Abstract
The distribution of lipoprotein(a) [Lp(a)] levels can differ dramatically across diverse racial/ethnic populations. The extent to which genetic variation in LPA can explain these differences is not fully understood. To explore this, 19 LPA tagSNPs were genotyped in 7,159 participants from the Third National Health and Nutrition Examination Survey (NHANES III). NHANES III is a diverse population-based survey with DNA samples linked to hundreds of quantitative traits, including serum Lp(a). Tests of association between LPA variants and transformed Lp(a) levels were performed across the three different NHANES subpopulations (non-Hispanic whites, non-Hispanic blacks, and Mexican Americans). At a significance threshold of p<0.0001, 15 of the 19 SNPs tested were strongly associated with Lp(a) levels in at least one subpopulation, six in at least two subpopulations, and none in all three subpopulations. In non-Hispanic whites, three variants were associated with Lp(a) levels, including previously known rs6919246 (p = 1.18×10−30). Additionally, 12 and 6 variants had significant associations in non-Hispanic blacks and Mexican Americans, respectively. The additive effects of these associated alleles explained up to 11% of the variance observed for Lp(a) levels in the different racial/ethnic populations. The findings reported here replicate previous candidate gene and genome-wide association studies for Lp(a) levels in European-descent populations and extend these findings to other populations. While we demonstrate that LPA is an important contributor to Lp(a) levels regardless of race/ethnicity, the lack of generalization of associations across all subpopulations suggests that specific LPA variants may be contributing to the observed Lp(a) between-population variance.
Introduction
Lipoprotein (a) [Lp(a)] levels have long been recognized as an independent risk factor for coronary artery disease (CAD)[1]–[3]. However, Lp(a) concentrations and their relationship with cardiovascular disease vary across races/ethnicities. The most notable example of this discrepancy is observed between populations of European- and African-decent. While the mean Lp(a) level is two- to threefold higher in blacks relative to whites[4], [5], elevated plasma Lp(a) levels have been reported to be associated with CAD in whites but have not been clearly demonstrated in blacks[6]–[10].
The epidemiology of Lp(a) in other US racial/ethnic populations, such as Mexican Americans, is not as well documented and often inconsistent. For example, compared to non-Hispanic whites, studies have shown Mexican Americans to have both higher[11] and lower[12] mean Lp(a) levels. The underlying cause(s) for these between-population differences has not been fully determined; however, there is evidence for the role of multiple, population-specific alleles in LPA [13], the gene that encodes for apolipoprotein(a) [apo(a)], which when bound to apolipoprotein B-100 and a low density lipoprotein (LDL)-like particle forms Lp(a).
Lp(a) levels not only vary dramatically across populations, they also have a remarkable inter-individual variability that ranges from barely detectable to greater than 250 nmol/l[14]. This inter-individual variability has a substantial genetic component. It has been determined that the apolipoprotein(a) gene is the major contributor to Lp(a) levels, accounting for more than 90% of the variance for that trait in European Americans[15].
Two types of genetic variants in LPA have been associated with Lp(a) levels: variations in the number of copies of the kringle IV-2 repeat and single nucleotide polymorphisms (SNPs). It has been estimated that the kringle IV-2 repeat alone explains 61–69% of the variability observed in Lp(a) levels in populations of European ancestry[15], [16]. In contrast, the kringle repeat appears to explain less of the variability (19–44%) in populations of African descent[17]–[19] and Mexican Americans (22–48%)[20], [21]. While the kringle IV-2 repeat polymorphism accounts for a large percentage of the variability of Lp(a) levels, the remaining variance has yet to be explained.
Recent studies have identified common SNPs in LPA as strongly associated with Lp(a) levels, explaining up to 36% of the trait variance in populations of European-descent[22]–[24]. While several studies have indicated certain SNPs are in substantial linkage disequilibrium (LD) with the kringle IV-2 repeat polymorphism[22], [23], evidence also exists that some SNPs are in relatively little LD with copy number variation in LPA [25] and may be independent contributors to Lp(a) levels. A recent genome-wide association study performed in a Hutterite population with kringle IV-2 repeat polymorphism data identified a SNP associated with Lp(a) levels independent of the kringle repeat, supporting the assumption that some common SNPs in LPA are independent of the kringle repeat polymorphisms (i.e., not in linkage disequilibrium)[24].
To date, relatively few studies have examined associations between LPA common SNPs and Lp(a) levels across multiple, diverse populations and no study has characterized the same panel of LPA common SNPs in populations of European-, African-, and Mexican-descent. To better characterize this genotype-phenotype relationship in more diverse populations, we have genotyped 19 European American and African American LPA tagSNPs in 7,159 participants from the Third National Health and Nutrition Examination Survey (NHANES III). NHANES III is a diverse, population-based cohort representing Americans of European-, African-, and Mexican-descent[26]. We report the significant association of LPA SNPs and Lp(a) levels in this diverse cohort and estimate the proportion of Lp(a) variance explained by these genetic variants.
Results
Population Characteristics
Characteristics of the NHANES III study participants are shown in Table 1. Genetic NHANES III included 2,631 non-Hispanic whites, 2,108 non-Hispanic blacks, and 2,073 Mexican Americans. As expected[27], the mean Lp(a) level in non-Hispanic blacks was 43.4 mg/dL (SD, 32.8 mg/dL), a twofold increase compared to non-Hispanic whites and a three-fold increase compared to Mexican Americans. Mexican Americans had significantly lower mean Lp(a) levels compared to whites (p<0.0001). Body mass index (BMI) was similar across all three populations (p = 0.093). Demographic variables age and sex, along with other blood lipid traits LDL-C, HDL-C, and triglycerides, differed significantly (p<0.0001) across populations.
Table 1. NHANES III study population characteristics.
| Variable | Non-Hispanic Whites(n = 2,631) | Non-Hispanic Blacks(n = 2,108) | Mexican Americans(n = 2,073) | P-value |
| Males, % | 40.0% | 42.6% | 49.4% | <0.0001 |
| Age (yr) | 50.2 (22.3) | 36.0 (18.3) | 37.1 (18.7) | <0.0001 |
| BMI (kg/m2) | 26.3 (5.6) | 27.3 (6.8) | 27.0 (5.6) | 0.093 |
| Lp(a) (mg/dL) | 20.3 (24.1) | 43.4 (32.8) | 14.9 (8.5) | <0.0001 |
| HDL-C (mg/dL) | 50.2 (15.6) | 53.8 (16.4) | 48.0 (13.1) | 0.112 |
| LDL-C (mg/dL) | 127.0 (38.0) | 118.8 (39.5) | 116.3 (34.1) | <0.0001 |
| TG (mg/dL) | 147.63 (116.8) | 108.8 (79.9) | 154.1 (121.2) | <0.0001 |
Study characteristics are shown for participants greater than 18 years of age who had non-missing Lp(a) levels. Samples sizes shown are the DNA samples available in Genetic NHANES III for each subpopulation. Values are shown as unweighted mean (SD). BMI, body mass index; Lp(a), lipoprotein(a); HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglycerides. P-values are based on one-way unweighted ANOVA.
