Abstract
Early embryogenesis in Drosophila melanogaster is controlled by maternal gene products, which are deposited in the egg during oogenesis. It is not well understood how maternal gene expression is controlled during germline development. pipsqueak (psq) is a complex locus that encodes several nuclear protein variants containing a PSQ DNA-binding domain and a BTB/POZ domain. Psq proteins are thought to regulate germline gene expression through epigenetic silencing. While psq was originally identified as a posterior-group gene, we show here a novel role of psq in embryonic terminal patterning. We characterized a new psq loss-of-function allele, psqrum, which specifically affects signaling by the Torso (Tor) receptor tyrosine kinase (RTK). Using genetic epistasis, gene expression analyses, and rescue experiments, we demonstrate that the sole function impaired by the psqrum mutation in the terminal system is an essential requirement for controlling transcription of the tor gene in the germline. In contrast, the expression of several other maternal genes, including those encoding Tor pathway components, is not affected by the mutation. Rescue of the psqrum terminal phenotype does not require the BTB/POZ domain, suggesting that the PSQ DNA-binding domain can function independently of the BTB/POZ domain. Our finding that tor expression is subject to dedicated transcriptional regulation suggests that different maternal genes may be regulated by multiple distinct mechanisms, rather than by a general program controlling nurse-cell transcription.
THE early steps of Drosophila embryonic development are under the control of a set of products supplied by the mother to the oocyte. The genes encoding these products are specifically transcribed in the female germline, which consists of cysts of 16 cells, the oocyte plus 15 auxiliary nurse cells. Thereafter, their RNAs or proteins are transported from the nurse cells into the oocyte (reviewed in Spradling 1993). However, while the role of these maternal genes is fundamental for early development, not much is known about how their expression is regulated.
Among those maternal products is a set of determinants responsible for the spatially restricted activation of early zygotic genes, which dictate the broad subdivisions of the future organism. Four different maternal systems are involved in setting up the body pattern of the embryo along the anterior–posterior and dorsoventral axes. One of these, the terminal system, is responsible for the specification of the terminal regions at both poles of the embryo (reviewed in Duffy and Perrimon 1994; Furriols and Casanova 2003). A central element in the terminal system is the product of the torso (tor) gene, a receptor tyrosine kinase (RTK) (Sprenger et al. 1989).
tor is transcribed in the nurse cells and its RNA is deposited in the oocyte, where it is thought to remain untranslated until fertilization. Upon translation, Tor protein accumulates ubiquitously at the blastoderm surface. However, it is activated only at the blastoderm poles by a mechanism that appears to be triggered by the localized cleavage of its ligand, the protein Trunk (Casanova et al. 1995; Casali and Casanova 2001). Upon activation, Tor triggers the Ras/Raf/MAPK signaling pathway, which downregulates a repressor complex containing the HMG domain protein Capicua (Cic) and the corepressor Groucho (Gro) (Paroush et al. 1997; Jimenez et al. 2000). As a result, two zygotic genes, tailless (tll) and huckebein (hkb), are specifically expressed at both embryonic poles. Mutations in components of the Tor signaling pathway give rise to embryos lacking the terminal regions (the terminal phenotype), whereas constitutive activation of the pathway leads to the opposite phenotype, in which terminal cell fates expand into central regions of the embryo (Klingler et al. 1988; Casanova and Struhl 1989; Furriols and Casanova 2003).
Not much is known about how the spatiotemporal patterns and levels of expression of the maternal genes are regulated. Here we describe a new role of pipsqueak (psq) in regulating tor transcription. psq is a widely expressed gene that by alternative splicing gives rise to several protein variants that share a PSQ DNA-binding motif (Weber et al. 1995; Horowitz and Berg 1996; Lehmann et al. 1998). Many psq mutant alleles have been recovered that show distinct embryonic and adult phenotypes. Our results now show that a particular set of psq allelic combinations gives rise to embryos with terminal defects. We demonstrate that these defects are due to a requirement of psq for tor expression in the germline. The specific transcriptional regulation of tor points to multiple and distinct regulatory mechanisms for different maternal genes, rather than a general mechanism for the regulation of nurse-cell transcription.
MATERIALS AND METHODS
Genetics:
The mutagenesis screen was previously described (Luschnig et al. 2004). The following fly stocks were used and are listed in FlyBase: dec1, FRTG13 dec+, hs-Flp122, psq0115, psq8109, psqD91, psqE34, psqE39, psqEP2011, psqF112, psqfs1, psqHK38, psqKG02404, psqKG00811, psqrev2, psqrev4, psqrev7, psqrev9, psqrev12, psqrev14, and torY9. The psqrum mutation was mapped to the cytological interval 47A−47B14 on the basis of noncomplementation of Df(2R)stan1, Df(2R)stan2, Df(2R)12, Df(2R)E3363, and Df(2R)47A and complementation of Df(2R)X1, Df(2R)X3, and Df(2R)en-A. The torY9 psqrum chromosome was generated by meiotic recombination. Double mutant psqrum; cic1 embryos were generated by inducing FRT82B cic1 homozygous germline clones (Chou and Perrimon 1996) in psqrum homozygous females. To rescue the psqrum mutation, males carrying either UASp-psq-1 or UASp-psq-2 and psqrum were crossed to females carrying psqrum and mat-α-tub67C-Gal4-VP16 (Hacker and Perrimon 1998). From this cross, females of the genotype UASp-psq; psqrum/psqrum; mat-α-tub67C-Gal4-VP16/+ were collected and their embryonic progeny were analyzed in cuticle preparations. The rescue experiment with UASp-tor was done accordingly, except that nos-Gal4-VP16 (Van Doren et al. 1998) was used as a Gal4 driver. Marked psqrum follicle cell clones were generated using the defective chorion method (Nilson and Schupbach 1998).
