Abstract
The histone protein family member X (H2AFX) is important in maintaining chromatin structure and genetic stability. Genetic variants in H2AFX may alter protein functions and thus cancer risk. In this case-control study, we genotyped four common single nucleotide polymorphisms (i.e., −1654A > G [rs643788], −1420G > A [rs8551], and −1187T > C [rs7759] in the H2AFX promoter region and 1057C > T [rs7350] in the 3′ untranslated region (UTR)) in 467 patients with sporadic breast cancer and 488 cancer-free controls. All female subjects were non-Hispanic whites aged ≤55 years. We found that significantly increased risk of breast cancer was associated with variant genotypes in the H2AFX promoter: adjusted odds ratio [OR] = 1.80, 95% confidence interval [CI] = 1.38–2.34 for −1654AG/GG; OR = 1.40, 95% CI = 1.07–1.83 for −1420GA/AA; and OR = 1.65, 95% CI = 1.26–2.16 for −1187TC/CC. Furthermore, the number of variant alleles in the promoter haplotypes was associated with increased risks of breast cancer in a dose-response manner (OR = 6.08, 95% CI = 3.25–11.38; OR = 6.83, 95% CI = 3.83–12.18; and OR = 23.61, 95% CI = 3.95–140.99 for one, two, and three variant alleles, respectively) (Ptrend < 0.0001). Age at onset of breast cancer significantly decreased as the number of variant alleles increased (Ptrend = 0.024). However, these effects were not observed in the 3′UTR 1057C > T polymorphism. Therefore, we believe that H2AFX promoter polymorphisms may contribute to the etiology of sporadic breast cancer in young non-Hispanic white women. Larger association studies and related functional studies are warranted to confirm these findings.
Keywords: Case–control, DNA repair, Double strand break, Genetic susceptibility, Molecular epidemiology
Introduction
In eukaryotes, to form the chromatin, DNA is packaged into nucleosomes, each having ~ 146 bp of DNA and four pairs of histone proteins (i.e., H2A, H2B, H3, and H4) that function in maintaining chromatin structure and genetic stability [1]. Each histone family is encoded by multigenes [2]. The H2AFX gene (also known as H2AX, H2A.X, and H2A/X; MIM# 601772) is the member X of the H2A family. In humans, H2AFX has been mapped to chromosome 11q23.2–q23.3 [3, 4], a region that is frequently lost in cancer (Mitelman Database of Chromosome Aberrations in Cancer: http://cgap.nci.nih.gov/Chromosomes/RecurrentAberrations). H2AFX spans 4 kb, contains one exon, and encodes a 143-amino acid protein (GenBank: DQ015918, or NM_002105).
H2AFX-deficient cells were found to be highly radio-sensitive and genomically unstable [5] and unable to form irradiation-induced foci because of impaired recruitment of double-strand break (DSB) repair proteins, such as NBS1, TP53BP1, and BRCA1 [6]. Mice deficient for both H2AFX and p53 had genomic instability and enhanced susceptibility to an early onset of various tumors [7, 8]. Further experiments showed that genomic instability and high radiosensitivity of the cells null for H2AFX could be reversed by transfection with the wild-type H2AFX [7]. Thus, H2AFX, as a genomic caretaker, is critical for the DSB repair.
Breast cancer, the most common cancer among women in the United States, has an etiologic association with DSBs [9–11]. Germline mutations in the BRCA1 and BRCA2 genes, which are involved in the DSB repair, predispose women to an early onset breast cancer, but mutations in BRCA1 and BRCA2 only contribute to ~5% of all breast cancers, mostly familial cases [12–14]. Nevertheless, the involvement of these two genes in DNA DSB repair provides some clue to the etiology of the remaining 95% of breast cancers. Ionizing radiation and other endogenous factors can cause DSBs [15, 16] that could increase risk for breast cancer [9–11, 17]. If any impairment exists in the H2AFX gene, it is possible that DSBs could not be repaired correctly or efficiently, leading to chromosomal breaks and genomic instability and thus predisposition to cancer.
H2AFX is highly polymorphic. We genotyped four tagging SNPs in an ongoing breast cancer case–control study [18] to test the hypothesis that H2AFX polymorphisms are associated with risk of sporadic breast cancer in women aged ≤55 years.
Materials and methods
Subjects
We capitalized on DNA samples that had been collected between August 1998 and August 2005 in an ongoing study of women ≤55 years old [18–20]. All subjects were non-Hispanic whites with incident primary breast cancer. These patients had registered and their diseases had been histopathologically confirmed at the Nellie B. Connally Breast Center at The University of Texas M. D. Anderson Cancer Center. No patients had treatment prior to entering the study. Of the 467 cases included in this study, 75 had ductal or lobular carcinoma in situ, and 392 had invasive carcinoma. According to the American Joint Committee on Cancer (AJCC) staging classifications, there were 75 cases of stage 0, 183 cases of stage I, 112 cases of stage IIA, 46 cases of stage IIB, 25 cases of stage IIIA, 6 cases of stage IIIB, 15 cases of stage IIIC, and 5 cases of stage IV. During the same time period, 488 cancer-free female controls were randomly recruited from among hospital visitors who were genetically unrelated to the cases or to each other and frequency-matched to the cases by age (±5 years).
After having signed an informed consent form, subjects were interviewed with a simple questionnaire to collect information on demographic characteristics, including smoking and alcohol use. Additional information that was only available for the cases included estrogen receptor / progesterone receptor expression, menopausal status, age at first full-term pregnancy, number of live births, use of hormone replacement or oral contraceptives, weight, height, and education level. Each subject donated a venous blood sample of ~30 mL, 1 mL of which was used for genomic DNA extraction. M. D. Anderson Cancer Center's institutional review board approved the research protocol.
