Abstract
Previous studies have suggested that transgene expression in plants can be affected by ploidy. Here we show that three different transgenes, a reporter transgene, an antisense transgene, and a hairpin RNA (hpRNA) transgene, are all expressed at a lower level in autotetraploid (4n) than in diploid (2n) Arabidopsis. RNA silencing of two endogenous genes was induced by the antisense and hpRNA transgenes and this silencing is significantly less effective in 4n than in 2n Arabidopsis; furthermore, the reduced silencing in 4n Arabidopsis correlated with reduced accumulation of silencing-inducer RNAs. Methylation analysis both of independent 2n and 4n transgenic lines and of 2n and 4n progeny derived from the same 3n transgenic parent, indicated that transgenes are more methylated in 4n than 2n Arabidopsis. These results suggest that transgenes are transcriptionally repressed in the 4n background, resulting in expression levels lower than in the 2n background. Transgenes designed to silence endogenous genes express lower concentrations of silencing-inducer RNAs in 4n Arabidopsis plants, resulting in less effective silencing of target genes than in 2n Arabidopsis plants.
POLYPLOIDIZATION, or whole genome duplication, occurs frequently in plants and is a major source of plant speciation (Stebbins 1966; Adams and Wendel 2005). Recent studies using newly formed synthetic auto- or allopolyploid plants have shown that polyploidization is associated with genome-wide changes in gene expression, and these changes appear to be controlled primarily by epigenetic mechanisms such as cytosine methylation and small RNAs (Comai et al. 2000; Kashkush et al. 2002; Wang et al. 2004; Xu et al. 2009; Yu et al. 2010).
In addition to changes in endogenous gene expression, polyploidization also affects the expression of transgenes. Transgenes in plants can be inactivated either transcriptionally, through inactivation of promoters by DNA methylation and histone deacetylation or postranscriptionally through sequence-specific degradation of mRNA (Matzke et al. 2002). A stably expressed hygromycin phosphotransferase (HPT) transgene in diploid Arabidopsis was subject to transcriptional inactivation when brought into triploid or tetraploid backgrounds (achieved either by crossing with 4n Arabidopsis or by chromosome doubling) (Mittelsten Scheid et al. 1996). This transcriptional inactivation was independent of transgene copy number and occurred solely as a consequence of a change in ploidy. A follow-up study demonstrated that the transcriptionally inactivated HPT allele could trans-inactivate a formerly active allele of the HPT transgene in tetraploid but not diploid Arabidopsis (Mittelsten Scheid et al. 2003). These findings suggest that transgenes are more prone to transcriptional inactivation in polyploids than in diploids.
The inactivated HPT allele is associated with both DNA methylation and heterochromatic histone modifications and can be reactivated only when both of these modifications are reversed (Mittelsten Scheid et al. 2003; Hetzl et al. 2007; Baubec et al. 2010). The authors proposed that DNA methylation and histone modifications cooperate to form a “double lock” on ploidy-associated transcriptional inactivation (Baubec et al. 2010), but how these mechanisms are initiated remains unknown.
Double-stranded RNA (dsRNA)-induced silencing, or RNA interference (RNAi), has become a powerful tool for knocking down gene expression in plants and animals (Wang and Waterhouse 2002; Hannon and Rossi 2004). During RNAi, dsRNA or hairpin RNA (hpRNA) is processed by Dicer, an RNase III-like enzyme, into 20–25 nt small interfering RNAs (siRNAs). These siRNAs are bound by Argonaute protein, guiding the Argonaute to cleave homologous single-stranded RNAs (Baulcombe 2004; Hannon and Rossi 2004). In plants, effective RNAi has been achieved mainly by expression of transgenes designed to express silencing-inducer RNAs (Eamens et al. 2008). Such transgene-induced RNAi is best studied in model plants such as Arabidopsis and rice, which are mostly diploids, yet many of the agriculturally important crops, such as wheat, cotton, and sugarcane, are polyploids.
The objective of the current work was to investigate whether or not the effectiveness of transgene-induced RNAi is altered by plant ploidy. The effectiveness of RNAi is expected to depend on the expression level of the silencing-inducer RNAs from the RNAi constructs. Thus, if ploidy alters the transcriptional activity of the RNAi transgenes (and hence the accumulation of silencing-inducer RNAs) it would also alter the effectiveness of target gene silencing. We investigated this possibility using diploid (2n) and autotetraploid (4n) Arabidopsis as model systems. By analyzing large numbers of independent 2n and 4n transgenic lines, and by comparing 2n and 4n progenies derived from the same triploid (3n) transgenic parents, we demonstrate that both the level of transgene expression and the effectiveness of transgene-induced RNAi are lower in 4n than in 2n plants. We also show that transgenes tend to be more methylated in 4n than in 2n Arabidopsis and this is likely to account for the relatively low levels of transgene expression and transgene-induced RNAi in 4n Arabidopsis.
MATERIALS AND METHODS
Plasmid constructs:
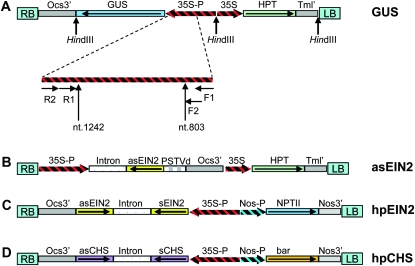
The β-glucuronidase (GUS) construct shown in Figure 1A was the same plasmid named as pCON in Chen et al. (2005). The hpCHS construct was previously described in Fusaro et al. (2006). To make the antisense construct, a 586-bp fragment of the EIN2 cDNA near the 5′ region was PCR amplified using primers 5′GCTGGATCCGGTACCTTGAATCCTACTCTGAG 3′ (forward) and 5′GAGATCGATCTCAGACTGACTCAGCA3′ (reverse), cloned into pGEM-T Easy (Promega), into which a BglII-digested full-length potato spindle tuber viroid (PSTVd) sequence was then inserted at the BamHI site. The asEIN2–PSTVd fusion sequence was then digested with XbaI and inserted at the XbaI site of pHannibal (Wesley et al. 2001), from which the 35S-intron-asEIN2-PSTVd-Ocs3′ fragment was excised with NotI and inserted into pWBVec2a (Wang et al. 1998), forming the final asEIN2 construct. For preparation of the hpEIN2 construct, a 911-bp fragment of EIN2 genomic DNA overlapping with the cDNA fragment was amplified using the same primers and cloned into pART7 (Gleave 1992), into which the PDK intron from pHannibal, and the EIN2 cDNA fragment from the asEIN2 construct, were inserted at the HindIII and XbaI sites, respectively. The 35S promoter-sense EIN2 genomic-intron-asEIN2-Ocs3′ fragment was then excised with NotI and inserted into pART27 (Gleave 1992), giving rise to the hpEIN2 construct.
Figure 1.—
Constructs used in this study. (A) The GUS overexpression construct. The striped bar below indicates the bisulphite PCR-amplified region. HindIII cleavage sites are indicated. (B) The EIN2 antisense construct. (C) The EIN2 hpRNA construct. (D) The chalcone synthase hpRNA construct. Ocs3′, Agrobacterium octopine synthase 3′ region; 35S-P, the cauliflower mosaic virus 35S promoter; 35S, a shorter version of the 35S promoter; Tml′, Agrobacterium tumor morphology large gene 3′ region; Nos-P and Nos3′, Agrobacterium nopaline synthase promoter and 3′ region, respectively; NPTII, neomycin phosphotransferase gene for kanamycin resistance; bar, phosphinothricin resistance gene; HPT, hygromycin phosphotransferase gene for hygromycin resistance; intron, the second intron of the Flaveria trinervia pyruvate orthophosphate dikinase (PdkA) gene; RB and LB, Agrobacterium T-DNA right and left borders, respectively.