Cuticle preparations:
Cuticle preparations were done as previously described (Luschnig et al. 2004).
Whole-mount in situ hybridization:
In situ hybridizations were performed using a standard protocol (Tautz and Pfeifle 1989). Digoxigenin–UTP-labeled antisense RNA probes were prepared from hkb, tll, and tor cDNAs. In situ hybridizations of wild-type and mutant embryos or ovaries were developed in parallel.
Antibody stainings:
Stainings were performed using standard protocols. The following antibodies were used: rabbit anti-Cic (Jimenez et al. 2000), mouse anti-HA (12CA5; Roche), mouse anti-dpERK (Cell Signaling Technologies), anti-β-galactosidase (559761; Cappel), and rabbit anti-Vasa (Gilboa and Lehmann 2004). Secondary antibodies were anti-rabbit Cy2, anti-mouse Cy3, and anti-rabbit-Cy5 at 1/300 (Jackson ImmunoResearch).
Microscopy:
Dark field photographs were taken using a Nikon Eclipse 80i microsocope with a Nikon digital camera DXM 1200F. Confocal images were obtained with a Leica SPE or an Olympus FV 1000 microscope.
Molecular characterization of the psqrum mutation:
Coding exons, exon–intron junctions, and 5′- and 3′-UTRs of the psq gene were sequenced in genomic DNA from psqrum homozygous flies and from flies carrying the parental chromosome (FRTG13 c px sp; Luschnig et al. 2004). Fragments of 400–600 bp were amplified and sequenced using various primers to obtain full multiple coverage of each fragment. Total RNA from mutant and wild-type ovaries was isolated using Trizol (Invitrogen). cDNA was synthesized using the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas), using both random hexamer primers and oligo-dT primers. We amplified cDNA from wild-type and mutant ovaries (and genomic DNA as a control) using primers located in the exons flanking the mutation. Fragments were purified, cloned into pGEM-T (Promega), and sequenced.
Real-time PCR:
Real-time PCR was performed with a Roche LightCycler 480 using the SYBR Green I method according to the manufacturer's instructions. Primers for tor, grk, ras, fs(1)ph, and for the rp49, alpha-Tubulin-84B, and act5C controls were designed on exon–exon junctions using Primer Express software (Applied Biosystems). We extracted total RNA from ovaries and prepared cDNAs using random hexamer primers. The nature of the PCR products was confirmed by melting curve analysis. All analyses were performed using the Relative Expression software tool (Pfaffl et al. 2002). Main steps of the automatic REST workflow are as follows: PCR efficiencies were calculated for every pair of primers by generating standard curves at increasing dilutions of cDNA (1:1, 1:5, 1:25, 1:125, and 1:625) and were used to correct raw data. rp49, alpha-tubulin-84B, and act5C were assumed to be equally expressed in wild-type and mutant ovaries and were used for normalization. A ratio between the normalized signals of tested genes in mutant and wild type was calculated and expressed as fold increase/decrease and statistically tested by a bootstrap test (10,000 randomizations).
Transgenic constructs:
UASp-psq-1 was generated by inserting the psq-1 cDNA (including 5′- and 3′-UTR sequences) as a NotI fragment from pHPT7-9 (Horowitz and Berg 1996) into pUASp (Rorth 1998). UASp-psq-2 was generated by amplifying the coding sequence of Psq-2 (corresponding to amino acids 420–1061 of Psq-1) from UASp-psq-1 using oligonucleotides Psq2-NotI-For (5′-tataGCGGCCGCatgactagtttaggcatgg) and pUASp-Rev (5′-tcaagctcctcgagttaacg). The PCR fragment was cut with NotI and inserted into pUASp. The resulting fragment retains the psq 3′-UTR, but lacks psq 5′-UTR sequences. The PCR-amplified part of the construct was confirmed by DNA sequencing. The tor–LacZ construct was generated by inserting a NotI genomic fragment of 3.5 kb from the tor promoter described in Furriols et al. (1998) into the C4PLZ transformation vector (Wharton and Crews 1993). UASp-tor was generated by cloning the tor cDNA (RE49094; DGRC) into pUASp. Transgenic flies were generated by P-element transformation. At least two independent insertions were analyzed for each construct.
RESULTS
rumpf is a new factor involved in Tor signaling:
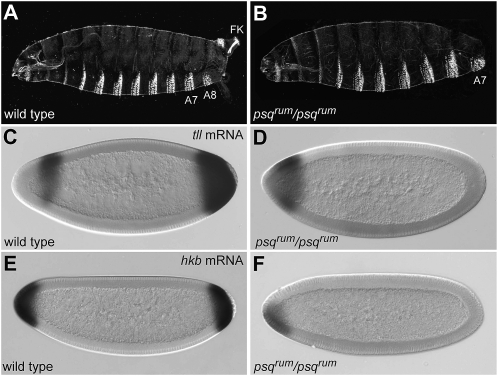
In a mutagenesis screen for new maternal genes (Luschnig et al. 2004) we found that embryos from 2R-225-5 homozygous mutant germline clones lacked cuticle structures derived from the anterior and posterior poles of the embryo (Figure 1, A and B), resembling the phenotype of terminal-class mutations. On the basis of this phenotype we named the locus rumpf (rum; German for “torso”). Homozygous rum flies show no obvious defects, but homozygous females produce embryos with terminal defects resembling those of rum germline clone-derived embryos. Although the cuticle phenotype of embryos lacking maternal rum function (from here on referred to as rum embryos) was somewhat variable and often showed deletions of one or more abdominal segments, all embryos completely lacked terminal structures (n = 100). The development of these structures depends on the terminal gap genes tll and hkb, whose expression at the posterior pole is solely dependent on Tor signaling. Posterior expression of tll and hkb was abolished in rum embryos, reminiscent of the phenotype of mutants lacking Tor signaling (Figure 1, C–F). These results indicate that activity of the Tor signaling pathway is severely compromised or completely abolished in rum embryos.