SNP selection
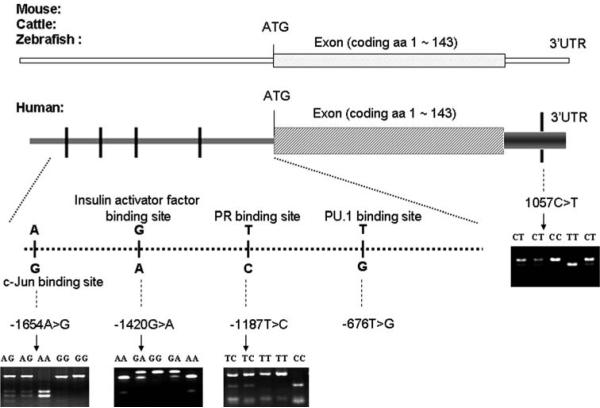
There were 14 single nucleotide polymorphisms (SNPs) that have been identified in its genomic region (i.e., from its 5′-flanking promoter region through the end of polyA in 3′ untranslated region [3′UTR]), according to the Environmental Genome Project (EGP) SNP database of the National Institute of Environmental Health Sciences (NIEHS) (http://egp.gs.washington.edu, accessed on December 11, 2006). These SNPs have also been reported in the Genbank dbSNP database (http://www.ncbi.nlm.nih.gov, accessed on December 11, 2006). Among the reported 14 SNPs, however, only five are common SNPs (i.e., with a minor allele frequency [MAF] ≥5%), four of these five being in the promoter region: −1654A > G [rs643788] (1654nt upstream to the initiation translation code ATG), −1420G > A [rs8551], and −1187T > C [rs7759], and −676T > G [rs2509851]. The fifth is in the 3′UTR, i.e., 1057C > T [rs7350] (Fig. 1).
Fig. 1.
Genomic structure, locations of common SNPs and possible binding sites in the H2AFX gene. Numbering is based on cDNA sequence (GenBank DQ015918). The nucleotide 5′ of the ATG-translation initiation codon is −1, +1 corresponds to the A of the ATG translation initiation codon in the reference sequence. The −676T > G locus was in complete LD with −1187T > C, thus, four SNPs (−1654A > G, −1420G > A, −1187T > C, and 1057C > T) were selected as the tagging SNPs. Bioinformatics analysis for the promoter SNPs: −1654G but not −1654A is the binding site for oncogene c-Jun; −1420G but not −1420A is a binging site fro insulin activator factor; −1187T but not −1187C has a binding site for progesterone receptor; and −676T (which completely linked with −1187T) but not −676G has a binding site for transcription factor PU.1
On the basis of genotyping data on 90 individuals from the NIEHS EGP-SNP resequencing database, we further calculated the D′-values and correlation coefficient (r2) for each pair of these five SNPs and found that the pair of −1187T > C and −676T > G was in complete linkage disequilibrium (LD) (D′ = 1.00 and r2 = 1.00). Thus, we included only four SNPs (−1654A > G, −1420G > A, −1187T > C, and 1057C > T) as the tagging SNPs, because they represented more than 95% of the haplotypes that are derived from all 14 SNPs, with . Figure 1 also shows the bioinformatics analysis of binding sites where the promoter SNPs are located.
Genotyping analysis
From each blood sample, the leukocyte cell pellet was obtained from the buffy coat by centrifuging 1 mL of the whole blood. Genomic DNA was extracted from the cell pellet using the DNA blood mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions. DNA purity and concentrations were determined by spectrophotometric measurement of absorbance at 260 and 280 nm.
The four H2AFX SNPs were genotyped using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method with the pairs of primers as listed in Table 1. For −1420G > A, a mismatched G replaced C at −2 bp from the polymorphic site of the sense primer to create an RsaI restriction site. Similarly, for 1057C > T, a mismatched TG replaced AA at +4 to +5 bp from the polymorphic site of the antisense primer to create a BclI restriction site. Polymerase chain reactions were performed with a PTC-200 DNA Engine Peltier thermal cycler (formerly MJ Research, Waltham, MA, USA). The four amplified fragments were then digested, respectively, by BccI, RsaI, BsiHKAI, and BclI (New England BioLabs, Beverly, MA, USA) and separated in 3% agarose gel (Fig. 1).
Table 1.
Polymerase chain reaction-restriction fragment length polymorphism genotypes of H2AFX SNPs
| Position and base change | Sense primer | Antisense primer | Annealing temperature (°C) | PCR products (bp) | Enzyme | Digested PCR products |
|---|---|---|---|---|---|---|
| −1654A>G (rs643788) | AACCTCACATT | TCTGGGACCA | 61 | 250 | BccI | AA: 119/100/31 |
| GCTCCTGCT | GAGAGAGAGG | AG: 219/119/100/31 | ||||
| GG: 219/31 | ||||||
| −1420G>A (rs8551) | ATCCTGGGCGT | ATGGAGAGGGA | 59 | 85 | RsaI | GG: 85 |
| TTCTTGCCCTGT | GAGAGTAGCAAG | GA: 85/62/23 | ||||
| AA: 62/23 | ||||||
| −1187T>C (rs7759) | CTGACTCTCAA | ATTGTCTCCATT | 61 | 150 | BsiHKAI | TT: 150 |
| GGAACCACTG | GAGAGCATGTG | TC: 150/96/54 | ||||
| CC: 96/54 | ||||||
| 1057C>T (rs7350) | TGGTGCTTAGCC | AGAAGGGGCGC | 61 | 93 | BclI | CC: 93 |
| CAGGACTTTCAG | CAGACCGTGATC | CT: 93/70/23 | ||||
| TT: 70/23 |
The genotyping results were evaluated by two researchers who were blinded to the subjects' case or control status. The genotype patterns were also confirmed by direct sequencing (data not shown). We randomly selected 96 samples (10.05% of 955 samples) for repeated assays of the four SNPs, and the results were 100% concordant.