Plant materials, transformation, crossing, and identification of 2n and 4n progeny:
The 2n and 4n Arabidopsis used for transformation were the diploid Arabidopsis thaliana ecotype Landsberg erecta (Ler) and a stable Ler tetraploid line obtained by chromosome doubling with colchicine treatment. Agrobacterium-mediated transformation was performed using the “floral dip” method described by Clough and Bent (1998). To select for transgenic lines, seed collected from Agrobacterium-infected plants was sterilized (Chen et al. 2005) and plated on MS medium containing 100 mg/liter of timentin plus appropriate selective agents [20 mg/liter of hygromycin for the GUS and asEIN2 constructs, 50 mg/liter of kanamycin for hpEIN2, and 5 mg/liter of phosphinothricin (PPT) for hpCHS]. Antibiotic or PPT-resistant seedlings were transferred to fresh MS plates containing the selective agents before being planted in soil. Crossing between 2n and 4n Arabidopsis was carried out by removing immature anthers from unopened flower buds in the 4n plants and fertilizing the stigma with pollen from freshly opened flowers of the 2n plants. To isolate 2n and 4n progeny from a 3n line, seed was collected from individual F2 plants and inspected for seed size (a 4n seed weighs ∼320 μg, ∼1.5 times bigger than a 2n seed that weighs ∼210 μg), and plants with uniformly larger- or smaller-sized seed were further checked for ploidy by counting chloroplast number in guard cells (Jacobs and Yoder 1989; Ho et al. 1990) from 10–12 stomata on the abaxial surface of leaves from each line using a confocal microscope, with 405 nm excitation generating autofluorescence from 450–550 nm to detect cell outlines, and 633 nm excitation generating strong chloroplast autofluorescence from 650 to 720 nm (a 4n guard cell has 8–12 chloroplasts, while a 2n cell has 4–6 chloroplasts). To select for homozygous or near-homozygous lines, individual F3 plants with relatively high transgene copy number were selected on the basis of semi-quantitative PCR using a primer pair for the GUS coding sequence and a primer pair for the Arabidopsis endogenous gene proteasome β-subunit G1 (primer sequences available upon request). Seed collected from the selected F3 plants was germinated on MS medium containing 20 mg/liter of hygromycin, and hygromycin-sensitive seedlings were further analyzed for GUS expression by staining with X-glucuronide (Jefferson et al. 1987).
Analysis of EIN2 silencing and GUS expression:
To assay for EIN2 silencing, seed was sterilized (Chen et al. 2005) and plated on MS medium containing 50 μg/liter of ACC (1-aminocyclopropane-1-carboxylic acid). The plates were sealed tightly with parafilm and incubated in total darkness at 4° for 2 days and then 22° for up to 2 weeks. Silencing was scored by visually comparing the elongation of hypocotyls with wild-type Arabidopsis. GUS activity was either quantitatively determined using the kinetic fluorimetric 4-methylumbelliferryl-β-glucuronide (MUG) assay (Chen et al. 2005) or visualized by histochemical staining of Arabidopsis seedlings with X-glucuronide (Jefferson et al. 1987). For T1 plants (the primary transformants), protein used for the MUG assay was extracted from 2–6 leaves of an individual plant, while for T2, 3n, and their progeny lines, protein was extracted from a pool of multiple (∼20–50) hygromycin-resistant sibling plants. MUG assays were performed on two different fluorometric machines with different parameter settings for the T1 plants and the rest of the materials, thereby giving different scales of readings and slope values. For the T1 plants (Figure 2A), the relative GUS activity represents slope value per 5 μg of protein, while for the rest, it is slope value per 1 μg of protein.
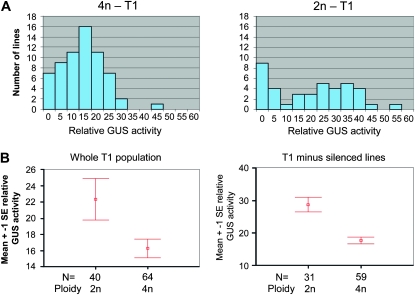
Figure 2.—
GUS activity in T1 transgenic lines determined by kinetic MUG assays at 37°. (A) Distribution of GUS expression levels for 4n and 2n T1 transgenic lines. The T1 population was heterozygous for the GUS transgene. (B) The overall level of GUS expression is significantly lower for the 4n population than the 2n population. (Left) Difference in mean GUS activity between the whole 2n and 4n populations in A. (Right) Difference after removing the “inactivated” or “silenced” lines (GUS activity <2). The relative GUS activity represents slope values per 5 μg of total plant protein.
DNA and RNA analysis:
DNA from the T2 GUS lines was isolated using the CTAB method described by Draper and Scott (1988). Total RNA from the T2 hpCHS lines was extracted using TRIzol reagent (Invitrogen) following the manufacturer's instruction. DNA and high molecular weight (HMW) RNA from all other samples was prepared following the phenol extraction method as previously described (Wang et al. 2008). Southern blot hybridization was performed as described (Wang et al. 2008) using a full-length octopine synthase terminator sequence as probe, which was excised from pART7 (Gleave 1992) with BamHI and NotI digestion, gel purified and radioactively labeled with α-32P using the Megaprimer DNA labeling kit (Amersham Biosciences). For Northern blot hybridization, 30 μg of total or HMW RNA was separated in 1.3% formaldehyde–agarose gel and blotted to a Hybond-N filter. The blot was hybridized (Wang et al. 2008) with 32P-labeled complementary RNA probes obtained by in vitro transcription using T7 or SP6 RNA polymerase and radioactive UTP from the respective sequences cloned into pGEM plasmids (Promega), including the 586 nt asEIN2 sequence, the full-length GUS coding sequence, and the bar gene sequence. For detection of chalcone synthase (CHS) siRNAs, ∼40 μg of total RNA samples was separated in 15% denaturing polyacrylamide gel, electroblotted and UV cross-linked onto Hybond-N filter, and hybridized with the same bar gene RNA probe following the procedure in Wang et al. (2008).
Methylation analysis using bisulphite PCR:
Bisulphite treatment of genomic DNA was carried out using the MethylEasy kit (Human Genetic Signatures) following the manufacturer's instruction. Approximately 5 μg of DNA from each sample was treated and purified with MethylEasy. Nested PCR was used to amplify the 35S promoter sequence from the bisulphite-treated DNA: the first round PCR was performed using the primers Top35S-F1 (5′TTAAGGTAAGTAATAGAGATTGGAGT3′) and Top35S-R2 (5′CTCCAAATAAAATAAACTTCCTTATATA3′), and the secondary PCR was performed with the nested primers Top35S-F2 (5′GAGATTGGAGTTTTTAAAAAGGTAGTT3′) and Top35S-R1 (5′TCAATAAAAATATCACATCAATCCACTT3′) (also see Figure 1A), using the same PCR cycles as described previously (Wang et al. 2001). These primers were designed to minimize biased amplification of methylated vs. unmethylated DNA: the last 13–19 nucleotides correspond to GUS sequences that contain no cytosines, so the primers bind to both bisulphite-converted (unmethylated) or unconverted (methylated) DNA. For sequencing analysis, the nested PCR product was gel purified using the UltraCleanTM 15 DNA purification kit (MO BIO Laboratories) and sequenced directly with BigDye 3.1 using one of the nested primers. To check the efficiency of bisulphite conversion, a 157-bp sequence of the chloroplast-encoded psaA protein gene was PCR amplified from the same bisulphite-treated DNA using the primers 5′ATGATGTTGTTAGAATTTYATATAGG3′ (forward) and 5′CATCATTTARCTATCRCAATTCTTT3′ (reverse).
Statistical analysis:
Populations were compared using a one- or two-tailed Welch's T-test (as specified) using Microsoft Excel 2002 (1-.3406.3501) SP 1. Where multiple measurements were taken of a single variable, the mean was taken as the score for analysis.
RESULTS
Expression of a β-glucuronidase transgene in 2n and 4n Arabidopsis:
Previous studies exploring how ploidy affects transgene expression have been based on the analysis of only a small number of transgenic lines. Here, diploid (2n) and tetraploid (4n) Arabidopsis plants were transformed with a construct expressing a GUS reporter gene (Figure 1A). Sixty-four 4n and forty 2n first generation transgenic lines (T1) were randomly selected and screened for GUS expression using fluorometric MUG assays.
The 4n and 2n transgenic populations showed a clear difference in the distribution of GUS expression levels (Figures 2A). The majority (89.0%) of the individual 4n lines had an intermediate level of GUS activity between 5 and 30 (arbitrary units, AU). In contrast, the 2n lines expressed a much wider range of GUS activity, with 15 lines (37.5%) showing an activity above >30 and 8 lines (20.0%) <2. A two-tailed T-test revealed a statistically significant difference (P = 0.036) between the mean GUS activity of the 2n (22.3 AU) and 4n (16.3 AU) transformants (Figure 2B, left panel; supporting information, Table S1A). The mean difference was 6.02 AU with a 95% C.I. of 0.4–11.6 AU. Furthermore, if the lines with GUS activity <2 AU were treated as “inactivated” or “silenced” and not included in the statistical analysis, the difference between the 2n and 4n transformants was even more striking. The mean GUS activity was 28.7 and 17.6 AU for the “active” 2n and 4n transformants, respectively, with a mean difference of 11.03 AU (95% C.I. 6.04–16.02; P < 0.001) (Figure 2B, right panel; Table S1B).