TagSNP allele frequencies are presented in Supplementary Table S1, by population. We calculated the Pearson correlation coefficient (r) between each of the three populations. Not surprisingly[28], [29], LPA allele frequencies observed in non-Hispanic whites were highly correlated with allele frequencies observed in Mexican Americans (r = 0.80). Also as expected[28]–[30], we observed weaker correlation between allele frequencies in non-Hispanic blacks compared with non-Hispanic whites (r = 0.60) and Mexican Americans (r = 0.48). Furthermore, compared with non-Hispanic whites, the proportion of SNPs that differed in allele frequency by more than ±0.10 was smaller in Mexican Americans (2/19 SNPs; 11%) than in blacks (11/19 SNPs; 58%).
We also compared the allele frequencies of these LPA SNPs in NHANES III to those in HapMap[31], [32] (Supplementary Table S2). Among the 12 LPA SNPs that overlapped this dataset and HapMap, we observed extremely high correlations (r≥0.99) in allele frequencies between non-Hispanic whites and HapMap CEU (US individuals of northern and western European ancestry) and between non-Hispanic blacks and both HapMap YRI (Yoruba from West Africa) and ASW (individuals with African ancestry from the Southwest USA). Mexican American allele frequencies were also very similar (r = 0.93) to those of HapMap MEX (individuals with Mexican ancestry in Los Angeles, California). Because Mexican Americans are a historically admixed population, a comparison with HapMap Asian populations was performed. The correlation between NHANES Mexican Americans and HapMap Han Chinese (HCB) and Japanese (JPT) was 0.77 and 0.78, respectively.
Haplotype frequencies were inferred for the 19 tagSNPs in LPA by NHANES III subpopulation. We observed eight common haplotypes (frequency >5%) in at least one subpopulation (Supplementary Table S3). While two haplotypes (#1 and #2) were common across all three populations, the remaining haplotypes were either common only to non-Hispanic blacks (#7 and #8), only non-Hispanic whites (#6), or shared between whites and Mexican Americans (#3, #4, #5). As expected[33], the majority of chromosomes from non-Hispanic whites (71.5%) and Mexican Americans (72.6%) were represented by common haplotypes inferred from LPA tagSNPs. Only approximately half of the chromosomes from non-Hispanics blacks (55.7%) were represented by common haplotypes, and the remaining half are scattered across rare haplotypes.
LPA SNP associations with Lp(a) levels
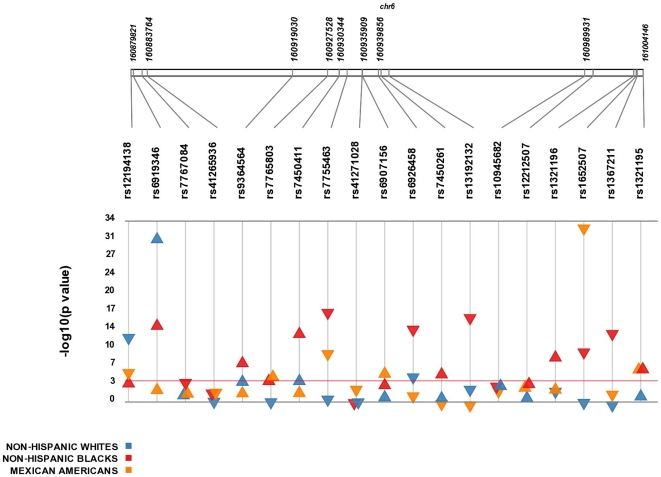
Each SNP was tested for an association with transformed Lp(a) levels. Results from this analysis are presented in Figure 1 and Table 2. After adjusting for age and sex, 15 of the 19 SNPs tested were significantly associated with Lp(a) levels in at least one subpopulation at p<0.0001, meeting the standard Bonferroni p-value threshold for multiple testing. Among non-Hispanic whites, we confirmed previous evidence of a strong association with rs6919346 (p = 1.2×10−30)[22], [24], which explained approximately 6% of the trait variance (R2 = 0.057) in our dataset. We also identified two novel associations with rs6926458 and rs12194138 (p = 5.3×10−6 and 2.1×10−13, respectively). To evaluate the combined effects of significantly associated variants, we calculated a continuous Genetic Risk Score (GRS) for each participant based on his or her total number of risk (i.e. Lp(a) increasing) alleles at each associated SNP. Based on the GRS, the additive effect of rs6919646, rs6926458, and rs12194138 explained 7% of the variation in transformed Lp(a) levels in non-Hispanic whites (Table 3).
Figure 1. Overview of association results between LPA SNPs and Lp(a) levels.
Plot showing the significance of all single-SNP associations with transformed Lp(a) levels. All results are unweighted adjusted for age and sex and are stratified by race/ethnicity. SNPs are plotted on top along the x axis in order from 5′ to 3′, and association with Lp(a) is indicated on the y axis (as −log10 p-value). Red line indicates p-value of 1×10−4. Direction of the triangle indicates direction of effect (β-coefficient).
Table 2. Associations between LPA SNPs and Lp(a) levels.
