Abstract
Enormous progress in mapping complex traits in humans has been made in the last 5 yr. There has been early success for prevalent diseases with complex phenotypes. These studies have demonstrated clearly that, while complex traits differ in their underlying genetic architectures, for many common disorders the predominant pattern is that of many loci, individually with small effects on phenotype. For some traits, loci of large effect have been identified. For almost all complex traits studied in humans, the sum of the identified genetic effects comprises only a portion, generally less than half, of the estimated trait heritability. A variety of hypotheses have been proposed to explain why this might be the case, including untested rare variants, and gene–gene and gene–environment interaction. Effort is currently being directed toward implementation of novel analytic approaches and testing rare variants for association with complex traits using imputed variants from the publicly available 1000 Genomes Project resequencing data and from direct resequencing of clinical samples. Through integration with annotations and functional genomic data as well as by in vitro and in vivo experimentation, mapping studies continue to characterize functional variants associated with complex traits and address fundamental issues such as epistasis and pleiotropy. This review focuses primarily on the ways in which genome-wide association studies (GWASs) have revolutionized the field of human quantitative genetics.
MANY phenotypes are quantitative in nature, and complex in etiology, with multiple environmental and genetic causes. The observation that complex traits cluster in relation to genetic relatedness suggests heritability, and advances in theoretical and experimental genetics, combined with analytical developments and high-throughput genomics, have provided an unprecedented view into the mode of inheritance and genetic architecture of complex traits.
Common diseases such as obesity, heart disease, type 2 diabetes mellitus, and others, have made Homo sapiens the most phenotypically studied organism. Genome-wide characterization of the levels and patterns of human genetic variation has enabled geneticists to interrogate this variation for association with complex phenotypes. In medical genetics, the ultimate objective is to identify causal functional variants and elucidate the mechanisms through which they exert their effects. Therefore, trait mapping studies can be considered hypothesis-generating exercises, helping to prioritize genes or genomic regions for further investigation. At the same time, trait mapping studies provide an overall description of genetic architecture: estimating heritability, the number of loci underlying phenotypic variation, and the distribution of effect sizes, as well as suggesting whether genetic interactions among loci (epistasis, see glossary in Table 1) or among traits (pleiotropy, see Table 1) exist.
TABLE 1.
Glossary of terms
| Term | Definition |
|---|---|
| Association mapping | a trait mapping approach that analyzes the co-occurrence of the trait and the marker in multiple individuals of a population; it relies on historical recombination. |
| Copy number variant | A segment of DNA that is present at variable copy number among individuals when compared to a diploid reference genome. Several publicly available catalogs of CNVs in the human genome are available (e.g., http://projects.tcag.ca/variation/; http://cnv.chop.edu/). |
| Epistasis | The phenomenon where the effect of a gene (or variant) on a trait is not independent of the effect on the trait of another gene (or variant). |
| Expression QTL (eQTL) | a region of the genome harboring a genetic variant contributing to gene expression variation. These are identified through association mapping (eQTL mapping) of variable transcription levels among individuals. |
| Fine mapping | a methodology to resolve locus boundaries. However the same LD that facilitates GWAS hampers fine mapping. In addition, multi-SNP analyses can help to refine the signal. Haplotype analyses using SNPs showing statistically independent association signals could reveal the existence of a single associated haplotype, refining the LD criterion for considering variants as putatively functional or could suggest multiple causal variants. Mapping in populations of different ancestry (that have different LD structure) may also help to refine a locus of interest. |
| Functional genomics | a field of study that combines genetics and molecular biology to understand the function of genes and other features of the genome. |
| Gene set enrichment analysis | A computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two states (e.g., different phenotypes or associated vs. nonassociated genetic variants). |
| Genetic architecture | (1) the number of loci affecting a complex trait, (2) the distribution of their effects, and (3) their interactions with each other (including dominance and epistatic effects) and with environmental variation. Although classically genetic architecture can be viewed as the decomposition of genetic variance for a trait into additive and interaction components, and (1) and (2) above are subsumed within additive genetic variance, in the GWAS era, we can directly investigate the number of loci and their distribution of effects. |
| Genetic heterogeneity | the phenomenon wherein different genetic variants underlie a single phenotypic trait in different individuals/families. |
| Imputation | a method to estimate genotypes of an individual at unmeasured SNP loci using a known reference panel of genetic variation and haplotypes derived from a sample of close ancestry. |
| Linkage mapping | a trait mapping approach that analyzes cosegregation of a trait with a marker in family pedigrees. |
| Mendelian disease | a disease in which mutations in a single gene cause the phenotype and for which the pattern of inheritance is very clear. |
| Meta-analysis | a term describing a wide variety of statistical procedures developed to pool and summarize results from multiple studies, thereby increasing statistical power. |
| Odds ratio (OR) | a statistic used to assess the likelihood of a particular outcome (e.g, disease state) if a certain factor (or allelic variant) is present. In the context of disease mapping, whether the odds ratio is greater than or less than one indicates whether a variant is a risk variant or protective, and the magnitude of the OR can be interpreted as the effect size. An OR of 1 indicates that the condition or event under study is equally likely in both groups. |
| Penetrance | the extent to which a genetic variant has an effect on individuals who carry it. High penetrance would refer to a Mendelian disease. |
| Pleiotropy | the phenomenon wherein a single gene or variant affects multiple phenotypes. |
| Prospective cohort | a set of individuals that is followed over time with repeated observational measurements taken throughout. The key feature is that traits studied at a subsequent point in time are being examined in a set of individuals that was not selected on the basis of that trait. |
| Systems genetics | holistic study of the effects of one or more genetic variants on the outcome (complex trait phenotype) and/or intermediate phenotypes such as gene expression profiles or other functional genomic or cell biological data. |
| The International HapMap Project | an international consortium to create a catalog of common genetic variation, primarily SNPs that occur in humans. The data reveal where genetic variants occur in the genome, how correlated they are with one another (linkage disequilibrium), and how they are distributed among individuals within and among populations. All data are publicly available; hapmap.ncbi.nlm.nih.gov/. |
| The 1000 Genomes Project | an international consortium-led project to fully sequence the genomes of at least 1000 people (now approximately 2000 people) of multiple ancestries from around the world. The project identifies SNPs as well as structural variants (rearrangements, deletions, or duplications of segments of the human genome). All data are publicly released as they become available; http://www.1000genomes.org/. |
Modern complex trait mapping in humans utilizes the linkage disequilibrium (LD, see Table 1)-based genome-wide association study (GWAS). GWAS involves correlating allele frequencies at each of several hundred thousand markers spaced throughout the genome with trait variation in a population-based sample (see box, case study of a GWAS: meta-analysis of six genome-wide association studies identifies seven new rheumatoid arthritis risk loci and below). GWAS is based on the premise that a causal variant is located on a haplotype, and therefore a marker allele in LD with the causal variant should show (by proxy) an association with a trait of interest. One of the advantages of the GWAS approach is that it is unbiased with respect to genomic structure and previous knowledge of the trait etiology, in contrast to candidate gene studies, where knowledge of the trait is used to identify candidate loci contributing to the trait of interest. Therefore, GWAS results hold the promise to reveal causal genes not previously suspected in disease etiology or indeed genetic effects of nongenic DNA regions, and GWASs hold the promise to estimate relatively complete genetic effects (additive and nonadditive) and pleiotropy in an unbiased way.
As of October 2010, 702 human GWASs have been published on 421 traits, the majority of medical relevance. The National Human Genome Research Institute at the National Institutes of Health updates weekly a catalog of published GWAS results (http://www.genome.gov/gwastudies; Hindorff et al. 2009; Johnson and O'Donnell 2009). There exist several hundred replicated disease-associated common single nucleotide polymorphisms (SNPs), and the list continues to grow. While much work remains to identify and characterize the full extent of the genetic contribution to human complex traits, human geneticists are now in a position to address fundamental questions of how and why complex traits vary among us. The large volume of results and systematic study of many traits makes review and interpretation of human GWAS results clearly relevant to all students of genetics today.
COMPLEX TRAIT GENETICS—KEY CONCEPTS
Following the rediscovery of Gregory Mendel's work in the early 20th century, a heated debate raged between “biometricians” and “Mendelians” as to the underlying mode of inheritance and genetic architecture of phenotypic traits. Focusing on continuous variation of characters within populations, biometricians Francis Galton and Karl Pearson developed statistical methods and concepts including correlation, regression, standard deviation, and principal components analysis to estimate the genetic component of phenotypic variance from the variance and covariance of traits and to further decompose genetic variance into additive and nonadditive components (Galton 1869, 1889, 1901; Pearson 1898). The Mendelian geneticists, including William Bateson and Hugo de Vries, were primarily interested in discrete traits and Mendel's laws of inheritance and worked to estimate the effects and modes of inheritance (e.g., dominance/recessivity) of strong allelic effects (Bateson 1902, 1909).
Ronald A. Fisher's (1918) important reconciliation that seemingly purely quantitative variation can be produced by the combined action of multiple genes, each inherited in a Mendelian fashion, opened the door to a unified theory of genetics. This, together with the discovery and understanding of genetic linkage (Bateson et al. 1905; Punnett 1909; Morgan 1911a; Morgan et al. 1915) and construction of the first linkage maps (Morgan 1911b; Sturtevant 1913, 1915), laid the foundation for genetic mapping and genetic analysis of quantitative traits.
Theoretical and experimental genetic studies of relatively simple quantitative traits, (Castle and Little 1910; East 1910; Altenburg and Muller 1920) led to the concept of the “polygene” (Thoday 1961), a set of loci underlying quantitative variation. Quantitative trait locus mapping, or QTL mapping, was first conceived by Sax (1923) and developed into a method for rigorous analysis of quantitative traits in experimental and natural populations. Of primary interest in the quantitative genetics of complex traits are the number of loci contributing to trait variance and the distribution of the magnitudes of their effects, topics that were widely debated by the early pioneers (e.g., Mather 1943; Waddington 1943; Mather and Jinks 1971; Thoday and Thompson 1976) and that are still relevant today.
A comprehensive understanding of the genetic architecture (see Table 1) of a complex trait includes quantification of heritability and partitioning of the genetic variance into additive and nonadditive components (Falconer and Mackay 1996; Lynch and Walsh 1998; Visscher 2008). Nonadditive genetic variance includes dominance and epistatic interactions of alleles within and between loci, as well as interactions between genes and environmental variation. Genetic correlation between traits, or pleiotropy, is also of interest when multiple traits are under study.