Statistical analysis
The χ2 tests were used to compare the distributions of demographic variables and selected risk factors between cases and controls. The Hardy–Weinberg equilibrium (p2 + 2pq + q2 = 1, where p is the frequency of the variant allele and q = 1−p) was tested by a goodness-of-fit χ2 test to compare the expected genotype frequencies with the observed frequencies in cancer-free controls. The association between case–control status and each SNP, measured by the odds ratio (OR) and its corresponding 95% confidence interval (CI), was estimated using an unconditional logistic regression model, with and without adjustment for age, smoking status (never versus ever), and alcohol drinking status (never versus ever). Logistic regression modeling was also used for the trend test.
The SAS/Genetics software program (Version 9.1, SAS Institute, Inc., Cary, NC, USA) was used to determine the LD of SNP pairs. Because the use of multiple SNPs may be a more efficient method of assessing genetic susceptibility to cancer associated with a candidate gene than an analysis of any single polymorphism, we used the Proc Haplotype procedure in the SAS/Genetics program and the PHASE program to reconstruct haplotypes in the promoter region on the basis of the observed genotypes and evaluated the associations between the haplotypes and breast cancer risk by logistic regression models. We further combined SNPs in the promoter region on the basis of the number of variant alleles within the genotypes and evaluated their associations with breast cancer risk. In the stratification analysis, we assessed the main effect of H2AFX in each subgroup. All statistical tests were two-sided, and P < 0.05 was considered statistically significant.
Results
Demographic characteristics of the study population
The analysis included 467 breast cancer cases and 488 cancer-free controls, and the characteristics of the study population have been described elsewhere [18]. The differences in the distributions of age, smoking status, and alcohol use between the cases and controls were not statistically significant (P = 0.935, 0.117, and 0.139, respectively) (data not shown). However, these variables were further adjusted for in the multivariate logistic regression model to control for any residual effect of possible confounding on the main effect of the selected H2AFX SNPs.
Distributions of H2AFX genotypes and association with risk of breast cancer
The genotype and allele distributions of H2AFX SNPs −1654A > G, −1420G > A, −1187T > C, and 1057C > T in the cases and controls are summarized in Table 2. The observed genotype frequencies of these SNPs were all in agreement with the Hardy–Weinberg equilibrium calculated for the controls (P = 0.710, 0.596, 0.608, and 0.160, respectively). The LD analysis of the controls showed that all polymorphisms were in incomplete LD with each other: −1654A > G in LD with −1420G > A, −1187T > C, and 1057C > T (D′ = 0.31, 0.50, and 0.44 and r2 = 0.03, 0.22, and 0.17, respectively); −1420G > A in LD with −1187T > C and 1057C > T (D′ = 0.77, and 0.80 and r2 = 0.20, and 0.22, respectively); and −1187T > C SNP in LD with 1057C > T (D′ = 0.65 and r2 = 0.42), suggesting that each SNP may have a relatively independent effect and also can be used to generate haplotypes for further analysis.
Table 2.
H2AFX genotypes and allele frequencies and logistic regression analysis for associations with risk of breast cancer
| Genotypesa | Cases n (%) | Controls n (%) | Pb | OR (95% CI)c |
|---|---|---|---|---|
| Total number of subjects | 467 | 488 | ||
| Total number of alleles | 934 | 976 | ||
| −1654A>G | ||||
| AA | 147 (31.5) | 220 (45.1) | <0.0001 | 1.00 (ref.) |
| AG | 236 (50.5) | 218 (44.7) | 1.63 (1.23–2.15) | |
| GG | 84 (18.0) | 50 (10.2) | 2.55 (1.69–3.84) | |
| P trend | <0.0001 | |||
| AG + GG | 320 (68.5) | 268 (54.9) | 1.80 (1.38–2.34) | |
| G allele | 0.433 | 0.326 | <0.0001 | |
| −1420G>A | ||||
| GG | 143 (30.6) | 186 (38.1) | 0.018 | 1.00 (ref.) |
| GA | 236 (50.5) | 235 (48.2) | 1.31 (0.98–1.74) | |
| AA | 88 (18.9) | 67 (13.7) | 1.72 (1.17–2.53) | |
| P trend | 0.005 | |||
| GA + AA | 324 (69.4) | 302 (61.9) | 1.40 (1.07–1.83) | |
| A allele | 0.441 | 0.378 | 0.005 | |
| −1187T>C | ||||
| TT | 141 (30.2) | 204 (41.8) | 0.001 | 1.00 (ref.) |
| TC | 252 (54.0) | 219 (44.9) | 1.66 (1.25–2.19) | |
| CC | 74 (15.8) | 65 (13.3) | 1.63 (1.10–2.43) | |
| P trend | 0.002 | |||
| TC + CC | 326 (69.8) | 284 (58.2) | 1.65 (1.26–2.16) | |
| C allele | 0.428 | 0.358 | 0.002 | |
| 1057C>T | ||||
| CC | 187 (40.0) | 206 (42.2) | 0.230 | 1.00 (ref.) |
| CT | 225 (48.2) | 211 (43.2) | 1.18 (0.90–1.54) | |
| TT | 55 (11.8) | 71 (14.6) | 0.86 (0.57–1.29) | |