Figure 1.—
psqrum impairs embryonic terminal development. (A and B) Cuticle phenotype of embryos from wild-type and psqrum homozygous females. psqrum embryos lack terminal structures (A8, abdominal segment 8; FK, Filzkörper). (C–F) Expression patterns of tailless (tll; C and D) and huckebein (hkb; E and F) in wild-type (C and E) and psqrum (D and F) early embryos. Expression patterns in psqrum mutants are reminiscent of those in embryos lacking Tor signaling.
rum is required for neither Tor receptor activation nor tll and hkb transcription:
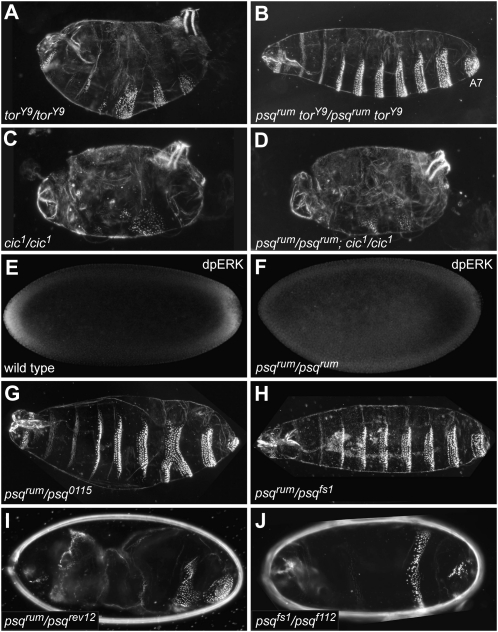
We asked at which level rum is interfering with the Tor pathway. The function of rum could be in either generating the terminally restricted signal in the somatic follicle cells or the transmission or interpretation of the signal. However, the germline-specific requirement excludes that the rum mutation affects a follicle cell function needed for Tor signaling. Consistent with this notion, large clones of rum mutant follicle cells do not affect terminal patterning of the embryo (supporting information, Figure S1). We asked whether rum acts upstream or downstream of the Tor RTK by testing epistatic relationships between rum and tor. Embryos from homozygous females carrying the tor gain-of-function mutation torY9 show expanded terminal regions at the expense of central body regions, as indicated by the deletion of several abdominal segments (Figure 2A). In contrast, all embryos from homozygous torY9 rum double mutant females show terminal defects resembling the rum single mutant phenotype (Figure 2B; n = 50). Thus, rum is epistatic to torY9.
Figure 2.—
psqrum acts upstream of MAPK and downstream of or at the level of the Tor receptor. (A) Embryos from torY9 homozygous females display ectopic activation of the Tor receptor as indicated by the deletion of several abdominal segments. (B) In contrast, embryos from torY9 psqrum double mutant females show terminal defects resembling the psqrum single mutant phenotype. (C) Embryos from cic1 homozygous females show a tor gain-of-function phenotype. (D) This phenotype is not modified in embryos from psqrum; cic1 homozygous germline clones. (E) MAPK activation is detected at the poles in wild-type embryos by a specific antibody that recognizes its diphosphorylated form (dpERK). (F) Such activation is not detected in psqrum mutants. (G–I) Heteroallelic combinations of psqrum with other psq alleles also give rise to terminal phenotypes. (J) A combination of other psq alleles besides psqrum also produces embryos showing terminal defects in addition to a lack of most abdominal segments.
In the absence of Tor signaling, a repressor complex containing Cic inhibits tll and hkb expression. In the absence of cic function, tll and hkb are derepressed and their respective expression domains extend into the middle of the embryo, resulting in a tor gain-of-function phenotype (Figure 2C; Jimenez et al. 2000). We observed the same tor gain-of-function phenotype in all embryos derived from rum; cic1 double mutant germline clones (Figure 2D; n = 40). Thus, cic is epistatic to rum. Finally, while MAPK/ERK phosphorylation is detected at the poles in the wild type (Gabay et al. 1997), it is not detected in rum embryos (Figure 2, E and F). Consequently, degradation of Cic protein at the poles is also abolished in rum embryos (data not shown). Altogether, these results indicate that rum acts upstream of MAPK and downstream of or at the level of the Tor receptor.