CASE-STUDY OF A GWAS:
META-ANALYSIS OF SIX GENOME-WIDE ASSOCIATION STUDIES IDENTIFIES SEVEN NEW RHEUMATOID ARTHRITIS RISK LOCI
RA is the most common autoimmune disease, characterized by chronic inflammation and destruction of the synovial joints later in life. In a recent study of RA risk (Stahl et al. 2010), six genome-wide association studies totaling over 5500 cases and over 20,000 controls were combined through meta-analysis. These studies included a new GWAS dataset of cases from the Brigham Rheumatoid Arthritis Sequential Study (Plenge et al. 2007) and shared controls genotyped on the Affymetrix SNPChip 6.0 platform, three datasets genotyped on the Illumina HumanHap 317K and 550K platforms (Plenge et al. 2007; Remmers et al. 2007; Gregersen et al. 2009), and the Wellcome Trust Case Control Consortium (2007) data including nonautoimmune disease cases as shared controls, genotyped on the Affymetrix 500K platform. All samples were of self-described European ancestry, with one sample set originating from Sweden, one from the United Kingdom, and four from North America. In an effort to minimize clinical heterogeneity of the samples, case samples were restricted to autoantibody positive RA, which is more severe than autoantibody negative RA, and for which previous genetic association studies have been much more productive (Raychaudhuri 2010).
Raw genotype data were acquired for the 22 autosomes and were filtered to remove poor-quality SNPs (high degree of missingness across individuals), to remove individuals that did not genotype well (high degree of missing data across SNPs), and to remove related individuals. PCA was applied to the genotype data to identify genetic outliers. Matching, based on the first five PCs, was used to remove excess controls in the more stratified North American (pan-European ancestry) datasets. Genome-wide imputation (Marchini et al. 2007) was used to infer genotypes at over 2.5 million SNPs in common across the studies. Logistic regression was used to test for association with case-control status in each GWAS dataset; the results were genomic-control corrected (Devlin and Roeder 1999) and then combined via inverse variance-weighted meta-analysis (de Bakker et al. 2008).
A q-q plot of the genome-wide distribution of results is presented in Figure 1, showing substantial departure from the null hypothesis of no association. This departure remained even after removing known RA risk-associated loci. Figure 2 presents a Manhattan plot of the statistical strength of association (−Log10P) across the autosomes, showing that several but not all previously known RA risk loci show strong association in these data (lack of replication is presumably due to lack of power; Stahl et al. 2010), and that several new loci exhibit strong associations. Thirty-four SNPs were tested for replication in additional samples, 10 of which achieved genome-wide significance in the combined analysis of over 41,000 case-control samples. These 10 SNPs represent three loci not previously known in any autoimmune disease, four loci previously implicated in other autoimmune diseases (Crohn's disease, systemic lupus erythematosis, and type 1 diabetes), and three loci previously implicated in RA risk: a celiac disease-associated AFF3 locus recently shown to be associated with RA (Barton et al. 2009), a more strongly associated SNP at the IL2RA locus (Barton et al. 2008), a new, independent SNP association at the CCL21 RA risk locus (Raychaudhuri et al. 2008).
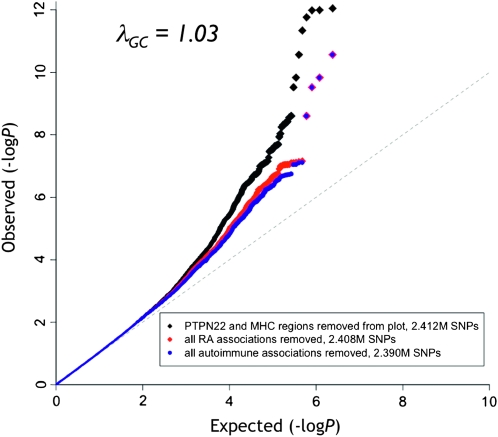
Figure 1.—
Q-Q plot of the RA GWAS meta-analysis (Stahl et al. 2010). Results for all SNPs excluding the strongly associated PTPN22 (chr1, 113.5–114.5 Mb) and MHC (chr6, 26–34 Mb) regions (which would otherwise dominate the tail of the distribution) are plotted in black. Results excluding SNPs in LD (r2 > 0.1) with previously known RA risk associations are plotted in red, showing that substantial association signal remains in the data. And results excluding SNPs in LD with validated autoimmune disease associations are plotted in blue, showing a degree of overlap between RA and related complex diseases. Genomic control λGC (scaled for 1000 cases and 1000 controls) for the data excluding PTPN22 and MHC (black) is shown in the inset. Image adapted from Stahl et al. 2010.
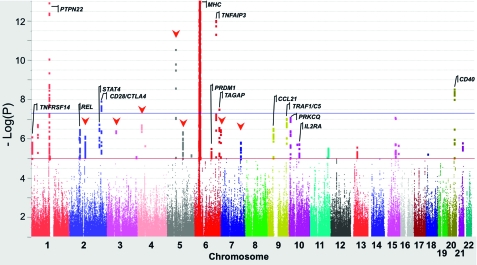
Figure 2.—
Manhattan plot for RA GWAS meta-analysis. Statistical strength of association (-Log10P) is plotted against genomic position with the 22 autosomal chromosomes in different colors. The blue horizontal line indicates the genome-wide significance threshold of P = 5 × 10−8; the red line is a threshold for “suggestive” association (P = 10−5). SNPs at 5 of 29 loci known from previous studies (gene symbols shown), and one of the 10 new loci identified in this study (marked by red triangles), achieved genome-wide significance in this meta-analysis (prior to the replication phase of the study). Over 200 SNPs representing 35 loci achieved P <10−5, versus roughly 10 expected by chance.
A total of 21 of 34 SNPs tested in the replication cohorts replicated with at least nominal significance, more than 10 times what would be expected by chance, suggesting that further studies with additional samples would lead to more validated RA risk loci. The q-q plot (Figure 1) also shows substantial departure from the null at even lower significance thresholds, which could be due to stratification or other weak, systematic bias in the data, but is also consistent with many more common variants weakly associated with RA risk.
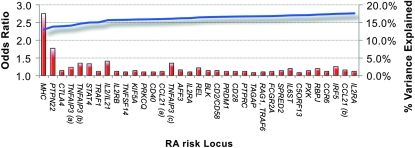
As with other recent GWAS discoveries, the loci validated in Stahl et al. (2010) have modest effect sizes (OR 1.1–1.3; Figure 3). On the basis of their ORs and allele frequencies, we can calculate the proportion of phenotypic variance explained in RA for each SNP under a liability threshold model (Falconer and Mackay 1996) and these can be assumed to sum to the total percentage of variance explained by validated RA risk alleles. Figure 3 shows that additional GWAS discoveries contribute little to the total variance explained, which seems to reach a plateau at 15–16% (Raychaudhuri 2010).
Figure 3.—
Validated RA risk alleles, their effect sizes (odds ratios, OR), and their percent variance explained. Modern understanding of the genetic etiology of RA has progressed quickly in the age of the GWAS, with scores of recently discovered risk alleles. As resources and technology have improved, our ability to discover alleles with more modest effect sizes has accelerated our ability to discover new risk alleles. On the other hand, with each discovered risk locus, the heritability (percent variance) explained is increasingly modest because new risk alleles' odds ratios are smaller while their frequencies in the population are comparable. Image courtesy of S. Raychaudhuri.
COMPLEX TRAIT MAPPING IN HUMANS
QTL mapping developed in large part as a methodology to uncover the genetic basis of quantitative traits in experimental crosses in model organisms and of traits of interest to animal and plant breeders and evolutionary geneticists. Early genetic mapping studies in humans utilized linkage mapping, a methodology that traces the transmission of phenotypes with genetic markers through pedigrees (reviewed in Ott 1991). In humans, linkage studies have been successful in identifying highly penetrant (see Table 1) genetic variants of large effect [odds ratio >100, (see Table 1)] underlying hundreds, if not thousands of Mendelian diseases (see Table 1; e.g., HTT gene in Huntington's disease, Gusella et al. 1983; CFTR gene in cystic fibrosis, Riordan et al. 1989).
In contrast to monogenic traits, complex traits have been more difficult to unravel using linkage approaches. Several common disease-predisposing variants that are associated with common disease variation were identified in early linkage/candidate gene studies, e.g., Factor VLeiden in deep venous thrombosis (Bertina et al. 1994), the APOEɛ-4 allele in Alzheimer's disease (Corder et al. 1993), and PPARγ in type 2 diabetes (Altshuler et al. 2000a). These observations were consistent with a “common disease/common variant” (CDCV) hypothesis, in which a disease phenotype results from the aggregate effects of polygenic variation, with causal variants present at high frequency in human populations. On the basis of these observations and on population genetic modeling of genetic variation (Fisher 1930; Haldane 1932; Wright 1969; Wright 1977; Wright 1978; Reich and Lander 2001), the CDCV hypothesis became a focus of human genetics at the turn of the 21st century (Lander 1996). Although it may seem paradoxical that deleterious alleles reach high frequency in a population, many complex disorders have a late age of onset and thus might not have been subject to strong purifying selection. Also, if a complex trait has an architecture wherein there are many loci with individually small effects on a trait (or fitness), or variants exhibit incomplete penetrance or pleiotropic effects, selection on any individual disease variant may have been only weakly deleterious, neutral, or indeed positive during human evolutionary history. An alternative, the common disease/rare variant (CDRV) hypothesis, posits that many genes/alleles with lower-frequency, higher-penetrance variants contribute to disease and is a straightforward extension to common diseases of the discoveries made for Mendelian disorders (Bodmer and Bonilla 2008). Population genetic modeling (Pritchard 2001) suggested that disease risk variants are likely to be mildly deleterious, have a high mutation rate, have a high total frequency, and exhibit extensive allelic heterogeneity. Therefore, Pritchard (2001) argued that the CDRV hypothesis is more consistent with human pathology and population biology than the CDCV hypothesis. These two contrasting views of the likely nature of variants underlying complex disease have very different implications for the likely success of different strategies for identifying the genetic basis underlying heritable complex disorders.
Risch and Merikangas (1996) considered the relative performance of different mapping methodologies for traits of various genetic architectures and demonstrated that linkage studies are well-powered to detect variants with large effects and high penetrance, but are underpowered for detection of variants of small effect. They determined that association mapping (see Table 1), a population-based alternative mapping approach, is especially well powered for mapping common variants (minor allele frequency, MAF > 0.05) of small effect size. In contrast to linkage analyses, association analyses test for a relationship between phenotypes and genotypes in large samples of “unrelated” individuals, assuming identity by state, where individuals of similar phenotype are assumed to share the same risk variants. Although the action of multiple factors (genetic or nongenetic), incomplete penetrance, and modest effects reduce analytical power, these limitations can be overcome with large sample sizes, except in the case of extensive allelic heterogeneity (Terwilliger and Weiss 1998). In addition, while linkage studies typically identify genomic regions of 5–10 Mb harboring tens to hundreds of genes, association studies are able to refine genomic loci to roughly 10–100 kb, often just a few genes, because many recombination events have occurred in the history of the population sample.
Human genetic variation and mapping tools:
Human genetic variation reflects demographic forces that occurred since the origin of anatomically modern humans around 100,000–200,000 yr ago and their migration out of Africa around 60,000 yr ago, including genetic drift, substructure, and migration, as well as genetic forces including mutation, recombination, and natural selection (Cavalli-Sforza et al. 1994; Cavalli-Sforza and Feldman 2003). Genetic differences between populations of different ancestry are modest: ancient polymorphisms that predate this migration are shared by all human populations and account for approximately 90% of human variants (Harris 1966; Li and Sadler 1991; Tishkoff and Verrelli 2003).