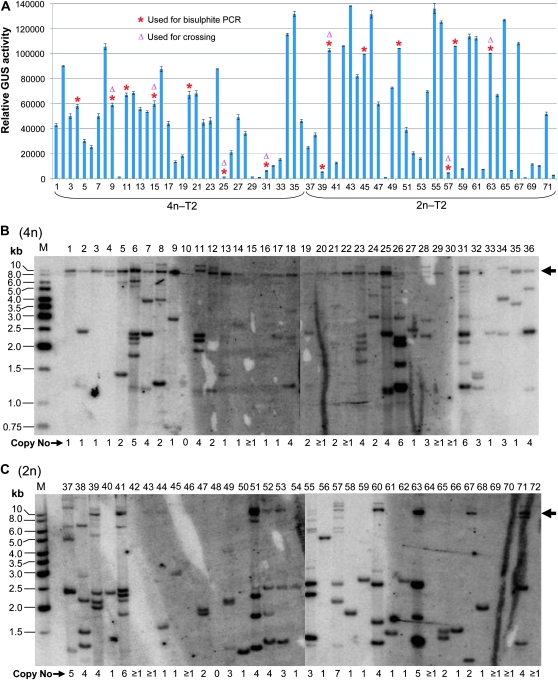
To further compare the 2n and 4n transgenic populations, 72 sec-generation (T2) lines derived from a subset of the T1 transformants were selected and analyzed for GUS expression and transgene insertion pattern. The pattern of GUS expression was maintained in the T2 generation: the majority of 4n lines showed intermediate levels of GUS activity compared to the much wider range seen in the 2n lines (Figure 3A). Southern blot hybridization showed that there was no clear difference in overall transgene copy number distribution between the 4n and 2n lines (Mann–Whitney U test, P = 0.90), estimated to be ranging from 1 to 7 copies based on the number of hybridization bands and a mean of 2.3 copies in both groups (Figure 3, B and C; Figure S1). Of note, the multiple-copy lines tended to show lower levels of GUS expression than the low-copy number lines, which was particularly true for the 4n population. These results suggested that the variation in GUS expression between the 4n and 2n populations was not caused by bias in transgene copy number distribution or differences in transgene dosage.
Figure 3.—
Molecular analysis of 36 4n or 2n T2 lines derived from a subset of the T1 transformants analyzed in Figure 2. The T2 lines were heterozygous for the GUS transgene. (A) GUS expression analysis. The relative GUS activity is slope values per 1 μg of total protein. The MUG assays for T2 plants and the rest of the samples shown in this article were determined using a different fluorometric machine, therefore giving a different scale of readings to the T1 samples in Figure 2. Error bars represent standard deviation of four measurements. Lines used in DNA methylation analysis (Figure 4) and in crossing for generating 3n progeny (Figure 5A) are marked with red asterisks and purple triangles, respectively. (B and C) Southern blot hybridization analysis. DNA was digested with HindIII and hybridized with the Ocs3′ fragment as a probe. The horizontal arrows indicate the high molecular weight band (∼10 kb in size) suggestive of cytosine methylation. The estimated transgene copy number is given below the blot. M, GeneRuler 1 kb DNA ladder (MBI Fermentas).
Methylation status of the GUS transgene in 2n and 4n Arabidopsis:
The majority of the 4n lines (32 of 36) contained a HMW band on Southern blots with HindIII digestion (Figure 3B), while relatively few 2n lines (10–12 out of 36) contained a similar band (Figure 3C). This band was not due to insufficient amounts of the HindIII enzyme in the digestion reactions: the DNA was uniformly digested across all 4n and 2n samples (Figure S2), and replicate experiments using different batches of DNA gave the same hybridization pattern (e.g., Figure 5G). This observation may be explained by the methylation sensitivity of HindIII: HindIII cuts at the sequence AAGCTT, but is inhibited by methylation of the cytosine residue. Thus, the HMW band may have been caused by incomplete digestion due to partial cytosine methylation at the HindIII sites, and the higher proportion of 4n lines displaying the HMW band suggests more widespread methylation of the transgene in 4n than 2n lines.
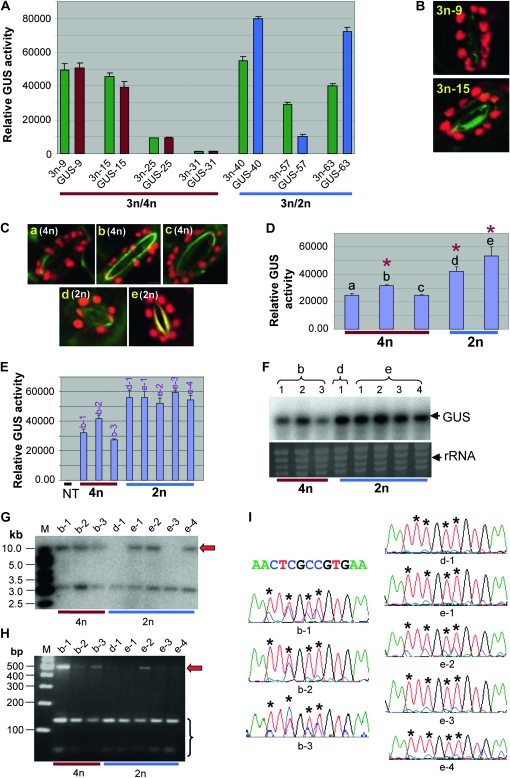
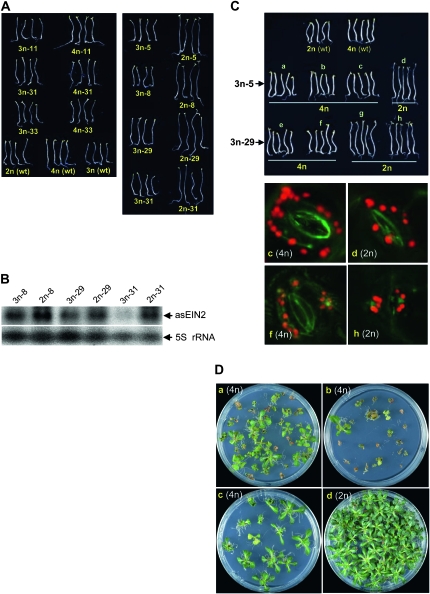
Figure 5.—
Expression and cytosine methylation analyses of 3n F1 GUS lines and the 2n and 4n progeny plants derived from the 3n line 3n-15. (A) Comparison of GUS expression between 3n plants and their parental 4n or 2n lines. These lines were heterozygous for the GUS transgene. Each pair of bars represents the mean value of GUS activity from three MUG assays of a parental 4n or 2n line (red or blue) and its 3n progeny (green). Error bars represent standard deviation. (B and C) Examples of visualization of chloroplasts in leaf guard cell confirming the 3n, 4n, or 2n ploidy level of the lines analyzed for GUS expression in A and D, respectively. (D) GUS expression analysis of 4n (a–c) and 2n (d and e) F3 progeny derived from the 3n F1 line 3n-15 in A. These F3 progeny plants were heterozygous for the GUS transgene. The asterisks indicate the three lines from which the F5 plants in E were derived. (E) GUS expression analysis of F5 lines derived from the F3 lines b, d, and e shown in D. These F5 lines were homozygous or near homozygous (see Table S2). NT, nontransgenic control. (F) Northern blot hybridization of GUS mRNA (upper) in the same F5 lines as shown in E, and ethidium bromide-stained rRNA as loading control (lower). (G) Southern blot hybridization analysis of the F5 lines. DNA was digested with HindIII and hybridized with Ocs3′. The red arrow indicates the high molecular weight band, suggesting partial resistance to HindIII digestion. M, GeneRuler 1 kb DNA marker. (H) MseI digestion of bisulphite PCR product for the F5 lines. M, GeneRuler 100 bp DNA marker. The red arrow indicates undigested PCR product, suggesting cytosine methylation. (I) Part of the sequencing trace files for the bisulphite PCR products of the F5 lines. As in Figure 4C, the four different nucleotides, A, T, C, and G, are shown in green, red, blue, and black, respectively. The red (T) and blue (C) mixed peaks indicate the presence of both bisulphite-converted and -unconverted cytosines. The original DNA sequence of the corresponding 13-bp region is given above. The asterisks indicate the position of cytosines.