rum is a mutation in the psq locus:
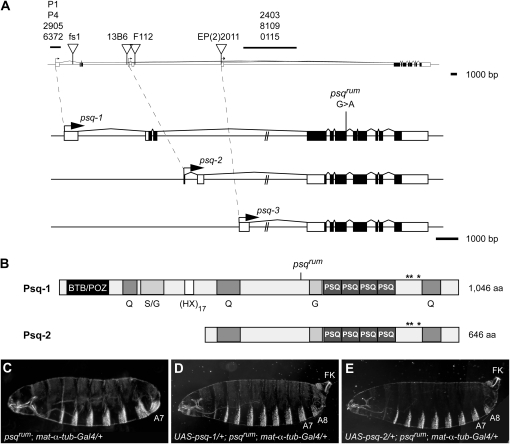
We mapped the rum mutation to the cytological interval 47A−47B14. Within this interval, two P-element insertions, EP(2)2011 and KG2404, failed to complement female sterility of the rum mutation. The two P-elements are inserted in the 48-kb spanning intron 3 of the psq gene (Figure 3A; Spradling et al. 1999; Bellen et al. 2004). psq is a large and complex locus, which, through the use of three alternative promoters and alternative splicing, gives rise to at least 12 different transcripts (FlyBase; Weber et al. 1995; Horowitz and Berg 1996; Ferres-Marco et al. 2006). The psq locus was originally identified on the basis of its maternal-effect posterior group phenotype (Schupbach and Wieschaus 1991; Siegel et al. 1993). However, several additional psq alleles show distinct embryonic or adult phenotypes. psq mutations include lethal alleles (0482; Horowitz and Berg 1996), semilethal alleles that cause rudimentary ovaries and a defect in R3/R4 photoreceptor specification (F112, E34, E39, D91; Weber et al. 1995), and viable female-sterile alleles causing defects in abdominal and dorsoventral patterning and in pole cell formation (fs1, I-30, HK38, 2403, 2905, 6372, P1, P4, and X1-30; Schupbach and Wieschaus 1991; Siegel et al. 1993; Horowitz and Berg 1996). However, a role of psq in terminal patterning has not been described. Complementation tests between rum and other psq alleles (summarized in Table S1) revealed complex interactions. Heteroallelic combinations between four psq alleles (fs1, 0115, KG02404, and rev12) and the rum mutation gave rise to embryonic terminal defects of variable strength and penetrance (Figure 2, G–I; Table S1). Interestingly, certain allelic combinations give rise to embryos that arrest early in development, while other alleles complement female sterility of the rum mutation. In addition, we found that embryos from transheterozygous females carrying a combination of two other recessive psq mutations (psqfs1/psqF112) also show terminal defects (Figure 2J). The complex genetic behavior of psq mutations suggests that the different Psq isoforms might mediate distinct functions during oogenesis and embryogenesis and that rum is a new psq allele that affects a requirement of psq in the Tor pathway.
Figure 3.—
psqrum is a mutation in the psq locus. (A) Genomic organization of the psq locus with P-element insertions indicated by triangles, or by horizontal bars for approximate positions, are shown on top. The three major classes of psq transcripts originating from the three alternative promoters are shown below. Coding exons are shown as solid boxes; noncoding exons and untranslated regions are shown as open boxes. The splice site mutation in psqrum is indicated. (B) Domain structure of two main Psq protein isoforms, Psq-1 and Psq-2. Note that only the long isoform Psq-1 contains the BTB/POZ domain, whereas both long and short isoforms contain the PSQ DNA-binding domain. Regions rich in glutamine (Q), glycine (G), or serine and glycine (S/G), and a repeat of 17 dipeptides of the structure (HX)17 are indicated. Asterisks denote potential MAPK phosphorylation sites (PXT/SP). The psqrum mutation is predicted to result in truncation of all Psq protein isoforms at the position marked by the vertical line above the Psq-1 protein. (C–E) Germline-specific expression of psq cDNAs rescues the terminal defects of psqrum embryos. A control psqrum embryo (C) lacks terminal structures, whereas expression of either psq-1 (D) or psq-2 (E) under the control of maternal Gal4 restores the formation of terminal structures. Note that maternal expression of psq-1 in psqrum embryos yielded a hatch rate of 90% (n = 200), while psq-2 expression yielded a hatch rate of 49% (n = 190). The unhatched embryos show variable terminal defects (data not shown).
Finally, to conclude whether the phenotype associated with the rum mutation is due to a loss of psq function, we generated transgenic flies expressing the longest psq transcript variant (psq-1; Figure 3; Horowitz and Berg 1996). Expression of this construct in the germline of rum females rescued the rum terminal defects in embryos (Figure 3, C and D). The majority (90%, n = 200) of the embryos hatched and developed into fertile adults. These results indicate that the terminal defects in rum embryos are due to a requirement of psq in the germline. We therefore refer to the rum mutation as psqrum from here on.
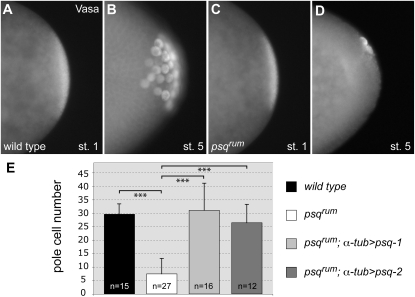
psqrum impairs pole cell formation:
psq was proposed to regulate posterior patterning and pole plasm formation through promoting the expression of Vasa (Vas) protein (Siegel et al. 1993). As for other psq alleles, psqrum embryos showed a severely reduced number of primordial germ cells (pole cells) (Figure 4). Wild-type embryos had on average 29.6 ± 3.75 (n = 15) Vas-positive pole cells, whereas psqrum embryos had only 7.4 ± 5.75 (n = 27) pole cells, with some embryos completely lacking pole cells. However, the mild posterior-patterning defects in psqrum embryos suggest that vas function in the mutants is nearly sufficient for normal abdominal patterning.
Figure 4.—
psqrum is required for pole cell formation. (A–D) Immunostaining with an anti-Vas antibody in embryos from wild-type (A and B) and psqrum (C and D) females. While Vas levels in early psqrum mutants appear similar to those of wild-type embryos, the number of pole cells is strongly reduced. (E) Quantification of reduction of pole cells in psqrum mutants. Expression of either psq-1 or psq-2 in the maternal germline restores pole cell formation in psqrum embryos. Mean pole cell numbers are not significantly different between psq-1 and psq-2 expressing psqrum embryos, as indicated by Student's t-test (P > 0.1). Error bars represent mean + SD. ***P < 0.001.