Botstein et al. (1980) pioneered the use of a large set of molecular markers for genetic analysis in humans. Physical maps (Hudson et al. 1995) provided scaffolding of the human genome, enabling positional cloning of quantitative trait loci. The Human Genome Project (Lander et al. 2001) stimulated many large-scale projects, including efforts to characterize the most abundant genetic variants in the human genome, single nucleotide polymorphisms (SNPs), which occur on average about every 1–2 kb between any two chromosomes. The combined efforts of The International SNP Consortium (TSC), the Human Genome Project, and other SNP discovery efforts (Altshuler et al. 2000b) led to the publication of the first genome-wide map of human genetic variation (Sachidanandam et al. 2001).
Observations from sequencing studies of individual loci and large-scale SNP data revealed that alleles of nearby SNPs tend to be strongly correlated with each other across individuals (Nickerson et al. 1998; Daly et al. 2001; Gabriel et al. 2002). That is, they are in strong LD and form limited numbers of haplotypes. In contrast to other species such as Drosophila, LD in humans does not decay gradually with distance. Rather, common genetic variation by and large is organized in “haplotype blocks,” local regions that have not been broken up by meiotic recombination, separated by recombination “hot spots” that occur every 100–200 kb (Daly et al. 2001; Reich et al. 2002; McVean and Cardin 2005). These observations provided the empirical foundation for the construction of a haplotype map of the human genome (see text below) for diverse populations, and these early studies demonstrated that LD patterns vary across populations (Gabriel et al. 2002; Reich et al. 2002; Rosenberg et al. 2002). Under the CDCV hypothesis, genetic variation organized into a limited number of haplotypes spanning a causal variant would be associated with disease, and relatively few “tag” SNPs chosen to represent the haplotypes would need to be genotyped in each haplotype block (de Bakker et al. 2005) to capture the effect.
The International HapMap Consortium (2003; see Table 1) characterized the patterns of common genetic variation within the human genome (over 3.1 million common SNPs MAF > 0.05; 25–35% of the total number of predicted common variants) were genotyped in 270 individuals from populations of African, Asian, and European ancestry. Subsequently, a subset of 1.6 million SNPs was genotyped in an additional 270 unrelated individuals of the original four populations plus nearly 900 additional individuals representing seven additional populations (Altshuler et al. 2010). HapMap's focus on common SNPs, exacerbated by discovery in small samples (Clark et al. 2005), means that it provides little information about patterns of variation for “rare” SNPs (MAF < 0.05). Also, structural variants including insertions/deletions (indels), inversions, and copy number variants (CNVs, see Table 1) were not directly surveyed (except as they are in LD with common SNPs, but see Redon et al. 2006).
To identify rare polymorphisms (MAF 0.001–0.05) and CNVs, and to localize CNV boundaries, the 1000 Genomes Project (1KG; see Table 1; Durbin et al. 2010) was launched in 2008 to sequence the genomes of over 2000 individuals. The 1KG Project is providing a catalog of low-frequency variants in the human genome, thus facilitating a next wave of GWAS to assess the role of variants with lower allele frequency.
Mapping complex traits with GWASs:
Technological advances have enabled cost-effective, ultra high-throughput SNP and CNV typing in large-scale, well-phenotyped sample collections, setting the stage for GWASs. (For recent reviews of GWAS methods, see Cardon and Bell 2001; Balding 2006; Hardy and Singleton 2009; Smith et al. 2009; for a recent example of a GWAS for rheumatoid arthritis risk, refer to box, case study of a GWAS: meta-analysis of six genome-wide association studies identifies seven new rheumatoid arthritis risk loci.)
In a GWAS, allele frequencies at thousands if not millions of loci are compared in individuals of varying phenotype (Cardon and Bell 2001). Defining the phenotype is an important consideration because phenotypic heterogeneity can reduce power (Ioannidis et al. 2009). Other complexities, including data quality per individual and per SNP, batch effects (Clayton et al. 2005), and relatedness among samples as well as genetic outliers (Price et al. 2006) must be accounted for to avoid systematic bias.
GWAS analysis tests for association of each SNP (state of the art is up to ∼10 million SNPs) with disease status or quantitative trait value in hundreds to tens of thousands of individuals. For quantitative traits, linear regression or Spearman's rank correlation is used to test each SNP for association between trait values and genotype. For categorical traits (e.g., case-control status or phenotypic extremes), chi-square or contingency table-based tests can be used in addition to logistic regression tests. Population stratification must be addressed in these analyses. Stratified analysis (e.g., using a Cochran–Mantel–Haentzel test), population structure covariates (e.g., inferred population assignments; Pritchard et al. 2000), or principal component analysis (PCA) eigenvectors (Price et al. 2006), or mixed model regression analysis (Aranzana et al. 2005; Aulchenko et al. 2007; Buckler et al. 2009) are approaches for dealing with cryptic population structure.
Assessment of GWAS results depends on the assumption that the vast majority of SNPs follow the null hypothesis, because relatively few of the tested genetic variants are expected to influence the trait of interest. Thus, the observed distribution of test statistics can be examined for signs of systematic bias. Devlin and Roeder (1999) suggested the use of a variance inflation factor (λGC), the ratio of the observed-to-expected median (chi-square) test statistic, and developed the widely used genomic control procedure wherein one calculates λGC and divides all of the test statistics by that factor. Test result distributions are often visualized in a quantile-quantile (q-q) plot of observed vs. expected test statistics or −log10(P) (see box, case study of a GWAS: meta-analysis of six genome-wide association studies identifies seven new rheumatoid arthritis risk loci; Figure 1).
Given the number of tests performed in a GWAS, multiple hypothesis testing is an important consideration, as was realized in the early days of QTL mapping in animal breeding (Neimann-Sorensen and Robertson 1961). Replication in independent samples is required for an association to be considered validated (same variant, trait, and direction of effect); standard practice is for a small set of the most significant variants from the “discovery phase” to be tested in an independent sample in a “replication phase” (multistage designs are also possible, e.g., Raychaudhuri et al. 2008). A consensus has emerged that a P-value less than 5 × 10−8 corresponds to genome-wide significance in a non-African population-based GWAS. This is a conservative Bonferroni correction based on roughly one million “effectively independent” common SNPs throughout the genome, given the pattern of linkage disequilibrium among common variants across the genome (Pe'er et al. 2008). A variation on this GWAS methodology—the Bayesian GWAS (Marchini et al. 2007; Servin and Stephens 2007; reviewed by Stephens and Balding 2009)—yields Bayes factors for SNP associations (analogous to a likelihood ratio of models with and without association, from which a posterior probability of association can be calculated given a prior probability of association), rather than P-values for the null hypothesis of no association.
The statistical power of a GWAS is a function of sample size, effect size, causal allele frequency, and marker allele frequency and its correlation with the causal variant. Because GWASs are underpowered to detect associations of modest effect sizes (odds ratio, OR of 1.1–1.5; Risch and Merikangas 1996; Spencer et al. 2009; Stahl et al. 2010), large population samples are required to detect variants of even moderate effect (OR 1.5–2). Meta-analyses (see Table 1) of independent GWASs for a trait reap the full benefit of GWASs that have already been performed, greatly increasing sample size and statistical power. When different GWASs use different genotyping array platforms, only a minority of the SNPs are in common. Recently, imputation (see Table 1) methods (reviewed in Li et al. 2009) have been developed to infer genotypes at untyped SNPs using a reference panel of more densely genotyped samples (e.g., HapMap2 data, 2.5 million SNPs, and early releases of 1KG data, ∼10 million SNPs). After imputation, GWAS results can be combined across multiple studies (meta-analysis methodology reviewed in de Bakker et al. 2008).
Notably, almost all GWASs to date have been conducted on populations of European descent, and almost none on populations of African descent. GWASs in alternative populations could identify population-specific associations with causal mutations that occurred after the migrations that established major ethnic populations, and may be especially important for rare variants. GWASs in alternative populations would also contribute to fine mapping (see Table 1), particularly if performed in populations of African descent, which have shorter LD stretches than non-African populations. Fine mapping across ethnicities is based on the idea that a SNP associated in multiple populations must be in LD with the causal variant in all populations (e.g., Udler et al. 2009) and assumes a single causal variant across populations and no population differences in disease etiology. Most GWAS signals have replicated across populations of different ethnicity (Waters et al. 2009, 2010; Teslovich et al. 2010), but in some cases differences between populations have been observed due to extreme allele frequency differences or lack of effect in one population vs. another (Kochi et al. 2009). Recently, it has been shown that under some circumstances, mapping in multiethnic cohorts can significantly increase power to detect associations, as genetic drift may elevate allele frequencies of some variants in different populations, thereby boosting statistical power to detect an association (Pulit et al. 2010). Thus, GWAS in additional populations are an important area of current and future research in medical genetics.
GWAS AND THEIR FINDINGS—IMPLICATIONS FOR GENETIC ARCHITECTURE
The first successful GWAS was of age-related macular degeneration, with ∼100,000 SNPs tested for association in 96 cases and 50 healthy controls (Klein et al. 2005), followed by GWASs for Crohn's Disease (Yamazaki et al. 2005), myocardial infarction (Ozaki and Tanaka 2005), inflammatory bowel disease (Duerr et al. 2006), and type 2 diabetes (Sladek et al. 2007). A landmark study by the Wellcome Trust Case Control Consortium (2007) (WTCCC) reported GWAS results for seven common diseases, including bipolar disorder (BD), coronary artery disease (CAD), Crohn's disease (CD), hypertension (HT), rheumatoid arthritis (RA), type I diabetes (T1D), and type II diabetes (T2D). For each disease, ∼500,000 SNP genotypes of 1500–2000 cases were compared to 3000 “shared” control samples. The study identified previously implicated risk loci, but, more important, revealed multiple new risk loci for some of the diseases. Interestingly, only one new association was found for CAD, and none were found for BD and HT, perhaps because of difficulties in defining disease phenotypes (heterogeneity in disease diagnosis and many underlying causes of disease), or perhaps due to differences in the genetic architecture of these diseases (e.g., fewer common variants with moderate to strong effects), making GWAS less powerful for these traits.
Following the success of the WTCCC study, the trickle of GWAS publications has become a flood. Below we describe results for a few well-studied traits, each with GWASs with >10,000 samples and genome-wide imputation (>2 million SNPs), representing a range of complex trait genetic architectures (at least in terms of common variants): T1D, involving early-onset autoimmune destruction of the insulin-producing β-cells of the pancreas, has a population prevalence of roughly 0.5% and estimated heritability of 88% (Hyttinen et al. 2003). GWAS of over 34,000 samples (Barrett et al. 2009a,b; Hakonarson et al. 2007, 2008; Cooper et al. 2008) have brought the list of validated T1D-risk associated loci to more than 40, explaining 80% of genetic variation of T1D [50% of which comes from the major histocompatability complex (MHC) region; Wei et al. 2009]. An example of another autoimmune disease with later onset but substantial overlap in etiology, RA, is given in box, case study of a GWAS: meta-analysis of six genome-wide association studies identifies seven new rheumatoid arthritis risk loci. Human height is highly heritable (∼80–90% heritability; Visscher 2008). A recent GWAS meta-analysis of nearly 180,000 individuals identified ∼200 loci that together explain ∼14% of height variation (Lango Allen et al. 2010). For other quantitative traits, including body mass index (Speliotes et al. 2010) and cholesterol (Kathiresan et al. 2008), GWASs of more than 240,000 and 22,000 individuals, respectively, have identified 32 and 18 loci that together explain only 2–4% and 5–6% of heritable variation.