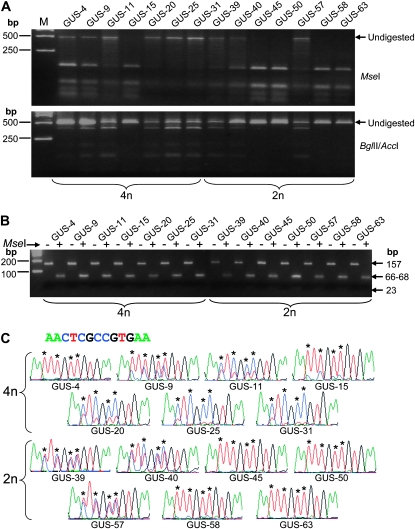
The GUS transgene construct contains only three HindIII sites, all of which are outside the transgene promoter (Figure 1A), so the pattern of HindIII digestion might not reflect the DNA methylation status of the GUS transcriptional unit, particularly in its promoter sequence. To investigate the methylation status, DNA samples from 7 each of the 4n and 2n T2 lines (indicated by asterisks in Figure 3A), composed of low- and high-copy number individuals, were treated with bisulphite—a process that converts unmethylated cytosines to uracils but does not affect methylated cytosines. A 442-bp sequence, from −108 to −549 of the 35S promoter including the B2–B5 domains important for the promoter activity (Figure 1A and Figure S3A) (Benfey and Chua 1990; Bhullar et al. 2007), was then PCR amplified with unbiased primers specific for the top strand. The PCR product was then digested with MseI or BglII/AccI and separated in an agarose gel. MseI recognizes the TTAA sequence that only occurs (14 times) in the bisulphite-converted PCR product of unmethylated DNA. BglII and AccI, on the other hand, recognize the AGATCT and GTCGAC sequences found only in the unconverted PCR product from methylated DNA (Figure S3B) and can therefore be used to corroborate the MseI digest result. To test the efficiency of bisulphite conversion, a 157-bp region of the Arabidopsis chloroplast gene encoding psaA protein (Figure S4, A and B), which is free of cytosine methylation, was PCR amplified from the same bisulphite-treated DNA and digested with MseI.
As shown in Figure 4A, PCR products from six of the 4n lines (GUS-4, -9, -11, -20, -25 and -31) and three of the 2n lines (GUS-39, -40, and -57) showed significant resistance to MseI digestion, indicating that the 35S promoter was methylated to various degrees in these lines. This was confirmed by the BglII/AccI digestion, which gave cleaved fragments indicating the presence of methylated cytosines. PCR products from the one remaining 4n line (GUS-15) and the four 2n lines (GUS-45, -50, -58, and -63) were almost completely digested by MseI but resistant to BglII/AccI digestion, indicating a near-complete absence of cytosine methylation. Bisulphite PCR product of the chloroplast DNA from all 14 samples was fully digested by MseI (Figure 4B; also Figure S4C for sequencing trace files that showed very little unconverted cytosine), indicating uniform and efficient bisulphite conversion. The restriction digests suggested that the 4n lines were more methylated than the 2n lines.
Figure 4.—
Cytosine methylation analysis using bisulphite PCR. (A) Bisulphite PCR product was digested with MseI (upper) or BglII/AccI (lower) and separated in 3% NuSieve 3:1 agarose gel. M, GeneRuler 1 kb DNA Ladder. (B) MseI digestion of bisulphite PCR product of a 157-bp region of the chloroplast-encoded psaA protein gene amplified from the same bisulphite-treated genomic DNA as for the 35S promoter sequence shown in A. Bisulphite PCR product was either treated (+) or untreated (−) with MseI and separated in 4% NuSieve agarose gel. Note that the DNA is fully digested by MseI for all samples, indicating uniform and efficient bisulphite conversion. (C) Part of the sequencing trace files for the bisulphite PCR products of the 35S promoter. The original DNA sequence of the corresponding 13-bp region is given above. The four different nucleotides, A, T, C, and G, are shown in green, red, blue, and black, respectively. The red and blue mixed peaks indicate the presence of both bisulphite-converted and unconverted cytosines. The asterisks indicate the positions of cytosines.
Sequencing of the bisulphite PCR product confirmed methylation differences between the 4n and 2n lines. Figure 4C shows a 13-bp region of the sequencing trace files that reflects the overall cytosine methylation status of the bisulphite-PCR product. This region (−479 to −491 of the 35S promoter; Figure S3A) contains four cytosine residues: two in CG context, one in CHG context (“H” representing A, C, or T), and one in CHH context (see the sequence on top of Figure 4C). The relative height between the cytosine peaks (blue) and the overlapping thymine peaks (red) should reflect the degree of cytosine methylation; a higher blue peak indicates stronger methylation and lower blue peaks, weaker methylation. Four of the seven 4n lines (GUS-11, GUS-20, GUS-25, and GUS-31) had the blue peaks dominating the red peaks, suggesting strong cytosine methylation. Two of these (GUS-25 and GUS-31) showed very low red thymine peaks at the cytosine positions, indicating almost complete methylation. The 4n lines GUS-9 and GUS-15 also showed significant amounts of unconverted cytosines (blue peaks) indicative of partial methylation. In contrast, only one 2n line, GUS40, exhibited a strong methylation pattern, with two lines (GUS-39 and GUS-57) showing an intermediate level of methylation (blue peaks = red peaks). Four of the 2n lines (GUS-45, GUS-50, GUS-58, and GUS-63) showed very little unconverted cytosines (blue peaks), indicating that they were almost completely unmethylated. This result is consistent with the restriction digestion result: both the frequency and degree of cytosine methylation were higher among the seven 4n lines than among the seven 2n lines analyzed.
There appeared to be an inverse correlation between the level of DNA methylation and that of gene expression (Figures 3A and 4). Despite this general trend, the 2n line GUS-40 had significant levels of cytosine methylation yet showed high-level GUS expression. Also, the 4n line GUS-4 showed very little methylation but had similar levels of GUS expression to the other four moderately expressing 4n lines (Figures 3A and 4). This variability may be due to the effect of differential insertion sites and copy numbers between lines, as this may affect the methylation and expression of the transgene.
Comparison of GUS transgene expression among 2n, 3n, and 4n plants containing identical transgene insertions:
We generated plants of different ploidy but containing identical transgene insertions. A subset of the 2n and 4n GUS lines shown in Figure 3A were crossed with wild-type 4n and 2n Arabidopsis, respectively, to generate triploid (3n) GUS transgenic lines (Figure 5, A and B). Both GUS-expressing (4n, GUS-9 and GUS-15; 2n, GUS-40 and GUS-63) and GUS-inactivated (4n, GUS-25 and GUS-31; 2n, GUS-57) lines were selected as the transgene donor lines.
There was no significant change in GUS expression between the 4n transgenic parental lines and their 3n progeny (P = 0.94, two-tailed Welch's T-Test). In contrast, the 2n parental lines and their 3n F1 progeny showed a clear difference in GUS expression (Figure 5A), however the small sample size did not allow statistical significance to be established. In the two high-expressor 2n lines (GUS-40 and GUS-63), the level of GUS expression decreased in the 3n progeny by ∼31 and 45%, respectively. Unlike the high expressors, the 3n F1 plants derived from the cross with the inactivated 2n line (GUS-57) showed a threefold increase in GUS expression, suggesting a partial release of inactivation.
Triploid (3n) Arabidopsis plants can be fertile and give rise to diploid (2n) and tetraploid (4n) progeny (Henry et al. 2007), and this was true for the 3n GUS lines. Therefore, from the 3n progeny of the cross between the single-copy 4n line (GUS-15) and wild-type 2n Arabidopsis, F2 plants were isolated with a 2n or 4n karyotype. This was done first by inspecting seed size from individual F2 plants followed by microscopic counting of chloroplast number in the guard cells (Figure 5C). As a result, three 4n and two 2n lines were isolated. Analysis of the F3 population of these lines showed that the level of GUS expression was lower in the three 4n lines than in the two 2n lines (Figure 5D), despite the two ploidy populations containing the same transgene locus.