Lack of terminal structures and pole cells in psqrum can be rescued by a Psq isoform lacking the BTB/POZ domain:
The psq locus encodes two major classes of proteins with different N termini (Figure 3B; Horowitz and Berg 1996). The long isoform Psq-1 (1046 aa) contains an N-terminal BTB/POZ domain and four repeats of the PSQ helix-turn-helix DNA-binding motif (Schwendemann and Lehmann 2002; Siegmund and Lehmann 2002). While the BTB/POZ domain provides a platform for protein–protein interactions (Perez-Torrado et al. 2006), the Psq repeats mediate binding to a GAGAG consensus DNA motif in vitro (Lehmann et al. 1998; Siegmund and Lehmann 2002). The short isoform Psq-2 (646 aa) is encoded by psq-2 and psq-3 class transcripts and lacks the N-terminal part of the protein harboring the BTB/POZ domain. However, the Psq repeats are common to all known Psq isoforms. To address whether Psq-1 and Psq-2 isoforms exhibit distinct functions with respect to Tor signaling, we generated a UAS-psq-2 transgene lacking the BTB/POZ domain. Germline-specific expression of this construct rescued the terminal defects in 49% (n = 190) of psqrum embryos (Figure 3E). While these embryos hatched and were morphologically normal, terminal patterning in the remaining unhatched embryos (51%, n = 190) was only partially rescued (data not shown). We cannot exclude that the partial rescuing activity of psq-2 transgenes may be due to differences in expression levels between UAS-psq-1 and UAS-psq-2 constructs. However, maternal expression of either UAS-psq-1 or UAS-psq-2 restored pole cell formation in psqrum embryos (Figure 4E). These findings suggest that the BTB/POZ domain is not strictly required for psq function in terminal signaling and pole cell formation. However, we cannot rule out that the transgenic Psq-2 protein might interact with and stabilize residual wild-type Psq proteins in psqrum mutants and thereby mediate the rescue in an indirect fashion.
The psqrum mutation alters the proper splicing of psq mRNAs:
To identify the molecular lesion that causes the psqrum mutation, we sequenced the annotated psq coding region, 5′- and 3′-untranslated regions (UTRs), and the reported intron/exon boundaries. We did not detect any change in the coding sequence and the putative regulatory regions analyzed. However, we found a G-to-A transition affecting a splice donor site at the junction between exon 6 and the following intron. By sequencing RT–PCR-amplified cDNA fragments, we confirmed that splicing at this site occurs in wild-type ovaries (Figure 5). The splicing is abnormal in psqrum mutants, as a new transcript is produced that escapes splicing and retains the mutated intron. However, splicing is not absolutely impaired in the mutant, as the wild-type variant is also detected (Figure 5). The new reading frame in the abnormal transcripts contains a stop codon 16 aa downstream of the nonspliced exon/intron junction. If translated, the resulting Psq proteins would be truncated and would lack the entire DNA-binding domain. Importantly, as all known psq transcripts share the mutated region, the mutation is likely to affect all Psq protein isoforms. Although the truncated Psq proteins could act in a dominant negative fashion, we consider more likely and consistent with the genetic data that psqrum is a loss-of-function mutation (see discussion). Thus, the psqrum phenotype appears to be caused by a reduction of the normal levels of one or more of the wild-type psq transcripts due to the aberrant splicing.
Figure 5.—
The psqrum mutation alters the proper splicing of psq mRNAs. Products from three independent PCRs from wild-type genomic DNA and RT–PCR from wild-type and psqrum ovaries with primers F and R at exons 6 and 8 of the psq gene. A genomic fragment of 622 bp and two cDNA fragments of ∼450 bp are detected in the wild type. Additional cDNA fragments of ∼600 bp are obtained from psqrum mutants. The diagram indicates the organization of the psq gene and the PCR products expected from amplification with primers F and R in conditions of normal and aberrant splicing. The point mutation in psqrum is indicated by an asterisk. The G in the splice donor site of the intron following exon 6 (in boldface type in the following sequence: CATATGGgtgagttg; intron sequence is lowercase) is mutated to A. Numbering of exons corresponds to their appearance in the psq–RA transcript variant (FlyBase).
psq is required for normal expression of tor mRNA:
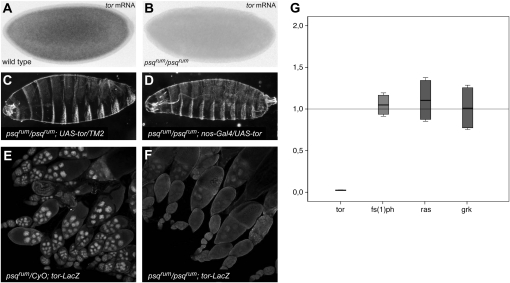
Since the psq locus encodes nuclear DNA-binding proteins, psq could influence Tor signaling by regulating the expression of one or more components of the pathway. On the basis of our genetic results, psq could be required for the expression of any of the genes that encode components acting between and including the Tor receptor and MAPK, i.e., Tor, Sos, Ras1, Raf, Dsor, and MAPK. The adaptor proteins Drk, Dos, and Dshc are less likely candidates, since mutation of any one of the corresponding genes does not lead to a complete loss of signaling activity, as observed in psqrum embryos (Luschnig et al. 2000). We measured steady-state transcript levels of ras1, raf, and dsor in wild-type and in psqrum ovaries by semiquantitative RT–PCR and found that they are detectable at apparently normal levels (data not shown). To analyze tor mRNA levels we performed quantitative real-time RT–PCR and found a strong reduction of tor mRNA levels in psqrum ovaries (46.7-fold mean reduction; P < 0.01; Figure 6G). Consistent with these results, strongly reduced tor mRNA levels were detected by in situ hybridization in psq embryos (psqrum and psqfs1/psqF112; Figure 6, A and B; Figure S2) and ovaries (psqfs1/psq0115; psqrev2/psq0297; and psqrum/psqrev4; data not shown). Conversely, transcript levels of gurken, fs(1)polehole, and ras1 were not significantly different between wild-type and psqrum ovaries (Figure 6G).