Distribution of effect sizes:
Mapping of complex traits in humans and model organisms shows that for the majority of traits studied, many loci contribute to the genetic component of trait variance. The distribution of effect sizes, however, is not completely consistent with the infinitesimal model of quantitative variation where many, many variants of small effect contribute to the trait (Fisher 1918; Bulmer 1980). Instead, for some traits, particularly the immune-related traits where the human leukocyte antigen (HLA) genes exert large effects, the trait architecture consists of a few loci of relatively large effect and many additional loci of very small effect, which is more consistent with Robertson's (1967, 1968) hypothesis of a roughly exponential distribution of effect sizes and consistent with models of adaptation by Fisher (1930), Kimura (1983), and Orr (1998). Among 531 genome-wide significant trait-SNP associations reported as of December 2008 (Hindorff et al. 2009), odds ratios range from 1.04 to 29.4 with first and third quartiles of 1.2 and 1.6. Thus, the vast majority of effect sizes identified to date are small (OR ≤ 1.5).
Gene–environment and gene–gene interactions:
Gene–environment (G × E) and gene–gene interactions may be ubiquitous aspects of complex trait genetics (Templeton 2000; Cordell 2009), due, for example, to genetic redundancy and evolutionary canalization (Waddington 1942; Gibson 2009). In humans, there are several well-documented interactions; for example, multiple studies have shown that the effects of FTO alleles [increasing body mass index by an amount equivalent to 1–1.5 kg body weight and increasing obesity risk by 30% (Frayling et al. 2007; Scuteri et al. 2007)] are attenuated by exercise (e.g., Vimaleswaran et al. 2005; Rampersaud et al. 2008). Most GWASs have not investigated G × E, primarily due to lack of data on environmental exposures. To facilitate testing for G × E, large prospective cohorts (see Table 1) are being established with robust, long-term quantification of environmental variables, e.g., the National Children's study in the United States (www.nationalchildrensstudy.gov) and the Avon Longitudinal Study of Parents and Children in the United Kingdom (www.bristol.ac.uk/alspac/).
Gene–gene or more specifically variant–variant interactions (epistasis) have been identified in model organisms (Phillips 2008; Tyler et al. 2009) and anecdotally in humans (Sing and Davignon 1985; Zerba et al. 2000; Small et al. 2002; Combarros et al. 2009), but to date have not been widely implicated as contributing to human complex trait variation (Hill et al. 2008). Analyses to detect epistatic interactions suffer from a substantially increased multiple testing burden that hampers detection and interpretation (reviewed by Cordell 2009) or have focused only on those SNPs with significant marginal effects (Barrett et al. 2008; Raychaudhuri et al. 2008).
Pleiotropy:
It has long been postulated that pleiotropy is ubiquitous (Caspari 1949). The existence of pleiotropic loci is well documented in model organisms; high-resolution mapping in Drosophila melanogaster, mouse, yeast and Arabidopsis thaliana has demonstrated that what at first appeared to be single QTL for multiple traits, were dissected into multiple variants, often with opposite effects (reviewed in Flint and Mackay 2009). It remains to be seen whether the same will hold true for humans. Several loci appear to have opposite effects across related diseases (Smyth et al. 2008; Maier et al. 2009), and could represent important checkpoints in the branching pathways that lead to the development of related but distinct diseases (Zhernakova et al. 2008). A few GWAS-discovered loci are associated with multiple diseases not previously thought to be related. The loci JAZF1 and TCF2 (or HNF1β) are associated with T2D as well as prostate cancer (Gudmundsson et al. 2007), suggesting that adaptive immunity and T cells play important roles in these different diseases. Interestingly, TCF2 variants contribute to susceptibility to prostate cancer, but are protective against T2D. Pleiotropy at the level of individual variants can influence the evolution of traits and populations (Barton and Keightley 2002; Mitchell-Olds et al. 2007; Roff and Fairbairn 2007) through both positive and negative selection. In humans, particularly non-African populations with relatively extensive LD, pleiotropy may very well turn out to operate at the level of both locus and individual variant, and could be an important contributor to common disease.
Natural selection on trait-associated variants:
The impact of natural selection on disease-associated variants is of keen interest in medical and population genetics (Di Rienzo 2006; Blekhman et al. 2008). Loci and variants underlying many traits, particularly those subject to geographical variation in selection (e.g., skin pigmentation and resistance to infectious agents), show population genetic evidence of the action of natural selection (Cavalli-Sforza et al. 1994; Sabeti et al. 2007; Coop et al. 2009; Pickrell et al. 2009). Genes underlying Mendelian disorders show strong evidence for purifying selection (Blekhman et al. 2008). In contrast, genes implicated in complex diseases do not show strong evidence of either purifying or positive selection (Blekhman et al. 2008). Recent genome scans for signals of positive natural selection based on long haplotypes and population differentiation, found enrichment of T2D risk loci (Pickrell et al. 2009; Chen et al. 2010), which may reflect its correlation with energy metabolism (Neel 1962). Barreiro and Quintana-Murci (2010) found that long-haplotype signatures of recent positive selection were enriched in SNPs associated with autoimmune but not other complex diseases; these risk alleles may have experienced recent positive selection through pleiotropic effects on immune-related phenotypes, including resistance to infectious disease and/or protection against other autoimmune diseases. Tests for selection on complex trait-associated loci or variants need further development and application, because predictions vary across diseases/traits and depend on the nature and degree of localization of the selection signal (Grossman et al. 2010), and they suffer because causal variants remain to be characterized for most complex trait associations. A few clear examples exist, however, for selection acting on individual variants. For example, the T1D- and celiac disease-associated risk allele of the SH2B3 locus shows evidence of selection (Pickrell et al. 2009; Zhernakova et al. 2010), perhaps due its greater response to bacterial infection (Zhernakova et al. 2010), and at the IFIH1 locus, associated with T1D risk and response to viral infection, selection appears to act on the haplotypes associated with infectious disease (Fumagalli et al. 2010). Thus, compelling anecdotal evidence of selection on disease-associated loci also suggests that pleiotropic effects, especially those relating to infectious disease, may be important determinants of selection.
The tests for positive selection described above, however, are based on detecting signals of selective sweeps at individual loci (Maynard Smith and Haigh 1974) and are not able to detect adaptive events that affected many alleles of small effect (Chevin et al. 2008; Hancock et al. 2010; Pritchard et al. 2010). Recently, Pritchard and Di Rienzo (2010) discussed “polygenic adaption,” which can occur by modest changes in allele frequency at multiple loci. Polygenic adaptation, an idea rooted in classical quantitative genetics, could be detected by combining trait-associated variants and then testing for a more significant correlation of their allele frequencies than expected under a neutral model (Pritchard et al. 2010).
Unexplained heritability remains:
Although GWASs have proven successful in identifying regions of the genome harboring variants that contribute to complex phenotypes and diseases, for most traits the effects of all associated loci account for a small proportion of the estimated heritability. With the exception of age-related macular degeneration and type 1 diabetes, for which collectively the proportion of heritability explained to date is approximately 50% and 80%, respectively (Klein et al. 2005; Maller et al. 2006; Barrett et al. 2009a), most complex disease variants identified to date together account for much less of the trait variance. Several proposed explanations for this “missing heritability” include (reviewed in Manolio et al. 2009; Eichler et al. 2010): (1) Effect sizes of associated variants may be underestimates due to incomplete linkage disequilibrium between causal variants and marker SNPs; (2) low-frequency polymorphisms (MAF 0.005–0.05) or rare variants (MAF < 0.005) that are not captured by current genotyping platforms, including CNVs, may contribute a portion of the unexplained heritability; (3) heritability may be overestimated (Slatkin 2009), with epistasis, epigenetics, and genotype–environment interactions contributing to trait heritability; and (4) many additional, currently undetected small effects may together comprise a significant contribution to heritability. Several of these ideas have stimulated the broadening of approaches taken to unravel complex traits.
The proportion of heritability attributable to common SNPs has been well characterized for many complex human traits, but not as extensively for CNVs. Despite several well-documented examples of CNV association with complex traits (e.g., deletion of CCL3L1 in HIV susceptibility; Gonzalez et al. 2005), common CNVs will not likely contribute substantially to unexplained heritability, as common CNVs are in LD with SNPs (Conrad et al. 2010; Craddock et al. 2010). However, it does appear that for some traits, including neurodevelopmental diseases such as schizophrenia (SCZ) (Stefansson et al. 2008; Walsh et al. 2008) and autism (Sebat et al. 2007; Wang et al. 2009), CNVs play a substantial role, so there is still interest in including CNVs in future GWASs.
Rare variants of large effect may explain a portion of the missing heritability. A heavy investment is being made to characterize low-frequency variants (MAF 0.001–0.05; e.g., through the 1KG Project), and commercial genotyping arrays will soon include newly ascertained low-frequency polymorphisms. A substantial number of causal variants could be very rare, possibly de novo or private to families, and these would need to be characterized through sequencing of clinical samples (Li and Leal 2009; Cirulli and Goldstein 2010), probably requiring specialized pooled-variant analysis strategies (Kryukov et al. 2009; Madsen and Browning 2009; Price et al. 2010). As sequencing technologies advance, whole-exome and whole-genome sequencing will become feasible for large numbers of individuals; initial studies (Choi et al. 2009; Lupski et al. 2010; Ng et al. 2010) are promising, though they present a practical challenge of accurate identification of the lowest frequency variants and distinguishing causal variants among so many.
Focused analysis of candidate genes and follow-up of loci identified through GWAS have demonstrated that some genes contain both common and rare variants associated with the trait [e.g., for lipid phenotypes (Cohen et al. 2005; Kotowski et al. 2006; Romeo et al. 2007), blood pressure (Ji et al. 2008), and type 1 diabetes], although the generality of these results will be resolved with empirical data. Dickson et al. (2010) demonstrated via simulations that trait association signals detected for common variants could, in fact, be caused by rare variants. But note that rare variants with very large effects (OR ∼10) would have already been identified through linkage studies, and despite many attempts, very few replicable linkages to complex diseases have been discovered (McCarthy 2002; Orozco et al. 2010). If rare variants influencing a trait are disproportionately located at the same loci as the common variants already identified, then targeted resequencing of regions revealed by GWAS will be a powerful approach (McCarthy 2009).