The F3 populations should be heterozygous for the GUS transgene, so the differential GUS expression shown in Figure 5D could be due to differences in transgene dosage between the 4n and 2n lines. To address this issue, homozygous or near-homozygous populations were isolated and used for expression analysis. Isolation of homozygous 2n lines was straightforward, as progeny from a heterozygous plant always gave a 3:1 segregation for GUS activity and hygromycin resistance while those from a homozygous plant invariably showed 100% activity and resistance. The segregation pattern is far more complex for 4n plants; thus, to isolate homozygous or near-homozygous 4n lines, semiquantitative PCR was performed to find individual F3 plants that contained a relatively high-copy number of the GUS transgene. We then screened the F4 and F5 populations derived from these F3 plants for hygromycin resistance and/or GUS activity. As a result, three F5 lines derived from the F3 line b (shown in Figure 5D) were isolated, which were homozygous or near homozygous (Table S2). The difference in GUS expression remained in these F5 populations (Figure 5, E and F), suggesting that it was not caused by gene dosage differences.
An unexpected observation during the isolation of homozygous lines was that a small proportion of the F5 4n plants (18 out of 1420) showed dramatic inactivation of the GUS transgene as indicated by low-intensity or localized GUS staining (Figure S5). Such GUS inactivation was not observed in any of the 2n plants analyzed. Taken together, these results suggested that the GUS transgene was transcriptionally less active and more prone to transcriptional inactivation in 4n than in 2n backgrounds.
To investigate whether DNA methylation differed between the 4n and 2n F5 lines, Southern blot hybridization of HindIII-digested DNA and MseI digestion of bisulphite-PCR product of the 35S promoter were performed as before. Both analyses showed an overall difference in cytosine methylation between the two populations. All three of the 4n lines showed significant levels of cytosine methylation, as indicated both by the HMW hybridizing band on the Southern blot (Figure 5G, lanes 2–4) and by the partial MseI digestion of the bisulphite-PCR product (Figures 5H, lanes 2–4). In contrast, two of the five 2n lines (d-1 and e-3) showed almost no methylation on the basis of the Southern blot (Figure 5G, lanes 5 and 8). The reduced methylation in the 2n lines was further indicated by MseI digestion of the bisulphite-PCR product: four 2n lines (including d-1 and e-3 plus e1 and e4) showed much reduced levels of cytosine methylation in the 35S promoter region compared to the three 4n lines (Figure 5H, compare lanes 5, 6, 8, and 9 with lanes 2–4). Sequencing of the bisulphite PCR product confirmed the MseI digestion result: one or more of the four cytosines shown displayed heavier methylation in the three 4n lines than for the five 2n lines (Figure 5I). Bisulphite PCR products of the 157-bp chloroplast DNA from all the eight samples were fully digested by MseI (Figure S6A; also see Figure S6B for sequencing trace files), indicating efficient and uniform bisulphite conversion. These results indicate that the methylation of the GUS transgene that occurred in the parental 4n line (GUS-15) was retained through 3n to its subsequent F5 4n progeny, but that this methylation was lost in some of the F5 2n progeny. Thus, methylation is more likely to be established and maintained in the 4n compared to the 2n genetic background.
Effect of ploidy on transgene-induced RNAi:
The experiments with the GUS transgene suggested that transgenes are expressed at lower levels in 4n than 2n Arabidopsis. Transgenes coding for silencing-inducer RNAs may show a similar difference in expression, resulting in reduced silencing of target genes in 4n plants. To investigate this, we transformed 2n and 4n Arabidopsis lines with one of three different silencing constructs: an antisense construct against the ETHYLENE INSENSITIVE 2 (EIN2) gene (Figure 1B), an hpRNA construct against EIN2 (Figure 1C), or an hpRNA construct against a CHS (Figure 1D) gene.
EIN2 and CHS were chosen as the target endogenous genes because silencing could be easily assayed. Silencing of the EIN2 gene can be evaluated by comparing the hypocotyl length of seedlings germinated in the dark on medium containing ACC. ACC inhibits hypocotyl growth, but EIN2-silenced seedlings have reduced sensitivity to ACC and grow longer hypocotyls than unsilenced or wild-type Arabidopsis, with the length of the hypocotyl reflecting the degree of EIN2 silencing. Silencing of the CHS gene results in the loss of the dark pigmentation (anthocyanin) in the seed coat, and the whiteness of the seed coat reflects the degree of CHS silencing. We included an antisense construct in addition to the more effective hpRNA constructs in the experiment because we expected that any difference in silencing between 2n and 4n transformants was more likely to be observed with the antisense construct due to its moderate efficacy.
Antisense transgene-induced silencing of EIN2:
Transformation with the antisense construct (asEIN2) resulted in 49 and 39 independent transformants of 2n and 4n backgrounds, respectively. Both the frequency and degree of EIN2 silencing were higher in the 2n than the 4n backgrounds: 21 of the 2n lines (42.8%) showed significant EIN2 silencing, while silencing was present in only eight of the 4n lines (20.5%) (Table 1 and Figure S7). The mean silencing score was 0.71 and 0.23 units in the 2n and 4n populations, respectively, and was statistically significant (Mann–Whitney, P = 0.036). Four of the 2n lines showed strong EIN2 silencing (2n:asEIN-5, -8, -29, and -31), but this was not seen in any 4n lines; and in four of the eight silenced 4n lines (4n:asEIN-13, -21, -33, and -39) only a small proportion (<10%) of T2 plants showed EIN2 silencing, with the rest showing wild-type phenotype (Table 1). This last observation suggested that the asEIN2 transgene is active in some siblings but inactive in the other siblings. Such variegation of the transgene occurred in a much smaller proportion of the 2n lines (2n:asEIN-22, -26, -48).
TABLE 1.
Antisense-mediated silencing of EIN2 in 2n and 4n Arabidopsis
| 2n:asEIN2 |
4n:asEIN2 |
||||
|---|---|---|---|---|---|
| Line | Silencing score | Line | Silencing score | Line | Silencing score |
| 2n:asEIN-2 | + | 2n:asEIN-26a | + | 4n:asEIN-11 | ++ |
| 2n:asEIN-3 | + | 2n:asEIN-29 | +++++ | 4n:asEIN-13a | + |
| 2n:asEIN-4 | + | 2n:asEIN-31 | +++ | 4n:asEIN-17 | + |
| 2n:asEIN-5 | +++ | 2n:asEIN-33 | + | 4n:asEIN-21a | + |
| 2n:asEIN-8 | ++++ | 2n:asEIN-34 | + | 4n:asEIN-28 | + |
| 2n:asEIN-9 | + | 2n:asEIN-35 | + | 4n:asEIN-31 | + |
| 2n:asEIN-11 | + | 2n:asEIN-38 | ++ | 4n:asEIN-33a | + |
| 2n:asEIN-14 | + | 2n:asEIN-41 | + | 4n:asEIN-39a | + |
| 2n:asEIN-17 | ++ | 2n:asEIN-45 | ++ | ||
| 2n:asEIN-22a | + | 2n:asEIN-48a | ++ | ||
| 2n:asEIN-25 | ++ | ||||
| % silenced line: 42.8% (21/49) | % silenced line: 20.5% (8/39) | ||||
Silencing was scored by visually comparing the hypocotyl length of transgenic seedlings with that of nontransgenic wild-type 2n and 4n seedlings germinated and grown on ACC medium in the dark.
Only a small proportion (<10%) of the T2 plants showed visible EIN2 silencing.
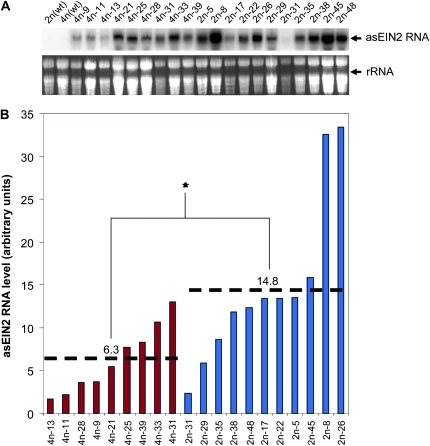
The levels of the asEIN2 RNA were lower overall in the 4n lines than in the 2n lines, and the majority of 4n lines showed intermediate levels, while the 2n lines showed a much wider range of asEIN2 expression (Figure 6). This result indicated that the asEIN2 transgene, like the GUS transgene, had reduced expression in the 4n background in comparison to the 2n background. It implies that the reduced efficiency of EIN2 silencing in the 4n background could be due to the reduced expression of the silencing-inducer antisense RNA.