| P trend | 0.917 | |||
| CT + TT | 280 (60.0) | 282 (57.8) | 1.10 (0.85–1.42) | |
| T allele | 0.359 | 0.362 | 0.891 |
The observed genotype frequency among the control subjects was in agreement with the Hardy–Weinberg equilibrium (p2 + 2pq + q2 = 1) (χ2 = 0.14, P = 0.710 for −1654A>G; χ2 = 0.28, P = 0.596 for −1420G>A; χ2 = 0.26, P = 0.608 for −1187T>C and χ2 = 1.97, P = 0.160 for 1057C>T)
Two-sided χ2 test for the distribution of either genotype or allele frequency
Adjusted for age, smoking, and alcohol use in logistic regression models
As shown in Table 2, the frequency distributions of the −1654A > G genotype and G variant allele significantly differed between the cases and the controls (P < 0.0001 for both). Logistic regression analysis showed that compared with the −1654AA genotype, −1654AG heterozygotes had a 1.63-fold increased risk of breast cancer (95% CI = 1.23–2.15), and the variant −1654GG homozygotes had a 2.55-fold increased risk of breast cancer (95% CI = 1.69–3.84). The carriers of G variant allele (i.e., AG + GG genotypes) had a 1.80-fold increased risk of breast cancer (95% CI = 1.38–2.34). There was a significant trend for an allele dose-effect on risk of breast cancer (Ptrend < 0.0001). For −1420G > A, the differences in the frequency distributions of genotype and A variant allele were also statistically significant between the cases and the controls (P = 0.018 and 0.005, respectively). Compared with the common −1420GG genotype, −1420GA heterozygotes had a borderline significantly increased risk of breast cancer (adjusted OR = 1.31, 95% CI = 0.98–1.74); and −1420AA variant homozygotes had a significantly increased risk of breast cancer (adjusted OR = 1.72, 95% CI = 1.17–2.53); the trend for an allele effect on cancer risk was significant (Ptrend = 0.005), and the carriers of A variant allele (i.e., GA + AA genotypes) had a 1.40-fold risk of breast cancer (95% CI = 1.07–1.83). Similarly, the frequency distributions of −1187T > C genotype and C variant allele differed significantly between the cases and the controls (P = 0.001 and 0.002, respectively). Compared with the common genotype −1187TT, the carriers of heterozygotes −1187TC and variant homozygotes −1187CC had significantly increased risk of breast cancer (adjusted OR = 1.66, 95% CI = 1.25–2.19; and OR = 1.63, 95% CI = 1.10–2.43, respectively); there was a significant trend for an allele dose-effect on risk of breast cancer (Ptrend = 0.002) and the carriers of C variant allele (i.e., TC + CC genotypes) had a 1.65-fold risk of breast cancer (95% CI = 1.26–2.16). However, for the 1057C > T SNP in the 3′UTR, the genotype and allele frequencies did not differ significantly between the cases and the controls (P = 0.230 and 0.891, respectively); consistently, there was no significant association between this SNP and risk of breast cancer. These data suggest that the SNPs in the H2AFX promoter region, but not in 3′UTR, are associated with risk of breast cancer.
H2AFX promoter haplotypes and risk of breast cancer
To determine the combined effects of the three promoter SNPs, we used both the SAS and PHASE programs to generate haplotypes based on the observed genotypes. As shown in Table 3, there were eight estimated haplotypes in the H2AFX promoter, of which five (AGT, AAT, AGC, GGC, and GAT) were ≥5%, representing 92.1% of chromosomes of the controls. The difference in the frequency distributions of haplotype alleles between the cases and controls was statistically significant (P < 0.0001). When the haplotype AGT without any variant allele was used as the reference, all other haplotypes (except for AAC) were associated with increased risk for breast cancer (Table 3). When the haplotypes were collapsed into four groups on the basis of the number of variant alleles, the difference in frequency distributions of these haplotype groups between the cases and controls was also statistically significant (P < 0.0001). Compared with the non-variant haplotype group, risk of breast cancer increased as the number of variant alleles increased in a dose-response manner (adjusted OR = 6.08, 95% CI = 3.25–11.38 for carriers of one variant allele; OR = 6.83, 95% CI = 3.83–12.18 for carriers of two variant alleles; and OR = 23.61, 95% CI = 3.95–140.99 for carriers of three variant alleles; Ptrend < 0.0001) (Table 3). We also calculated the post-hoc powers for each haplotype in Table 3 by using the online "Power and Sample Size Program" (http://biostat.mc.vanderbilt.edu/twiki/bin/view/Main/PowerSampleSize). Most of the estimated ORs of the haplotypes had sufficient statistical power (0.8) except for the haplotypes AAC and GAC (data not shown).
Table 3.