The distribution of effect sizes for common variants affecting human complex traits is highly skewed toward small effect sizes, and the true distribution is likely even more skewed than the empirical distribution, as GWASs are underpowered to detect small effects. The identification of additional loci of small effect will be partially addressed through meta-analysis of multiple GWAS, but given stringent significance thresholds, it is unlikely that GWAS will ever be powered to identify the full spectrum of small effects. Several recent analytical approaches have been developed to test whether common variants of extremely small effect size might contribute en masse to trait variation. This approach was successfully applied to SCZ, suggesting a highly polygenic model of common variants with small effects, together explaining approximately 35% of population variance in disease (Purcell et al. 2009), still a minority of the estimated 80% heritability of SCZ. Interestingly, Purcell et al. (2009) demonstrated that the same variants contributing to schizophrenia risk also play a role in bipolar disorder. Recently, Yang et al. (2010b) estimated that 67% of the heritability of human height could be explained by a polygenic model. A proportion of the estimated heritability remains unaccounted for in both of these traits, but the model nevertheless accounts for far more than that of known, validated associations. These approaches are unable to identify specific variants contributing to trait variation. Models with many genetic variables obscure the estimation of marginal effects, so that although an overall effect can be inferred, the effects of individual variants cannot be identified with any accuracy (Gibson 2010). Despite this, the approach has practical value; a polygenic analysis defines a large set of variants of which an unknown subset affect phenotype, that together represent real underlying biology. Thus information-based study of the set of variants as a whole may hold the greatest promise of GWAS to dissect human complex traits (see PROMISE OF GWAS—SYSTEMS GENETICS OF COMPLEX TRAITS).
THE PROGRESS OF GWAS—MEDICAL GENETIC DISCOVERY
GWAS discoveries promise to provide understanding of mechanisms of disease etiology and disease pathogenesis. Associated loci not previously suspected of a role in disease etiology have suggested unexpected biology, such as CFH and other genes of the complement pathway (inflammatory pathway regulation) involved in macular degeneration (Klein et al. 2005), and genes of the autophagy pathway (degradation of intracellular components in lysosomes, induced by bacterial infection) involved in inflammatory bowel disease (Rioux et al. 2007; Cho et al. 2008; Mathew 2008). Such genes and pathways would not have been tested in candidate gene studies. GWAS of related diseases often reveal an overlapping genetic basis, and the overlaps promise to shed light on disease mechanisms. Several so-called “autoimmunity” loci are associated with multiple autoimmune diseases, including the HLA genes of the MHC region, and other genes involved in both innate and adaptive immunity (Maier and Hafler 2008; Zhernakova et al. 2008). Among cancers, the 8q24 “gene desert” region, has been found to harbor common variants associated with bladder, breast, colon, ovarian, and prostate cancers (reviewed in Ghoussaini et al. 2008), whereas most other cancer GWAS discoveries are disease specific (reviewed in Easton and Eeles 2008).
The associated SNPs identified through GWAS are unlikely to be the functional variants themselves. Rather, they serve as markers for an underlying haplotype containing the functional variant, but for which the complete pattern of sequence variation is unknown. Hindorff et al. (2009) reported bioinformatic analyses of a recent tally of trait-associated SNPs, providing clues as to the types of genetic variants contributing to complex trait variation. Coding regions were overrepresented (11% of trait-associated SNPs vs. 2% of the genome sequence) among GWAS hits, 43% of associations were located in intergenic regions (outside of promoters and transcribed regions), and 45% were located in introns. Nonsynonymous codons and promoter regions were significantly enriched for trait-associated SNPs, while intergenic regions were significantly underrepresented. Trait-associated SNPs may be preferentially located in genic regions, but they can lie anywhere in the genome, including in gene deserts (e.g., the prostate cancer locus at 8q24; Yeager et al. 2007 and the Crohn's disease locus at 5p13; Wellcome Trust Case Control Consortium 2007). The current picture of gene-centric functional variation is driven by effects of common variants; it remains to be seen whether the pattern will hold once a wider range of allele frequencies and effect sizes have been characterized.
From a medical genetics perspective, the ultimate goal of GWAS is to identify the causal variants underlying validated trait-SNP associations and to characterize their functional effects. In practice the most strongly associated variant at a locus identified through GWAS is presumed to be in LD with the causal, functional variant and becomes the focus for follow-up studies. Fine mapping of an associated locus, followed by deep resequencing of the associated region in samples of interest identifies all possible functional variants, and then a variety of bioinformatic and genomic approaches are used to prioritize variants for experimental studies to verify the functional consequences of putative functional variants (see below).
Examples of experimentally confirmed functional variants underlying validated GWAS hits are accumulating, and they reveal a variety of functional mechanisms underlying trait variation. The IRF5 locus includes variants that disrupt intron splicing, decrease mRNA transcript stability, and delete part of the interferon regulating factor (IRF) protein (Graham et al. 2007), explaining independent associations with systemic lupus erythmatosis (Sigurdsson et al. 2005; Graham et al. 2006), inflammatory bowel disease (Dideberg et al. 2007), and RA (Stahl et al. 2010). Allele-specific chromatin remodeling affecting the expression of several genes in the ORMDL3 locus region (Verlaan et al. 2009) explains its association with asthma (Moffatt et al. 2007), Crohn's disease (Barrett et al. 2008), and T1D (Barrett et al. 2009a). At a locus associated with elevated LDL-cholesterol levels in the blood and myocardial infarction, a common nonprotein-coding variant was found to create a transcription factor binding site that alters the expression of the SORT1 gene in the liver (Musunuru et al. 2010). In another recent study, the largest GWAS meta-analysis to date of blood lipid traits, Teslovich et al. (2010) identified 59 distinct gene variants and validated the biological significance of three of the novel genes in mice, holding promise for therapeutics.
THE PROMISE OF GWAS—SYSTEMS GENETICS OF COMPLEX TRAITS
If polygenic analysis is required to understand the genetic basis of complex traits, this leads to a systems biology perspective in which many perturbations of a complex network contribute to the outcome of complex trait phenotype in ways that may not be possible to disentangle on a per-variant basis. A systems genetics (see Table 1) approach is thus needed, in which large sets of genetic variants and/or genes are analyzed together, genetic data are integrated with external functional data types, and the results inform the biology of the complex trait directly (reviewed in Mackay et al. 2009). Systems genetics is perhaps the culmination of the classic quantitative genetics perspective, where patterns of phenotypic and genetic covariance shed light on complex trait biology directly, but fueled with GWAS and functional genomic (see Table 1) data.
Functional annotation of the genome can shed light on mechanisms of trait biology. One common approach is to determine whether trait-associated variants cluster into groups of specific biological functions more than would be expected by chance, e.g., for gene ontology (GO) terms. Large-scale databases integrate various types of data from the literature to build pathways, and commercial and public tools exist to facilitate access [e.g., Ingenuity (http://www.ingenuity.com); Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/)]).
Recently, GWASs have been conducted on mRNA levels, themselves quantitative traits, in expression QTL (eQTL) mapping studies (see Table 1) (e.g., Dixon et al. 2007; Goring et al. 2007; Stranger et al. 2007a,b; Dimas et al. 2009). Several studies show that complex trait-associated variants overlap with eQTL variants (e.g., Emilsson et al. 2008; Nica et al. 2010), helping to prioritize a gene and mechanism for functional follow-up. Furthermore, a recent study of GWAS associations reported that on a global scale, GWAS-identified variants are significantly more likely to be eQTL than minor-allele-frequency-matched SNPs chosen from high-throughput GWAS platforms (Nicolae et al. 2010). Although some eQTL datasets are publicly available (e.g., http://www.sanger.ac.uk/resources/software/genevar/; Yang et al. 2010a), few are from primary cell types, and available datasets may not be relevant to some traits or diseases, and few studies have performed both complex trait GWAS and eQTL mapping in the same individuals. To provide the scientific community with a resource to facilitate large-scale analyses, the National Institutes of Health recently launched the Genotype-Tissue Expression (GTeX) project (http://nihroadmap.nih.gov/GTEx/index.asp) to provide a publicly available catalog of tissue-specific gene expression profiles and eQTL.
Similarly, other data types such as methylation/acetylation, protein–protein interactions, and miRNA regulatory networks, can be integrated with GWAS results. Gene set enrichment analysis (GSEA; see Table 1) (Subramanian et al. 2005) and related analyses (Lage et al. 2008) have identified correlated expression profiles of trait-associated genes across experiments and tissues in several diseases. High-confidence protein–protein interactions have successfully identified candidate genes within linkage/association intervals on the basis of their protein products' interactions with those implicated in similar diseases (Lage et al. 2007).
Integrative analyses have thus far focused largely on validated SNPs (Lage et al. 2007; Lango Allen et al. 2010; Nicolae et al. 2010) and provide an accurate but incomplete picture of the genetic system underlying complex traits. As future GWASs bring the numbers of validated SNPs from a few tens to >100 for complex diseases, these analyses will become a gold standard for comparison with systems genetics approaches based on broader sets of variants (e.g., from polygenic analysis, GWAS, or sequencing studies) to provide insights into complex trait biology directly.
The Bayesian GWAS framework is appropriate for using external biological and functional genomics-based information to inform prior probabilities of SNP association (Stephens and Balding 2009). Leveraging independent, functional knowledge to establish priors should be straightforward, based on odds ratios of the external data in validated trait-associated SNPs, but remains a key challenge for the development of Bayesian GWAS methods because of their heterogeneity and potential bias. While exciting approaches for combining heterogeneous data are being developed (Lage et al. 2008; Huttenhower et al. 2009; Lee et al. 2009; Battle et al. 2010), these issues must be taken into consideration in the design and interpretation of truly integrative systems genetics analyses.
CONCLUSION
Genome-wide association studies in humans have already proven a resounding success in providing a framework for unraveling the genetic basis of complex traits. The results have provided unprecedented views into the contribution of common variants to complex traits, illuminated genome function, and have opened new possibilities for the development of therapeutic interventions. Trait architecture conforms to a roughly exponential distribution of effect sizes: the majority of common complex trait-associated variants studied thus far have modest effects (OR < 2), and for most traits, substantial heritability remains to be explained. Identifying the genetic basis of the remaining trait variance will require additional discoveries, particularly of rare trait-associated variants and better characterization of genetic modes of action and interaction and refined estimates of heritability. DNA sequencing will play a key role in the next generation of GWAS, through candidate locus resequencing in large cohorts, whole-exome sequencing, and eventually whole-genome sequencing of large numbers of individuals. Also, integration of functional biological knowledge into association analyses promises to point directly to putative functional variants. Importantly, identification of causal variants and expansion of these studies into populations of diverse ancestry will facilitate further biological understanding and population genetics of complex traits. Continued accelerated pace of discovery of medically important trait-associated variants in humans will depend on implementation of new technologies and analytic approaches to integrate diverse data types, but also, critically, on the lessons learned from the burst of discovery that has been the result of the first round of genome-wide association studies in humans.