| SNPs | Non-Hispanic Whitesn = 2,397 | Non-Hispanic Blacksn = 1,711 | Mexican Americansn = 1,749 | ||||||
| β(95% CI) | R2 | P-value | β(95% CI) | R2 | P-value | β(95% CI) | R2 | P-value | |
| rs1321196 | −0.13(−0.22, −0.05) | 0.0036 | 0.0026 | 0.21(0.14, 0.28) | 0.0193 | 1.88×10−8 | 0.12(0.02, 0.22) | 0.0031 | 0.0204 |
| rs1321195 | 0.05(−0.07, 0.18) | 0.0003 | 0.3863 | 0.53(0.31, 0.75) | 0.0123 | 3.03×10−6 | 0.40(0.23, 0.56) | 0.0127 | 3.49×10−6 |
| rs1367211 | −0.004(−0.10, 0.09) | 0.0000 | 0.9330 | −0.27(−0.34, −0.20) | 0.0345 | 3.67×10−14 | −0.15(−0.26, 0.04) | 0.0039 | 0.0092 |
| rs1652507 | −0.05(−0.16, 0.05) | 0.0005 | 0.3295 | −0.45(−0.59, −0.32) | 0.0247 | 1.06×10−10 | −0.54(−0.63, −0.46) | 0.0858 | 5.44×10−34 |
| rs6907156 | 0.21(−0.51, 0.92) | 0.0001 | 0.5722 | 0.15(0.05, 0.25) | 0.0053 | 0.0031 | 0.73(0.39, 1.06) | 0.0103 | 2.14×10−5 |
| rs6919346 | 0.61(0.51, 0.71) | 0.0565 | 1.18×10−30 | 0.75(0.56, 0.94) | 0.0344 | 2.16×10−14 | 0.18(0.02, 0.33) | 0.0029 | 0.0248 |
| rs6926458 | −0.23(−0.32, −0.13) | 0.0087 | 5.29×10−6 | −0.45(−0.57, −0.34) | 0.0364 | 5.90×10−15 | −0.15(−0.28, 0.03) | 0.0032 | 0.0189 |
| rs7755463 | −0.47(−0.99, 0.06) | 0.0014 | 0.0823 | −0.33(−0.40, −0.25) | 0.0449 | 4.01×10−18 | −0.89(−1.16. −0.62) | 0.0230 | 2.14×10−10 |
| rs7767084 | 0.07(−0.05, 0.18) | 0.0006 | 0.2467 | −0.41(−0.61, −0.21) | 0.0097 | 5.88×10−5 | 0.10(−0.03, 0.23) | 0.0015 | 0.1187 |
| rs9364564 | 0.19(0.08, 0.29) | 0.0049 | 0.0007 | 0.33(0.21, 0.46) | 0.0156 | 2.29×10−7 | 0.11(−0.02, 0.24) | 0.0016 | 0.1053 |
| rs12212507 | 0.03(−0.15, 0.21) | 0.0001 | 0.7633 | 0.63(0.23, 1.03) | 0.0063 | 0.0019 | 0.57(0.14, 1.00) | 0.0037 | 0.0093 |
| rs13192132 | −0.14(−0.23, −0.06) | 0.0043 | 0.0011 | −0.43(−0.53, −0.33) | 0.0425 | 3.85×10−17 | −0.003(−0.11, 0.10) | 0.0000 | 0.9508 |
| rs10945682 | 0.12(0.04, 0.21) | 0.0033 | 0.0041 | −0.14(−0.21, −0.06) | 0.0079 | 0.0003 | −0.16(−0.26, −0.06) | 0.0055 | 0.0022 |
| rs12194138 | −0.41(−0.52, −0.30) | 0.0229 | 2.05×10−13 | 0.34(0.12, 0.56) | 0.0020 | 0.0014 | −0.47(−0.66, −0.28) | 0.0149 | 7.97×10−7 |
| rs7450261 | 0.10(−1.29, 1.48) | 0.0000 | 0.8907 | 0.35(0.18, 0.51) | 0.0108 | 2.84×10−5 | −0.33(−1.34, 0.68) | 0.0003 | 0.5176 |
| rs7450411 | 0.19(0.08, 0.30) | 0.0051 | 0.0005 | 0.37(0.27, 0.47) | 0.0308 | 7.37×10−13 | 0.12(−0.02, 0.25) | 0.0017 | 0.0910 |
| rs7765803 | −0.05(−0.14, 0.04) | 0.0005 | 0.2471 | 0.13(0.06, 0.20) | 0.0071 | 0.0005 | 0.21(0.10, 0.31) | 0.0089 | 8.54×10−5 |
| rs41265936 | −0.80(−2.04, 0.44) | 0.0007 | 0.2037 | −0.22(−0.37, −0.06) | 0.0047 | 0.0054 | −0.88(−1.48, −0.28) | 0.0046 | 0.0039 |
| rs41271028 | −0.45(−1.22, 0.32) | 0.0006 | 0.2556 | −0.06(−0.18, 0.07) | 0.0007 | 0.3608 | −0.66(−1.06, −0.27) | 0.0061 | 0.0011 |
The association of LPA SNPs with log transformed Lp(a) levels is shown by a regression coefficient (beta, β) and 95% confidence interval (CI) for each SNP, adjusted for age and sex. Measures of variance explained (R2) are provided for each SNP based on unadjusted regressions. Significant associations (P-value<0.0001) are in bold.
Table 3. Additive effects of LPA risk alleles on Lp(a) levels.
| Non-Hispanic Whites | Non-Hispanic Blacks | Mexican Americans | |
| Total n | 2269 | 1605 | 1665 |
| No. SNPs used in GRS | 3 | 12 | 6 |
| β (95% CI) | 0.36 (0.31–0.42) | 0.11 (0.09–0.13) | 0.34 (0.29–0.39) |
| P-value | <10−35 | <10−34 | <10−41 |
| R2 | 0.07 | 0.09 | 0.11 |
The amount of variance explained (R2) in transformed Lp(a) levels by the Genetic Risk Score (GRS) is displayed, along with the regression coefficient (beta, β) and 95% confidence interval (CI) for each association.
Mexican Americans had twice the number of significant associations compared with non-Hispanic whites, with six SNPs associated with transformed Lp(a) levels at p<0.0001. One SNP in particular, rs1652507, was strongly associated at p = 5.44×10−34 and had the largest effect size of all the associations (R2 = 0.086). Two of the six associated SNPs (rs1321195 and rs7765803) have previously been associated with Lp(a) in a cohort of Europeans[22]. The joint effect of all six associated SNPs, as measured by the GRS, explained 11% of the variance in Lp(a) trait distribution observed in Mexican Americans.
Of the three subpopulations, non-Hispanic blacks had the greatest number of significant associations at p<0.0001 with 12 SNPs. Each associated SNP contributed 1% to 4.5% of the trait variance, with the additive effect of the SNPs contributing up to 9% of the total variance in Lp(a) levels. Five of the 12 associated SNPs (rs1321195, rs1652507, rs6919346, rs6926458, and rs7755463) were also associated in one of the two other racial/ethnic groups, non-Hispanic whites or Mexican Americans, and the directions of the effect (beta) were consistent across the associated subpopulations.
LPA risk allele distribution
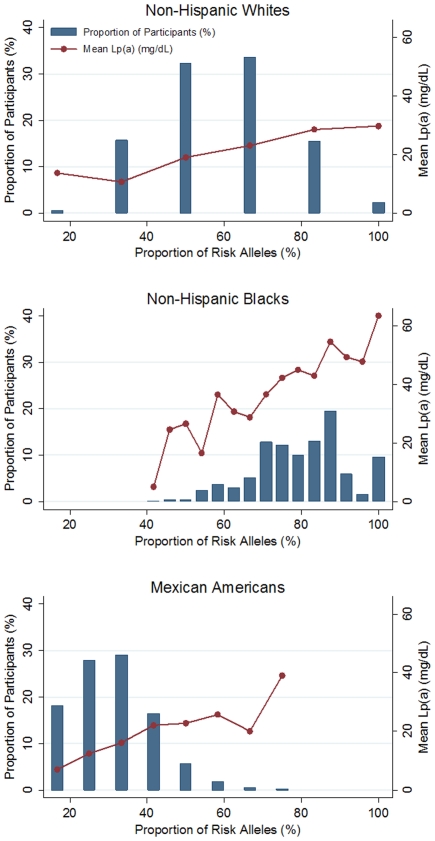
The proportion of risk alleles (i.e. the total number of risk alleles divided by the number of risk alleles possible) was examined across all NHANES subpopulations (Figure 2). In general, the distributions differed greatly among non-Hispanic whites, non-Hispanic blacks, and Mexican Americans. In non-Hispanic whites, the proportion of risk alleles followed a normal distribution and the average (mean) number of risk alleles was 3.5 out of the possible six risk alleles (58.3%). In contrast, the distribution of risk alleles was skewed to the left in non-Hispanic blacks and to the right in Mexican Americans (Figure 2). The average number of risk alleles in non-Hispanic black participants was 17 out of 24 possible risk alleles (70.8%) while the average number in Mexican American participants was 5.5 out of the 12 possible risk alleles (45.8%). Overall, non-Hispanic blacks had the largest genetic burden of all three subpopulations defined by these alleles, with 99.0% percent of participants possessing greater than 50% of the possible risk alleles. This genetic burden is significantly greater than that carried by non-Hispanic whites (51.4%, p<0.001) or Mexican Americans (2.7%, p<0.001). Figure 2 also illustrates mean Lp(a) levels in participants with various proportions of risk alleles. As expected, mean Lp(a) is higher in participants with a greater proportion of risk alleles, again reflecting the role that these variants may play in contributing to both between- and within-population Lp(a) trait variation.