Frequency distribution of haplotypes and combined genotypes in the promoter region of H2AFX and association with risk of breast cancer
| Cases n (%) | Controls n (%) | OR (95% CI)a | OR (95% CI)a | |
|---|---|---|---|---|
| H2AFX haplotype | 934 | 976 | ||
| Number of variant alleles in haplotypesb | ||||
| 0 (AGT) | 90 (9.6) | 240 (24.5) | 1.00 (ref.) | 1.00 (ref.) |
| 1 (AAT) | 296 (31.7) | 284 (29.1) | 5.44 (2.74–10.79) | 6.08 (3.25–11.38) |
| 1 (AGC) | 131 (14.0) | 118 (12.1) | 6.79 (3.21–14.38) | |
| 1 (GGT) | 70 (7.5) | 48 (4.9) | 11.05 (3.74–32.66) | |
| 2 (AAC) | 13 (1.4) | 16 (1.6) | 3.58 (0.60–21.28) | 6.83 (3.83–12.18) |
| 2 (GGC) | 231 (24.7) | 201 (20.7) | 6.07 (3.28–11.23) | |
| 2 (GAT) | 78 (8.4) | 56 (5.7) | 12.71 (4.73–34.19) | |
| 3 (GAC) | 25 (2.6) | 13 (1.4) | 23.61 (3.95–140.99) | 23.61 (3.95–140.99) |
| Trend test | P<0.0001 | |||
| Combined H2AFX genotypes | 467 | 488 | ||
| Number of variant alleles in genotypesc | ||||
| 0 | 8 (1.7) | 46 (9.4) | 1.00 (ref.) | 1.00 (ref.) |
| 1 | 47 (10.1) | 107 (21.9) | ||
| 2 | 141 (30.2) | 117 (24.0) | 3.33 (2.24–4.96) | 3.24 (2.29–4.59) |
| 3 | 202 (43.3) | 179 (36.7) | 3.15 (2.17–4.57) | |
| 4 | 65 (13.9) | 37 (7.6) | 5.02 (3.03–8.32) | 5.02 (3.03–8.32) |
| 5 | 3 (0.6) | 2 (0.4) | ||
| 6 | 1 (0.2) | 0 (0) | ||
| Trend test | P<0.0001 |
ORs were adjusted for age, smoking, and alcohol use in logistic regression models
Order of SNPs: H2AFX-1654A>G, −1420G>A, and −1187T>C and the variant alleles were −1654G, −1420A, and −1187C; the number represents the count of variant alleles within each haplotype
Genotype combinations of three SNPs in the promoter region of H2AFX gene; The number represents the number of variants alleles
Combined H2AFX genotypes and risk of breast cancer
To further determine the combined effect of the three promoter SNPs, we also assessed the associations between risk of breast cancer and the number of promoter variant alleles (i.e., −1654G, −1420A or −1187C) among the combined genotypes, which were divided into three groups for further stratification analysis. The difference in the frequency distribution of these three groups between cases and controls was significant (P < 0.0001). When carriers of zero-to-one variant alleles were used as the reference, breast cancer risk increased as the number of variant alleles increased in a dose-response manner (adjusted OR = 3.24, 95% CI = 2.29–4.59 for two-to-three variant alleles and adjusted OR = 5.02, 95% CI = 3.03–8.32 for four-to-six variant alleles; Ptrend < 0.0001) (Table 3).
Stratification analysis of H2AFX promoter genotypes and risk of breast cancer
We further performed a stratification analysis of the associations between the number of H2AFX variant alleles and risk of breast cancer in subgroups by age, smoking and alcohol drinking status, and patient subgroups (i.e., tumor estrogen and progesterone receptor status and tumor stage). The main effect of the H2AFX genotypes was evident in each subgroup, and the risk increased with the number of variant alleles increased in all subgroups in a dose-response manner (Ptrend < 0.001 for all subgroups). However, the altered risk was more pronounced in those aged ≤45 years (OR = 9.02; 95% CI = 4.09–19.88) and in ever-drinkers (OR = 9.24; 95% CI = 4.34–19.68) who had the genotypes with four-to-six variant alleles, compared with those who had the genotypes that contained zero-to-one variants (Table 4). In the case-only analysis, we did not find any evidence of interactions between H2AFX variants and ER/PR status, tumor histology and stages, and subjects' clinical characteristics (i.e., menopausal status, age at first full-term pregnancy, number of live births, and use of hormone replacement or oral contraceptives) (data not shown).
Table 4.
Stratification analysis of associations between H2AFX genotypes and risk of breast cancer
| Number of variant alleles in H2AFX |
Adjusted OR 95% CIa | Ptrendb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases |
Controls |
|||||||||
| 0 or 1 | 2 or 3 | 4–6 | 0 or 1 | 2 or 3 | 4–6 | 0 or 1 | 2 or 3 | 4–6 | ||
| Age (years) | ||||||||||
| ≤45 | 18 (9.1) | 146 (73.3) | 35 (19.6) | 73 (34.4) | 123 (58.0) | 16 (7.6) | 1.00 | 4.87 (2.75–8.63) | 9.02 (4.09–19.88) | <0.001 |
| 46–55 | 37 (13.8) | 197 (73.5) | 34 (12.7) | 80 (29.0) | 173 (62.7) | 23 (8.3) | 1.00 | 2.46 (1.58–3.84) | 3.31 (1.69–6.47) | <0.001 |
| Smoking | ||||||||||
| Never | 31 (11.1) | 209 (75.2) | 38 (13.7) | 79 (29.7) | 165 (62.0) | 22 (8.3) | 1.00 | 3.16 (1.98–5.03) | 4.17 (2.10–8.28) | <0.001 |
| Ever | 24 (12.7) | 134 (70.9) | 31 (16.4) | 74 (33.3) | 131 (59.0) | 17 (7.7) | 1.00 | 3.30 (1.94–5.58) | 5.68 (2.65–12.15) | <0.001 |
| Drinking | ||||||||||
| Never | 25 (11.6) | 165 (76.7) | 25 (11.6) | 64 (25.8) | 161 (64.9) | 23 (9.3) | 1.00 | 2.64 (1.58–4.41) | 2.77 (1.33–5.77) | <0.001 |
| Ever | 30 (11.9) | 178 (70.6) | 44 (17.5) | 89 (37.1) | 135 (56.3) | 16 (6.7) | 1.00 | 3.84 (2.40–6.17) | 9.24 (4.34–19.68) | <0.001 |
| Estrogen receptorc | ||||||||||
| Negative | 17 (13.3) | 84 (65.6) | 27 (21.1) | 153 (31.4) | 296 (60.6) | 39 (8.0) | 1.00 | 2.49 (1.42–4.37) | 7.05 (3.39–14.69) | <0.001 |
| Positive | 34 (11.6) | 222 (76.1) | 36 (12.3) | 1.00 | 3.40 (2.25–5.14) | 4.07 (2.25–7.37) | <0.001 | |||
| Progesterone receptorc | ||||||||||
| Negative | 22 (12.5) | 123 (69.9) | 31 (17.6) | 153 (31.4) | 296 (60.6) | 39 (8.0) | 1.00 | 2.85 (1.73–4.69) | 5.94 (3.03–11.62) | <0.001 |
| Positive | 29 (11.9) | 183 (75.0) | 32 (13.1) | 1.00 | 3.31 (2.13–5.14) | 4.26 (2.28–7.93) | <0.001 | |||
| Stage | ||||||||||
| 0 | 7 (9.3) | 58 (77.4) | 10 (13.3) | 153 (31.4) | 296 (60.6) | 39 (8.0) | 1.00 | 4.19 (1.86–9.44) | 5.38 (1.89–15.29) | <0.001 |
| 1 | 23 (12.6) | 136 (74.3) | 24 (13.1) | 1.00 | 3.04 (1.86–4.95) | 4.15 (2.08–8.29) | <0.001 | |||
| 2 | 20 (12.7) | 114 (72.1) | 24 (15.2) | 1.00 | 3.01 (1.79–5.05) | 4.97 (2.46–10.04) | <0.001 | |||
