Abstract
In many organisms, meiotic recombination occurs preferentially at a limited number of sites in the genome known as hotspots. In the fission yeast Schizosaccharomyces pombe, simple sequence motifs determine the location of at least some, and possibly most or all, hotspots. Recently, we showed that a large number of different sequences can create hotspots. Among those sequences we identified some recurring motifs that fell into at least five distinct families, including the well-characterized CRE family of hotspots. Here we report the essential sequence for activity of two of the novel hotspots, the oligo-C and CCAAT hotspots, and identify associated trans-acting factors required for hotspot activity. The oligo-C hotspot requires a unique 8-bp sequence, CCCCGCAC, though hotspot activity is also significantly affected by adjacent nucleotides. The CCAAT hotspot requires a more complex and degenerate sequence, including the originally identified seven nucleotide CCAATCA sequence at its core. We identified transcription factors, the CCAAT-binding factor (CBF) and Rst2, which are required specifically for activity of the CCAAT hotspots and oligo-C hotspots, respectively. Each of these factors binds to its respective motifs in vitro. However, unlike CRE, the sequence required for hotspot activity is larger than the sequence required for binding, suggesting the involvement of additional factors.
MEIOSIS is an essential process in the life cycle of sexually reproducing organisms. It differs from mitosis in that two successive cell divisions follow a single round of chromosome replication, resulting in the production of four haploid products from a single diploid precursor cell. The production of haploid cells occurs at the first meiotic division, during which homologous chromosomes from each parent pair and segregate to opposite poles. Prior to segregation, homologous chromosomes exchange genetic information through recombination. In both Saccharomyces cerevisiae and Schizosaccharomyces pombe, and probably other organisms, recombination is initiated by the formation of double strand DNA breaks (DSBs) (Gerton et al. 2000; Mahadevaiah et al. 2001; Cromie et al. 2007). In many organisms, including humans, mice, and both the budding and fission yeasts, these DSBs occur preferentially at so-called hotspots (Petes 2001; Kauppi et al. 2004; Nishant and Rao 2005). Although the distribution of DSBs can vary widely across the genome, the distribution of interhomolog crossovers, at least in S. pombe, is considerably more uniform due to the preferential selection of the sister chromatid for DSB repair at strong DSB hotspots (Hyppa and Smith 2010).
The factors determining the position of DSB hotspots have been the subject of research for many years, during which some common themes have emerged. Transcription factor binding, which may serve as a mechanism to open chromatin structure prior to initiation of DSBs, has been implicated in the activity of several hotspots in both the fission and budding yeasts. In S. cerevisiae, the HIS4 gene contains a hotspot whose activity requires binding of the Bas1, Bas2, and Rap1 transcription factors (White et al. 1993). Bas1 is also associated with elevated levels of DSBs at four other sites in the S. cerevisiae genome. However, this is not a simple relationship, as Bas1 binding also appears to represses DSB formation at nine other sites (Mieczkowski et al. 2006). Meiotic DSB hotspots occur primarily in promoter-containing intergenic regions (IGRs) (Baudat and Nicolas 1997; Gerton et al. 2000). This observation suggests that, similar to HIS4, other hotspots require the binding of transcription factors, since transcription factors are expected to bind primarily in gene promoters, although their binding outside these regions has not been thoroughly investigated. However, other than the Bas1 target sequence, no other simple sequence motifs producing recombination hotspots have been identified in S. cerevisiae.
DSBs in the fission yeast S. pombe also occur primarily in IGRs, particularly in large IGRs (Cromie et al. 2007). Many of these hotspots are sites of production of noncoding RNAs (Wahls et al. 2008), which is again consistent with the binding of transcription factors at those sites, though a causative relationship has not been established. One well-characterized hotspot in S. pombe, M26 (also referred to as CRE), requires a specific sequence, ATGACGT, and heterodimeric transcription factor, Atf1-Pcr1, for hotspot activity (Schuchert et al. 1991; Kon et al. 1997). The M26 motif generates a hotspot at many, but not all, sites in the S. pombe genome. This indicates that local DNA sequence alone is not sufficient for generating hotspots (Ponticelli and Smith 1992; Steiner and Smith 2005), which has also been demonstrated in S. cerevisiae; for example the insertion of hotspot DNA into different chromosomal contexts does not always create a new hotspot (Haring et al. 2003).
In mammals, transcription factor binding may also be a factor in regulating the locations of recombination hotspots (Zhang et al. 2004), but this has not been demonstrated, and the importance of factors other than local sequence motifs, which may include epigenetic modifications, trans-acting factors, and/or chromatin structure, is indicated by observations of sex-specific recombination rates (Kong et al. 2002) and changes in hotspot activity without changes to local sequence (Neuman and Jeffreys 2006). However, interest in the general importance of specific motifs has continued due to observations of local base substitutions associated with changes in hotspot activity (Jeffreys et al. 1998; Jeffreys and Neumann 2002, 2005) and the discovery of a specific 13-bp motif, CCNCCNTNNCCNC, which may be necessary for up to 40% of human recombination hotspots (Myers et al. 2008). More recently, it was shown that this motif is recognized by the PRDM9 methylase (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010), which trimethylates lysine 4 of histone H3 (H3K4me3), providing an interesting link with recombination in yeast, since H3K4 trimethylation was also shown to mark the location of some hotspots in S. cerevisiae (Borde et al. 2009).
In S. pombe, we identified a large number of unique sequences that create meiotic recombination hotspots (Steiner et al. 2009). Many of these sequences contained common motifs that could be grouped into five distinct families, including the previously identified CRE family of hotspots (Fox et al. 2000). For two of the novel hotspots, the oligo-C and CCAAT motifs, we were able to identify potential transcription factors that may be required for their activity. In this report, we characterized the essential sequence of those hotspots and identify transcription factors required for their hotspot activity.
MATERIALS AND METHODS
Strains and genetic procedures:
All strains used in this study are listed in Table 1. Strains were constructed by linear transformation of WS129, which contains a ura4+-kanMX6 construct inserted in the ade6 gene (Steiner et al. 2009). Transforming DNA was generated by overlap-extension PCR (Vallejo et al. 1995) using wild-type (ade6+) genomic DNA as template and outside primers oWS202: 5′-ACGAACATCATTAAGCGCGAAGCG-3′ and oWS203: 5′-ACGCATGAGTTGTGGAAGTCGAGA-3′ and inside primers containing the desired mutations. Transformation was performed using lithium acetate as previously described (Steiner et al. 2009). All transformants were verified by sequencing and Southern blot hybridization.
TABLE 1.