Figure 6.—
Expression analysis of 4n and 2n asEIN2 lines. (A) Northern blot hybridization using sense EIN2 RNA as probe (top). (Bottom) Loading control showing ethidium bromide-stained rRNA bands. (B) Relative intensity of the hybridizing signals. For quantifying the hybridizing signals, the intensity of the GUS mRNA band was measured using the Multi Gauge software (Fujifilm) and calibrated against the intensity of the ethidium bromide-stained rRNA band. The mean value of three measurements was taken as the score for each line, and the score was used to perform a one-tailed Welch's T-test. The two dashed lines indicate the mean value of the 4n and 2n samples (6.3 and 14.8, respectively). *The difference between 4n and 2n is statistically significant (P < 0.01).
It is important to note that the steady-state RNA level detected by the Northern blot hybridization does not necessarily reflect the transcriptional activity of the asEIN2 transgene in all lines: post-transcriptional silencing of the transgene itself could reduce the accumulation of the asEIN2 RNA in some lines. If transcriptional inactivation of the asEIN2 transgene occurred, this would result in reduced transgene RNA production and thus reduced silencing of the target endogenous EIN2 gene. If instead siRNA-induced post-transcriptional silencing of the transgene occurred, the siRNAs produced would target not only the transgene itself but also the transgene's target: endogenous EIN2. Thus, EIN2 silencing would be maintained, though there would be little detectable transgene RNA. This could explain the relatively strong silencing observed in a small number of lines (e.g., 2n:asEIN-31) that had low levels of asEIN RNA.
To compare EIN2 silencing in plants with different ploidy but containing identical asEIN2 transgene insertions, a subset of the 2n and 4n asEIN2 lines were crossed with wild-type 4n and 2n Arabidopsis, respectively, to generate 3n transgenic lines (Figure 7A). From two of these 3n F1 plants, derived from crosses with the two strongly silenced 2n parental lines (2n-5 and 2n-29), we isolated four F3 lines each that had a 2n or 4n karyotype on the basis of seed size and chloroplast number of leaf guard cells (e.g., Figure 7C and Figure S8).
Figure 7.—
Assay for EIN2 silencing induced by the EIN2 antisense construct (asEIN2). (A) EIN2 silencing in F1 3n plants and their parental 4n (left) or 2n lines (right). Within each panel the plants on the left are 3n lines and those on the right the 4n or 2n parental lines. Seed was plated on MS medium containing ACC and allowed to germinate in darkness. Five seedlings from each line were then photographed. Longer hypocotyls in comparison to wild-type Arabidopsis indicate EIN2 silencing. (B) Northern blot hybridization detection of EIN2 antisense RNAs in three of the 3n lines and their 2n parental lines. (C) EIN2 silencing in 4n and 2n progeny derived from two of the 3n lines, 3n-5 and 3n-29. Only the 2n progeny displayed observable EIN2 silencing. The ploidy level was confirmed by visualization of chloroplasts in the leaf guard cells (lower). (D) Hygromycin sensitivity of offspring from 3n-5 plants in C. A large proportion of seedlings of the 4n progeny lines a and b shown in C died or showed little growth on medium containing 20 mg/liter of hygromycin, indicating silencing of the hygromycin resistance selectable marker gene in the asEIN2 transgene. This is in contrast to the seedlings of the 2n progeny line d derived from the same 3n parent, which showed vigorous growth on hygromycin medium, indicating active expression of the selectable marker gene.
As shown in Figure 7A, EIN2 silencing was similar between the three 4n parental lines and their 3n progeny (left panel). In contrast, EIN2 silencing in the 2n parental lines was almost completely released when the same asEIN2 transgene insertions were brought into the 3n background (Figure 7A, right panel) and this release correlated with a reduction in the asEIN2 transcript level (Figure 7B). These results bear a striking resemblance to the observation in GUS transgene expression, which showed no clear difference between 4n parents and their 3n progeny but strong downregulation when the GUS transgene was transferred from 2n to 3n backgrounds (Figure 5A).
Of the progeny derived from the two 3n F1 lines, only the 2n progeny displayed appreciable EIN2 silencing, while none of the 4n progeny (having the same asEIN2 transgene insertions) showed visible EIN2 silencing (Figure 7C). Taken together, these results suggest that antisense-mediated silencing is less efficient in 4n than in 2n backgrounds, and this is likely due to reduced expression of the antisense transgene. When the F3 asEIN2 plants were grown on hygromycin medium a significant proportion of plants of the 4n lines a and b in Figure 7C exhibited hygromycin sensitivity (Figure 7D), suggesting that the asEIN2 construct had become inactive in these plants. In contrast, the 2n F3 line d derived from the same parental line 3n-5, grew vigorously on hygromycin, indicating stable expression of the hygromycin resistance gene. This is similar to the observation with the F5 GUS plants, where only the 4n plants were subject to GUS transgene inactivation (Figure S5) and again suggests that transgene inactivation is more common in the 4n than the 2n background.
hpRNA transgene-induced RNAi:
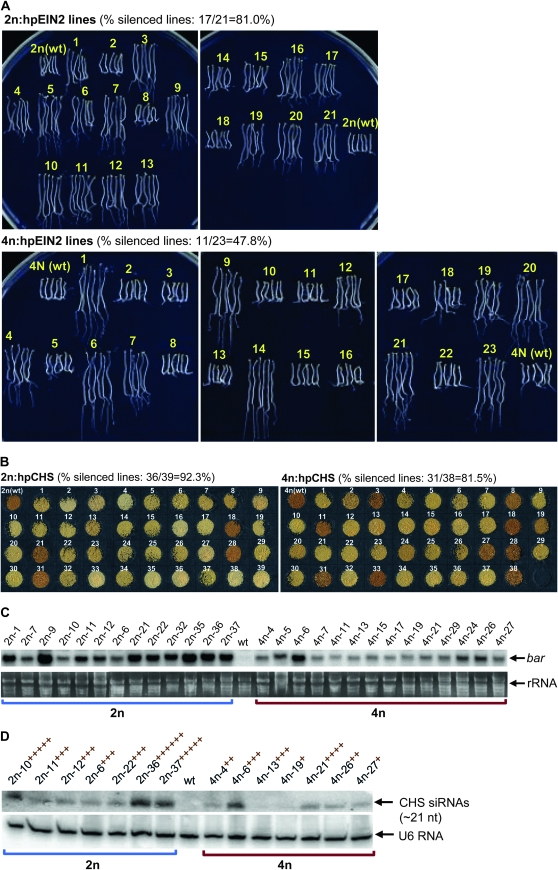
As hpRNA transgenes are more potent than antisense transgenes at inducing RNAi in plants, it was expected that strong silencing would occur in both 2n and 4n plants. Indeed, strong silencing of EIN2 and CHS were observed in both 2n and 4n Arabidopsis backgrounds (Figure 8, A and B). Nevertheless, comparisons of two relatively large 2n and 4n transgenic populations revealed a clear difference in the efficacy of hpRNA transgene-induced RNAi.
Figure 8.—
hpRNA transgene-induced silencing in 2n and 4n Arabidopsis. (A) Assay for EIN2 silencing in 2n and 4n Arabidopsis transformed with the EIN2 hpRNA construct. Longer hypocotyls indicate EIN2 silencing. (B) Pictures of seed from 2n and 4n Arabidopsis transformed with the chalcone synthase hpRNA construct. The reduction or loss of the brown pigmentation indicates CHS silencing. (C) Northern blot hybridization analysis of 2n and 4n hpCHS lines using 32P-labeled bar antisense RNA as a probe. The lower panel is ethidium bromide-stained rRNA as a loading control. (D) Detection of siRNAs derived the hpCHS transgene in a subset of the 2n and 4n hpCHS lines. The number of + symbols indicates degrees of CHS silencing. The lower panel is U6 RNA hybridized with a 32P-labeled antisense oligo as a loading control.