Figure 6.—
psqrum is required for normal expression of tor in the germline. (A and B) tor in situ hybridization in wild-type embryo (A) and psqrum mutant (B). In psqrum mutants, tor mRNA levels are strongly reduced. (C and D) GAL4-driven expression of tor is sufficient to rescue the terminal phenotype in a psqrum homozygous background. (E) A tor–lacZ construct gives rise to LacZ protein in the nurse cells (note nuclear accumulation because of a nuclear localization signal in the construct). (F) LacZ accumulation from the tor–lacZ construct is abolished in homozygous psqrum ovaries. (G) tor mRNA is 46.7-fold reduced in psqrum ovaries compared to the wild type, as detected by real-time PCR. In contrast, mRNA levels of fs(1)ph, ras, and grk are not affected. The y-axis indicates relative expression levels as fold change between wild-type and psqrum ovaries.
A further confirmation that Tor is the critical component of the pathway affected in psqrum mutants comes from the observation that tor expression driven by the Gal4/UAS system (Brand and Perrimon 1993) is sufficient to rescue the terminal phenotype of psqrum mutants (Figure 6, C and D). Importantly, this indicates that none of the remaining signaling components downstream of the Tor receptor are significantly impaired in psqrum mutants. Finally, to support that psqrum alters tor at the transcriptional level, we analyzed the expression of a lacZ reporter gene under the control of the tor promoter. While the reporter is clearly detected in the nurse cells in wild-type ovaries, no signal was detected in ovaries from psqrum females (Figure 6, E and F). Altogether, these data point to a specific and essential role of psq in the regulation of tor transcription during oogenesis.
DISCUSSION
We report here a particular role of psq in the regulation of tor transcription in the Drosophila germline. However, only some psq mutations show a terminal phenotype. In addition, these results also highlight a distinct mode of transcriptional control of the tor gene, different from other maternal genes, including those that encode other members of the Tor signaling pathway. Below we discuss these issues and their implications.
The role of psq in tor transcriptional regulation:
psq has functions in many developmental stages and presumptive null mutants are lethal (Weber et al. 1995; Horowitz and Berg 1996). Here, we report that a set of psq mutations unveils a specific role in tor transcription. Why is this phenotype only observed associated with these particular psq mutations? Among the psq alleles that allow adult survival, strong mutations block oogenesis at early stages (Siegel et al. 1993; Weber et al. 1995; Horowitz and Berg 1996). Thus, in those cases, an early requirement in oogenesis would mask a later requirement for tor transcription. Psq proteins are present in multiple isoforms (Weber et al. 1995; Horowitz and Berg 1996; Ferres-Marco et al. 2006). Only a few psq alleles have been molecularly characterized and among those many are due to transposon insertions (Weber et al. 1995; Horowitz and Berg 1996; Spradling et al. 1999; Schwendemann and Lehmann 2002; Thibault et al. 2004). Therefore, it is difficult on the basis of the molecular analysis of the psq mutations to assign distinct functions to the isoforms generated by the different transcripts.
A number of reasons argue for the psqrum mutation unveiling a physiological function of psq in tor transcription, rather than the rum phenotype being a neomorphic effect caused by a special truncated Psq protein. First, the terminal phenotype of the psqrum mutation is observed in homozygosity, as well as in trans-heterozygous combinations with several other psq loss-of-function alleles. Second, the terminal phenotype of the psqrum mutation arises in association with a mild posterior phenotype, a well-known psq loss-of-function phenotype. Third, a transheterozygous combination of other psq loss-of-function alleles also produces embryos showing reduced tor expression and terminal defects. And finally, expression of psq rescues the psqrum terminal phenotype.
The psqrum mutation causes a decrease in the wild-type splicing at one specific site. Nevertheless, since all known isoforms share this splicing, we cannot infer whether a particular isoform is responsible for tor transcriptional regulation. However, the rescue experiments indicate that both a long Psq isoform containing the BTB/POZ domain and a short isoform lacking this domain are capable of providing the psq function controlling tor transcription that is missing in psqrum mutants. Thus, on the one hand, the BTB/POZ domain appears to be dispensable for this psq function, but on the other hand, a long isoform can substitute for a short isoform, arguing against separate functions of these psq isoforms in the context of Tor signaling. Thus, an overall decrease of many psq isoforms in the psqrum mutant could be affecting tor transcription.
In this regard, it is worth considering together the terminal and posterior defects associated with the psqrum mutation. tor is affected more strongly than vas in psqrum mutants, while the opposite is true for other psq mutants (e.g., psqHK38, psq2403, and psqfs1; Schupbach and Wieschaus 1991; Siegel et al. 1993; Horowitz and Berg 1996), in which vas is strongly affected, but not tor, according to their cuticle phenotype. These data argue against a simple model in which tor and vas transcription would be impaired below different thresholds of psq activity.