Strains
| Strain | Genotype | ade6 mutationa | Positionb |
|---|---|---|---|
| WS4 | h− lys4-95 | ||
| WS105 | h+ ade6-469 ura4-D18 his7-366 | AGATGCCTTGAGGTGT | 1460–1475 |
| WS129 | h90 ade6-4001c ura4-D18 leu1-32 (pWS35) | ||
| WS224 | h90 ade6-4002c ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATTT | 125–140 |
| WS242 | h90 ade6-4032 ura4-D18 leu1-32 pWS35 | CGTACTCCAATCAATTT | 125–140 |
| WS243 | h90 ade6-4033 ura4-D18 leu1-32 pWS35 | CGTGCTCCAATCAATTT | 125–140 |
| WS244 | h90 ade6-4034 ura4-D18 leu1-32 pWS35 | CGTTCTCCAATCAATTT | 125–140 |
| WS246 | h90 ade6-4036 ura4-D18 leu1-32 pWS35 | CGTCGTCCAATCAATTT | 125–140 |
| WS247 | h90 ade6-4037 ura4-D18 leu1-32 pWS35 | CGTCTTCCAATCAATTT | 125–140 |
| WS248 | h90 ade6-4038 ura4-D18 leu1-32 pWS35 | CGTCCACCAATCAATTT | 125–140 |
| WS249 | h90 ade6-4039 ura4-D18 leu1-32 pWS35 | CGTCCCCCAATCAATTT | 125–140 |
| WS250 | h90 ade6-4040 ura4-D18 leu1-32 pWS35 | CGTCCGCCAATCAATTT | 125–140 |
| WS251 | h90 ade6-4041 ura4-D18 leu1-32 pWS35 | CGTCCTACAATCAATTT | 125–140 |
| WS252 | h90 ade6-4042 ura4-D18 leu1-32 pWS35 | CGTCCTGCAATCAATTT | 125–140 |
| WS253 | h90 ade6-4043 ura4-D18 leu1-32 pWS35 | CGTCCTTCAATCAATTT | 125–140 |
| WS254 | h90 ade6-4044 ura4-D18 leu1-32 pWS35 | CGTCCTCAAATCAATTT | 125–140 |
| WS255 | h90 ade6-4045 ura4-D18 leu1-32 pWS35 | CGTCCTCGAATCAATTT | 125–140 |
| WS256 | h90 ade6-4046 ura4-D18 leu1-32 pWS35 | CGTCCTCTAATCAATTT | 125–140 |
| WS257 | h90 ade6-4047 ura4-D18 leu1-32 pWS35 | CGTCCTCCCATCAATTT | 125–140 |
| WS258 | h90 ade6-4048 ura4-D18 leu1-32 pWS35 | CGTCCTCCGATCAATTT | 125–140 |
| WS259 | h90 ade6-4049 ura4-D18 leu1-32 pWS35 | CGTCCTCCTATCAATTT | 125–140 |
| WS261 | h90 ade6-4051 ura4-D18 leu1-32 pWS35 | CGTCCTCCAGTCAATTT | 125–140 |
| WS262 | h90 ade6-4052 ura4-D18 leu1-32 pWS35 | CGTCCTCCATTCAATTT | 125–140 |
| WS263 | h90 ade6-4053 ura4-D18 leu1-32 pWS35 | CGTCCTCCAAACAATTT | 125–140 |
| WS264 | h90 ade6-4054 ura4-D18 leu1-32 pWS35 | CGTCCTCCAACCAATTT | 125–140 |
| WS265 | h90 ade6-4055 ura4-D18 leu1-32 pWS35 | CGTCCTCCAAGCAATTT | 125–140 |
| WS266 | h90 ade6-4056 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATAAATTT | 125–140 |
| WS267 | h90 ade6-4057 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATGAATTT | 125–140 |
| WS268 | h90 ade6-4058 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATTAATTT | 125–140 |
| WS269 | h90 ade6-4059 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCCATTT | 125–140 |
| WS270 | h90 ade6-4060 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCGATTT | 125–140 |
| WS271 | h90 ade6-4061 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCTATTT | 125–140 |
| WS272 | h90 ade6-4062 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCACTTT | 125–140 |
| WS273 | h90 ade6-4063 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAGTTT | 125–140 |
| WS274 | h90 ade6-4064 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCATTTT | 125–140 |
| WS275 | h90 ade6-4065 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAAATT | 125–140 |
| WS276 | h90 ade6-4066 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAACTT | 125–140 |
| WS277 | h90 ade6-4067 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAAGTT | 125–140 |
| WS278 | h90 ade6-4068 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATAT | 125–140 |
| WS279 | h90 ade6-4069 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATCT | 125–140 |
| WS280 | h90 ade6-4070 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATGT | 125–140 |
| WS284 | h− ura4-D18 leu1-32 his7-366 php2Δ∷kanc | 125–140 | |
| WS344 | h90 ade6-4050 ura4-D18 leu1-32 pWS35 | CGTCCTCCACTCAATTT | 125–140 |
| WS345 | h90 ade6-4035 ura4-D18 leu1-32 pWS35 | CGTCATCCAATCAATTT | 125–140 |
| WS346 | h90 ade6-4080 ura4-D18 leu1-32 pWS35 | AGTCCTCCAATCAATTT | 125–140 |
| WS348 | h90 ade6-4081 ura4-D18 leu1-32 pWS35 | GGTCCTCCAATCAATTT | 125–140 |
| WS350 | h90 ade6-4082 ura4-D18 leu1-32 pWS35 | TGTCCTCCAATCAATTT | 125–140 |
| WS352 | h90 ade6-4083 ura4-D18 leu1-32 pWS35 | CATCCTCCAATCAATTT | 125–140 |
| WS354 | h90 ade6-4084 ura4-D18 leu1-32 pWS35 | CCTCCTCCAATCAATTT | 125–140 |
| WS356 | h90 ade6-4085 ura4-D18 leu1-32 pWS35 | CTTCCTCCAATCAATTT | 125–140 |
| WS358 | h90 ade6-4086 ura4-D18 leu1-32 pWS35 | CGACCTCCAATCAATTT | 125–140 |
| WS360 | h90 ade6-4087 ura4-D18 leu1-32 pWS35 | CGCCCTCCAATCAATTT | 125–140 |
| WS362 | h90 ade6-4088 ura4-D18 leu1-32 pWS35 | CGGCCTCCAATCAATTT | 125–140 |
| WS382 | h90 ade6-4099c ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTGAd | 128–143 |
| WS424 | h− ade6-4099 ura4-D18 leu1-32 hsr1Δ∷hygBe | TTGAACCCCGCACTGA | 128–143 |
| WS427 | h+ ade6-469 ura4-D18 his7-366 rst2Δ∷ura4+f | AGATGCCTTGAGGTGT | 1460–1475 |
| WS429 | h− ade6-4099 rsv1Δ∷kang | TTGAACCCCGCACTGA | 128–143 |
| WS430 | h− ade6-4099 ura4-D18 leu1-32 | TTGAACCCCGCACTGA | 128–143 |
| WS431 | h− ade6-4099 ura4-D18 leu1-32 scr1∷ura4+h | TTGAACCCCGCACTGA | 128–143 |
| WS433 | h+ ade6-469 ura4-D18 his7-366 rsv1Δ∷kan | AGATGCCTTGAGGTGT | 1460–1475 |
| WS435 | h+ ade6-469 ura4-D18 his7-366 hsr1Δ∷hygB | AGATGCCTTGAGGTGT | 1460–1475 |
| WS438 | h− ade6-4099 ura4-D18 leu1-32 rst2Δ∷ura4+ | TTGAACCCCGCACTGA | 128–143 |
| WS448 | h+ ade6-469 ura4-D18 his7-366 scr1∷ura4+ | AGATGCCTTGAGGTGT | 1460–1475 |
| WS452 | h− ade6-M26 ura4-D18 leu1-32 rst2Δ∷ura4+ | TTGATGGATGACGTGA | 128–143 |
| WS454 | h− ura4-D18 rst2Δ∷ura4+ | ||
| WS460 | h90 ade6-4109 ura4-D18 leu1-32 pWS35 | TTAAACCCCGCACTGA | 128–143 |
| WS461 | h90 ade6-4110 ura4-D18 leu1-32 pWS35 | TTCAACCCCGCACTGA | 128–143 |
| WS462 | h90 ade6-4111 ura4-D18 leu1-32 pWS35 | TTTAACCCCGCACTGA | 128–143 |
| WS463 | h90 ade6-4112 ura4-D18 leu1-32 pWS35 | TTGCACCCCGCACTGA | 128–143 |