Figure 2. Distribution of LPA risk alleles in non-Hispanic whites, non-Hispanic blacks, and Mexican Americans.
Plots showing the frequency distributions of the proportion of risk alleles in the three NHANES III subpopulations. Proportion of risk alleles was calculated by dividing the total number of LPA risk alleles (i.e. the GRS, see Materials and Methods) by the total number of possible risk alleles in each population, multiplied by 100%. Mean Lp(a) values are also plotted for each corresponding proportion.
Discussion
In this study, we identified several variants in the LPA gene that are strongly associated with Lp(a) levels in a diverse epidemiologic study. More specifically, three SNPs in non-Hispanic whites, twelve SNPs in non-Hispanic blacks, and six SNPs in Mexican Americans were strongly associated at p<0.0001. While no single LPA variant was significantly associated in all three racial/ethnic groups, six SNPs were significantly associated in two subpopulations and the directions of effects were consistent.
Most previously published studies characterizing the relationship between Lp(a) and LPA have focused on the effects of the kringle IV-2 copy number polymorphism. More recently, a genome-wide association study in Hutterites identified one SNP in LPA (rs6919346) that associated with Lp(a) levels, independent of kringle IV-2 copy number[24]. Subsequent studies have found this variant to be independently associated with increased Lp(a) levels in European Caucasians[22], [23] and South Asians and Chinese[23]. In our study, the same allele (G) was also strongly associated with increased trait levels not only in non-Hispanic whites (β = 0.61, p = 1.18×10−30) but also in non-Hispanic blacks (β = 0.75, p = 2.16×10−14). In contrast, the association in Mexican Americans was much less robust (p = 0.02), but the effect trended in the same direction (β = 0.18). This intronic tagSNP is not in linkage disequilibrium (LD) with any genotyped SNP (Supplementary Figure S1), nor with the kringle IV repeat[25]. As others have suggested[24], rs6919346 may be tagging the causal variant or, due to the fact that it resides in a CRE-binding site, may play a role in gene expression.
It is interesting to note that while we did replicate the association between Lp(a) levels and rs6919346, we did not necessarily replicate the associations reported recently for rs10945682 and rs7765803[23]. LPA rs10945682 was not associated with Lp(a) levels in NHANES III at the significant threshold of p<0.0001 (Table 2). Furthermore, while the direction of effect in non-Hispanic whites was consistent with that observed for Europeans and Asians studied by Lanktree et al (taking into account the coded allele), the direction of effect was opposite in the non-Hispanic black and Mexican American subpopulations in NHANES III. LPA rs7765803 was not associated with Lp(a) levels in non-Hispanic whites (p = 0.2471) while it was strongly associated in European and Asian populations in Lanktree et al. Finally, the data reported here are not consistent with the linkage disequilibrium data reported by Lanktree et al. LPA rs10945682 and rs6919346 are reported to be in the same linkage disequilibrium block[23] but the LD calculated in our non-Hispanic white samples and in HapMap CEU suggests there is little LD (r2 = 0.06 and 0.03 in non-Hispanic whites and CEU, respectively) between the two SNPs. It is possible that this discrepancy can be explained by unidentified population substructure or by the use of different LD measures, but this is unclear from the literature and requires further investigation.
As alluded to above, the relationship between LPA tagSNPs and Lp(a) levels may represent a direct (i.e. causal) or indirect (i.e. proxy for true causal variant) relationship. The latter situation most likely applies to the majority of SNPs genotyped in this study. Of the 19 LPA SNPs, 17 are located in introns, and the two nonsynonymous SNPs (rs7765803 and rs41265936, Supplementary Table S1) are not predicted to alter protein function using SIFT[34]. Additional studies are needed to determine if these variants regulate LPA expression in vivo. However, since apo(a) is present only in humans, Old World primates, and the hedgehog, resources for these studies are limited to transgenic mice and rabbits as models[35].
In an attempt to evaluate the joint effect of significantly associated variants, a genetic risk score (GRS) was calculated. Based on this GRS, these variants together explained 7%, 9%, and 11% of the variance in Lp(a) levels in non-Hispanic whites, non-Hispanic blacks, and Mexican Americans, respectively. In comparison to the effect attributed to the kringle repeat region based on previous studies[15], [16], [20], [21], [28], [36]–[38], the effect of these SNPs is considerably small.
This study has several strengths and limitations. The greatest strength is the use of a large and diverse population. While there have been several studies of LPA SNPs and its association with Lp(a) that have included both European and African descent populations, no single study, to our knowledge, has also included Mexican Americans genotyped for the same LPA SNPs. This latter point cannot be under emphasized as the Hispanic or Latino population is the fastest growing minority population in the United States yet remains relatively underrepresented in genetic association studies[39].
A limitation is that the method of measuring serum Lp(a) levels in NHANES III does not account for apo(a) isoform size. While accurate measurement of apo(a) isoform is ideal, the reliability of the Lp(a) measurement used here has been adequately demonstrated[26]. Furthermore, there is no generally accepted laboratory procedure or national standardization program for Lp(a) measurement, which may help to explain the lack of generalizabilty across studies[14].
A second major limitation is that NHANES III does not have data on kringle repeat size for each participant. Several methods are used to measure kringle repeat size such as Southern blot[28] and quantitative PCR[40], neither of which can be used in NHANES III DNA samples given investigators are aliquoted limited amounts of DNA from crude cell lysates. Without these data, it is unclear if the associations between LPA SNPs and Lp(a) levels reported here are independent of the KIV-2 copy number variant, which has a well-established, large effect on Lp(a) levels.