As Psq is thought to repress gene expression through epigenetic silencing (Huang et al. 2002; Ferres-Marco et al. 2006), Psq could activate tor expression indirectly, through the repression of a still unidentified tor repressor. Alternatively, Psq could activate tor expression directly. Indeed, genetic interaction studies suggest that psq and Trithorax-like (Trl) act together in transcriptional activation as well as in transcriptional silencing of homeotic genes (Schwendemann and Lehmann 2002).
Distinct regulation of tor transcription in the germline:
Not much is known about how transcription is regulated in Drosophila nurse cells. One possibility is that spatially and temporally coexpressed genes share a common mode of transcriptional regulation. Indeed, enrichment of specific core motifs in the promoters of genes with female germline expression (Fitzgerald et al. 2006) is consistent with such a hypothesis. The multiple effects of psq mutations during oogenesis might argue for such a general role of psq. However, and in spite of psq's multiple requirements in the germline, tor transcription appears to be distinctly regulated. A similar case appears to apply to bcd transcription, which was found to be specifically controlled by Serendipity-δ (Sry-δ), a zinc finger protein. sry-δ null alleles block oogenesis, and only a particular genetic combination revealed the specific requirement of sry-δ for bcd transcription (Payre et al. 1994). Thus, both psq and sry-δ have a basic function in oogenesis, probably through the transcriptional control of other germline genes, and a specific function in the control of tor and bcd, respectively. This similarity is particularly intriguing considering the peculiarities of early Drosophila embryogenesis and the fact that anterior patterning by bcd seems to be restricted to Diptera (Casci 2000) and that tor-dependent terminal patterning appears in Diptera and Coleoptera, but not in Hymenoptera (Wilson and Dearden 2009). Thus, the regulation of bcd and tor transcription by a specific function of more general germline transcription factors might be related to their particular recruitment to embryonic patterning. Interestingly, in the case of terminal patterning, tor transcription appears to be regulated independently from that of genes encoding the other elements of the signaling pathway (e.g., Ras and Raf), as induced expression of tor is sufficient to rescue the psqrum terminal phenotype. However, the only essential function of these other elements of the Tor pathway in the oocyte is to transmit the Tor signal, as indicated by the phenotype of mutant germline clones (Ambrosio et al. 1989). Thus, in the absence of tor activity, these products appear to be silent in the Drosophila germline. Altogether, these data suggest a possible multiple-step way to acquire new regulatory mechanisms in a given set of cells. This possibility appears particularly suggestive in the light of recent results pointing to Tor as the receptor for prothoracicotropic hormone (PTTH), which stimulates the production of the molting hormone ecdysone (Rewitz et al. 2009). Could this be a more ancient role of tor that would subsequently have been recruited for embryonic terminal patterning in some insects? Such a scenario appears to apply to the Toll signaling pathway, which shares some similarities with the Tor pathway (Casanova et al. 1995), and has a widely conserved function in immunity in many animals (Leulier and Lemaitre 2008) and whose components are also transcribed by the Drosophila female germline to specify the embryonic dorsoventral pattern.
Acknowledgments
We thank Celeste Berg, Maria Dominguez, Ruth Lehmann, Marek Mlodzik, Ze'ev Paroush, Trudi Schüpbach, Alexander Schwendemann, Daniel St Johnston, and the Bloomington Stock Center for fly stocks and reagents. We thank Marta Casado, Jose de las Heras, Xavier Franch, and Nicolás Martín for help and experimental advice, and Michalis Averof for comments on the manuscript. We thank colleagues in our laboratories for discussion. S.L. thanks Christian Lehner for his support. Work in J.C.'s laboratory is supported by grants from the Generalitat de Catalunya and the Spanish Ministerio de Ciencia e Innovación (BFU2009-07629 and Consolider CSD2007-00008). Work in S.L.'s laboratory is supported by the Swiss National Science Foundation, the German Research Foundation, the Julius Klaus-Stiftung Zürich, and the Kanton Zürich.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.121624/DC1.