| WS464 | h90 ade6-4113 ura4-D18 leu1-32 pWS35 | TTGGACCCCGCACTGA | 128–143 |
| WS465 | h90 ade6-4114 ura4-D18 leu1-32 pWS35 | TTGTACCCCGCACTGA | 128–143 |
| WS466 | h90 ade6-4115 ura4-D18 leu1-32 pWS35 | TTGACCCCCGCACTGA | 128–143 |
| WS467 | h90 ade6-4116 ura4-D18 leu1-32 pWS35 | TTGAGCCCCGCACTGA | 128–143 |
| WS468 | h90 ade6-4117 ura4-D18 leu1-32 pWS35 | TTGATCCCCGCACTGA | 128–143 |
| WS469 | h90 ade6-4118 ura4-D18 leu1-32 pWS35 | TTGAAACCCGCACTGA | 128–143 |
| WS470 | h90 ade6-4119 ura4-D18 leu1-32 pWS35 | TTGAAGCCCGCACTGA | 128–143 |
| WS471 | h90 ade6-4120 ura4-D18 leu1-32 pWS35 | TTGAATCCCGCACTGA | 128–143 |
| WS472 | h90 ade6-4121 ura4-D18 leu1-32 pWS35 | TTGAACACCGCACTGA | 128–143 |
| WS473 | h90 ade6-4122 ura4-D18 leu1-32 pWS35 | TTGAACGCCGCACTGA | 128–143 |
| WS474 | h90 ade6-4123 ura4-D18 leu1-32 pWS35 | TTGAACTCCGCACTGA | 128–143 |
| WS475 | h90 ade6-4124 ura4-D18 leu1-32 pWS35 | TTGAACCACGCACTGA | 128–143 |
| WS476 | h90 ade6-4125 ura4-D18 leu1-32 pWS35 | TTGAACCGCGCACTGA | 128–143 |
| WS477 | h90 ade6-4126 ura4-D18 leu1-32 pWS35 | TTGAACCTCGCACTGA | 128–143 |
| WS478 | h90 ade6-4127 ura4-D18 leu1-32 pWS35 | TTGAACCCAGCACTGA | 128–143 |
| WS479 | h90 ade6-4128 ura4-D18 leu1-32 pWS35 | TTGAACCCGGCACTGA | 128–143 |
| WS480 | h90 ade6-4129 ura4-D18 leu1-32 pWS35 | TTGAACCCTGCACTGA | 128–143 |
| WS481 | h90 ade6-4130 ura4-D18 leu1-32 pWS35 | TTGAACCCCACACTGA | 128–143 |
| WS482 | h90 ade6-4131 ura4-D18 leu1-32 pWS35 | TTGAACCCCCCACTGA | 128–143 |
| WS483 | h90 ade6-4132 ura4-D18 leu1-32 pWS35 | TTGAACCCCTCACTGA | 128–143 |
| WS484 | h90 ade6-4133 ura4-D18 leu1-32 pWS35 | TTGAACCCCGAACTGA | 128–143 |
| WS485 | h90 ade6-4134 ura4-D18 leu1-32 pWS35 | TTGAACCCCGGACTGA | 128–143 |
| WS486 | h90 ade6-4135 ura4-D18 leu1-32 pWS35 | TTGAACCCCGTACTGA | 128–143 |
| WS487 | h90 ade6-4136 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCCCTGA | 128–143 |
| WS488 | h90 ade6-4137 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCGCTGA | 128–143 |
| WS489 | h90 ade6-4138 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCTCTGA | 128–143 |
| WS490 | h90 ade6-4139 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCAATGA | 128–143 |
| WS491 | h90 ade6-4140 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCAGTGA | 128–143 |
| WS492 | h90 ade6-4141 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCATTGA | 128–143 |
| WS493 | h90 ade6-4142 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACAGA | 128–143 |
| WS494 | h90 ade6-4143 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACCGA | 128–143 |
| WS495 | h90 ade6-4144 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACGGA | 128–143 |
| WS496 | h90 ade6-4145 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTAA | 128–143 |
| WS497 | h90 ade6-4146 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTCA | 128–143 |
| WS498 | h90 ade6-4147 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTTA | 128–143 |
| WS499 | h90 ade6-4148 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTGC | 128–143 |
| WS500 | h90 ade6-4149 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTGG | 128–143 |
| WS501 | h90 ade6-4150 ura4-D18 leu1-32 pWS35 | TTGAACCCCGCACTGT | 128–143 |
| WS594 | h90 ade6-4194 ura4-D18 leu1-32 pWS35 | TAGAACCCCGCACTGA | 128–143 |
| WS595 | h90 ade6-4195 ura4-D18 leu1-32 pWS35 | TCGAACCCCGCACTGA | 128–143 |
| WS596 | h90 ade6-4196 ura4-D18 leu1-32 pWS35 | TGGAACCCCGCACTGA | 128–143 |
| WS597 | h90 ade6-4197 ura4-D18 leu1-32 pWS35 | ATGAACCCCGCACTGA | 128–143 |
| WS598 | h90 ade6-4198 ura4-D18 leu1-32 pWS35 | CTGAACCCCGCACTGA | 128–143 |
| WS599 | h90 ade6-4199 ura4-D18 leu1-32 pWS35 | GTGAACCCCGCACTGA | 128–143 |
| WS602 | h90 ade6-4200 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATTA | 125–140 |
| WS603 | h90 ade6-4201 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATTC | 125–140 |
| WS604 | h90 ade6-4202 ura4-D18 leu1-32 pWS35 | CGTCCTCCAATCAATTG | 125–140 |
The nucleotide sequence within ade6 for the indicated alleles. Insertions are underlined. Nucleotide substitutions are shown in boldface type. Red font indicates a single nucleotide substitution relative to either the ade6-4002 or ade6-4099 alleles.
The nucleotide positions of the ade6 sequence shown in the third column. Nucleotide positions are numbered from the beginning of the open reading frame.
The ade6-4099 allele, as well as all the other alleles based on ade6-4099, also contain the closely linked stop mutation, A121T, not shown.
Homothallic crosses (Figure 1) were performed by growing cells in NBL [0.67% yeast nitrogen base without amino acids (Difco)] supplemented with 100 μg/ml adenine and 50 μg/ml uracil at 32° to saturation. One milliliter of each culture was washed once in 1 ml 0.85% NaCl, resuspended in 50 μl of 0.85% NaCl, and plated onto sporulation agar (Gutz et al. 1974) supplemented with 100 μg/ml adenine and 50 μg/ml uracil (SPA +AU). These plates were incubated for 2 days at 25° to allow for mating and sporulation. For heterothallic crosses (Figure 2) strains were grown in YEL (Gutz et al. 1974) supplemented with adenine, uracil, leucine, lysine, and histidine (5S). rst2 mutants were grown in liquid EMM2 medium (Nurse 1975) with the same supplements, which produced a greater yield of viable spores compared to YEL. A total of 0.5 ml of each strain in the cross was mixed together, washed with 0.85% NaCl, and sporulated on SPA +5S. Asci were digested overnight in 1 ml of water containing 5 μl glusulase (Perkin-Elmer) followed by the addition of 450 μl of 95% ethanol for 15 min to kill remaining vegetative cells. The free spores were washed once in 1 ml of water and resuspended in 1 ml of water.
