletion of Blimp1 in all B cells (Blimp1CD19KO) or GC B cells (Blimp1Cγ1KO), respectively. Consistent with previous data (Shapiro-Shelef et al., 2003), Blimp1CD19KO B cells failed to differentiate into immunoglobulin-secreting plasma cells, both in vivo and after ex vivo stimulation (Figure S5 and data not shown). On the contrary, immunized Blimp1Cγ1KO mice had selective impairment in GC-derived IgG1 but not in IgM production, consistent with a specific block in GC-dependent plasma cell differentiation (Figure S5). The Blimp1CD19KO mice also showed a significant increase of the fraction of B220+PNAhi GC B cells and CD21+CD23− extra-follicular marginal zone (MZ) B cells (Figure 5A). Although we cannot formally exclude that a decrease in the CD23+CD21− follicular (FO) B cell compartment can contribute to the increase in the MZ B cell fraction, the observation that FO B cell development is normal in an analogous Blimp1 knockout model (Shapiro-Shelef et al., 2003) suggests that the accumulation of both GC and MZ B cells is due to a block in terminal B cell differentiation.
Figure 5. Blimp1 B cell conditional knockout mice develop lymphoproliferative disorders.
(A) Representative flow cytometric analysis of splenic B cell suspensions isolated from mice of the indicated genotypes and stained for CD21/CD23 (top) and B220/PNA (bottom). A significant increase in both GC (B220+PNAhi) and extra follicular, MZ (CD21+CD23−) B cell subpopulations can be seen in the Blimp1CD19KO animals, as compared to their control littermates. Data are quantitated on the right (mean ± SD; n=3). (B) Spleen/body weight ratio in mice of the indicated genotypes, analyzed between 10–16 months of age. Splenomegaly was defined as an increase in the spleen/body weight ratio above 0.7% (dashed line). Solid lines indicate the mean value in each genotype. (C) Frequency of lymphoproliferative disorders in the three cohorts shown in (B). MZBCH=marginal zone B cell hyperplasia; LPD=lymphoproliferative disease; DLBCL=diffuse large B cell lymphoma; p values (student’s t-test) are provided if significant (<0.05). See also Figure S5 and Table S3.
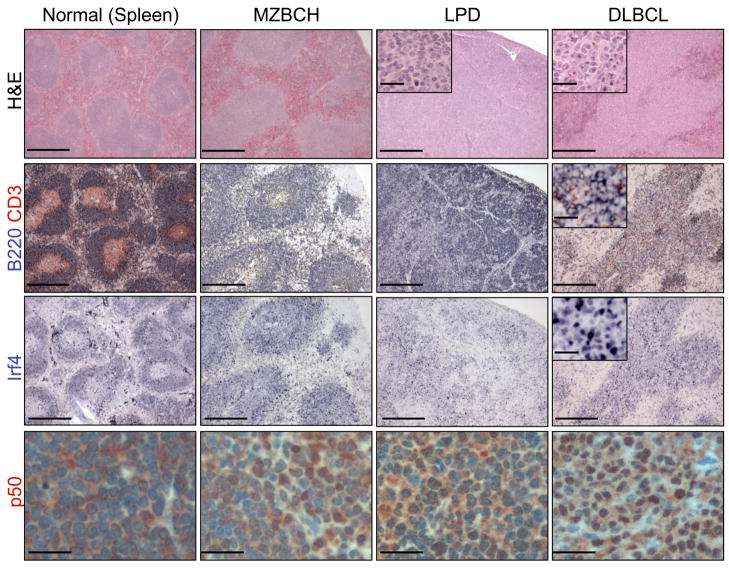
Macroscopic examination of animals sacrificed between 10–16 months of age showed the presence of splenomegaly, suggestive of lymphoproliferation, in most Blimp1CD19KO mice (90%, n=34/38) (Figure 5B). Subsequent histological examination of the lymphoid organs revealed that ~68% (n=26/38) of the animals had developed a variety of lymphoproliferative disorders, including MZ B cell hyperplasia (MZBCH; n=14), characterized by the expansion of small cells surrounding the B cell follicles; lymphoproliferative disease (LPD; n=8), defined by the initial obliteration of the follicular architecture; and DLBCL (n=4), characterized by a complete effacement of the tissue architecture due to infiltrating large cells (Figures 5C and 7). These disorders were of B cell origin, as they stained positive for the pan-B cell marker B220 (Figure 7). Furthermore, analysis of the rearranged Ig genes demonstrated a clonal origin of the disease in all DLBCL and LPD tested, as well as in 5/14 (36%) MZBCH (Figure S6A). Most of the clonal lymphoproliferations carried somatically hypermutated immunoglobulin genes (71%, n=10/14), documenting the tumor derivation from a cell that had transited the GC (Table S3).