The amount of linkage disequilibrium, or lack thereof, between the KIV-2 region and other LPA variants is a controversial issue. Previous studies have reported strong LD between the KIV-2 alleles and SNPs in or around LPA [22], [41]–[43]. In contrast, additional studies indicate the lack of strong LD[24], [28]. More specifically, the tagSNPs genotyped in this study had been selected from a previous study[28] that provided data on kringle IV-2 repeat size, and no strong LD (r2>0.80) was found for any of the SNPs tested[28]. However, there was moderate LD (r2 = 0.45 in European American and r2 = 0.57 in African American samples) between kringle repeat sizes 10 and 14 and LPA SNPs 74970 and rs41271028, respectively[28]. LPA 74970 was not genotyped here. LPA rs41271028 was genotyped here but was not significantly associated with Lp(a) levels in any of the three subpopulations after correction for multiple testing (Table 2). Thus, the tagSNPs genotyped here and significantly associated with Lp(a) levels after correction for multiple testing are not in high or moderate LD with specific kringle repeat sizes examined in the original dataset reported by Crawford et al[28]. Further studies are needed in NHANES and other large datasets to characterize the full spectrum of LPA genetic variation and its impact on Lp(a) levels in diverse populations.
Another limitation of this study is that only approximately 30–35% of the LDSelect “bins” for European Americans and African Americans are represented by tagSNPs as many LPA SNPs failed assay design or genotyping in NHANES III. And, tagSNPs selection was limited to common variation, leaving rarer variation such as LPA rs10455872 (<5% MAF) untested. Thus, much of the genetic variation in LPA and its association with Lp(a) levels in these populations remains to be explored. Furthermore, tagSNPs were not selected specifically for the Mexican American subpopulation. At the time of tagSNPs selection, HapMap 3 Mexican American samples were not available at the time, and it was unclear which populations should be used for tagSNPs selection to adequately represent this admixed population. It is important to note that, however, that while our tagSNPs selection process may have been biased for populations of European- and African-descent, the allele frequencies observed in NHANES III Mexican Americans were very similar to that of non-Hispanic whites. Furthermore, our lack of Mexican American specific tagSNPs does not undermine the observation that there is an excess of significant variants associated only in non-Hispanic blacks compared to non-Hispanic whites.
Because of these strengths, and despite these limitations, we have taken an important step in understanding how LPA genetic variants contribute to Lp(a) levels in a diverse population. One of the major findings of our study was that there were notably more significant associations between Lp(a) and LPA SNPs in non-Hispanic blacks compared to non-Hispanic whites and Mexican Americans. Moreover, nearly half of these associations were exclusive to non-Hispanic blacks. Our results suggest that between-population differences in Lp(a) levels can be explained, in part, by multiple population-specific cis-acting variants in LPA. While the role of multiple trans-acting factors in Lp(a) trait distribution has been disputed[44]–[46] and cannot be ruled out, our results reaffirm the need for more comprehensive studies of the effects of LPA variants in large, diverse populations.
Materials and Methods
Ethics Statement
All procedures were approved by the CDC Ethics Review Board and written informed consent was obtained from all participants. This candidate gene association study was approved by the CDC Ethics Review Board (protocols #2003-08 and #2006-11) and the University of Washington's Institutional Review Board (IRB #23667; HSRC D committee). Because no identifying information was accessed by the investigators, this study was considered exempt from Human Subjects by Vanderbilt University's Institutional Review Board (IRB #061062; HS2 committee).
Study Population
Ascertainment of the Third National Health and Nutrition Examination Survey (NHANES III) and method of DNA collection have been previously described[47]–[49] and so will only be briefly described here. The National Health and Nutrition Examination Surveys are cross-sectional surveys conducted by the National Center for Health Statistics (NCHS) at the Centers for Disease Control and Prevention (CDC). NHANES III was conducted between 1988–1990 (phase 1) and 1991–1994 (phase 2)[50], [51]. Like all the NHANES, NHANES III is a complex survey design that over-sampled minorities (non-Hispanic blacks and Mexican Americans), the young, and the elderly. All NHANES have interviews that collect demographic, socioeconomic, dietary, and health-related data. Also, all NHANES study participants undergo a detailed medical examination at a central location known as the Mobile Examination Center (MEC). The medical examination includes the collection of physiological measurements by CDC medical personnel and blood and urine samples for laboratory tests. Beginning with phase 2 of NHANES III, DNA samples were collected from study participants aged 12 years and older.
Laboratory Measures
Serum total cholesterol, triglycerides, and HDL cholesterol were measured using standard enzymatic methods. LDL cholesterol was calculated using the Friedewald equation, with missing values assigned for samples with triglyceride levels greater than 400 mg/dl. Serum Lp(a) levels were measured immunochemically by enzyme-linked immunosorbant assay (ELISA) (Strategic Diagnostics, Newark, DE), which does not have cross reactivity with plasminogen or LDL and is non-sensitive to apo(a) size heterogeneity[26]. Quality control measures of the Lp(a) assay have been described elsewhere and the reliability of this Lp(a) measurement has been adequately demonstrated [26].
SNP Selection and Genotyping
Single nucleotide polymorphisms (SNPs) were selected from SeattleSNPs data on European Americans (n = 23) and African Americans (n = 24) re-sequenced for SNP discovery as previously described[25]. Briefly, tagSNPs were chosen for genotyping in both populations separately using LDSelect[52] at minor allele frequency (MAF) >5% and r2>0.80. At the time of tagSNPs selection (2006), LPA variation data was not available for Mexican Americans or other Hispanic reference samples. Forty-nine SNPs were considered for genotyping, 35 SNPs were targeted for genotyping, and 20 were successfully genotyped. Genotyping was performed using the Illumina GoldenGate assay (as part of a custom 384 OPA) by the Center for Inherited Disease Research (CIDR) through the National Heart Lung and Blood Institute's Resequencing and Genotyping Service. A display of the chromosomal locations of all 20 LPA SNPs, along with their relative locations to the 5′ untranslated region (represented by rs1800769) and the kringle repeat (represented by rs9457952 and rs9457986, which flank the kringle repeat), is presented in Supplementary Figure S2.
Genotyping call rates and tests of Hardy Weinberg Equilibrium stratified by self-reported race/ethnicity were calculated for all genotyped LPA SNPs (Supplementary Table S1). The average genotyping call rate for all 20 SNPs was 95.9%. SNP rs4073498 was out of Hardy Weinberg Equilibrium (HWE; p<0.01) in all three racial/ethnic groups and was therefore excluded from all analyses as mandated by CDC. Five additional SNPs (rs1321195, rs1652507, rs7755463, rs7450261, and rs41265936) were found to be out HWE in one subpopulation but were carried forward in the analysis. In addition to these quality control metrics, we genotyped blinded duplicates as required by CDC, and all SNPs reported here passed quality control metrics required by CDC. All genotype data reported here were deposited into the NHANES III Genetic database and are available for secondary analysis through CDC.