Figure 1.—
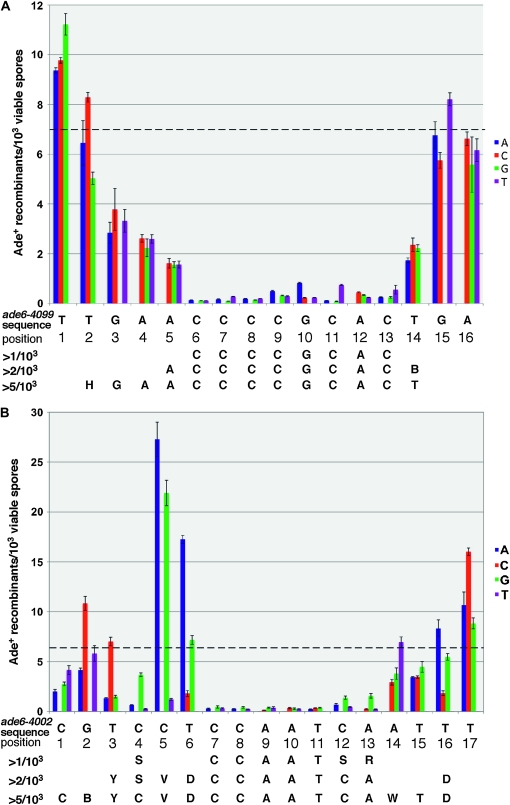
Essential sequence for the oligo-C (ade6-4099) and CCAAT (ade6-4002) hotspots. The frequency of ade6+ recombinants is shown for each nucleotide substitution at the indicated position. The relevant sequence of the ade6-4099 (A) and ade6-4002 (B) alleles is shown immediately beneath each graph. Each bar represents the frequency of recombination (±1 SEM) observed in strains containing single nucleotide substitutions relative to the sequences shown. The sequence necessary to achieve recombination levels >1, 2, or 5 Ade+/103 spores is indicated for each of the two hotspots. The average recombination observed for the unsubstituted hotspots is indicated by the dashed lines. A minimum of three crosses was performed for each value shown.
Figure 2.—
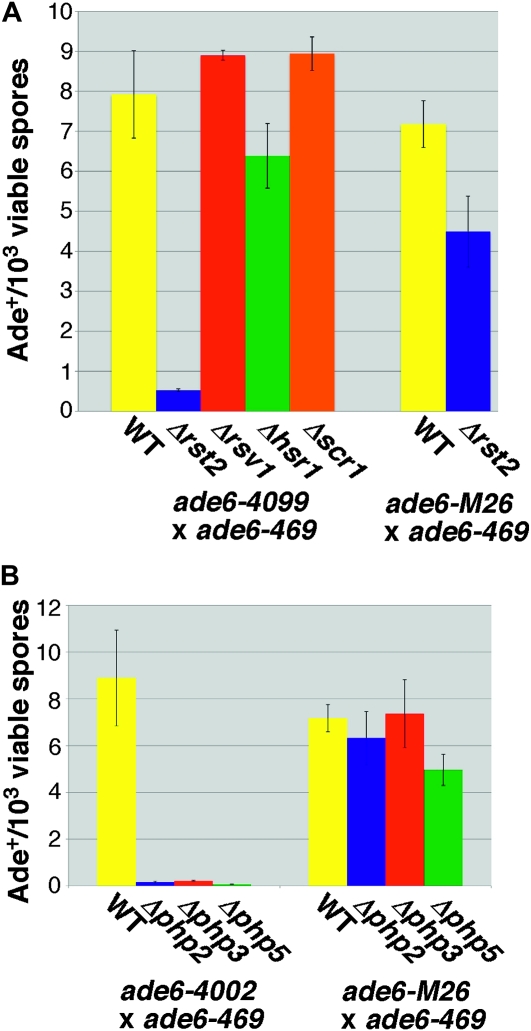
Specific transcription factors are required for the oligo-C (A) and CCAAT (B) hotspots. Bars represent the frequency of recombination to ade6+ ±1 SEM for strains of the indicated genotype. A minimum of three crosses was performed to obtain each value.
Each of the homothallic strains shown in Table 1 contains the plasmid pWS35, which carries a 613-bp fragment of the ade6 gene (Steiner et al. 2009). Thus, the chromosomal copy of ade6 can recombine with the plasmid to become ade6+ during meiosis. This chromosome X plasmid recombination has previously been shown to be an accurate measure of hotspot activity in comparison to chromosome X chromosome recombination (Ponticelli and Smith 1989; Steiner et al. 2009). The frequency of ade6+ recombinants for both homothallic and heterothallic crosses was determined by plating appropriate dilutions of spores onto YEA +5S (total spores) and YEA +4SG (Ade+ spores) as previously described (Steiner and Smith 2005). YEA +4SG is the same as YEA +5S except it contains 100 μg/ml guanine in place of adenine, which inhibits growth of adenine auxotrophs.
Electrophoretic mobility shift assays:
For gel shift assays involving the CCAAT hotspot and related sequences (Figure 3), probes were generated by PCR using primers oWS302: 5′-TTGGGCCGAATGATGGTAGAG-3′ and oWS303: 5′-GCCAAGGCATCAGTGTTAATATG-3′, Taq polymerase (New England Biolabs), and the appropriate genomic DNA template (Table 1). The resulting 206-bp products were purified and end labeled using T4-polynucleotide kinase and γ-32P-ATP(Sambrook and Russell 2001). Whole cell protein extracts were prepared from strains WS4 (wild type) and WS284 (php2Δ∷kan) as previously described (Steiner and Smith 2005). Binding reactions were performed in a final volume of 10 μl at room temperature for 20 min. Each reaction contained 0.2 ng of probe, 10 μg protein extract, 0.2 μg poly dI-dC (Sigma), 12% glycerol, 12 mm HEPES (pH 7.9), 0.6 mm EDTA, and 0.6 mm DTT. All ingredients except the probe were mixed and allowed to preincubate for 5 min prior to addition of the probe. Binding reactions were electrophoresed in a 0.5× TBE (Sambrook and Russell 2001) 5% polyacrylamide gel at 50 volts for 2 hr. Following electrophoresis, gels were transferred to Whatman 3MM paper, vacuum dried at 80° for 45 min, and analyzed using a Personal Molecular Imager (BioRad).
Figure 3.—
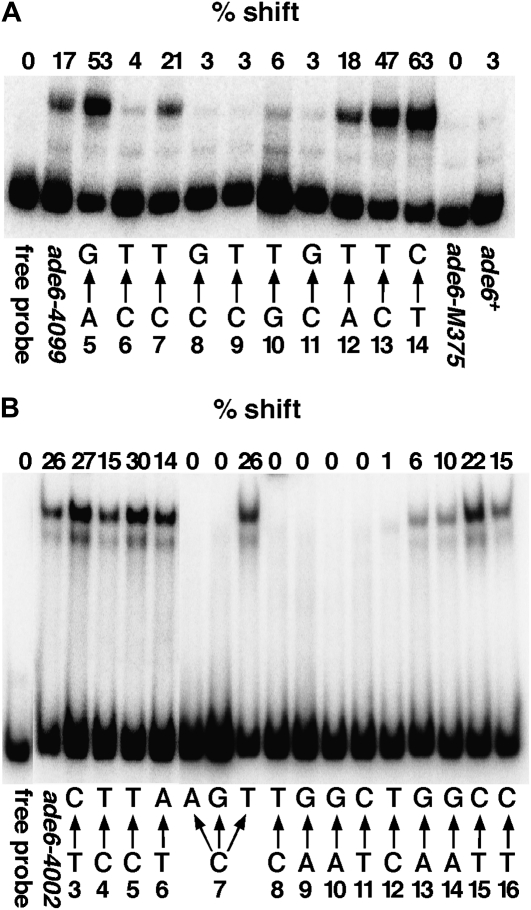
Sequences required for transcription factor binding are less stringent than sequences required for hotspot activity. EMSA assays were performed as described in materials and methods. The relevant portions of the ade6-4099 (A) and ade6-4002 (B) alleles are shown beneath each figure. Numbers correspond to nucleotide positions shown in Figure 1. The percentage of probe experiencing a mobility shift is indicated above each lane. The specific nucleotide substitution tested is indicated beneath each lane by arrows, for example an A→G substitution at position 5. Each probe was tested two or three times with similar results.
