Abstract
Raman spectroscopy (RS) and optical coherence tomography (OCT) are powerful tools for optical analysis of tissues with mutually complementary strengths and limitations. OCT excels at visualizing tissue microstructure but lacks molecular specificity, while RS can relay tissue biochemical composition but typically cannot relate microstructure. Previous implementations of combined RS-OCT have utilized a common sample arm while maintaining independent RS and OCT detection arms. We present the design and application of an integrated RS-OCT instrument with a common detection arm for both RS and OCT. The detector is a spectrograph capable of sequential detection of the 855-nm OCT signal and the Raman scatter generated by a 785-nm source. The capabilities of the instrument are demonstrated ex vivo in the calvaria and retina of rodents, as well as in vivo in human skin.
Keywords: Raman spectroscopy, optical coherence tomography, tissue characterization, bone, retina, skin
Introduction
Optical imaging and spectroscopy have both been applied to the noninvasive characterization of tissues. Imaging techniques such as optical coherence tomography (OCT)1 excel at relaying images of tissue microstructure, while spectroscopic methods such as Raman spectroscopy (RS)2 are capable of probing the molecular composition of tissue with excellent specificity. The ability of OCT to perform real-time cross sectional imaging with micrometer-scale resolution has been utilized for both quantitative and qualitative assessment of tissues in a wide range of applications. For example, quantitative measurements of retinal nerve fiber layer thickness can provide valuable information for glaucoma assessment,3 while qualitative analysis of the esophageal epithelium can identify characteristic features of Barrett’s dysplasia.4 Although visualization of tissue microstructure is often sufficient to characterize tissue type, different structural features can often have a fairly similar appearance in OCT despite having different underlying molecular makeups.5, 6, 7 This limitation results from the fact that OCT images are simply maps of reflectivity and do not directly reveal the molecular composition of the sample. RS, on the other hand, can generate in-elastic scattering spectra with sharp spectral features corresponding to the vibrational modes of biological molecules intrinsic to the sample. RS has demonstrated the ability to characterize the molecular features of pathology in a number of tissues, including the cervix,8 skin,9 breast,10 and gastrointestinal (GI) tract.11 In contrast to OCT, the primary limitation of RS is that the weak nature of in-elastic scattering precludes rapid spectral imaging over a large spatial area. Clearly, characterization of both morphology and biochemical composition could compensate for the limitations of both RS and OCT, and allow for a more complete analysis of tissues. For example, the detection of early dental caries has already been identified as a potential application where the mutual benefit of morphological and biochemical characterization with OCT and RS can be beneficial.12 This work suggested that the mutually complementary strengths and limitations of RS and OCT are well suited for integration into a single instrument. The realization of such an instrument would allow data collected from the two modalities to augment one another, and could advance the biomedical applications of RS and OCT beyond what is possible with either technique independently.
The most straightforward approach for combining RS and OCT into a single instrument consists of integrating the sampling optics while maintaining independent detection hardware. To date, the two reports of instruments combining RS and OCT have pursued the common sample arm approach. The first system combined a time-domain OCT engine using a 1310-nm source and rapid-scanning optical delay reference arm with a 785-nm RS system.13 This instrument demonstrated the ability of RS-OCT to perform in-vivo analysis and evaluated highly scattering tissues such as the breast and skin. Specifically, the instrument demonstrated the benefits of RS-OCT by: 1. utilizing OCT to guide Raman spectral acquisition of small (< 500 μm) regions of irregular tissue, and 2. utilizing RS to characterize the biochemical composition of ambiguous structures within an OCT image. A second RS-OCT instrument combined a Fourier-domain OCT system with a 855-nm broadband source and a spectrometer-based detection system (i.e., spectral-domain OCT) with a 633-nm RS system.14 The advantage of previously reported RS-OCT systems is that the use of independent detection arms allows hardware configurations for each technique to be optimized independently. The drawback, however, is that such configurations require extensive instrumentation that may not be necessary if it were possible to further integrate the two modalities. Since both RS and OCT can be performed with systems that incorporate a spectrograph and charge-coupled device (CCD) for detection,15, 16 it is possible that a streamlined instrument with a common detection arm can be realized with appropriate design considerations. The primary challenges in the design of a common-detector RS-OCT system are: 1. selection of the appropriate light sources, 2. spectrograph design, and 3. selection of appropriate detector architecture.
We present the design of an integrated system capable of performing RS-OCT with both a common sample arm and detection arm. In addition, we demonstrate the ability of this instrument to characterize the morphological and biochemical features of different tissues, demonstrate the capabilities of the instrument for both ex-vivo and in-vivo analysis, and discuss the observed strengths and limitations of the device.