Statistical Methods
Analyses were performed for each self-reported race/ethnicity separately. Quality control measures were implemented in PLINK[53]. Tests of association were performed using SAS version 9.1 and were limited to participants greater than 18 years of age who had non-missing Lp(a) levels regardless of fasting status. Each genetic variant was tested for association with ln(Lp(a)+1) levels (a transformation that approximated normality) using linear regression assuming an additive genetic model. Analyses were performed adjusted for age and sex, and results were plotted using Synthesis-View[54], [55]. Data were accessed remotely from the CDC's Research Data Center (RDC) in Hyattsville, Maryland using Analytic Data Research by Email (ANDRE). Statistical significance was defined as p<0.0001, which represents the Bonferroni corrected p-value [p = 0.0008 = 0.05/(20 SNPs × 3 populations)]. Using STATA 10.1, the frequency of risk alleles was compared between populations using Pearson's chi-squared test. Pair-wise linkage disequilibrium (r2) was calculated using the Genome Variation Server provided by SeattleSNPs (http://gvs.gs.washington.edu/GVS/). Haplotypes were inferred by SAS/Genetics using the expectation-maximization algorithm in each subpopulation separately.
To account for selection and non-response biases, the National Center for Health Statistics provides a weighting methodology, which has been described elsewhere[56]. We performed tests of association both unweighted (using SAS version 9.1) and weighted (using SUDAAN). The results did not differ appreciably; therefore, unweighted results are presented here and only select weighted results are presented in Supplementary Table S4.
Genetic Risk Score Calculation
The Genetic Risk Score (GRS) was calculated for every participant, respective to each population separately, using SNPs that were associated with transformed Lp(a) levels at p<0.0001. We used a count method and assumed each SNP to be independently associated with increased levels of Lp(a). Assuming an additive genetic model for each SNP, a value of 2 was given to individuals who were homozygous for the “risk” allele (i.e. the allele associated with increased levels of transformed Lp(a) levels). Values of 1 and 0 were given to genotypes containing 1 or 0 copies of the risk allele, respectively. The GRS was calculated summing the number of risk alleles at each locus. Participants with incomplete genotype data at any SNP used in the GRS were excluded from analysis. Linear regression, with continuous GRS as the independent variable, was used to evaluate the joint effects (R2) of associated genetic variants for Lp(a) trait variation. A weighted GRS (WGRS) was also calculated by multiplying each β-coefficient from adjusted tests of association by the number of risk alleles, and then summing the products. Compared to the GRS, the results of the WGRS do not appreciably differ (Supplementary Table S5); therefore, GRS was used for the main analyses in the paper.
Supporting Information
Pair-wise linkage disequilibrium (r2) calculated for 19 LPA SNPs in non-Hispanic whites (A), non-Hispanic blacks (B), and Mexican Americans (C) in NHANES III.
(DOC)
Location of genotyped LPA SNPs relative to the kringle repeat region and a SNP in the 5′ untranslated region. Synthesis-View[1] was used to plot the 20 LPA SNPs genotyped in this study. Three other SNPs not genotyped in this study are also represented in this plot within the boxes: rs1800769 (which represents a 5′ UTR SNP genotyped by Rainwater et al 1997[2]) and rs9457986 and rs9457952, which flank the kringle repeat. Chromosomal locations are based on genome build 36.
(DOC)
SNP location and genotyping quality control metrics, stratified by race/ethnicity.
(DOC)
Frequency of LPA variants in HapMap samples.
(DOC)
LPA common haplotypes and haplotype frequencies. Only haplotypes with frequencies >5% in at least one population are displayed. Alleles are ordered based on chromosomal location (5′ to 3′). Frequencies >5% are in bold.
(DOC)
Associations between LPA SNPs and Lp(a) levels, weighted for selection and non-response biases. The association of LPA SNPs with log transformed Lp(a) levels is shown by a regression coefficient (beta, β) and 95% confidence interval (CI) for each SNP, adjusted for age and sex. Measures of variance explained (R2) are provided for each SNP based on unadjusted regressions. Significant associations (P-value<0.0001) are in bold.
(DOC)
Additive effects of LPA alleles associated with increased Lp(a) levels. The amount of variance explained (R2) in transformed Lp(a) levels by the Weighted Genetic Risk Score (WGRS) is displayed, along with the median WGRS score, WGRS interquartile range (IQR), regression coefficient (beta, β) and 95% confidence interval (CI) for each association.
(DOC)
Acknowledgments
We would like to thank Dr. Geraldine McQuillan and Jody McLean for their help in accessing the Genetic NHANES III data. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
Footnotes
Competing Interests: MJR and JDS own stock in Illumina, Inc. To MJR and JDS's knowledge, the remaining authors have no conflicts of interest to disclose. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: Genotyping services were provided by the Johns Hopkins University under federal contract number (N01-HV-48195) from the National Heart, Lung, and Blood Institute (NHLBI). This work was funded, in part, by NIH grants U01 HL66682 (DAN) and U01 HL66642 (DAN). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bennet A, Di Angelantonio E, Erqou S, Eiriksdottir G, Sigurdsson G, et al. Lipoprotein(a) Levels and Risk of Future Coronary Heart Disease: Large-Scale Prospective Data. Arch Intern Med. 2008;168:598–608. doi: 10.1001/archinte.168.6.598. [DOI] [PubMed] [Google Scholar]
- 2.Berglund L, Ramakrishnan R. Lipoprotein(a): An Elusive Cardiovascular Risk Factor. Arterioscler Thromb Vasc Biol. 2004;24:2219–2226. doi: 10.1161/01.ATV.0000144010.55563.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danesh J, Collins R, Peto R. Lipoprotein(a) and coronary heart disease. Meta-analysis of prospective studies. Circulation. 2000;102:1082–1085. doi: 10.1161/01.cir.102.10.1082. [DOI] [PubMed] [Google Scholar]