Gel shift assays involving the oligo-C hotspot and related sequences were performed as described above with the following differences. Probes were labeled with T4 DNA polymerase and α-32P-dCTP (Sambrook and Russell 2001). The purified DNA binding domain of the Rst2 protein (Rst2ZF) was utilized in place of whole cell protein extracts. This protein was overexpressed in Escherichia coli and purified as described (Kunitomo et al. 2000). Briefly, protein expression was induced in strain Rosetta 2(DE3)pLacI (Novagen) carrying the plasmid pET-Rst2ZF, which encodes the two zinc finger motifs of Rst2 along with a histidine tag. One hundred milliliters of these cells was centrifuged and resuspended in 6 ml of His-bind buffer (0.5 m NaCl, 20 mm Tris-HCl, 5 mm imidazole, pH 7.9). Cells were snap frozen and thawed in an ice water bath twice, followed by intermittent sonication on ice to disrupt the cells. The His-tagged Rst2ZF protein was purified using His-bind resin as described by the manufacturer (Novagen). The purified protein was concentrated by ultrafiltration and resuspended in 50 mm KCl at a concentration of 1 mg/ml. Ten-microliter binding reactions were performed in 50 mm KCl containing 2 μg Rst2ZF, 1 μg poly dI-dC, and 1 ng probe for 20 min at room temperature. These reactions were electrophoresed at room temperature in 5% polyacrylamide gels containing 4 mm Tris, 190 mm glycine, pH 8.0 for 90 min at 4 mA constant current.
Bioinformatics:
We used the hotspot map reported by Hyppa et al. (2008), interpreting their data as indicating a hotspot where the hybridization ratio was elevated above background, on the basis of the statistical method used by the authors, in either of their replicate experiments with rad50S mutant strains. By these criteria there were 440 hotspots. These hotspots included 4781 probes covering 1.31 Mb, or ∼10.4% of the 12.57 Mb of sequenced genome. This definition probably included some weaker hotspots not reported by Cromie et al. (2007), since the DSB map in that report showed only 700 kb of hotspot DNA. However, false positive results are unlikely to explain the discrepancy, because we defined DSB hotspots using the program ChIPOT1e (v1.0; with a P-value cutoff of 0.001), which is designed to reduce noise by using sliding windows wide enough to cover several probes (Buck et al. 2005).
We used an online version of MEME Bailey and Elkan (1994) for motif detection; URL: http://meme.nbcr.net/meme/cgi-bin/meme.cgi.
RESULTS
Note: the following standard degenerate nucleotide letter designations are used: R = A or G; Y = C or T; S = C or G; W = A or T; B = C, G, or T; D = A, G, or T; H = A, C, or T; V = A, C, or G; N = any nucleotide.
The oligo-C hotspot:
The oligo-C hotspot (ade6-4099) was originally constructed on the basis of a partially degenerate 11-bp motif, DACCCCGCACD, identified in a screen for recombination hotspots (Steiner et al. 2009). To determine whether this entire sequence is required for hotspot activity, we systematically mutagenized each nucleotide in and around the motif. Figure 1A shows that mutation of any nucleotide in the motif reduces recombination; however the central eight nucleotides (positions 6–13) are most critical for activity of this hotspot. Given that the S. pombe genome is 36% GC (Egel 2004) and contains ∼12.6 Mb of sequenced DNA, one would predict ∼25 occurrences of this GC-rich 8-nucleotide sequence, on the basis of random nucleotide association. However, it is found only 10 times, making it modestly underrepresented in the genome. This underrepresentation is similar to the 7-bp M26 motif, which is also approximately threefold underrepresented (285 occurrences vs. 769 predicted). Three of the 8 nucleotide oligo-C motifs coincide with meiotic DSBs (Hyppa et al. 2008). Thus, the oligo-C hotspot may not contribute significantly to the overall frequency of meiotic recombination in S. pombe. However, these numbers may be an underestimate since variations of this motif, which may function by a similar mechanism, also create hotspots (Steiner et al. 2009).
A search of the TransFac database suggested that the oligo-C motif is a target for the MIG1 transcription factor of S. cerevisiae, a Cys2His2 zinc finger protein (Badis et al. 2008). The homologous proteins MIG1, MIG2, and MIG3 of S. cerevisiae are all reported to bind the same sequence, CCCCGCA (Badis et al. 2008), which matches perfectly the first seven nucleotides of the oligo-C hotspot. The four strongest orthologs of MIG1 in S. pombe are scr1, rsv1, hsr1, and rst2, so we tested deletions in each gene for their effect on the activity of the hotspot. Of the four, only deletion of rst2 had a significant effect, decreasing recombination ∼15-fold. By comparison, the M26 hotspot showed only a modest, 1.6-fold reduction, suggesting that the effect on recombination is specific to the oligo-C hotspot (Figure 2A).
The Rst2 protein is required for full expression of the ste11 and fbp1 genes involved in sexual development and gluconeogenesis, respectively (Kunitomo et al. 2000; Higuchi et al. 2002). As Ste11 is a crucial transcription factor responsible for regulating the expression of many genes involved in mating and meiosis, rst2 mutants show similar meiotic defects (Kunitomo et al. 2000). We were unable to detect any binding of the radiolabeled oligo-C probes by electrophoretic mobility shift assays (EMSAs) using whole cell extracts, so we used the purified N-terminal DNA binding domain of Rst2 described by Kunitomo et al. (2000). Using purified protein, we observed binding to the oligo-C motif (ade6-4099; Figure 3A).
Because the ade6-M26 hotspot of S. pombe shows a strict correlation between binding of its associated transcription factor, Atf1-Pcr1, and hotspot activity (Wahls and Smith 1994), we tested whether a similar correlation were also true of the oligo-C hotspot. Surprisingly, we observed only an imperfect correlation (Figures 1A and 3A). Though an eight-nucleotide sequence is required for hotspot activity (positions 6–13; Figure 1A), only five nucleotides (positions 6, 8, 9, 10, and 11) showed a significant decrease in binding relative to the unsubstituted ade6-4099 allele. In some cases, (positions 5, 13, and 14), nucleotide substitutions actually increased binding. Thus, we conclude that binding of Rst2, at least on naked DNA, is necessary but not sufficient for activity of the oligo-C hotspot.
Kunitomo et al. (2000) observed binding of Rst2 to a sequence similar to the ade6-4099 hotspot, CCCCTCAT, which differs at positions 10 and 13 (Figures 1A and 3A). We observed only weak binding with a G→T substitution at position 10. However, the C→T substitution at position 13, though inactive as a hotspot, bound strongly to Rst2ZF (Figure 3A), which could potentially offset the reduced binding observed with the single G→T substitution at position 10 and explain this apparent discrepancy.
The CCAAT hotspot:
The CCAAT hotspot was originally identified as a 7-bp motif, CCAATCA, which appeared multiple times among a pool of ∼400 larger sequences containing hotspots. A single base insertion was used to construct this motif in the ade6 gene, creating a hotspot of recombination (Steiner et al. 2009). Although this result was consistent with a requirement for only the 7-bp motif indicated, systematic mutagenesis revealed that a larger degenerate sequence is actually required, at least at the position in ade6 tested here. Figure 1B shows that the CCAAT hotspot is strongly dependent on the originally identified 7-bp motif (positions 7–13); however, neighboring nucleotides also significantly affect activity. Recombination frequencies >0.1% (approximately one-sixth the level of the unmutagenized hotspot) require a degenerate 10-bp sequence, SNNCCAATSR, but higher levels of recombination have increasingly stringent sequence requirements. For example, recombination >0.2%, requires at least a degenerate 14-bp sequence, YSVDCCAATCANND. However, since nucleotide substitutions even at position 1 (Figure 1B) are still significantly below the level of the unmutagenized hotspot, the influence of even more distant nucleotides cannot be excluded. A few nucleotide substitutions even resulted in significant increases in hotspot activity, for example at positions 2, 5, 6, and 17. At least in one case, this is probably due to the formation of another hotspot motif; for example, the C→A substitution at position 5 (Figure 1B) produces the complement of the M26 motif, ACGTCAT (the A to the left of position 1 is not shown). These results contrast with both the oligo-C and M26 hotspots, which show only modest effects of substitutions >3 bp from the central core sequence (Figure 1A and Schuchert et al. 1991).