| 3–4 | 5 (9.8) | 35 (68.6) | 11 (21.6) | 1.00 | 3.70 (1.41–9.68) | 9.26 (2.96–28.93) | <0.001 | |||
Compared with 0 or 1 allele. ORs were adjusted for age, smoking, and alcohol use in logistic regression models within each stratum
Trend tests for breast cancer risk, with number of variant alleles in each stratum
Estrogen and progesterone receptor findings were only available for 374 cases
Age at onset of breast cancer and variants of H2AFX
Because an early age at onset of cancer is a feature of genetic susceptibility, we determined the relationship between age at onset of breast cancer and the number of variant alleles in the combined H2AFX promoter genotypes in the 467 cases. A Pearson correlation analysis showed that age at onset decreased significantly as the number of variant alleles in the combined genotypes increased (r = −0.10, P = 0.024) in these cases. We assigned patients to one of three groups on the basis of the number of variant alleles in the combined genotypes and found that the age at onset (mean ± SD) was 47.2 ± 4.7 years in 55 patients with zero-to-one variant alleles, but was significantly lower in 343 patients with two-to-three (45.2 ± 6.8; P = 0.010) and 69 patients with four-to-six variant alleles (44.7 ± 6.7; P = 0.017) (data not shown).
Discussion
In this hospital-based case–control study, we determined the associations between four common tagging SNPs of H2AFX (−1654A > G, −1420G > A, −1187T > C, and 1057C > T) and risk of sporadic breast cancer in non-Hispanic white women aged ≤55 years. We found that the minor variant genotypes of each SNP and haplotypes with minor alleles in the H2AFX promoter region were associated with an increased risk of breast cancer in an allele dose-response manner. A stratification analysis showed that this increased risk was more evident in those aged ≤45 years; furthermore, the increased number of variant alleles was associated with decreased age at onset of breast cancer. These results suggest that genetic variants in the H2AFX promoter region may play an important role in the etiology of breast cancer, particular in women aged ≤45 years in this study population.
Few studies have investigated the role of H2AFX variants in the development of cancer. The coding region of H2AFX was first sequenced in 101 unrelated breast cancer patients from 101 hereditary breast cancer families, but no variants or mutations in the exons of H2AFX were identified [21]. Similarly, no variants in the coding region of H2AFX were detected in the cancer cells of 68 leukemia and 44 lymphoma patients [22]. The results of these studies suggest that the coding region of H2AFX is genetically conservative and that genetic variants outside the coding region, environmental factors, or gene-environment interaction may play an important role in the development of malignancies. It is known that the promoter and 3′UTR regions of a gene have functions in regulating transcription, mRNA stability, and gene production [23] and that SNPs are the common forms of genetic variations in these core and proximal promoter regions [24], and thus searching for functional variants in the promoter, 3′UTR or regulatory regions of the gene [25–27] becomes important. Recently, Novik et al. reported that the promoter SNP −417G > A (rs2509049) in H2AFX has a protective association with non-Hodgkin lymphoma [28]. Our finding that an increased risk of breast cancer was associated with variants in the promoter region of H2AFX provides an additional support for the notion that SNPs in the H2AFX promoter region are biologically important.
Since the effects of the H2AFX promoter SNPs on the gene expression in tumor tissues are not clarified and it is likely that the phosphorylation of H2AFX may be affected by these promoter SNPs, we further performed bioinformatics analysis with the TESS program (http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ=SEA-FR-QueryS) to understand possible biological roles of these SNPs. We found that the variant −1654G allele, but not −1654A allele, creates a new binding site of oncogene c-Jun and that this variant may function in inducing oncogenic transformation [29], proliferation, and angiogenesis of breast cancer [30]. In addition, the −1420G allele, but not the variant −1420A allele, has a binding site of insulin activator factor, although the functions of this binding site remain unclear [31]. Because insulin resistance, diabetes, and obesity related to insulin resistance or hyperinsulinemia have been shown to be associated with increased risk of breast cancer [32, 33], we postulated that the −1420A variant allele may cause loss of possible protective effects of insulin activator factor due to the loss of its binding site. Furthermore, the −1187T allele, but not the variant −1187C allele, has a binding site of progesterone receptor. It has been shown that the lack of progesterone receptor is a risk factor for breast cancer and is associated with poor survival [34, 35]. This hormone has also been shown to protect against mammary tumorigenesis in rats [36]; therefore, it is likely that the −1187C variant allele may cause loss of the protection from normally available hormones in breast cells. Finally, the −676T (which completely linked with −1187T) allele, but not −676G (which completely linked with −1187C), has a binding site for the transcription factor PU.1, which is a member of the ets transcription factor family that activates gene transcription. Studies have shown that PU.1 is required for normal blood cell development, and abnormalities of the PU.1 gene can lead to leukemia [37–39]. Taken together, the results of bioinformatics analysis suggest that the presence of genetic variants in the H2AFX promoter region predicts risk of developing breast cancer and is consistent with the findings in the present study.