- 4.The Emerging Risk Factors Collaboration. Lipoprotein(a) Concentration and the Risk of Coronary Heart Disease, Stroke, and Nonvascular Mortality. JAMA: The Journal of the American Medical Association. 2009;302:412–423. doi: 10.1001/jama.2009.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guyton JR, Dahlen GH, Patsch W, Kautz JA, Gotto AM., Jr Relationship of plasma lipoprotein Lp(a) levels to race and to apolipoprotein B. Arterioscler Thromb Vasc Biol. 1985;5:265–272. doi: 10.1161/01.atv.5.3.265. [DOI] [PubMed] [Google Scholar]
- 6.Heiss G, Schonfeld G, Johnson JL, Heyden S, Hames CG, et al. Black-white differences in plasma levels of apolipoproteins: the Evans County Heart Study. Am Heart J. 1984;108:807–814. doi: 10.1016/0002-8703(84)90676-8. [DOI] [PubMed] [Google Scholar]
- 7.Moliterno DJ, Jokinen EV, Miserez AR, Lange RA, Willard JE, et al. No association between plasma lipoprotein(a) concentrations and the presence or absence of coronary atherosclerosis in African-Americans. Arterioscler Thromb Vasc Biol. 1995;15:850–855. doi: 10.1161/01.atv.15.7.850. [DOI] [PubMed] [Google Scholar]
- 8.Sharrett AR, Ballantyne CM, Coady SA, Heiss G, Sorlie PD, et al. Coronary heart disease prediction from lipoprotein cholesterol levels, triglycerides, lipoprotein(a), apolipoproteins A-I and B, and HDL density subfractions: The Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 2001;104:1108–1113. doi: 10.1161/hc3501.095214. [DOI] [PubMed] [Google Scholar]
- 9.Sorrentino MJ, Vielhauer C, Eisenbart JD, Fless GM, Scanu AM, et al. Plasma lipoprotein (a) protein concentration and coronary artery disease in black patients compared with white patients. Am J Med. 1992;93:658–662. doi: 10.1016/0002-9343(92)90199-l. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasan SR, Dahlen GH, Jarpa RA, Webber LS, Berenson GS. Racial (black-white) differences in serum lipoprotein (a) distribution and its relation to parental myocardial infarction in children. Bogalusa Heart Study. Circulation. 1991;84:160–167. doi: 10.1161/01.cir.84.1.160. [DOI] [PubMed] [Google Scholar]
- 11.Kamboh MI, Rewers M, Aston CE, Hamman RF. Plasma apolipoprotein A-I, apolipoprotein B, and lipoprotein(a) concentrations in normoglycemic Hispanics and non-Hispanic whites from the San Luis Valley, Colorado. Am J Epidemiol. 1997;146:1011–1018. doi: 10.1093/oxfordjournals.aje.a009229. [DOI] [PubMed] [Google Scholar]
- 12.Haffner SM, Gruber KK, Morales PA, Hazuda HP, Valdez RA, et al. Lipoprotein(a) concentrations in Mexican Americans and non-Hispanic whites: the San Antonio Heart Study. Am J Epidemiol. 1992;136:1060–1068. doi: 10.1093/oxfordjournals.aje.a116571. [DOI] [PubMed] [Google Scholar]
- 13.Chretien JP, Coresh J, Berthier-Schaad Y, Kao WH, Fink NE, et al. Three single-nucleotide polymorphisms in LPA account for most of the increase in lipoprotein(a) level elevation in African Americans compared with European Americans. J Med Genet. 2006;43:917–923. doi: 10.1136/jmg.2006.042119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcovina SM, Koschinsky ML, Albers JJ, Skarlatos S. Report of the National Heart, Lung, and Blood Institute Workshop on Lipoprotein(a) and Cardiovascular Disease: recent advances and future directions. Clin Chem. 2003;49:1785–1796. doi: 10.1373/clinchem.2003.023689. [DOI] [PubMed] [Google Scholar]
- 15.Boerwinkle E, Leffert CC, Lin J, Lackner C, Chiesa G, et al. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J Clin Invest. 1992;90:52–60. doi: 10.1172/JCI115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boomsma DI, Knijff P, Kaptein A, Labeur C, Martin NG, et al. The effect of apolipoprotein(a)-, apolipoprotein E-, and apolipoprotein A4- polymorphisms on quantitative lipoprotein(a) concentrations. Twin Res. 2000;3:152–158. doi: 10.1375/136905200320565436. [DOI] [PubMed] [Google Scholar]
- 17.Ali S, Bunker CH, Aston CE, Ukoli FA, Kamboh MI. Apolipoprotein A kringle 4 polymorphism and serum lipoprotein (a) concentrations in African blacks. Hum Biol. 1998;70:477–490. [PubMed] [Google Scholar]
- 18.Kraft HG, Lingenhel A, Pang RW, Delport R, Trommsdorff M, et al. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: the distribution of null alleles is non-random. Eur J Hum Genet. 1996;4:74–87. doi: 10.1159/000472175. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt K, Kraft HG, Parson W, Utermann G. Genetics of the Lp(a)/apo(a) system in an autochthonous Black African population from the Gabon. Eur J Hum Genet. 2006;14:190–201. doi: 10.1038/sj.ejhg.5201512. [DOI] [PubMed] [Google Scholar]
- 20.Chiu L, Hamman RF, Kamboh MI. Apolipoprotein A polymorphisms and plasma lipoprotein(a) concentrations in non-Hispanic Whites and Hispanics. Hum Biol. 2000;72:821–835. [PubMed] [Google Scholar]
- 21.Rainwater DL, Kammerer CM, Vandeberg JL, Hixson JE. Characterization of the genetic elements controlling lipoprotein(a) concentrations in Mexican Americans. Evidence for at least three controlling elements linked to LPA, the locus encoding apolipoprotein(a). Atherosclerosis. 1997;128:223–233. doi: 10.1016/s0021-9150(96)05993-x. [DOI] [PubMed] [Google Scholar]
- 22.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 23.Lanktree MB, Anand SS, Yusuf S, Hegele RA. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 2010;3:39–46. doi: 10.1161/CIRCGENETICS.109.907642. [DOI] [PubMed] [Google Scholar]
- 24.Ober C, Nord AS, Thompson EE, Pan L, Tan Z et al. Genome-wide association study of plasma lipoprotein(a) levels identifies multiple genes on chromosome 6q. J Lipid Res. 2009;50:798–806. doi: 10.1194/jlr.M800515-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crawford DC, Peng Z, Cheng JF, Boffelli D, Ahearn M, et al. LPA and PLG sequence varaition and kringle IV-2 copy number in two populations. Hum Hered. 2008;66:199–209. doi: 10.1159/000143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Center for Disease Control and Prevention. The Third National Health and Nutrition Examination Survey, 1988-94. 1996. Reference manuals and reports (CD ROM). US Department of Health and Human Services National Center for Health Statistics.