Figure 7. Blimp1 B cell conditional knockout mice develop DLBCL with an activated phenotype and constitutive NF-κB activation.
Representative spleen and lymph node sections from Blimp1 knock-out mice presenting with a spectrum of lymphoproliferative disorders, including MZBCH, LPD, and overt DLBCL, as compared to a wild-type control (see results for a detailed histological description, and Figure S6 for additional data). Tissues were stained with hematoxylin and eosin (H&E) or immunostained with antibodies against the B220 pan-B cell marker, IRF4 and the NF-κB subunit p50, as indicated (Scale bar, 1250 μm; inset, 125 μm; scale is 50 μm for p50 stain).
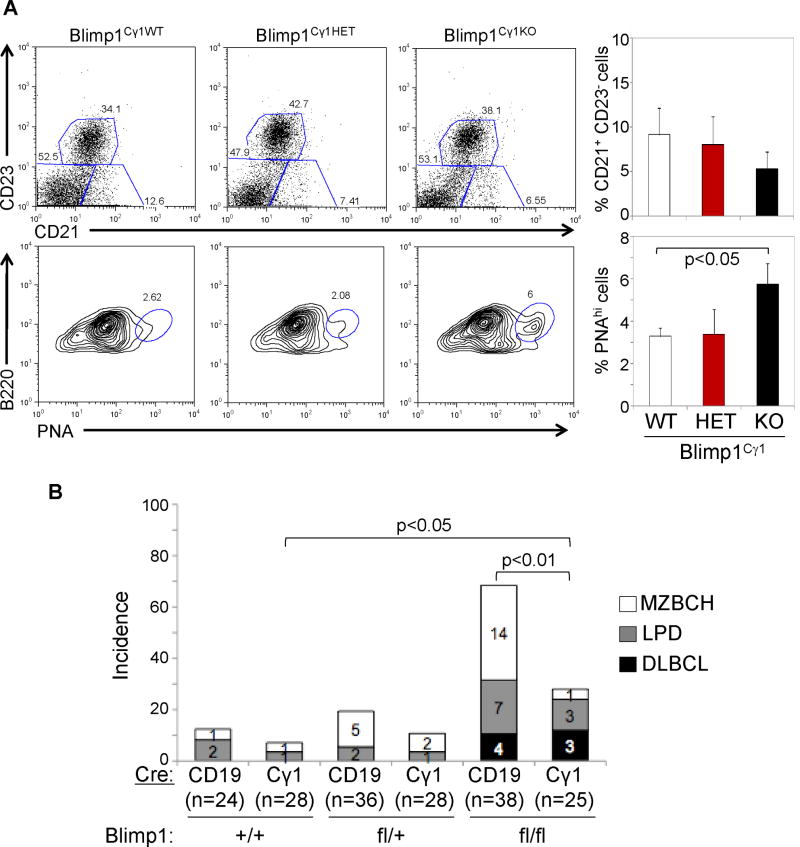
Similar to the Blimp1CD19KO mice, Blimp1Cγ1KO mice showed a 2-fold increase in the proportion of GC B cells after immunization (Figure 6A). However, these animals did not display an increase in the MZ B cell compartment, consistent with the restriction of Blimp1 deletion to GC B cells. When analyzed between 12–18 months of age, Blimp1Cγ1KO mice did not show splenomegaly nor increased incidence of MZBCH, yet they developed LPD and DLBCL at similar penetrance with respect to the Blimp1CD19KO model (Figure 6B), demonstrating that deletion of Blimp1 in the GC alone was sufficient to promote lymphomagenesis.