References
- Altenburg, E., and H. J. Muller, 1920. The Genetic Basis of Truncate Wing,—an Inconstant and Modifiable Character in Drosophila. Genetics 5(1): 1–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler, D., J. N. Hirschhorn, M. Klannemark, C. M. Lindgren, M. C. Vohl et al., 2000. a The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 26 76–80. [DOI] [PubMed] [Google Scholar]
- Altshuler, D., V. J. Pollara, C. R. Cowles, W. J. Van Etten, J. Baldwin et al., 2000. b An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407 513–516. [DOI] [PubMed] [Google Scholar]
- Altshuler, D. M., R. A. Gibbs, L. Peltonen, E. Dermitzakis, S. F. Schaffner et al., 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana, M. J., S. Kim, K. Zhao, E. Bakker, M. Horton et al., 2005. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 1 e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko, Y. S., D. J. de Koning and C. Haley, 2007. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding, D. J., 2006. A tutorial on statistical methods for population association studies. Nat. Rev. Genet. 7 781–791. [DOI] [PubMed] [Google Scholar]
- Barreiro, L. B., and L. Quintana-Murci, 2010. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat. Rev. Genet. 11 17–30. [DOI] [PubMed] [Google Scholar]
- Barrett, J. C., S. Hansoul, D. L. Nicolae, J. H. Cho, R. H. Duerr et al., 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J. C., D. G. Clayton, P. Concannon, B. Akolkar, J. D. Cooper et al., 2009. a Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett, J. C., J. C. Lee, C. W. Lees, N. J. Prescott, C. A. Anderson et al., 2009. b Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41 1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, A., W. Thomson, X. Ke, S. Eyre, A. Hinks et al., 2008. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum. Mol. Genet. 17 2274–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, A., S. Eyre, X. Ke, A. Hinks, J. Bowes et al., 2009. Identification of AF4/FMR2 family, member 3 (AFF3) as a novel rheumatoid arthritis susceptibility locus and confirmation of two further pan-autoimmune susceptibility genes. Hum. Mol. Genet. 18 2518–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3 11–21. [DOI] [PubMed] [Google Scholar]
- Bateson, W., 1902. Mendel's Principles of Heredity: A Defence. Cambridge University Press, Cambridge, UK.
- Bateson, W, 1909. Mendel's Principles of Heredity. Cambridge University Press, Cambridge UK.
- Bateson W, E. R. Saunders, and R. C. Punett, 1905. Experimental studies in the physiology of heredity. Reports to the Evolution Committee of the Royal Society 2 1–55, 80–99. [Google Scholar]
- Battle, A, M. C. Jonikas, P. Walter, J. S. Weisman and D. Koller, 2010. Automated identification of pathways from quantitative genetic interaction data. Mol. Syst. Biol. 6 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertina, R. M., B. P. Koeleman, T. Koster, F. R. Rosendaal, R. J. Dirven et al., 1994. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature 369 64–67. [DOI] [PubMed] [Google Scholar]
- Blekhman, R., O. Man, L. Herrmann, A. R. Boyko, A. Indap et al., 2008. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer, W., and C. Bonilla, 2008. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 40 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein, D., R. L. White, M. Skolnick and R. W. Davis, 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 314–331. [PMC free article] [PubMed] [Google Scholar]
- Buckler, E. S., J. B. Holland, P. J. Bradbury, C. B. Acharya, P. J. Brown et al., 2009. The genetic architecture of maize flowering time. Science 325 714–718. [DOI] [PubMed] [Google Scholar]
- Bulmer, M., 1980. The Mathematical Theory of Quantitative Genetics. Clarendon Press, Oxford.
- Cardon, L. R., and J. I. Bell, 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2 91–99. [DOI] [PubMed] [Google Scholar]
- Caspari, E., 1949. A synopsis of contemporary evolutionary thinking. Evolution 3 377. [DOI] [PubMed] [Google Scholar]
- Castle, W. E., and C. C. Little, 1910. On a modified Mendelian ratio among yellow mice. Science 32 868–870. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza, L. L., P. Menozzi and A. Piazza, 1994. The History and Geography of Human Gene. Princeton University Press, Princeton, NJ.
- Cavalli-Sforza, L. L., and M. W. Feldman, 2003. The application of molecular genetic approaches to the study of human evolution. Nat. Genet. 33(Suppl): 266–275. [DOI] [PubMed] [Google Scholar]
- Chen, H., N. Patterson and D. Reich, 2010. Population differentiation as a test for selective sweeps. Genome Res. 20 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin, L. M., S. Billiard and F. Hospital, 2008. Hitchhiking both ways: effect of two interfering selective sweeps on linked neutral variation. Genetics 180 301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H. S., T. J. Byun, S. B. Ahn, T. Y. Kim, C. S. Eun et al., 2008. A case of familial Crohn's disease observed in a parent and his offspring. Korean J. Gastroenterol. 52 247–250. [PubMed] [Google Scholar]
- Choi, M., U. I. Scholl, W. Ji, T. Liu, I. R. Tikhonova et al., 2009. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 106 19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli, E. T., and D. B. Goldstein, 2010. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 11 415–425. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. J. Hubisz, C. D. Bustamante, S. H. Williamson and R. Nielsen, 2005. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15 1496–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, D. G., N. M. Walker, D. J. Smyth, R. Pask, J. D. Cooper et al., 2005. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 37 1243–1246. [DOI] [PubMed] [Google Scholar]
- Cohen, J., A. Pertsemlidis, I. K. Kotowski, R. Graham, C. K. Garcia et al., 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37 161–165. [DOI] [PubMed] [Google Scholar]
- Combarros, O., M. Cortina-Borja, A. D. Smith and D. J. Lehmann, 2009. Epistasis in sporadic Alzheimer's disease. Neurobiol. Aging 30 1333–1349. [DOI] [PubMed] [Google Scholar]
- Conrad, D. F., D. Pinto, R. Redon, L. Feuk, O. Gokcumen et al., 2010. Origins and functional impact of copy number variation in the human genome. Nature 464 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop, G., J. K. Pickrell, J. Novembre, S. Kudaravalli, J. Li et al., 2009. The role of geography in human adaptation. PLoS Genet. 5 e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, J. D., D. J. Smyth, A. M. Smiles, V. Plagnol, N. M. Walker et al., 2008. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 40 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, H. J., 2009. Detecting gene-gene interactions that underlie human diseases. Nat. Rev. Genet. 10(6): 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corder, E. H., A. M. Saunders, W. J. Strittmatter, D. E. Schmechel, P. C. Gaskell et al., 1993. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 261 921–923. [DOI] [PubMed] [Google Scholar]
- Craddock, N., M. E. Hurles, N. Cardin, R. D. Pearson, V. Plagnol et al., 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, M. J., J. D. Rioux, S. F. Schaffner, T. J. Hudson and E. S. Lander, 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29 229–232. [DOI] [PubMed] [Google Scholar]
- de Bakker, P. I., M. A. Ferreira, X. Jia, B. M. Neale, S. Raychaudhuri et al., 2008. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17 R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker, P. I., R. Yelensky, I. Pe'er, S. B. Gabriel, M. J. Daly et al., 2005. Efficiency and power in genetic association studies. Nat. Genet. 37 1217–1223. [DOI] [PubMed] [Google Scholar]
- Devlin, B., and K. Roeder, 1999. Genomic control for association studies. Biometrics 55 997–1004. [DOI] [PubMed] [Google Scholar]
- Di Rienzo, A., 2006. Population genetics models of common diseases. Curr. Opin. Genet. Dev. 16 630–636. [DOI] [PubMed] [Google Scholar]
- Dickson, S. P., K. Wang, I. Krantz, H. Hakonarson and D. B. Goldstein, 2010. Rare variants create synthetic genome-wide associations. PLoS Biol. 8 e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg, V., G. Kristjansdottir, L. Milani, C. Libioulle, S. Sigurdsson et al., 2007. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum. Mol. Genet. 16 3008–3016. [DOI] [PubMed] [Google Scholar]
- Dimas, A. S., S. Deutsch, B. E. Stranger, S. B. Montgomery, C. Borel et al., 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, A. L., L. Liang, M. F. Moffatt, W. Chen, S. Heath et al., 2007. A genome-wide association study of global gene expression. Nat. Genet. 39 1202–1207. [DOI] [PubMed] [Google Scholar]
- Duerr, R. H., K. D. Taylor, S. R. Brant, J. D. Rioux, M. S. Silverberg et al., 2006. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin, R. M., G. R. Abecasis, D. L. Altshuler, A. Auton, L. D. Brooks et al., 2010. A map of human genome variation from population-scale sequencing. Nature 467 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East, E. M., 1910. A Mendelian interpretation of variation that is apparently continuous. Am. Nat. 44 65–82. [Google Scholar]
- Easton, D. F., and R. A. Eeles, 2008. Genome-wide association studies in cancer. Hum. Mol. Genet. 17 R109–R115. [DOI] [PubMed] [Google Scholar]
- Eichler, E. E., J. Flint, G. Gibson, A. Kong, S. M. Leal et al., 2010. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 11 446–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson, V., G. Thorleifsson, B. Zhang, A. S. Leonardson, F. Zink et al., 2008. Genetics of gene expression and its effect on disease. Nature 452 423–428. [DOI] [PubMed] [Google Scholar]
- Falconer, D., and T. Mackay, 1996. Introduction to Quantitative Genetics. Longman, New York. [DOI] [PMC free article] [PubMed]
- Fisher, R., 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans. Roy. Soc. Edin. 52 399–433. [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Flint, J., and T. F. Mackay, 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayling, T. M., N. J. Timpson, M. N. Weedon, E. Zeggini, R. M. Freathy et al., 2007. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316 889–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli, M., R. Cagliani, S. Riva, U. Pozzoli, M. Biasin et al., 2010. Population genetics of IFIH1: ancient population structure, local selection and implications for susceptibility to type 1 diabetes. Mol. Biol. Evol. 27 2555–2566. [DOI] [PubMed] [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296 2225–2229. [DOI] [PubMed] [Google Scholar]
- Galton, F., 1869. Hereditary Genius: an Inquiry Into Its Laws and Consequences. Macmillan, London.
- Galton, F., 1889. Natural Inheritance. Macmillan, London.