Silencing of both EIN2 and CHS was less frequent in the 4n background than in the 2n background (Figure 8, A and B; Table S3 and Table S4). Figure 8B suggests that, in addition to the reduced frequency of hpRNA transgene-induced silencing of the CHS gene, the degree of silencing was also lower in the 4n background than in the 2n background. A number of the 2n hpCHS lines had extremely light seed coat color (e.g., 2n:hpCHS-2, -4, -16, -33, -35, -36, -38, and -39), but none of the 4n lines showed such extreme loss of pigmentation (Figure S9A). Quantification of the seed color intensity confirmed that the seed of the 4n population was as a whole darker than that of the 2n lines (Figure S10). This was not due to a difference in the target CHS gene expression, as a RT–PCR analysis showed no increase in CHS transcript abundance in the 4n Arabidopsis (Figure S9C). Three of the 4n lines (4n:hpCHS-2, 4n:hpCHS-31, and 4n:hpCHS-38) had a mixture of light- and dark-colored seed (Figure S9B), suggesting transgene variegation in these lines. In the hpEIN2 transgenic lines, silencing was less frequent in the 4n lines than in the 2n lines (Figure 8A and Table S3), but it was difficult to quantitatively compare the degree of EIN2 silencing between the strongly silenced 2n and 4n lines, because the larger 4n seed allows for more elongation of hypocotyls on ACC medium in darkness.
It was considered that a relative reduction in transgene dosage due to increased chromosome number in the 4n background could have led to reduced RNAi efficiency in these plants. Nonetheless, segregation analyses of the hpRNA lines suggest that this was not the case: the majority of the unsilenced 4n lines actually contained relatively high copy numbers of the hpEIN2 or hpCHS transgenes as indicated by the high segregation ratio for kanamycin (Kan) or phosphinothricin (PPT) resistance (Table S3 and Table S4). On the other hand, the 4n lines with a lower copy number tended to be the more strongly silenced lines. Thus, there appeared to be an inverse correlation between transgene copy number and the level of hpRNA transgene-induced RNAi in the 4n Arabidopsis. In contrast to the high-copy number 4n lines (most of which showed no silencing of the target genes), many of the high-copy number 2n lines showed various degrees of target gene silencing, including strong silencing (Table S3 and Table S4). As was the case with the 4n plants, the low-copy number 2n lines tended to be among the most strongly silenced.
A subset of the hpCHS lines was analyzed with Northern blot hybridization. The expression level of hpCHS RNA is difficult to determine as it is the substrate for Dicer, so instead the expression of the PPT resistance gene bar (part of the hpCHS construct) and the level of hpCHS-derived siRNAs were measured. As shown in Figures 8C, the overall expression levels of bar were lower in the 4n lines than the 2n lines. This ploidy-dependent expression difference was reminiscent of that seen in the GUS and antisense EIN2 transgenes (Figures 3A and 6) and suggests that the hpRNA transgenes are less expressed in 4n than in 2n backgrounds. Northern blot hybridization of siRNAs showed a similar trend: as a whole the 4n lines accumulated lower levels of siRNAs than the 2n lines analyzed (Figure 8D). The abundance of the siRNAs correlated relatively well (though not perfectly) with the degree of CHS silencing. Taken together, these results suggested that, like the antisense transgene, hpRNA transgenes are expressed at lower levels, and hence induce less effective RNAi in 4n than in 2n backgrounds.
DISCUSSION
In this article we provide evidence that transgenes are expressed at a lower level in an autotetraploid background than in a diploid background. The three transgenes tested, namely the GUS sense transgene, the EIN2 antisense transgene, and the CHS hpRNA transgene, all showed a lower overall level of expression in 4n than 2n Arabidopsis.
Transgenes can be inactivated at both transcriptional and post-transcriptional levels, and transcriptional repression in plants is often associated with increased cytosine methylation especially in the promoter region of a gene (Matzke et al. 2002; Baulcombe 2004). Our analysis of the GUS transgenic lines indicated that transgenes have more widespread cytosine methylation in the 4n background than in the 2n background (Figures 3 and 4). In addition to this, we analyzed the offspring of a single 3n GUS transgenic parent for methylation status of the transgenic DNA: the transgene was more methylated in the 4n than the 2n progeny, and GUS expression levels showed an inverse correlation with the degree of DNA methylation in the transgene promoter (Figure 5, E–I). These results suggest that the reduction of transgene expression in 4n Arabidopsis is due to reduced transgene transcription rather than post-transcriptional RNA degradation. This is consistent with the previous observation of ploidy-associated transcriptional inactivation of an HPT transgene that is also associated with increased DNA methylation (Mittelsten Scheid et al. 1996).
Despite the higher overall level of transgene expression, the 2n GUS transgenic population contained a larger proportion of lines (8 out of 40) than the 4n population (4 out of 64) that showed extremely low levels (or inactivation) of GUS expression (Figure 2). Also, an inactive GUS transgene in a 2n line became significantly reactivated when introduced into a 3n background by crossing with wild-type 4n Arabidopsis (Figure 5A). A possible explanation for this seeming paradox is that the GUS transgene in these lines is transcribed at excessive levels, triggering post-transcriptional silencing or cosuppression (Schubert et al. 2004); the generally higher levels of transgene transcription in the 2n background would cause more frequent cosupression, resulting in a higher proportion of GUS-silenced lines than in the 4n background.
While the expression and methylation analyses suggested that transgenes are transcriptionally repressed in the 4n lines, most of these lines showed an intermediate level of transgene expression. This differs from the conventionally defined transcriptional gene silencing associated with dramatic reduction in transgene expression (Matzke et al. 2002). A transgenic study in animals may provide an explanation for the transcriptional repression in 4n Arabidopsis: Robertson et al. (1995) showed that the expression of a globin transgene within all mouse lines is heterocellular, with individual cells either showing no expression of the transgene at all or expressing it at a high level, characteristic of a particular line. The authors found that although the number of transgene-expressing cells varies greatly between different transgenic lines, within a transgenic line, individual mice have strikingly similar numbers of expressing cells. Thus, the variation in total transgene activity between lines is due mainly to differences in the percentage of cells that actively express the transgene. The authors called the phenomenon position-dependent variegation, since the degree of variation appears to depend on transgene integration site but not transgene copy number.
The following observations raise the possibility that similar position-dependent transgene variegation may occur in Arabidopsis plants: among the asEIN2 and hpCHS transgenics, some lines gave rise to both silenced and unsilenced progeny, more frequently in the 4n than the 2n populations; of the F5 GUS siblings derived from a 3n parent, a small number of 4n plants showed strong GUS inactivation, while this was not observed in the 2n plants of the same parent (the rest of the siblings showing active GUS expression; Figure S5); digestion of the bisulphite PCR product by MseI appeared to yield either fully cut or fully uncut DNA but not intermediate fragments (Figures 4A and 5H); and the HindIII digestion of genomic DNA also appeared to primarily give either fully cut or fully uncut but not intermediate hybridizing bands (Figures 3, B and C, and 5G). These appear to suggest that DNA from individual cells of a particular transgenic line is either fully unmethylated or fully methylated. Thus, the transgenic Arabidopsis lines may contain a mixture of transcriptionally active and transcriptionally inactive cells, and the overall transgene expression levels may reflect the percentage of the cells that are actively expressing the transgene.
Our results show that RNAi induced by antisense and hairpin RNA transgenes is less effective in 4n than in 2n Arabidopsis. This reduced RNAi effectiveness appears to be due to reduced expression of the silencing-inducer RNAs. With the EIN2 antisense transgene, no strongly silenced lines could be recovered from the 4n population. Furthermore, transgene-induced EIN2 silencing in the 2n lines was largely lost when the antisense transgene was introduced into the 3n background by crossing with wild-type 4n Arabidopsis. This strong effect of ploidy on antisense-induced silencing is probably due to the dependence of this silencing method on the level of antisense transgene expression.
With the two hpRNA constructs, strong RNAi of the two endogenous target genes could be achieved in both the 2n and 4n Arabidopsis, indicating that the RNAi technology can be successfully applied to knocking down gene expression in polyploidy plants. However, the proportion of plants showing strong RNAi was significantly lower in the 4n populations. Furthermore, comparing the strongly silenced 4n and 2n populations, the 4n lines appear to show a reduced intensity of target gene silencing. This result appears to be in conflict with a recent report indicating that hpRNA transgene-induced silencing is not affected by ploidy change (Pignatta et al. 2008). In their report the authors selected four single-copy 2n hpCHS lines with various degrees of CHS silencing, converted them into 4n plants by colchicine treatment, and then compared CHS silencing between the 2n and 4n populations. They detected no significant changes in the mRNA level of the target CHS gene or the accumulation of its biosynthetic product anthocyanin between the 2n lines and their respective 4n counterparts. The authors concluded that transgene-induced silencing is not affected by a change in ploidy. We cannot explain the discrepancy between our result and that of Pignatta et al. (2008), although some possible sources of differences may be identified. For one, Pignatta et al. (2008) did not examine the epigenetic status and expression of the hpCHS transgene, which could have undergone changes during colchicine-induced chromosome doubling but which were not sufficient to cause a significant change in target gene silencing. In our study, random transgenic populations were examined, which showed a clear difference between 2n and 4n populations primarily in the frequency of silencing, while in the reported study, the authors used selected single-copy lines that might be inherently stable in regard to transgene expression and transgene-induced target gene silencing. Ploidy-dependent epigenetic changes are often progressive (Adams and Wendel 2005), and it would be interesting to investigate whether the difference in hpRNA transgene-induced RNA silencing becomes more prominent in subsequent generations of the 2n and 4n lines in the two studies.