Figure 6. Conditional deletion of Blimp1 in the GC promotes lymphomagenesis.
(A) Representative flow cytometric analysis of the CD21+CD23− MZ B cell and B220+PNAhi GC B cell compartments in mice of the indicated genotypes, analyzed ten days after SRBC immunization (left). Data from three mice per genotype are quantitated on the right (mean ± SD). (B) Percentage of mice developing lymphoproliferative disorders in the CD19-Cre and Cγ1-Cre models, sacrificed between 10–18 months of age. See also Table S4.
In both models, the majority of the DLBCL were Irf4-positive (86%, n=6/7) (Figure 7) and negative for GC and differentiation markers (i.e., Bcl6, PNA and CD138) (Figure S6B), consistent with the phenotype of a post-GC, activated, pre-plasmablastic B cell (Falini et al., 2000). Interestingly, these tumors also displayed nuclear NF-κB p50 (Figure 7) indicative of constitutive NF-κB activation, as observed in human ABC-DLBCL (Davis et al., 2001). Sequencing analysis of three common targets of genetic lesions associated with constitutive NF-κB activity in the human disease (Tnfaip3/A20, Card11 and Cd79b) did not reveal the presence of mutations in six DLBCL cases analyzed (data not shown), suggesting that constitutive NF-κB activation is necessary for ABC-DLBCL pathogenesis, but is induced by different mechanisms in this model.
Over time, Blimp1CD19KO mice showed significantly reduced survival, such that at 15 months of age only 20% of the animals were still alive (Figure S6C). Interestingly, the majority of these mice had an expansion of the bronchus-associated lymphoid tissue (BALT) (79%, n=30/38) consistent with the diagnosis of BALT hyperplasia (BALT-H), in the presence (n=21/30) or absence (n=9/30) of other lymphoproliferative disorders (Figure S6D). Immunostaining of the BALT-H demonstrated an expansion of both Pax5+ B cells and CD3+ T cells within a relatively preserved B-T cell architecture (Figure S6E), suggestive of a benign lymphoproliferation. In contrast, Blimp1Cγ1KO mice did not show reduced survival nor suffered from BALT-H (Figure S6C and D), suggesting that the BALT-H results from the expansion of Blimp1-deleted, extra-follicular mature B cells (see Table S4 for a comparison of the phenotypes from the two models). Taken together, these data provide conclusive evidence for BLIMP1 as a bona fide tumor suppressor gene in ABC-DLBCL.
DISCUSSION
Heterogeneous mechanisms of BLIMP1 inactivation
This study demonstrates that BLIMP1 is inactivated in ABC-DLBCL through a variety of genetic means. Moreover to truncating mutations and deletions, we describe a mechanism by which missense mutations inactivate BLIMP1 by either destabilizing the protein or impairing its trans-repression activity. Moreover, we show that transcriptional silencing of BLIMP1 via deregulating BCL6 translocations represents an alternative mechanism to inactivate its function. The mutually exclusive nature of BLIMP1 and BCL6 lesions in DLBCL is consistent with the current model for plasma cell differentiation, whereby IRF4-mediated transcriptional repression of BCL6 leads to de-repression of BLIMP1 and subsequent GC exit (Klein and Dalla-Favera, 2008; Saito et al., 2007); thus, structural alterations of either gene should play analogous roles in blocking terminal differentiation. In addition, these lesions will likely contribute to lymphomagenesis through additional, distinct mechanisms. This possibility is supported by the fact that the IμHABCL6 mouse model of DLBCL does not phenocopy the Blimp1 conditional-knockout mouse model, as the IμHABCL6 mice have both an increased incidence and heterogeneity of DLBCL (Cattoretti et al., 2005). Since BCL6 is known to regulate multiple cellular functions, including DNA damage responses (Phan and Dalla-Favera, 2004; Ranuncolo et al., 2007; Ranuncolo et al., 2008), signal transduction (Basso et al., 2010; Juszczynski et al., 2009) and cell cycle progression (Phan et al., 2005; Shaffer et al., 2000), it is possible that ABC-DLBCL cases carrying BCL6 lesions have distinct phenotypic traits.