Our study was hospital-based, and the controls selected may not represent the general population. However, the genotype frequency distributions in our study population were similar to those reported for other populations. For example, the frequencies of minor alleles (i.e., the −1654G, −1420A, −1187C, and 1057T alleles) among the 488 non-Hispanic white women controls in our study were 0.33, 0.38, 0.36, and 0.36, respectively, which are similar to those reported for the white subjects included in EGP_CEPH-PANEL of the Genebank dbSNP (http://www.ncbi.nlm.nih.gov/SNP) (0.36, 0.41, 0.43, and 0.41, respectively) and the general American population included in the NIEHS database (http://egp.gs.washington.edu/) (0.37, 0.42, 0.32, and 0.29, respectively). Compared to some published association studies of breast cancer, our study size may not be large enough. However, we calculated the statistical power for detecting risks associated with the promoter haplotypes in this association study and achieved a 90% study power (two-sided test, α = 0.001) to detect the minimum OR of 2.1 for carriers of two-to-three variant alleles (which occurred at a frequency of 29.4% in the controls). Another limitation of our study is the lack of reproductive information for the control subjects, which excluded the evaluation of their modification of risk associated with the H2AFX polymorphisms. One final concern is that because we restricted the subjects to non-Hispanic whites aged ≤55 years, it is uncertain whether our findings can be generalizable to other subpopulations.
In conclusion, in this hospital-based case–control study of sporadic breast cancer, we found significant associations between genetic variants in the promoter of H2AFX and risk of breast cancer in non-Hispanic white women aged ≤55 years, especially in those aged ≤45 years. Age at onset of breast cancer significantly decreased as the number of variant alleles in the H2AFX promoter region increased. Our results suggest that polymorphisms in the H2AFX promoter region contribute to the etiology of sporadic breast cancer. Larger, preferably population-based case–control studies, as well as well-designed mechanistic studies, are needed to validate our findings.
Acknowledgments
This study was supported in part by National Institutes of Health grants CA 108364 (L.E.W.), ES11740 (Q.W.), and CA 16672 (M. D. Anderson Cancer Center). We thank Margaret Lung and Leanel Fairly for their assistance in recruiting the subjects; Luo Wang, Ping Xiong, Yawei Qiao, Kejing Xu, Zhensheng Liu, Zhaozheng Guo, Jianzhong He, and Yinyan Li for their laboratory assistance; and Ann Sutton for scientific editing.
Abbreviations
- CI
Confidence interval
- DSB
Double-strand break
- LD
Linkage disequilibrium
- H2AFX
H2A histone familymember X
- OR
Odds ratio
- PCR-RFLP
Polymerase chain reaction-restriction fragment length polymorphism
- SNP
Single nucleotide polymorphism
- UTR
Untranslated region
Footnotes
Present Address: J. Lu The Institute for Chemical Carcinogenesis, Guangzhou Medical College, 195 Dongfengxi Rd., Guangzhou, Guangdong, P.R. China
Reference
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 3.Ivanova VS, Hatch CL, Bonner WM. Characterization of the human histone H2A.X gene. Comparison of its promoter with other H2A gene promoters. J Biol Chem. 1994;269:24189–24194. [PubMed] [Google Scholar]
- 4.Ivanova VS, Zimonjic D, Popescu N, Bonner WM. Chromosomal localization of the human histone H2A.X gene to 11q23.2-q23.3 by fluorescence in situ hybridization. Hum Genet. 1994;94:303–306. doi: 10.1007/BF00208289. [DOI] [PubMed] [Google Scholar]
- 5.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114:371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 9.Ralhan R, Kaur J, Kreienberg R, Wiesmuller L. Links between DNA double strand break repair and breast cancer: accumulating evidence from both familial and nonfamilial cases. Cancer Lett. 2007;248:1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhang C, Naftalis E, Euhus D. Carcinogen-induced DNA double strand break repair in sporadic breast cancer. J Surg Res. 2006;135:120–128. doi: 10.1016/j.jss.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 12.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90:978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 13.Rosen EM, Fan S, Pestell RG, Goldberg ID. BRCA1 gene in breast cancer. J Cell Physiol. 2003;196:19–41. doi: 10.1002/jcp.10257. [DOI] [PubMed] [Google Scholar]
- 14.Ting NS, Lee WH. The DNA double-strand break response pathway: becoming more BRCAish than ever. DNA Repair (Amst) 2004;3:935–944. doi: 10.1016/j.dnarep.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 15.Little JB. Radiation carcinogenesis. Carcinogenesis. 2000;21:397–404. doi: 10.1093/carcin/21.3.397. [DOI] [PubMed] [Google Scholar]
- 16.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufmann WK, Filatov L, Oglesbee SE, Simpson DA, Lotano MA, McKeen HD, Sawyer LR, Moore DT, Millikan RC, Cordeiro-Stone M, Carey LA. Radiation clastogenesis and cell cycle checkpoint function as functional markers of breast cancer risk. Carcinogenesis. 2006;27:2519–2527. doi: 10.1093/carcin/bgl103. [DOI] [PubMed] [Google Scholar]