The negative effect of high ploidy on transgene expression and transgene-induced RNAi suggested by our study has practical implications. For instance, to obtain transgenic polyploid lines with high levels of transgene product or efficient transgene-induced RNAi, one might need to generate relatively large numbers of transgenic lines. To possibly make matters worse, a polyploid species might be particularly difficult to transform using conventional vectors because the selectable marker genes could be expressed poorly in the poplyploid background, resulting in inefficient selection of stable transformants.
Our study was carried out using a newly formed synthetic autotetraploid Arabidopsis line, where we observed ploidy-induced transcriptional repression similar to the previously reported HPT transgene silencing (Mittelsten Scheid et al. 1996, 2003). It remains to be investigated whether or not such ploidy-dependent reduction of transgene expression as well as transgene-induced RNAi occurs in naturally occurring autopolyploid plants or in allopolyploids, which comprise many of the important crop species. Rapid and widespread inactivation of endogenous genes has been reported to occur in newly formed allopolyploid Arabidopsis and wheat (Liu et al. 1998; Comai et al. 2000). Interestingly, genes that are inactivated due to allopolyploidy are often duplicated genes with repeated DNA elements (Comai et al. 2000; Adams and Wendel 2005), which, like transgenes, are often the target of epigenetic silencing in plants (Pikaard et al. 2008). This suggests that not only are transgene expression and transgene-induced RNAi generally affected by ploidy changes, but transgenes could become a useful tool for investigating ploidy-induced changes of endogenous gene expression.
Acknowledgments
We thank Limin Wu, Jing Chen, Sameer Tiwari, and Wenwu Guo for technical assistance; Ian Greaves for providing bisulphite PCR primers for the chloroplast DNA; Carl Davies for photography, and Peter Waterhouse and Jean Finnegan for support and helpful discussions. We would also like to thank the two anonymous reviewers for their constructive comments. T.E.F. and D.S were funded by Commonwealth Scientific and Industrial Research Organization summer studentships; M.B.W. was partly supported by an Australian Research Council future fellowship (FT0991956).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124370/DC1.
References
- Adams, K. L., and J. F. Wendel, 2005. Polyploid and genome evolution in plants. Curr. Opin. Plant Biol. 8 135–141. [DOI] [PubMed] [Google Scholar]
- Baubec, T., H. Q. Dinh, A. Pecinka, B. Rakic, W. Rozhon, et al. 2010. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 22 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D., 2004. RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Benfey, P. N., and N. H. Chua, 1990. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250 959–966. [DOI] [PubMed] [Google Scholar]
- Bhullar, S., S. Datta, S. Advani, S. Chakravarthy, T. Gautam, et al. 2007. Functional analysis of cauliflower mosaic virus 35S promoter: re-evaluation of the role of subdomains B5, B4 and B2 in promoter activity. Plant Biotechnol. J. 5 696–708. [DOI] [PubMed] [Google Scholar]
- Chen, S., C. A. Helliwell, L. M. Wu, E. S. Dennis, N. Upadhyaya et al., 2005. A novel T-DNA vector design conducive for selection of transgenic lines with simple integration and stable transgene expression. Funct. Plant Biol. 32 671–681. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Comai, L., A. P. Tyagi, K. Winter, R. Holmes-Davis, S. H. Reynolds et al., 2000. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell 12 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, J., and R. Scott, 1988. The isolation of plant nucleic acids, pp. 199–236 in Plant Genetic Transformation and Gene Expression: A Laboratory Manual, edited by J. Draper, R. Scott, P. Armitage and R. Walden. Alden Press, Oxford, UK.
- Eamens, A., M. B. Wang, N. A. Smith and P. M. Waterhouse, 2008. RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol. 147 456–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro, A.F., L. Matthew, N. A. Smith, S. J. Curtin, J. Dedic-Hagan et al., 2006. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7 1168–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A. P., 1992. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20 1203–1207. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J., and J. J. Rossi, 2004. Unlocking the potential of the human genome with RNA interference. Nature. 431 371–378. [DOI] [PubMed] [Google Scholar]
- Henry, I. M., B. P. Dilkes and L. Comai, 2007. Genetic basis for dosage sensitivity in Arabidopsis thaliana. PLoS Genet. 3 e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzl, J., A. M. Foerster, G. Raidl and O. Mittelsten Scheid, 2007. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51 526–536. [DOI] [PubMed] [Google Scholar]
- Ho, L., Y. Wan, J. M. Widholm and A. L. Rayburn, 1990. The use of stomatal chloroplast number for rapid determination of ploidy level in maize. Plant Breed. 105 203–210. [Google Scholar]
- Jacobs, J., and J. I. Yoder, 1989. Ploidy levels in transgenic tomato plants determined by chloroplast number. Plant Cell Rep. 7 662–663. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., T. A. Kavanagh and M. W. Bevan, 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levvy, 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B., J. M. Vega and M. Feldman, 1998. Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41 535–542. [DOI] [PubMed] [Google Scholar]
- Matzke, M. A., W. Aufsatz, T. Kanno, M. F. Mette and A. J. Matzke, 2002. Homology-dependent gene silencing and host defense in plants. Adv. Genet. 46 235–275. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., L. Jakovleva, K. Afsar, J. Maluszynska and J. Paszkowski, 1996. A change of ploidy can modify epigenetic silencing. Proc. Natl. Acad. Sci. USA 93 7114–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelsten Scheid, O., K. Afsar and J. Paszkowski, 2003. Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34 450–454. [DOI] [PubMed] [Google Scholar]
- Pignatta, D., B. Dilkes, T. Wroblewski, R. W. Michelmore and L. Comai, 2008. Transgene-induced gene silencing is not affected by a change in ploidy level. PLoS One 3 e3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard, C.S., J. R. Haag, T. Ream and A. T. Wierzbicki, 2008. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 13 390–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, G., D. Garrick, W. Wu, M. Kearns, D. Martin et al., 1995. Position-dependent variegation of globin transgene expression in mice. Proc. Natl. Acad. Sci. USA 92 5371–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, D., B. Lechtenberg, A. Forsbach, M. Gils, S. Bahadur et al., 2004. Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16 2561–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G.L., 1966. Chromosomal variation and evolution. Science 152 1463–1469. [DOI] [PubMed] [Google Scholar]
- Wang, J., L. Tian, A. Madlung, H. S. Lee, M. Chen et al., 2004. Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167 1961–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., N. A. Smith, E. S. Dennis, P. M. Waterhouse, P. J. Unrau et al., 2008. Synthesis of complementary RNA by RNA-dependent RNA polymerases in plant extracts is independent of an RNA primer. Funct. Plant Biol. 35 1091–1099. [DOI] [PubMed] [Google Scholar]
- Wang, M.B., and P. M. Waterhouse, 2002. Application of gene silencing in plants. Curr. Opin. Plant Biol. 5 146–150. [DOI] [PubMed] [Google Scholar]
- Wang, M. B., Z. Y. Li, P. R. Matthews, N. M. Upadhyaya and P. M. Waterhouse, 1998. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hort. 461 401–407. [Google Scholar]
- Wang, M. B., V. Wesley, E. J. Finnegan, N. A. Smith and P. M. Waterhouse, 2001. Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA 7 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V., C. A. Helliwell, N. A. Smith, M. B. Wang, D. T. Rouse et al., 2001. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Xu, Y., L. Zhong, X. Wu, X. Fang and J. Wang, 2009. Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta 229 471–483. [DOI] [PubMed] [Google Scholar]
- Yu, Z., G. Haberer, M. Matthes, T. Rattei, K. F. Mayer et al., 2010. Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107 17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]