Despite IRF4 expression, one-third of ABC-DLBCL lacked BLIMP1 protein expression in the absence of BLIMP1 or BCL6 structural alterations. Analysis of a limited number of cases ruled out the possibility that BLIMP1 promoter hypermethylation was responsible for the absence of Blimp1 expression, suggesting that additional unknown genetic lesions may contribute to blocking terminal differentiation. In our series, amplification of SPIB or PAX5, two genes that are also known to repress BLIMP1 (Mora-Lopez et al., 2007; Schmidlin et al., 2008), did not seem to be involved (data not shown). However, genome wide approaches may be necessary to determine whether structural alterations in other genes and/or microRNAs, such as those recently described to regulate BLIMP1 expression (Leucci et al., 2010; Malumbres et al., 2009; Nie et al., 2008; West et al., 2009; Zhang et al., 2009), play a role in preventing BLIMP1 protein expression in this subset.
Association of BLIMP1 inactivation with other lesions
The long latency and the clonality of DLBCL in the Blimp1 conditional-knockout mouse models indicate that oncogenic events affecting other pathways cooperate with BLIMP1 inactivation to promote a full neoplastic phenotype and to cause DLBCL. Since the NF-κB transcriptional complex is known to be constitutively active in the majority of ABC-DLBCL, genetic alterations within this pathway may be important contributors to lymphomagenesis. Interestingly, concurrent inactivation of BLIMP1 and mutations within NF-κB pathway components are found in a significant fraction of human ABC-DLBCL (data not shown). Furthermore, the DLBCLs that develop in Blimp1 conditional-knockout mice display evidence of constitutive NF-κB activation. Thus, future in vivo studies should address the cooperative role of these two pathways in lymphomagenesis.
BLIMP1 as a tumor suppressor gene
In the present study, we have provided converging evidence from human genetics, functional studies and mouse models that BLIMP1 is a bona fide tumor suppressor gene in ABC-DLBCL. One major role for BLIMP1 inactivation in lymphomagenesis is to block terminal differentiation and to modulate the expression of genes involved in cell cycle progression. This notion is supported by our observation that introduction of BLIMP1 into a DLBCL cell line leads to G1 cell cycle arrest, as well as by previous work showing that activated Blimp1 knockout mouse B cells have increased proliferative capacity (Shapiro-Shelef et al., 2003). Thus, the expansion of GC and non-GC MZ B cells observed in our mice may be in part due to the enhanced proliferation of Blimp1-null activated B cells failing to undergo terminal differentiation. The Blimp1CD19KO and Blimp1Cγ1KO mice will provide useful pre-clinical models to explore additional pathways contributing to lymphomagenesis.
EXPERIMENTAL PROCEDURES
Tumor samples and classification
Primary biopsies from 139 newly diagnosed DLBCL patients were obtained from the archives of the Departments of Pathology at Columbia University and Weill Cornell Medical College, after approval by the respective Institutional Review Boards [Exempt Human Subject Research of anonymized/de-identified existing pathological specimens, under regulatory guideline 45 CFR 46.101(b)(4)]. For a description of the 19 DLBCL cell lines used, see Supplementary Experimental Procedures. Samples were classified by gene expression profile analysis into the ABC or GCB subtypes as previously described (Compagno et al., 2009). Cases that could not be profiled due to the lack of material or low percentage of tumor cells in the biopsy were classified into GC and non-GC DLBCL based on expression of CD10, BCL6 and IRF4 (Hans et al., 2004).