Unlike the oligo-C motif, the CCAATCA motif is abundant in the S. pombe genome. Of the 1408 occurrences of the motif, 172 (12%) fall within hotspots (Table 2), which is significant (P < 0.01), on the basis of the binomial distribution with 10.4% of the genome occurring in hotspots. In terms of frequency per unit distance, this corresponds to an excess of 17% in hotspots compared to the genome as a whole. Given that the CCAATCA motif occurs very frequently in the genome, it is reasonable to expect that some, but not all, of its 172 occurrences within hotspots are coincidental. With 10.4% of the genome present in regions enriched for DSBs, one would predict ∼146 (1408 × 0.104) chance occurrences within hotspots, with a 95% confidence interval of 124–169 chance occurrences. Subtracting these values from 172, this suggests that the CCAATCA sequence is responsible for 3–48 of the 440 DSB hotspots in the genome.
TABLE 2.
CCAATCA and derivative motifs show significant association with DSB hotspots in the genome
| Motif | Total number in genome | Number within hotspots | Fraction within hotspots (see text) | Fractional enrichment within hotspotsa |
|---|---|---|---|---|
| CCAAT | 30,442 | 3086 | 0.101 | −0.03 |
| YCAATC | 14,147 | 1528** | 0.108 | 0.04 |
| CCAATC | 6413 | 679* | 0.106 | 0.02 |
| DCCAATC | 4237 | 469** | 0.111 | 0.06 |
| VDCCAATC | 2637 | 295* | 0.112 | 0.08 |
| DCCAATCA | 1114 | 134** | 0.120 | 0.16b |
| CCAATCA | 1408 | 172** | 0.122 | 0.17b |
| DCCAATCANND | 887 | 112** | 0.126 | 0.21b |
| SVDCCAATC | 938 | 121** | 0.129 | 0.24b |
| CCAATCANND | 1119 | 145*** | 0.130 | 0.25b |
| VDCCAATCA | 652 | 87** | 0.133 | 0.28b |
| YSVDCCAATC | 479 | 67** | 0.140 | 0.34b |
| VDCCAATCANND | 524 | 76*** | 0.145 | 0.39b |
| SVDCCAATCA | 218 | 33** | 0.151 | 0.46b |
| SVDCCAATCANND | 170 | 30** | 0.176 | 0.70bc |
| YSVDCCAATCA | 100 | 18** | 0.180 | 0.73b |
| YSVDCCAATCANND | 83 | 16** | 0.193 | 0.85bc |
| ATGACGTd | 285 | 63*** | 0.221 | 1.12 |
*P < 0.05; **P < 0.01; ***P < 0.001; probability that the number of motifs falling within hotspots is due to chance. Probabilities are based on the binomial distribution formula with 10.4% of the genome found within DSB hotspots (see text).
(Observed fraction –0.104)/0.104.
P < 0.05; χ2 probability that the increased fraction of the indicated motif found within hotspots is not different from the five-base CCAAT motif.
P < 0.05; χ2 probability that the increased fraction of the indicated motif found within hotspots is not different from the seven-base CCAATCA motif.
The M26 motif is provided for comparison.
For comparison, 63 of the 285 appearances of the M26 heptamer (22%) lie within hotspots. M26 also shows considerably greater enrichment in hotspots compared with the genome as a whole (112%), suggesting that a higher proportion of M26 motifs, compared to CCAATCA motifs, create hotspots. By the same reasoning as above, it can be estimated that M26 is responsible for 23–43 hotspots in the genome. Thus, in spite of its considerably weaker association with hotspots, the CCAATCA motif, due to its abundance in the genome, may, nevertheless, be responsible for a number of DSB hotspots comparable to that of M26. It should be noted that our estimate for the number of M26-responsible hotspots in S. pombe is lower than another recent estimate (Wahls and Davidson 2010), given our assumption that some M26–DSB associations are coincidental.
Of course, our analysis of the CCAAT hotspot within ade6 indicated that nucleotides flanking the CCAATCA heptamer are also critical for activity of that hotspot (Figure 1B). Inclusion of one or more of these flanking nucleotides significantly increases the association with genome-wide hotspots (Table 2). For example, the sequence YSVDCCAATCANND is found 83 times in the genome, 16 of which are associated with breaks. This corresponds to a frequency excess of 85% in hotspots compared to the genome as a whole, which is significantly greater than the 7-base CCAATCA sequence alone, and closer to the enrichment observed for M26. Surprisingly, the 5-base CCAAT sequence alone, which is essential in most organisms for binding to the CCAAT-binding factor CBF (see below) is modestly underrepresented in hotspots (Table 2).
Binding of the CCAAT-binding factor to the CCAAT hotspot:
Given the sequence of the CCAAT hotspot, we tested whether the well-characterized CBF was required for activity of this hotspot. The CBF in S. pombe is a heterotrimer composed of the Php2, Php3, and Php5 subunits (McNabb et al. 1997). As in S. cerevisiae, the CBF in S. pombe is required mainly for the synthesis of mitochondrial proteins. Thus, mutations in any of the genes encoding subunits of the CBF result in disruption of the CBF complex and the inability to grow on nonfermentable carbon sources (Olesen et al. 1991; Mercier et al. 2006). Deletion of any of the three subunits of the CBF resulted in a >40-fold decrease in recombination of the CCAAT hotspot (Figure 2B and Steiner et al. 2009). This same decrease in recombination was not observed for ade6-M26, suggesting the CBF is required specifically for the CCAAT hotspot.
Previous studies of CCAAT-binding factors from other organisms have shown that binding has an almost absolute requirement for the 5-nucleotide CCAAT sequence (Mantovani 1998, 1999), though binding was also significantly influenced by nucleotides adjacent to that motif (Dorn et al. 1987; Hatamochi et al. 1988; Kim and Sheffery 1990). Given the results of our recombination analysis, we tested whether the sequence required for high levels of recombination is also required for binding of the CBF. Figure 3 shows that the sequence required for binding in vitro is considerably smaller than the region required for hotspot activity. Surprisingly, we found that either a C or a T nucleotide at position 7 (Figure 3B) sufficed for binding of the CBF in vitro. (The presence of the T substitution at this position was confirmed and retested with the same result; data not shown). Furthermore, little or no binding of the CBF was observed for the C→T mutation at position 12. Thus, the minimal sequence for binding of the S. pombe CBF in vitro appears to be YCAATC. However, given that we did not test every nucleotide substitution within this sequence, additional degeneracies are possible. As expected, no binding of the CCAAT hotspot sequence was observed using protein extracts obtained from a php2 deletion strain (data not shown), indicating that the CBF is responsible for the observed mobility shifts.
Other genomic features associated with hot vs. nonhot CCAATCA motifs:
We used the software MEME to investigate whether hotspot-associated occurrences of either the CCAATCA or M26 motifs shared common flanking sequences, but found no obvious patterns, suggesting that any influence of particular flanking nucleotides is context dependent. This has previously been shown for M26 (Steiner and Smith 2005), and it is likely also true for CCAATCA, since the most active form of that motif found in ade6, CBYCVDCCAATCAWTD (Figure 1B), is not even found in the S. pombe genome.