- Ghoussaini, M., H. Song, T. Koessler, A. A. Al Olama, Z. Kote-Jarai et al., 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J. Natl. Cancer Inst. 100 962–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, G., 2009. Decanalization and the origin of complex disease. Nat. Rev. Genet. 10 134–140. [DOI] [PubMed] [Google Scholar]
- Gibson, G., 2010. Hints of hidden heritability in GWAS. Nat. Genet. 42 558–560. [DOI] [PubMed] [Google Scholar]
- Gonzalez, E., H. Kulkarni, H. Bolivar, A. Mangano, R. Sanchez et al., 2005. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307 1434–1440. [DOI] [PubMed] [Google Scholar]
- Goring, H. H., J. E. Curran, M. P. Johnson, T. D. Dyer, J. Charlesworth et al., 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 39 1208–1216. [DOI] [PubMed] [Google Scholar]
- Graham, R. R., S. V. Kozyrev, E. C. Baechler, M. V. Reddy, R. M. Plenge et al., 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 38 550–555. [DOI] [PubMed] [Google Scholar]
- Graham, R. R., C. Kyogoku, S. Sigurdsson, I. A. Vlasova, L. R. Davies et al., 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc. Natl. Acad. Sci. USA 104 6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen, P. K., C. I. Amos, A. T. Lee, Y. Lu, E. F. Remmers et al., 2009. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet. 41 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, S. R., I. Shylakhter, E. K. Karlsson, E. H. Byrne, S. Morales et al., 2010. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science 327 883–886. [DOI] [PubMed] [Google Scholar]
- Gudmundsson, J., P. Sulem, V. Steinthorsdottir, J. T. Bergthorsson, G. Thorleifsson et al., 2007. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 39 977–983. [DOI] [PubMed] [Google Scholar]
- Gusella, J. F., N. S. Wexler, P. M. Conneally, S. L. Naylor, M. A. Anderson et al., 1983. A polymorphic DNA marker genetically linked to Huntington's disease. Nature 306 234–238. [DOI] [PubMed] [Google Scholar]
- Hakonarson, H., S. F. Grant, J. P. Bradfield, L. Marchand, C. E. Kim et al., 2007. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature 448 591–594. [DOI] [PubMed] [Google Scholar]
- Hakonarson, H., H. Q. Qu, J. P. Bradfield, L. Marchand, C. E. Kim et al., 2008. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes 57 1143–1146. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1932. The Causes of Evolution. Longmans, Green & Co., London.
- Hancock, A. M., G. Alkorta-Aranburu, D. B. Witonsky and A. Di Rienzo, 2010. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365 2459–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy, J., and A. Singleton, 2009. Genomewide association studies and human disease. N. Engl. J. Med. 360 1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, H., 1966. Enzyme polymorphisms in man. Proc. R. Soc. B 164 298–310. [DOI] [PubMed] [Google Scholar]
- Hill, W. G., M. E. Goddard and P. M. Visscher, 2008. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 4 e1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff, L. A., P. Sethupathy, H. A. Junkins, E. M. Ramos, J. P. Mehta et al., 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, T. J., L. D. Stein, S. S. Gerety, J. Ma, A. B. Castle et al., 1995. An STS-based map of the human genome. Science 270 1945–1954. [DOI] [PubMed] [Google Scholar]
- Huttenhower, C, K. T. Mutungu, N. Indik, W. Yang, M. Schroeder et al., 2009. Detailing regulatory networks through large scale data integration. Bioinformatics 25 3267–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyttinen, V., J. Kaprio, L. Kinnunen, M. Koskenvuo and J. Tuomilehto, 2003. Genetic liability of type 1 diabetes and the onset age among 22,650 young Finnish twin pairs: a nationwide follow-up study. Diabetes 52 1052–1055. [DOI] [PubMed] [Google Scholar]
- International Hapmap Consortium, 2003. The International HapMap Project. Nature 426 789–796. [DOI] [PubMed] [Google Scholar]
- Ioannidis, J. P., G. Thomas and M. J. Daly, 2009. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, W., J. N. Foo, B. J. O'Roak, H. Zhao, M. G. Larson et al., 2008. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat. Genet. 40 592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. D., and C. J. O'Donnell, 2009. An open access database of genome-wide association results. BMC Med. Genet. 10 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan, S., O. Melander, C. Guiducci, A. Surti, N. P. Burtt et al., 2008. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 40 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge.
- Klein, R. J., C. Zeiss, E. Y. Chew, J. Y. Tsai, R. S. Sackler et al., 2005. Complement factor H polymorphism in age-related macular degeneration. Science 308 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochi, Y., A. Suzuki, R. Yamada and K. Yamamoto, 2009. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J. Autoimmun. 32 158–162. [DOI] [PubMed] [Google Scholar]
- Kotowski, I. K., A. Pertsemlidis, A. Luke, R. S. Cooper, G. L. Vega et al., 2006. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 78 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov, G. V., A. Shpunt, J. A. Stamatoyannopoulos and S. R. Sunyaev, 2009. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl. Acad. Sci. USA 106 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage, K., N. T. Hansen, E. O. Karlberg, A. C. Eklund, F. S. Roque et al., 2008. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc. Natl. Acad. Sci. USA 105 20870–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage, K., E. O. Karlberg, Z. M. Storling, P. I. Olason, A. G. Pedersen et al., 2007. A human phenome-interactome network of protein complexes implicated in genetic disorders. Nat. Biotechnol. 25 309–316. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., 1996. The new genomics: global views of biology. Science 274 536–539. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody et al., 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Lango Allen, H., K. Estrada, G. Lettre, S. I. Berndt, M. N. Weedon et al., 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. I., A. M. Dudley, D. Drubin, P. A. Silver, N. J. Krogan et al., 2009. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 5 e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and S. M. Leal, 2009. Discovery of rare variants via sequencing: implications for the design of complex trait association studies. PLoS Genet. 5 e1000481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. H., and L. A. Sadler, 1991. Low nucleotide diversity in man. Genetics 129 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., C. Willer, S. Sanna and G. Abecasis, 2009. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10 387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski, J. R., J. G. Reid, C. Gonzaga-Jauregui, D. Rio Deiros, D. C. Chen et al., 2010. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 362 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mackay, T. F., E. A. Stone and J. F. Ayroles, 2009. The genetics of quantitative traits: challenges and prospects. Nat. Rev. Genet. 10 565–577. [DOI] [PubMed] [Google Scholar]
- Madsen, B. E., and S. R. Browning, 2009. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 5 e1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, L. M., and D. A. Hafler, 2008. The developing mosaic of autoimmune disease risk. Nat. Genet. 40 131–132. [DOI] [PubMed] [Google Scholar]
- Maier, L. M., C. E. Lowe, J. Cooper, K. Downes, D. E. Anderson et al., 2009. IL2RA genetic heterogeneity in multiple sclerosis and type 1 diabetes susceptibility and soluble interleukin-2 receptor production. PLoS Genet. 5 e1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller, J., S. George, S. Purcell, J. Fagerness, D. Altshuler et al., 2006. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 38 1055–1059. [DOI] [PubMed] [Google Scholar]
- Manolio, T. A., F. S. Collins, N. J. Cox, D. B. Goldstein, L. A. Hindorff et al., 2009. Finding the missing heritability of complex diseases. Nature 461 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini, J., B. Howie, S. Myers, G. McVean and P. Donnelly, 2007. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39 906–913. [DOI] [PubMed] [Google Scholar]
- Mather, K., and J. L. Jinks, 1971. Biometrical Genetics: The Study of Continuous Variation. Chapman and Hall, London.
- Mather, K., 1943. Polygene in development. Nature 151 560. [Google Scholar]
- Mathew, C. G., 2008. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat. Rev. Genet. 9 9–14. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and J. Haigh, 1974. The hitch-hiking effect of a favourable gene. Genet. Res. 23 23–35. [PubMed] [Google Scholar]
- McCarthy, M. I., 2002. Susceptibility gene discovery for common metabolic and endocrine traits. J. Mol. Endocrinol. 28 1–17. [DOI] [PubMed] [Google Scholar]
- McCarthy, M. I., 2009. Exploring the unknown: assumptions about allelic architecture and strategies for susceptibility variant discovery. Genome Med. 1 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean, G. A., and N. J. Cardin, 2005. Approximating the coalescent with recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell-Olds, T., J. H. Willis and D. B. Goldstein, 2007. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat. Rev. Genet. 8 845–856. [DOI] [PubMed] [Google Scholar]
- Moffatt, M. F., M. Kabesch, L. Liang, A. L. Dixon, D. Strachan et al., 2007. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature 448 470–473. [DOI] [PubMed] [Google Scholar]
- Morgan, T. H., 1911. a The Origin of Nine Wing Mutations in Drosophila. Science 33 496–499. [DOI] [PubMed] [Google Scholar]
- Morgan, T. H., 1911. b Random Segregation Versus Coupling in Mendelian Inheritance. Science 34 384. [DOI] [PubMed] [Google Scholar]
- Morgan, T. H., A. H. Sturtevant, H. J. Muller and C. B. Bridges, 1915. The Mechanism of Mendelian Heredity. Henry Holt, New York.
- Musunuru, K., A. Strong, M. Frank-Kamenetsky, N. E. Lee, T. Ahfeldt et al., 2010. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 466 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel, J. V., 1962. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet. 14 353–362. [PMC free article] [PubMed] [Google Scholar]
- Neimann-Sorensen, A., and A. Robertson, 1961. The association between blood groups and several production characteristics in three Danish cattle breeds. Acta Agr. Scand. 11 163–196. [Google Scholar]
- Ng, S. B., K. J. Buckingham, C. Lee, A. W. Bigham, H. K. Tabor et al., 2010. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42 30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica, A. C., S. B. Montgomery, A. S. Dimas, B. E. Stranger, C. Beazley et al., 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 6 e1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D. A., S. L. Taylor, K. M. Weiss, A. G. Clark, R. G. Hutchinson et al., 1998. DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nat. Genet. 19 233–240. [DOI] [PubMed] [Google Scholar]
- Nicolae, D. L., E. Gamazon, W. Zhang, S. Duan, M. E. Dolan et al., 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6 e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco, G., J. C. Barrett and E. Zeggini, 2010. Synthetic associations in the context of genome-wide association scan signals. Hum. Mol. Genet. 19 R137–R144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1998. Testing natural selection versus genetic drift in phenotypic evolution using Quantitative Trait Locus data. Genetics 149 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott, J., 1991. Analysis of Human Genetic Linkage, Ed. 2. Johns Hopkins University Press, Baltimore.
- Ozaki, K., and T. Tanaka, 2005. Genome-wide association study to identify SNPs conferring risk of myocardial infarction and their functional analyses. Cell. Mol. Life. Sci. 62 1804–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, K., 1898. Mathematical contributions to the theory of evolution. On the Law of Ancestral Heredity. Proc. R. Soc. London 62 386–412. [Google Scholar]
- Pe'er, I., R. Yelensky, D. Altshuler and M. J. Daly, 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 32 381–385. [DOI] [PubMed] [Google Scholar]
- Phillips, P. C., 2008. Epistasis: the essential role of gene interactions in the structure and evolution of genetic systems. Nat. Rev. Genet. 9 855–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, J. K., G. Coop, J. Novembre, S. Kudaravalli, J. Z. Li et al., 2009. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19 826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge, R. M., C. Cotsapas, L. Davies, A. L. Price, P. I. de Bakker et al., 2007. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet. 39 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. L., N. J. Patterson, R. M. Plenge, M. E. Weinblatt, N. A. Shadick et al., 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38 904–909. [DOI] [PubMed] [Google Scholar]
- Price, A. L., G. V. Kryukov, P. I. de Bakker, S. M. Purcell, J. Staples et al., 2010. Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet. 86 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., 2001. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 69 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., and A. Di Rienzo, 2010. Adaptation–not by sweeps alone. Nat. Rev. Genet. 11 665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens, N. A. Rosenberg and P. Donnelly, 2000. Association mapping in structured populations. Am. J. Hum. Genet. 67 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., J. K. Pickrell and G. Coop, 2010. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20 R208–R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit, S. L., B. F. Voight and P. I. de Bakker, 2010. Multiethnic genetic association studies improve power for locus discovery. PLoS One 5 e12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punnett, R. C., 1909. Mendelism. Wilshire, New York.