Mutational and copy number analyses of the BLIMP1 gene
Genomic DNA from 158 DLBCL samples was extracted according to standard methods and used for amplification of the BLIMP1 coding exons as previously described (Pasqualucci et al., 2006). Purified amplicons were sequenced directly from both strands and compared to the corresponding germline sequence (NM_001198) using the Mutation Surveyor Version 2.41 software (Soft Genetics, State College, PA). Mutations were confirmed on independent PCR products and their somatic origin was verified by analysis of paired normal DNA, where available. In addition, all mutations were verified in available databases of germline variants (NCBI dbSNP, Build 130; Ensembl; Watson genome sequence). Copy number analysis of BLIMP1 was performed using a combination of fluorescent in situ hybridization, single nucleotide polymorphism profiling arrays using the Affymetrix Genome-Wide Human SNP Array 6.0 platform (Affymetrix, Santa Clara, CA, USA) and quantitative copy number PCR (see supplemental methods for additional details).
Transient transfections/Luciferase reporter assays
293T cells were transiently transfected with equimolar amounts (200 ng) of wild-type and mutant pCMV-HA-BLIMP1 vectors via the calcium phosphate precipitation method, as described (Gu et al., 1993). For analysis of protein stability, transfected 293T cells were treated with 50 μg/mL cycloheximide (Sigma-Aldrich) for 2, 4, and 8 hours before harvesting for protein analysis. For luciferase reporter assays, 293T cells were co-transfected with 10 ng of wild-type or mutant pCMV-HA-BLIMP1 vectors, 10 ng of CIITA-luc reporter construct and 0.1 ng of the TK-RL Renilla reporter as control for transfection efficiency (Promega, Madison, WI). Cells were harvested 48 hours after transfection, and the Dual Luciferase Reporter Assay (Promega) was performed according to the manufacturer’s instructions, in three independent experiments.
Cell proliferation and cell cycle assays
Cellular proliferation was measured by an MTT assay (Roche) using 10,000 cells/well in duplicate in two independent experiments. To assess the cell cycle profile, 60 hours after infection with the indicated lentiviral supernatants, BJAB cells were labeled with bromodeoxyuridine (BrdU) for 30 minutes before harvesting. BrdU incorporation and DNA content were analyzed with the APC BrdU flow kit (Becton Dickinson) (see supplemental methods for a detailed transduction protocol).
BCL6 knockdown
A previously validated BCL6 lentiviral shRNA construct (Basso et al., 2010) was used to knockdown BCL6 expression in RCK8 B cells. Control and BCL6 shRNA lentiviruses were produced and transduced into RCK8 B cells as described. Twenty-four hours after the second round of infection, cells were selected with 1μg/mL puromycin for 5.5 days, and live cells were purified by MACS using the Dead Cell Removal Kit (Miltenyi Biotec).
Ex vivo plasma cell differentiation assay
Primary mouse B cells were purified from the spleen of 8-week old Blimp1CD19KO mice using the Mouse B cell Isolation Kit (Miltenyi Biotec, Auburn, CA) and cultured in RPMI-1640 medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 U/ml streptomycin, 55 μm β-mercaptoethanol and 10 μg/mL lipopolysaccharides (Sigma-Aldrich). For the complementation assay, cells were transduced after one day with PINCO retroviral vectors expressing wild-type HA-BLIMP1 or its derivatives, as described. The percentage of CD138+ B220− plasma cells in the transduced (GFP+) population was determined 48 and 72 hours after the second round of infection by flow cytometric analysis, using anti-CD138-APC (Pharmingen, San Jose, CA) and anti-B220-PerCP (Pharmingen) antibodies. As control, splenic B cells from wild-type mice were treated in parallel with LPS and analyzed for differentiation one day earlier than transduced cells.