- 18.Lu J, Wei Q, Bondy ML, Li D, Brewster A, Shete S, Yu TK, Sahin A, Meric-Bernstam F, Hunt KK, Singletary SE, Ross MI, Wang LE. Polymorphisms and haplotypes of the NBS1 gene are associated with risk of sporadic breast cancer in non-Hispanic white women < = 55 years. Carcinogenesis. 2006;27:2209–2216. doi: 10.1093/carcin/bgl077. [DOI] [PubMed] [Google Scholar]
- 19.Shi Q, Wang LE, Bondy ML, Brewster A, Singletary SE, Wei Q. Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis. 2004;25:1695–1700. doi: 10.1093/carcin/bgh167. [DOI] [PubMed] [Google Scholar]
- 20.Xiong P, Bondy ML, Li D, Shen H, Wang LE, Singletary SE, Spitz MR, Wei Q. Sensitivity to benzo(a)pyrene diol-epoxide associated with risk of breast cancer in young women and modulation by glutathione S-transferase polymorphisms: a case-control study. Cancer Res. 2001;61:8465–8469. [PubMed] [Google Scholar]
- 21.Monteiro AN, Zhang S, Phelan CM, Narod SA. Absence of constitutional H2AX gene mutations in 101 hereditary breast cancer families. J Med Genet. 2003;40:e51. doi: 10.1136/jmg.40.4.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh SH, Rosenquist R. Absence of H2AX gene mutations in B-cell leukemias and lymphomas. Leukemia. 2005;19:464. doi: 10.1038/sj.leu.2403651. [DOI] [PubMed] [Google Scholar]
- 23.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 24.Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O'Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- 25.Ponomarenko JV, Merkulova TI, Vasiliev GV, Levashova ZB, Orlova GV, Lavryushev SV, Fokin ON, Ponomarenko MP, Frolov AS, Sarai A. rSNP_Guide, a database system for analysis of transcription factor binding to target sequences: application to SNPs and site-directed mutations. Nucleic Acids Res. 2001;29:312–316. doi: 10.1093/nar/29.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim Biophys Acta. 2006;1762:17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 27.De Gobbi M, Viprakasit V, Hughes JR, Fisher C, Buckle VJ, Ayyub H, Gibbons RJ, Vernimmen D, Yoshinaga Y, de Jong P, Cheng JF, Rubin EM, Wood WG, Bowden D, Higgs DR. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 28.Novik KL, Spinelli JJ, Macarthur AC, Shumansky K, Sipahimalani P, Leach S, Lai A, Connors JM, Gascoyne RD, Gallagher RP, Brooks-Wilson AR. Genetic variation in H2AFX contributes to risk of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2007;16:1098–1106. doi: 10.1158/1055-9965.EPI-06-0639. [DOI] [PubMed] [Google Scholar]
- 29.Vogt PK. Jun, the oncoprotein. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 30.Vleugel MM, Greijer AE, Bos R, van der Wall E, van Diest PJ. c-Jun activation is associated with proliferation and angiogenesis in invasive breast cancer. Hum Pathol. 2006;37:668–674. doi: 10.1016/j.humpath.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Whelan J, Cordle SR, Henderson E, Weil PA, Stein R. Identification of a pancreatic beta-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol Cell Biol. 1990;10:1564–1572. doi: 10.1128/mcb.10.4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose DP, Komninou D, Stephenson GD. Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev. 2004;5:153–165. doi: 10.1111/j.1467-789X.2004.00142.x. [DOI] [PubMed] [Google Scholar]
- 33.Lipscombe LL, Goodwin PJ, Zinman B, McLaughlin JR, Hux JE. Increased prevalence of prior breast cancer in women with newly diagnosed diabetes. Breast Cancer Res Treat. 2006;98:303–309. doi: 10.1007/s10549-006-9166-3. [DOI] [PubMed] [Google Scholar]
- 34.Colditz GA, Rosner BA, Chen WY, Holmes MD, Hankinson SE. Risk factors for breast cancer according to estrogen and progesterone receptor status. J Natl Cancer Inst. 2004;96:218–228. doi: 10.1093/jnci/djh025. [DOI] [PubMed] [Google Scholar]
- 35.Grann VR, Troxel AB, Zojwalla NJ, Jacobson JS, Hershman D, Neugut AI. Hormone receptor status and survival in a population-based cohort of patients with breast carcinoma. Cancer. 2005;103:2241–2251. doi: 10.1002/cncr.21030. [DOI] [PubMed] [Google Scholar]
- 36.Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, D'Cruz CM, Chodosh LA. Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy. Cancer Res. 2006;66:6421–6431. doi: 10.1158/0008-5472.CAN-05-4235. [DOI] [PubMed] [Google Scholar]
- 37.Li SL, Schlegel W, Valente AJ, Clark RA. Critical flanking sequences of PU.1 binding sites in myeloid-specific promoters. J Biol Chem. 1999;274:32453–32460. doi: 10.1074/jbc.274.45.32453. [DOI] [PubMed] [Google Scholar]
- 38.Marecki S, McCarthy KM, Nikolajczyk BS. PU.1 as a chromatin accessibility factor for immunoglobulin genes. Mol Immunol. 2004;40:723–731. doi: 10.1016/j.molimm.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 39.Mueller BU, Pabst T, Osato M, Asou N, Johansen LM, Minden MD, Behre G, Hiddemann W, Ito Y, Tenen DG. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]