- 27.Marcovina SM, Albers JJ, Wijsman E, Zhang ZH, Chapman NH, et al. Differences in Lp(a) concentrations and apo(a) polymorphs between black and white Americans. J Lipid Res. 1996;37:2569–2585. [PubMed] [Google Scholar]
- 28.Crawford DC, Peng Z, Cheng JF, Boffelli D, Ahearn M, et al. LPA and PLG sequence variation and kringle IV-2 copy number in two populations. Hum Hered. 2008;66:199–209. doi: 10.1159/000143403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bamshad M. Genetic influences on health: does race matter? JAMA. 2005;294:937–946. doi: 10.1001/jama.294.8.937. [DOI] [PubMed] [Google Scholar]
- 30.Carlson CS, Eberle MA, Rieder MJ, Smith JD, Kruglyak L, et al. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat Genet. 2003;33:518–521. doi: 10.1038/ng1128. [DOI] [PubMed] [Google Scholar]
- 31.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 32.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, et al. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 35.Boffa MB, Marcovina SM, Koschinsky ML. Lipoprotein(a) as a risk factor for atherosclerosis and thrombosis: mechanistic insights from animal models. Clin Biochem. 2004;37:333–343. doi: 10.1016/j.clinbiochem.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Ali S, Bunker CH, Aston CE, Ukoli FA, Kamboh MI. Apolipoprotein A kringle 4 polymorphism and serum lipoprotein (a) concentrations in African blacks. Hum Biol. 1998;70:477–490. [PubMed] [Google Scholar]
- 37.Kraft HG, Lingenhel A, Pang RW, Delport R, Trommsdorff M, et al. Frequency distributions of apolipoprotein(a) kringle IV repeat alleles and their effects on lipoprotein(a) levels in Caucasian, Asian, and African populations: the distribution of null alleles is non-random. Eur J Hum Genet. 1996;4:74–87. doi: 10.1159/000472175. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt K, Kraft HG, Parson W, Utermann G. Genetics of the Lp(a)/apo(a) system in an autochthonous Black African population from the Gabon. Eur J Hum Genet. 2006;14:190–201. doi: 10.1038/sj.ejhg.5201512. [DOI] [PubMed] [Google Scholar]
- 39.Choudhry S, Seibold MA, Borrell LN, Tang H, Serebrisky D, et al. Dissecting complex diseases in complex populations: asthma in latino americans. Proc Am Thorac Soc. 2007;4:226–233. doi: 10.1513/pats.200701-029AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lanktree MB, Rajakumar C, Brunt JH, Koschinsky ML, Connelly PW, et al. Determination of lipoprotein(a) kringle repeat number from genomic DNA: copy number variation genotyping using qPCR. J Lipid Res. 2009;50:768–772. doi: 10.1194/jlr.D800050-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraft HG, Haibach C, Lingenhel A, Brunner C, Trommsdorff M, et al. Sequence polymorphism in kringle IV 37 in linkage disequilibrium with the apolipoprotein (a) size polymorphism. Hum Genet. 1995;95:275–282. doi: 10.1007/BF00225193. [DOI] [PubMed] [Google Scholar]
- 42.Luke MM, Kane JP, Liu DM, Rowland CM, Shiffman D, et al. A polymorphism in the protease-like domain of apolipoprotein(a) is associated with severe coronary artery disease. Arterioscler Thromb Vasc Biol. 2007;27:2030–2036. doi: 10.1161/ATVBAHA.107.141291. [DOI] [PubMed] [Google Scholar]
- 43.Ogorelkova M, Kraft HG, Ehnholm C, Utermann G. Single nucleotide polymorphisms in exons of the apo(a) kringles IV types 6 to 10 domain affect Lp(a) plasma concentrations and have different patterns in Africans and Caucasians. Hum Mol Genet. 2001;10:815–824. doi: 10.1093/hmg/10.8.815. [DOI] [PubMed] [Google Scholar]
- 44.Barkley RA, Brown AC, Hanis CL, Kardia SL, Turner ST, et al. Lack of genetic linkage evidence for a trans-acting factor having a large effect on plasma lipoprotein[a] levels in African Americans. J Lipid Res. 2003;44:1301–1305. doi: 10.1194/jlr.M300163-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Mooser V, Scheer D, Marcovina SM, Wang J, Guerra R, et al. The Apo(a) gene is the major determinant of variation in plasma Lp(a) levels in African Americans. Am J Hum Genet. 1997;61:402–417. doi: 10.1086/514851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholz M, Kraft HG, Lingenhel A, Delport R, Vorster EH, et al. Genetic control of lipoprotein(a) concentrations is different in Africans and Caucasians. Eur J Hum Genet. 1999;7:169–178. doi: 10.1038/sj.ejhg.5200290. [DOI] [PubMed] [Google Scholar]
- 47.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, et al. Genetic Variation Is Associated With C-Reactive Protein Levels in the Third National Health and Nutrition Examination Survey. Circulation. 2006;114:2458–2465. doi: 10.1161/CIRCULATIONAHA.106.615740. [DOI] [PubMed] [Google Scholar]
- 48.Chang Mh, Lindegren ML, Butler MA, Chanock SJ, Dowling NF, et al. Prevalence in the United States of Selected Candidate Gene Variants: Third National Health and Nutrition Examination Survey, 1991-1994. Am J Epidemiol. 2009;169:54–66. doi: 10.1093/aje/kwn286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinberg KK, Sanderlin KC, Ou C-Y, Hannon WH, McQuillan GM, et al. DNA banking in epidemiologic studies. Epidemiol Rev. 1997;19:156–162. doi: 10.1093/oxfordjournals.epirev.a017938. [DOI] [PubMed] [Google Scholar]
- 50.Centers for Disease Control and Prevention. Plan and Operation of the Third National Health and Nutrition Examination Survey, 1988-94. 2004. Bethesda, MD.
- 51.Centers for Disease Control and Prevention. Third National Health and Nutrition Examination Survey, 1988-94, Plan and Operations Procedures Manuals 1996.
- 52.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pendergrass S, Dudek SM, Roden DM, Crawford DC, Ritchie MD. Visual integration of results from a large DNA biobank (biovu) using synthesis-view. Pac Symp Biocomput. 2011;265-275 doi: 10.1142/9789814335058_0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pendergrass S, Dudek S, Crawford D, Ritchie M. Synthesis-View: visualization and interpretation of SNP association results for multi-cohort, multi-phenotype data and meta-analysis. BioData Mining. In press. 2010 doi: 10.1186/1756-0381-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohr SL. Pacific Grove, Calif: Duxbury Press; 1999. Sampling: Design and Analysis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pair-wise linkage disequilibrium (r2) calculated for 19 LPA SNPs in non-Hispanic whites (A), non-Hispanic blacks (B), and Mexican Americans (C) in NHANES III.
(DOC)
Location of genotyped LPA SNPs relative to the kringle repeat region and a SNP in the 5′ untranslated region. Synthesis-View[1] was used to plot the 20 LPA SNPs genotyped in this study. Three other SNPs not genotyped in this study are also represented in this plot within the boxes: rs1800769 (which represents a 5′ UTR SNP genotyped by Rainwater et al 1997[2]) and rs9457986 and rs9457952, which flank the kringle repeat. Chromosomal locations are based on genome build 36.
(DOC)
SNP location and genotyping quality control metrics, stratified by race/ethnicity.
(DOC)
Frequency of LPA variants in HapMap samples.
(DOC)
LPA common haplotypes and haplotype frequencies. Only haplotypes with frequencies >5% in at least one population are displayed. Alleles are ordered based on chromosomal location (5′ to 3′). Frequencies >5% are in bold.
(DOC)
Associations between LPA SNPs and Lp(a) levels, weighted for selection and non-response biases. The association of LPA SNPs with log transformed Lp(a) levels is shown by a regression coefficient (beta, β) and 95% confidence interval (CI) for each SNP, adjusted for age and sex. Measures of variance explained (R2) are provided for each SNP based on unadjusted regressions. Significant associations (P-value<0.0001) are in bold.
(DOC)
Additive effects of LPA alleles associated with increased Lp(a) levels. The amount of variance explained (R2) in transformed Lp(a) levels by the Weighted Genetic Risk Score (WGRS) is displayed, along with the median WGRS score, WGRS interquartile range (IQR), regression coefficient (beta, β) and 95% confidence interval (CI) for each association.
(DOC)