Mice
Blimp1CD19KO and Blimp1Cγ1KO mice and control littermates were generated on a mixed C57BL/6:129Sv background by crossing Blimp1fl/+ mice with Blimp1CD19HET or Blimp1Cγ1HET mice respectively, followed by offspring intercrossing. Genotyping was performed by PCR analysis and the protocol is available upon request. Animals were monitored for tumor incidence and survival biweekly over a period of 10–18 months, and sacrificed for analysis when visibly ill or at the end of the study, according to protocols approved by the Columbia University Institutional Animal Care and Use Committee. Flow cytometric analysis of B and T cell lymphoid compartments and histological examination of mouse tissues were also performed at 3, 12 and 15 months of age (n=3–5 mice per genotype). A detailed description of the methods is available in the accompanying supplemental material. Kaplan-Meier event-free survival curves were generated using the GraphPad Prism 5 software (GraphPad Sofware, La Jolla, CA), and statistical significance was calculated using the long-rank (Mantel-Cox) test. The student’s t-test (unpaired, two-tailed) was used to assess whether differences in the incidence of lymphoproliferative disorders were significant in knockout mice compared to control wild-type littermates.
SIGNIFICANCE.
ABC-DLBCL, the less curable subtype of DLBCL, has the molecular footprint of a tumor that likely arose from post-germinal centre B cells undergoing plasma cell differentiation. Our results show that the pathway controlling terminal differentiation is inactivated in these tumors by a heterogeneous set of mutually exclusive genetic lesions, leading to genetic or epigenetic inactivation of the master plasma cell regulator BLIMP1. The observation that conditional B cell deletion of Blimp1 in the mouse promotes DLBCL with features of human ABC-DLBCL demonstrates that the plasma cell differentiation pathway is critically involved in suppressing lymphomagenesis and provides conclusive evidence for BLIMP1 as a tumor suppressor. Mice with B cell specific deletion of Blimp1 may represent faithful models of pre-clinical interest for this disease.
Supplementary Material
Acknowledgments
We thank U. Klein, D. Dominguez-Sola and M. Saito for helpful discussions; A. Grunn for help with the sequencing analysis; J. Piskurich for providing the CIITA luciferase reporter, and G. Bornkamm for the bicistronic expression cassette. We also thank the Flow Cytometry, Transgenic Mouse and Genomics Shared Resources of the Herbert Irving Comprehensive Cancer Center. This work was supported by N.I.H. Grants CA-092625 and CA-37295, by a Specialized Center of Research grant from the Leukemia&Lymphoma Society, and by the Stewart Trust Fund. L.P. is on leave from the University of Perugia.
Footnotes
The authors declare no conflicts of interest.