- Purcell, S. M., N. R. Wray, J. L. Stone, P. M. Visscher, M. C. O'Donovan et al., 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersaud, E., B. D. Mitchell, T. I. Pollin, M. Fu, H. Shen et al., 2008. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch. Intern. Med. 168 1791–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri, S., 2010. Recent advances in the genetics of rheumatoid arthritis. Curr. Opin. Rheumatol. 22 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri, S., E. F. Remmers, A. T. Lee, R. Hackett, C. Guiducci et al., 2008. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet. 40 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon, R., S. Ishikawa, K. R. Fitch, L. Feuk, G. H. Perry et al., 2006. Global variation in copy number in the human genome. Nature 444 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D. E., and E. S. Lander, 2001. On the allelic spectrum of human disease. Trends Genet. 17 502–510. [DOI] [PubMed] [Google Scholar]
- Reich, D. E., S. F. Schaffner, M. J. Daly, G. McVean, J. C. Mullikin et al., 2002. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32 135–142. [DOI] [PubMed] [Google Scholar]
- Remmers, E. F., R. M. Plenge, A. T. Lee, R. R. Graham, G. Hom et al., 2007. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med. 357 977–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan, J. R., J. M. Rommens, B. Kerem, N. Alon, R. Rozmahel et al., 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science 245 1066–1073. [DOI] [PubMed] [Google Scholar]
- Rioux, J. D., R. J. Xavier, K. D. Taylor, M. S. Silverberg, P. Goyette et al., 2007. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch, N., and K. Merikangas, 1996. The future of genetic studies of complex human diseases. Science 273 1516–1517. [DOI] [PubMed] [Google Scholar]
- Robertson, A., 1967. The Nature of Quantitative Genetic Variation. University of Wisconsin Press, Madison, WI.
- Robertson, A., 1968. The Spectrum of Genetic Variation. Syracuse University Press, Syracuse, NY.
- Roff, D. A., and D. J. Fairbairn, 2007. The evolution of trade-offs: where are we? J. Evol. Biol. 20 433–447. [DOI] [PubMed] [Google Scholar]
- Romeo, S., L. A. Pennacchio, Y. Fu, E. Boerwinkle, A. Tybjaerg-Hansen et al., 2007. Population-based resequencing of ANGPTL4 uncovers variations that reduce triglycerides and increase HDL. Nat. Genet. 39 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A., J. K. Pritchard, J. L. Weber, H. M. Cann, K. K. Kidd et al., 2002. Genetic structure of human populations. Science 298 2381–2385. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., P. Varilly, B. Fry, J. Lohmueller, E. Hostetter et al., 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam, R., D. Weissman, S. C. Schmidt, J. M. Kakol, L. D. Stein et al., 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409 928–933. [DOI] [PubMed] [Google Scholar]
- Sax, K., 1923. The association of size differences with seed coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuteri, A., S. Sanna, W. M. Chen, M. Uda, G. Albai et al., 2007. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 3 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J., B. Lakshmi, D. Malhotra, J. Troge, C. Lese-Martin et al., 2007. Strong association of de novo copy number mutations with autism. Science 316 445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin, B., and M. Stephens, 2007. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson, S., G. Nordmark, H. H. Goring, K. Lindroos, A. C. Wiman et al., 2005. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am. J. Hum. Genet. 76 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing, C. F., and J. Davignon, 1985. Role of the apolipoprotein E polymorphism in determining normal plasma lipid and lipoprotein variation. Am. J. Hum. Genet. 37 268–285. [PMC free article] [PubMed] [Google Scholar]
- Sladek, R., G. Rocheleau, J. Rung, C. Dina, L. Shen et al., 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445 881–885. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., 2009. Epigenetic inheritance and the missing heritability problem. Genetics 182 845–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, K. M., L. E. Wagoner, A. M. Levin, S. L. Kardia and S. B. Liggett, 2002. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N. Engl. J. Med. 347 1135–1142. [DOI] [PubMed] [Google Scholar]
- Smith, J. G., J. K. Lowe, S. Kovvali, J. B. Maller, J. Salit et al., 2009. Genome-wide association study of electrocardiographic conduction measures in an isolated founder population. Kosrae. Heart Rhythm 6 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, D. J., J. D. Cooper, J. M. Howson, N. M. Walker, V. Plagnol et al., 2008. PTPN22 Trp620 explains the association of chromosome 1p13 with type 1 diabetes and shows a statistical interaction with HLA class II genotypes. Diabetes 57 1730–1737. [DOI] [PubMed] [Google Scholar]
- Speliotes, E. K., C. J. Willer, S. I. Berndt, K. L. Monda, G. Thorleifsson et al., 2010. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 42 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, C. C., Z. Su, P. Donnelly and J. Marchini, 2009. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 5 e1000477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E. A., S. Raychaudhuri, E. F. Remmers, G. Xie, S. Eyre et al., 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson, H., D. Rujescu, S. Cichon, O. P. Pietilainen, A. Ingason et al., 2008. Large recurrent microdeletions associated with schizophrenia. Nature 455 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, M., and D. J. Balding, 2009. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 10 681–690. [DOI] [PubMed] [Google Scholar]
- Stranger, B. E., M. S. Forrest, M. Dunning, C. E. Ingle, C. Beazley et al., 2007. a Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B. E., A. C. Nica, M. S. Forrest, A. Dimas, C. P. Bird et al., 2007. b Population genomics of human gene expression. Nat. Genet. 39 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant, A.H., 1913. The Linear Arragement of Six Sex Linked Factors in in Drosophila, as Shown by their Mode of Association. J. Exp. Zool. 14 43–59. [Google Scholar]
- Sturtevant, A. H., 1915. The behavior of the chromosomes as studied through linkage. Verlag Gerb. Borntraeger, Berlin 234–287.
- Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert et al., 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton, AR, 2000. Epistasis and complex traits, pp. 41–57 in Epistasis and the Evolutionary Process. Edited by J. Wolf, B. Brodie III and M. Wade. Oxford University Press, New York.
- Terwilliger, J. D., and K. M. Weiss, 1998. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr. Opin. Biotechnol. 9 578–594. [DOI] [PubMed] [Google Scholar]
- Teslovich, T. M., K. Musunuru, A. V. Smith, A. C. Edmondson, I. M. Stylianou et al., 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoday, J. M., 1961. Location of polygenes. Nature 191 368–370. [Google Scholar]
- Thoday, J. M., and J. N. Thompson, Jr., 1976. The number of segregating genes implied by continuous variation. Genetica 46 335–344. [Google Scholar]
- Tishkoff, S. A., and B. C. Verrelli, 2003. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu. Rev. Genomics Hum. Genet. 4 293–340. [DOI] [PubMed] [Google Scholar]
- Tyler, A. L., F. W. Asselbergs, S. M. Williams and J. H. Moore, 2009. Shadows of complexity: what biological networks reveal about epistasis and pleiotropy. Bioessays 31 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler, M. S., K. B. Meyer, K. A. Pooley, E. Karlins, J. P. Struewing et al., 2009. FGFR2 variants and breast cancer risk: fine-scale mapping using African American studies and analysis of chromatin conformation. Hum. Mol. Genet. 18 1692–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlaan, D. J., B. Ge, E. Grundberg, R. Hoberman, K. C. Lam et al., 2009. Targeted screening of cis-regulatory variation in human haplotypes. Genome Res. 19 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaleswaran, K. S., V. Radha, S. Ghosh, P. P. Majumder, R. Deepa et al., 2005. Peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene polymorphisms and their relationship to Type 2 diabetes in Asian Indians. Diabet. Med. 22 1516–1521. [DOI] [PubMed] [Google Scholar]
- Visscher, P. M., 2008. Sizing up human height variation. Nat. Genet. 40 489–490. [DOI] [PubMed] [Google Scholar]
- Waddington, C. H., 1942. Canalization of development and the inheritance of acquired characters. Nature 150 563–565. [Google Scholar]
- Waddington, C. H., 1943. Polygenes and oligogenes. Nature 151 394. [Google Scholar]
- Walsh, T., J. M. McClellan, S. E. McCarthy, A. M. Addington, S. B. Pierce et al., 2008. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320 539–543. [DOI] [PubMed] [Google Scholar]
- Wang, K., H. Zhang, D. Ma, M. Bucan, J. T. Glessner et al., 2009. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, K. M., L. Le Marchand, L. N. Kolonel, K. R. Monroe, D. O. Stram et al., 2009. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol. Biomarkers Prev. 18 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, K. M., D. O. Stram, M. T. Hassanein, L. Le Marchand, L. R. Wilkens et al., 2010. Consistent association of type 2 diabetes risk variants found in Europeans in diverse racial and ethnic groups. PLoS Genet. 6 e1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, Z., K. Wang, H. Q. Qu, H. Zhang, J. Bradfield et al., 2009. From disease association to risk assessment: an optimistic view from genome-wide association studies on type 1 diabetes. PLoS Genet. 5 e1000678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium, 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1969. Evolution and the Genetics of Populations, Vol. 2, The Theory of Gene Frequencies. University of Chicago Press, Chicago.
- Wright, S., 1977. Evolution and the Genetics of Populations, Vol. 3, Experimental Results and Evolutionary Deductions. University of Chicago Press, Chicago.
- Wright, S., 1978. Evolution and the Genetics of Populations, Vol. 4, Variation Within and Between Natural Populations. University of Chicago Press, Chicago.
- Yamazaki, K., D. McGovern, J. Ragoussis, M. Paolucci, H. Butler et al., 2005. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum. Mol. Genet. 14 3499–3506. [DOI] [PubMed] [Google Scholar]
- Yang, T. P., C. Beazley, S. B. Montgomery, A. S. Dimas, M. Gutierrez-Arcelus et al., 2010. a Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics 26 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., B. Benyamin, B. P. McEvoy, S. Gordon, A. K. Henders et al., 2010. b Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager, M., N. Orr, R. B. Hayes, K. B. Jacobs, P. Kraft et al., 2007. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 39 645–649. [DOI] [PubMed] [Google Scholar]
- Zerba, K. E., R. E. Ferrell and C. F. Sing, 2000. Complex adaptive systems and human health: the influence of common genotypes of the apolipoprotein E (ApoE) gene polymorphism and age on the relational order within a field of lipid metabolism traits. Hum. Genet. 107 466–475. [DOI] [PubMed] [Google Scholar]
- Zhernakova, A., E. M. Festen, L. Franke, G. Trynka, C. C. van Diemen et al., 2008. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am. J. Hum. Genet. 82 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhernakova, A., C. C. Elbers, B. Ferwerda, J. Romanos, G. Trynka et al., 2010. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am. J. Hum. Genet. 86 970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]