We also looked for additional features that might correlate with active vs. inactive occurrences of the hotspot-associated motifs CCAATCA, YSVDCCAATCA, YSVDCCAATCANND, and M26 (ATGACGT). All of these motifs are much more likely to be associated with hotspots in noncoding regions (NCRs) than in coding sequence (CDS) DNA. Although there is a significant bias for hotspots to occur in NCRs (66% of total hotspots), this bias is even greater for hotspot-associated motifs and does not reflect the general distribution of those motifs in the genome (Table 3). We found that NCRs containing a hotspot and at least one motif are on average about threefold larger than NCRs with hotspots and no motif. However, this trend toward motifs being present in larger NCRs was even greater when the analysis was applied to nonhot regions only, so it is not indicative of motif activity. Given that hotspot activity is often concentrated in promoter regions, we also asked whether motifs within hotspots were closer to transcriptional start sites (TSS) than nonhot motifs, but we found the distance between motif and TSS to be larger on average in hot than in nonhot regions for all motifs except YSVDCCAATCA, and the difference in this case was marginal (<3%).
TABLE 3.
Comparison of hotspot association in coding vs. noncoding regions
| Total motifs |
Hotspot-associated motifs |
|||
|---|---|---|---|---|
| Motif | CDS | NCR | CDS | NCR |
| CCAATCA | 65% (915) | 35% (493) | 38% (65) | 62% (107) |
| YSVDCCAATCA | 50% (50) | 50% (50) | 17% (3) | 83% (15) |
| YSVDCCAATCANND | 51% (42) | 49% (41) | 19% (3) | 81% (13) |
| M26 (ATGACGT) | 45% (128) | 55% (157) | 22% (14) | 78% (49) |
CDS, coding sequence; NCR, noncoding region. The actual numbers used for determination of percentages are shown in parentheses. A total of 66% of the nucleotides covered by all 440 hotspots are found in noncoding sequence.
Another feature previously found to correlate with recombination hotspots in both S. cerevisiae and humans is polypurine tract (PPT) density (Kong et al. 2002; Bagshaw et al. 2006), and we found this also to be true in S. pombe. The density of PPTs of at least 15 bp is 2.35-fold greater in hot than nonhot NCRs (P < 10−51 by Mann–Whitney test), and this enrichment is even greater within 1 kb of each of the four hotspot-associated motifs considered (P < 0.014 by Mann–Whitney test; Table 4). Since PPTs can form non-B-DNA structures under physiological conditions (Ohno et al. 2002), we speculate that this correlation could reflect an activity of PPTs to render chromosome structure permissive to the binding of hotspot-activating proteins. However, any effect they may have must operate over some distance, since there is no PPT within 100 bp of a hotspot-associated YSVDCCAATCA motif.
TABLE 4.
Polypurine tracts are significantly enriched near hot noncoding regions and hotspot-associated motifs
| Mean % of nucleotides within 1 kb of hotspot-associated motifs composed of PPTs ≥15 bpa |
|||
|---|---|---|---|
| Motif | Hot NCRs | Nonhot NCRs | Pc |
| Any motif or noneb | 1.408 | 0.598 | <6.76 × 10−52 |
| CCAATCA | 2.289 | 0.718 | <3.21 × 10−8 |
| M26 | 2.232 | 0.710 | <1.75 × 10−5 |
| YSVDCCAATCA | 3.550 | 1.259 | <0.00218 |
| YSVDCCAATCANND | 4.402 | 1.210 | <0.0134 |
PPT, polypurine tract; NCR, noncoding region.
For example, if 3% of nucleotides in a 2-kb region are composed of PPTs ≥15 bp, they could be found in a single 60-bp PPT, three 20-bp PPTs, or other combinations, provided the minimum PPT length is 15 bp.
Mean % of nucleotides within hotspots composed of PPTs ≥15 bp regardless of motif presence or absence.
For each hotspot-associated motif, the P-values are for the comparison between PPT densities in hot NCRs regardless of motif presence and PPT densities within 1 kb of motifs in hotspots (Mann–Whitney test).
DISCUSSION
Here we have characterized two new sequence motifs that act as meiotic recombination hotspots. Like the previously characterized M26 (CRE) hotspot, both the oligo-C and CCAAT hotspots show an obvious nucleotide sequence requirement. The oligo-C hotspot requires an eight-nucleotide sequence with relatively well-defined borders, though immediately adjacent nucleotides also affect hotspot activity (Figure 1A). In contrast, the CCAAT hotspot has considerably less well-defined borders. Though there is an obvious requirement for a continuous and specific seven-nucleotide sequence, CCAATCA, substitutions at least six nucleotides distant from this motif show significant effects on recombination levels. These results indicate a large and degenerate sequence is required for hotspot activity, at least at this position within ade6.
We identified the transcription factors Rst2 and the CBF as essential trans-acting factors required for activity of the oligo-C and CCAAT hotspots, respectively. In this respect both of these hotspots conform to the paradigm established by the M26 (CRE) hotspot, which also requires a transcription factor, Atf1-Pcr1, for activity. However the M26 hotspot shows a much stricter correlation between hotspot activity and the ability to bind Atf1-Pcr1 in vitro (Wahls and Smith 1994; Steiner and Smith 2005). In contrast, the sequence required for activity of both the oligo-C and CCAAT hotspots is more extensive than the sequence required for binding of their respective factors. Though hotspot activity is not observed in the absence of binding, binding (sometimes strong binding) is observed in the absence of hotspot activity. This result implies that each of these hotspots requires one or more additional factors for activity, at least at the locations tested here. These could be cofactors that interact directly with Rst2 or the CBF or bind independently in adjacent intervals.
The oligo-C hotspot sequence identified here is found only 10 times in the sequenced portion of the S. pombe genome, only 3 of which are associated with DSB hotspots. However, the seven-base CCAATCA motif is abundant in the genome and is significantly enriched within meiotic DSBs. Notably, this enrichment increases when the flanking nucleotides required for hotspot activity within ade6 are included (Table 2). This implies that those flanking nucleotides also influence hotspot activity in other regions, though it is not clear whether they are required in all contexts. Compared to M26, the CCAATCA motif is less strongly associated with DSB hotspots; however, because it is much more abundant than M26, both motifs may be responsible for comparable numbers of DSBs in the genome. These two motifs alone are likely to be responsible for 26–91 (6–21%) of the 440 DSB hotspots observed in the genome.
Acknowledgments
We thank Jürg Baehler, Kunihiro Ohta, Masayuki Yamamoto, and the Yeast Genetic Resource Center of Japan for supplying strains and reagents used in this study, and we thank Marlo Brown for assistance with statistical analyses. This work was supported by a National Institutes of Health grant, R15GM078065 (to W.W.S.).
References
- Badis, G., E. T. Chan, H. v. Bakel, L. Pena-Castillo, D. Tillo et al., 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw, A. T. M., J. P. W. Pitt and N. J. Gemmell, 2006. Association of poly-purine/poly-pyrimidine sequences with meiotic recombination hotspots. BMC Genomics 7 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T. L., and C. Elkan, 1994 Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology, AAAI Press, Menlo Park, CA, pp. 28–36. [PubMed]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat, F., J. Buard, C. Grey, A. Fledel-Alon, C. Ober et al., 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde, V., N. Robine, W. Lin, S. Bonfils, V. Geli et al., 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 28 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M. J., A. B. Nobel and J. D. Lieb, 2005. ChIPOTle: a user-friendly tool for the analysis of ChIP-chip data. Genome Biol. 6 R97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., C. Wilkinson, S. Watt, C. J. Penkett, W. M. Toone et al., 2008. Multiple pathways differentially regulate global oxidative stress responses in fission yeast. Mol. Biol. Cell 19 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., R. W. Hyppa, H. P. Cam, J. A. Farah, S. I. S. Grewal et al., 2007. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLOS Genet. 3 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn, A., J. Bollekens, A. Staub, C. Benoist and D. Mathis, 1987. A mulitiplicity of CCAAT Box-Binding Proteins. Cell 50 863–872. [DOI] [PubMed] [Google Scholar]
- Egel, R. (Editor), 2004. The Molecular Biology of Schizosaccharomyces pombe: Genetics, Genomics, and Beyond. Springer-Verlag, Berlin.