Accession Numbers: The expression data reported in this paper have been deposited in the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database (Series Accession Number GSE12195).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abramson JS, Shipp MA. Advances in the biology and therapy of diffuse large B cell lymphoma: moving toward a molecularly targeted approach. Blood. 2005;106:1164–1174. doi: 10.1182/blood-2005-02-0687. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–5471. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- Basso K, Saito M, Sumazin P, Margolin AA, Wang K, Lim WK, Kitagawa Y, Schneider C, Alvarez MJ, Califano A, Dalla-Favera R. Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood. 2010;115:975–984. doi: 10.1182/blood-2009-06-227017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, Hao Z, Waisman A, Egert A, Ghitza D, Rajewsky K. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci U S A. 2006;103:7396–7401. doi: 10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, Mo T, Murty VV, Dalla-Favera R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B cell-receptor signalling in diffuse large B cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, Pacini R, Alunni C, Natali-Tanci L, Ugolini B, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–2092. [PubMed] [Google Scholar]
- Gaidano G, Hauptschein RS, Parsa NZ, Offit K, Rao PH, Lenoir G, Knowles DM, Chaganti RS, Dalla-Favera R. Deletions involving two distinct regions of 6q in B cell non-Hodgkin lymphoma. Blood. 1992;80:1781–1787. [PubMed] [Google Scholar]
- Gu W, Cechova K, Tassi V, Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci U S A. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL, Lynch JC, Armitage JO, Warnke RA, Alizadeh AA, et al. The t(14;18) defines a unique subset of diffuse large B cell lymphoma with a germinal center B cell gene expression profile. Blood. 2002;99:2285–2290. doi: 10.1182/blood.v99.7.2285. [DOI] [PubMed] [Google Scholar]
- Iqbal J, Greiner TC, Patel K, Dave BJ, Smith L, Ji J, Wright G, Sanger WG, Pickering DL, Jain S, et al. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B cell lymphoma. Leukemia. 2007;21:2332–2343. doi: 10.1038/sj.leu.2404856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Sanger WG, Horsman DE, Rosenwald A, Pickering DL, Dave B, Dave S, Xiao L, Cao K, Zhu Q, et al. BCL2 translocation defines a unique tumor subset within the germinal center B cell-like diffuse large B cell lymphoma. Am J Pathol. 2004;165:159–166. doi: 10.1016/s0002-9440(10)63284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszczynski P, Chen L, O’Donnell E, Polo JM, Ranuncolo SM, Dalla-Favera R, Melnick A, Shipp MA. BCL6 modulates tonic BCR signaling in diffuse large B cell lymphomas by repressing the SYK phosphatase, PTPROt. Blood. 2009;114:5315–5321. doi: 10.1182/blood-2009-02-204362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- Klein U, Dalla-Favera R. Germinal centres: role in B cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008a;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008b;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Onnis A, Cocco M, De Falco G, Imperatore F, Giuseppina A, Costanzo V, Cerino G, Mannucci S, Cantisani R, et al. B cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int J Cancer. 2010;126:1316–1326. doi: 10.1002/ijc.24655. [DOI] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–599. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, et al. Molecular profiling of diffuse large B cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- Mora-Lopez F, Reales E, Brieva JA, Campos-Caro A. Human BSAP and BLIMP1 conform an autoregulatory feedback loop. Blood. 2007;110:3150–3157. doi: 10.1182/blood-2007-05-092262. [DOI] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, Chadburn A, Tuschl T, Knowles DM, Tam W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, Sano M, Barton SC, Obukhanych T, Nussenzweig M, Tarakhovsky A, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B cell lymphoma. Blood. 2003;101:2914–2923. doi: 10.1182/blood-2002-11-3387. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Lin KI, Lin Y, Wang Y, Ting JP, Calame K. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B cells. Blood cells, molecules & diseases. 2008;41:95–99. doi: 10.1016/j.bcmd.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–862. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, Pernis A, Pasqualucci L, Dalla-Favera R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, et al. The molecular signature of mediastinal large B cell lymphoma differs from that of other diffuse large B cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- Schmidlin H, Diehl SA, Nagasawa M, Scheeren FA, Schotte R, Uittenbogaart CH, Spits H, Blom B. Spi-B inhibits human plasma cell differentiation by repressing BLIMP1 and XBP-1 expression. Blood. 2008;112:1804–1812. doi: 10.1182/blood-2008-01-136440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002a;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Staudt LM. Lymphoid malignancies: the dark side of B cell differentiation. Nat Rev Immunol. 2002b;2:920–932. doi: 10.1038/nri953. [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Sunyaev S, Ramensky V, Koch I, Lathe W, 3rd, Kondrashov AS, Bork P. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, et al. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. Embo J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Lista F, Lo Coco F, Knowles DM, Offit K, Chaganti RS, Dalla-Favera R. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, et al. Patterns of microRNA expression characterize stages of human B cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.