References
- Ambrosio, L., A. P. Mahowald and N. Perrimon, 1989. Requirement of the Drosophila raf homologue for torso function. Nature 342 288–291. [DOI] [PubMed] [Google Scholar]
- Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson et al., 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Casali, A., and J. Casanova, 2001. The spatial control of Torso RTK activation: a C-terminal fragment of the Trunk protein acts as a signal for Torso receptor in the Drosophila embryo. Development 128 1709–1715. [DOI] [PubMed] [Google Scholar]
- Casanova, J., and G. Struhl, 1989. Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev. 3 2025–2038. [DOI] [PubMed] [Google Scholar]
- Casanova, J., M. Furriols, C. A. McCormick and G. Struhl, 1995. Similarities between trunk and spatzle, putative extracellular ligands specifying body pattern in Drosophila. Genes Dev. 9 2539–2544. [DOI] [PubMed] [Google Scholar]
- Casci, T., 2000. Evo-devo. A head with no torso. Nat. Rev. Genet. 1 9. [DOI] [PubMed] [Google Scholar]
- Chou, T. B., and N. Perrimon, 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy, J. B., and N. Perrimon, 1994. The torso pathway in Drosophila: lessons on receptor tyrosine kinase signaling and pattern formation. Dev. Biol. 166 380–395. [DOI] [PubMed] [Google Scholar]
- Ferres-Marco, D., I. Gutierrez-Garcia, D. M. Vallejo, J. Bolivar, F. J. Gutierrez-Aviño et al., 2006. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439 430–436. [DOI] [PubMed] [Google Scholar]
- FitzGerald, P. C., D. Sturgill, A. Shyakhtenko, B. Oliver and C. Vinson, 2006. Comparative genomics of Drosophila and human core promoters. Genome Biol. 7 R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols, M., and J. Casanova, 2003. In and out of Torso RTK signalling. EMBO J. 22 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furriols, M., A. Casali and J. Casanova, 1998. Dissecting the mechanism of torso receptor activation. Mech. Dev. 70 111–118. [DOI] [PubMed] [Google Scholar]
- Gabay, L., R. Seger and B. Z. Shilo, 1997. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development 124 3535–3541. [DOI] [PubMed] [Google Scholar]
- Gilboa, L., and R. Lehmann, 2004. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr. Biol. 14 981–986. [DOI] [PubMed] [Google Scholar]
- Hacker, U., and N. Perrimon, 1998. DRhoGEF2 encodes a member of the Dbl family of oncogenes and controls cell shape changes during gastrulation in Drosophila. Genes Dev. 12 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz, H., and C. A. Berg, 1996. The Drosophila pipsqueak gene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development 122 1859–1871. [DOI] [PubMed] [Google Scholar]
- Huang, D.-H., Y.-L. Chang, C.-C. Yang, I.-C. Pan and B. King, 2002. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol. Cell. Biol. 22 6261–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, G., A. Guichet, A. Ephrussi and J. Casanova, 2000. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 14 224–231. [PMC free article] [PubMed] [Google Scholar]
- Klingler, M., M. Erdelyi, J. Szabad and C. Nusslein-Volhard, 1988. Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature 335 275–277. [DOI] [PubMed] [Google Scholar]
- Lehmann, M., T. Siegmund, K. G. Lintermann and G. Korge, 1998. The pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273 28504–28509. [DOI] [PubMed] [Google Scholar]
- Leulier, F., and B. Lemaitre, 2008. Toll-like receptors–taking an evolutionary approach. Nat. Rev. Genet. 9 165–178. [DOI] [PubMed] [Google Scholar]
- Luschnig, S., J. Krauss, K. Bohmann, I. Desjeux and C. Nusslein-Volhard, 2000. The Drosophila SHC adaptor protein is required for signaling by a subset of receptor tyrosine kinases. Mol. Cell 5 231–241. [DOI] [PubMed] [Google Scholar]
- Luschnig, S., B. Moussian, J. Krauss, I. Desjeux, J. Perkovic et al., 2004. An F1 genetic screen for maternal-effect mutations affecting embryonic pattern formation in Drosophila melanogaster. Genetics 167 325–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson, L. A., and T. Schupbach, 1998. Localized requirements for windbeutel and pipe reveal a dorsoventral prepattern within the follicular epithelium of the Drosophila ovary. Cell 93 253–262. [DOI] [PubMed] [Google Scholar]
- Paroush, Z., S. M. Wainwright and D. Ish-Horowicz, 1997. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development 124 3827–3834. [DOI] [PubMed] [Google Scholar]
- Payre, F., M. Crozatier and A. Vincent, 1994. Direct control of transcription of the Drosophila morphogen bicoid by the serendipity delta zinc finger protein, as revealed by in vivo analysis of a finger swap. Genes Dev. 8 2718–2728. [DOI] [PubMed] [Google Scholar]
- Perez-Torrado, R., D. Yamada and P. A. Defossez, 2006. Born to bind: the BTB protein-protein interaction domain. Bioessays 28 1194–1202. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M. W., G. W. Horgan and L. Dempfle, 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30 e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz, K. F., N. Yamanaka, L. I. Gilbert and M. B. O'Connor, 2009. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326 1403–1405. [DOI] [PubMed] [Google Scholar]
- Rorth, P., 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78 113–118. [DOI] [PubMed] [Google Scholar]
- Schupbach, T., and E. Wieschaus, 1991. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendemann, A., and M. Lehmann, 2002. Pipsqueak and GAGA factor act in concert as partners at homeotic and many other loci. Proc. Natl. Acad. Sci. USA 99 12883–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, V., T. A. Jongens, L. Y. Jan and Y. N. Jan, 1993. pipsqueak, an early acting member of the posterior group of genes, affects vasa level and germ cell-somatic cell interaction in the developing egg chamber. Development 119 1187–1202. [DOI] [PubMed] [Google Scholar]
- Siegmund, T., and M. Lehmann, 2002. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev. Genes Evol. 212 152–157. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., 1993. Germline cysts: communes that work. Cell 72 649–651. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., D. Stern, A. Beaton, E. J. Rhem, T. Laverty et al., 1999. The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger, F., L. M. Stevens and C. Nusslein-Volhard, 1989. The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature 338 478–483. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Thibault, S. T., M. A. Singer, W. Y. Miyazaki, B. Milash, N. A. Dompe et al., 2004. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36 283–287. [DOI] [PubMed] [Google Scholar]
- Van Doren, M., A. L. Williamson and R. Lehmann, 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8 243–246. [DOI] [PubMed] [Google Scholar]
- Weber, U., V. Siegel and M. Mlodzik, 1995. pipsqueak encodes a novel nuclear protein required downstream of seven-up for the development of photoreceptors R3 and R4. EMBO J. 14 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton, Jr., K. A., and S. T. Crews, 1993. CNS midline enhancers of the Drosophila slit and Toll genes. Mech. Dev. 40 141–154. [DOI] [PubMed] [Google Scholar]
- Wilson, M. J., and P. K. Dearden, 2009. Tailless patterning functions are conserved in the honeybee even in the absence of Torso signaling. Dev. Biol. 335 276–287. [DOI] [PubMed] [Google Scholar]