- Fox, M. E., T. Yamada, K. Ohta and G. R. Smith, 2000. A family of CRE-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics 156 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz, H., H. Heslot, U. Leupold and N. Loprieno, 1974. Schizosaccharomyces pombe, pp. 395–446 in Handbook of Genetics, edited by R. C. King. Plenum Press, New York.
- Haring, S., G. Halley, A. Jones and R. Malone, 2003. Properties of natural double-strand-break sites at a recombination hotspot in Saccharomyces cerevisiae. Genetics 165 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatamochi, A., P. T. Golumbek, E. V. Schaftingen and B. D. Crombrugghe, 1988. CCAAT DNA binding factor consisting of two different components that are both required for DNA binding. J. Biol. Chem. 263 5940–5947. [PubMed] [Google Scholar]
- Higuchi, T., Y. Watanabe and M. Yamamoto, 2002. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fisision yeast. Mol. Biol. Cell 22 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, K., C. S. Hoffman and K. Ohta, 2006. Reciprocal nuclear shuttling of two antagonizing Zn finger proteins modulates Tup family corepressor function to repress chromatin remodeling. Eukaryot. Cell 5 1980–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa, R. W., G. A. Cromie and G. R. Smith, 2008. Indistinguishable landcapes of meiotic DNA breaks in rad50+ and rad50S strains of fission yeast revealed by a novel rad50+ recombination intermediate. PLoS Genet. 4 e1000267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyppa, R. W., and G. R. Smith, 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys, A. J., J. Murray and R. Neumann, 1998. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol. Cell 2 267–273. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., and R. Neumann, 2002. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat. Genet. 31 267–271. [DOI] [PubMed] [Google Scholar]
- Jeffreys, A. J., and R. Neumann, 2005. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 14 2277–2287. [DOI] [PubMed] [Google Scholar]
- Kauppi, L., A. J. Jeffreys and S. Keeney, 2004. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 5 413–424. [DOI] [PubMed] [Google Scholar]
- Kim, C. G., and M. Sheffery, 1990. Physical characterization of the purified CCAAT transcription factor, α-CP1. J. Biol. Chem. 265 13362–13369. [PubMed] [Google Scholar]
- Kon, N., M. D. Krawchuk, B. G. Warren, G. R. Smith and W. P. Wahls, 1997. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 94 13756–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31 241–247. [DOI] [PubMed] [Google Scholar]
- Kunitomo, H., T. Higuchi, Y. Iino and M. Yamamoto, 2000. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11+ gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 11 3205–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah, S. K., J. M. A. Turner, F. Baudat, E. P. Rogakou, P. deBoer et al., 2001. Recombinational DNA double strand breaks in mice precede synapsis. Nat. Genet. 27 271–276. [DOI] [PubMed] [Google Scholar]
- Mantovani, R., 1998. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 26 1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, R., 1999. The molecular biology of the CCAAT-binding factor NF-Y. Gene 239 15–27. [DOI] [PubMed] [Google Scholar]
- Mata, J., A. Wilbrey and J. Bahler, 2007. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 8: R217.211–217.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNabb, D. S., K. A.-S. Tseng and L. Guarente, 1997. The Saccharomyces cerevisiae Hap5p homolog from fission yeast reveals two conserved domains that are essential for assembly of heterotetrameric CCAAT-binding factor. Mol. Cell. Biol. 17 7008–7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, A., B. Pelletier and S. Labbe, 2006. A transcription factor cascade involving Fep1 and the CCAAT-binding factor Php4 regulates gene expression in response to iron deficiency in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 5 1866–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., M. Dominska, M. J. Buck, J. L. Gerton, J. D. Lieb et al., 2006. A global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 26 1014–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, S., C. Freeman, A. Auton, P. Donnelly and G. McVean, 2008. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat. Genet. 40 1124–1129. [DOI] [PubMed] [Google Scholar]
- Myers, S., R. Bowden, A. Tumian, R. E. Bontrop, C. Freeman et al., 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, R., and A. J. Jeffreys, 2006. Polymorphism in the activity of human crossover hotspot independent of local DNA sequence variation. Hum. Mol. Genet. 15:1401–1411. [DOI] [PubMed] [Google Scholar]
- Nishant, K. T., and M. R. S. Rao, 2005. Molecular features of meiotic recombination hot spots. Bioessays 28 45–56. [DOI] [PubMed] [Google Scholar]
- Nurse, P., 1975. Genetic control of cell size at cell division in yeast. Nature 256 547–551. [DOI] [PubMed] [Google Scholar]
- Ohno, M., T. Fukagawa, J. S. Lee and T. Ikemura, 2002. Triplex-forming DNAs in the human interphase nucleus visualized in situ by polypurine/polypyrimidine DNA probes and antitriplex antibodies. Chromosoma 111 201–213. [DOI] [PubMed] [Google Scholar]
- Olesen, J. T., J. D. Fikes and L. Guarente, 1991. The Schizosaccharomyces pombe homolog of Saccharomyces cerevisiae HAP2 reveals selective and stringent conservation of the small essential core protein domain. Mol. Cell. Biol. 11 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov, E. D., P. M. Petkov and K. Paigen, 2010. Prdm9 controls activation of mammalian recombinatoin hotspots. Science 327 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes, T. D., 2001. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2 360–370. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1989. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli, A. S., and G. R. Smith, 1992. Context dependence of a eukaryotic recombination hotspot. Proc. Natl. Acad. Sci. USA 89 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., and D. Russell, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schuchert, P., M. Langsford, E. Käslin and J. Kohli, 1991. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 10 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, W. W., and G. R. Smith, 2005. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics 169 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, W. W., E. M. Steiner, A. R. Girvin and L. E. Plewik, 2009. Novel nucleotide sequence motifs that produce hotspots of meiotic recombinatoin in Schizosaccharomyces pombe. Genetics 182 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo, A. N., R. J. Pogulis and L. R. Pease, 1995. Mutagenesis by PCR, pp. 603–612 in PCR Primer: A Laboratory Manual. Edited by C. W. Dieffenbach and G. S. Dveksler. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Wahls, W. P., and G. R. Smith, 1994. A heteromeric protein that binds to a meiotic homologous recombination hotspot: correlation of binding and hotspot activity. Genes Dev. 8 1693–1702. [DOI] [PubMed] [Google Scholar]
- Wahls, W. P., and M. K. Davidson, 2010. Discrete DNA sites regulate global distribution of meiotic recombination. Trends Genet. 26 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls, W. P., E. R. Siegel and M. K. Davidson, 2008. Meiotic recombination hotspots of fission yeast are directed to loci that express non-coding RNA. PLOS One 3 e2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M. A., M. Dominska and T. D. Petes, 1993. Transcription factors are required for the meiotic recombination hotspot at the HIS4 locus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 90 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., F. Li, J. Li, M. Q. Zhang and X. Zhang, 2004. Evidence and characteristics of putative human alpha recombination hotspots. Hum. Mol. Genet. 13 2823–2828. [DOI] [PubMed] [Google Scholar]