Materials and Methods
To realize an integrated RS-OCT system with a common detector, the initial concern is selecting light sources that result in spectral overlap of the Raman scatter spectrum and the OCT bandwidth. RS of tissue has been reported at wavelengths from the ultraviolet17 to 1064 nm in the infrared.18 Typically, sources in the near-infrared are preferred because tissue autofluorescence is reduced. However, Raman scattering intensity and detector responsivity also typically decrease with increasing wavelength. A wavelength-stabilized external cavity 785-nm diode laser (Sacher Lasertechnik Group, Marburg, Germany) was selected as the Raman source, because tissue autofluorescence is minimized without increasing the wavelength so much as to significantly reduce the collected Raman signal intensity. OCT can also be performed over a range of wavelengths in the near-infrared; however, it is typically performed near wavelengths at 830 or 1310 nm. The additional benefit of the 785-nm RS source is that the resultant “fingerprint” region for organic molecules, which ranges from 500- to 2000-cm−1 relative wavenumbers, spans the wavelength band from 815 to 930 nm (500 to 2000 cm−1) and nicely overlaps a spectral range where OCT sources are readily available. The selected OCT source (Exalos, Incorporated, Zurich, Switzerland) is centered at 855 nm with a -3-dB bandwidth of 40 nm and a full spectral width ranging from approximately 800 to 900 nm, and is thus well suited for integration with the 785-nm RS source. It should be noted that although RS of tissues is also commonly performed with an 830 nm source, the corresponding fingerprint region (866 to 995 nm) does not overlap well with any readily available broadband light sources suitable for OCT that the authors are aware of. In addition, the quantum efficiency of silicon detectors in the 830-nm fingerprint region is inferior to that of the 785-nm fingerprint region, which is beneficial for both modalities.
After selection of the appropriate sources, it is critical that the design of the detection hardware facilitate both RS and OCT of tissues. Detectors capable of exceptionally high sensitivity coupled with a resistance to etaloning are necessary to acquire Raman spectra from tissues, due to the weak nature of Raman scattering and the relatively intense tissue autofluorescence background. These detectors are typically back-illuminated, deep-depletion CCD arrays with cooling mechanisms in place to limit the dark noise. Although the detectors used in OCT also clearly benefit from low noise, they differ in that they are typically front-illuminated CCD arrays with a large dynamic range and high-speed readout rates. Due to the stringent detector requirements of RS, a back-illuminated, deep-depletion, thermoelectrically cooled 1024×256 CCD camera (Newton 920-BRDD, Andor Technology, Belfast, Northern Ireland) typically used for RS was selected. This detector is capable of a 2.5-MHz pixel readout rate and 26×26-μm pixels with a full well capacity = 180,600 e−∕pixel. The readrate and dynamic range of the camera are similar to the specifications of detectors typically employed for OCT, while it is also sensitive enough for detection of tissue Raman signals.
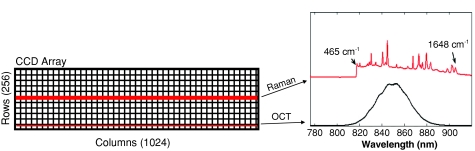
The detector is coupled to an f∕1.8 imaging spectrograph (magnification = 1.13) with a 785-nm Raman holographic transmission grating (Kaiser Optical Systems, Ann Arbor, Michigan). The spectrograph is configured to cover a wavelength range from 780 to 920 nm, which spans the full bandwidth of the OCT source, and thus does not adversely affect the theoretical axial resolution limit for imaging. The spectral range here corresponds to Raman shifts up to 1870 cm−1, which is sufficient to nearly cover the entire range of features arising from tissue. Figure 1 illustrates the detector illumination and the spectral overlap of the Raman fingerprint region and the OCT source spectrum.
Figure 1.
Illustration of the CCD array illumination for RS and OCT, along with the spectral overlap of an example Raman spectrum (acetaminophen) and the OCT source.
In addition to spectral coverage, it is critical that sufficient spectral resolution is achieved for both RS and OCT. To identify the sharp spectral features of tissue Raman signals, a spectral resolution of at least 10 cm−1 is necessary. In spectral domain OCT, the maximum depth range is inversely related to the spectral resolution.16 Therefore, it is important that the spectral resolution be sufficiently high to allow the depth range to exceed the optical penetration depth of the imaging light source, which is generally 1 to 2 mm in the tissue. The instrument we have developed collects light in separate 100-μm multimode (RS) and 6-μm single-mode (OCT) fibers, which are coupled to the spectrograph with a custom input adapter, and illuminates different rows of the CCD. The resulting spectral resolution is 0.141 nm for OCT, and 7 cm−1 for RS. The OCT spectral resolution was back-calculated from the calibrated axial scan range. The scan range was measured to be 1.27 mm in air by translating a mirror mounted on a calibrated micrometer stage through the sample arm optical path. The spectral resolution (δλ) can be calculated as δλ = λcenter2∕4 ⋅ n ⋅ zmax,16 where λcenter is the spectrograph center wavelength, n is the index of refraction, and zmax is the maximum unaliased scan range. Calibration of the spectrograph is performed with the atomic emission lines of a neon-argon lamp.
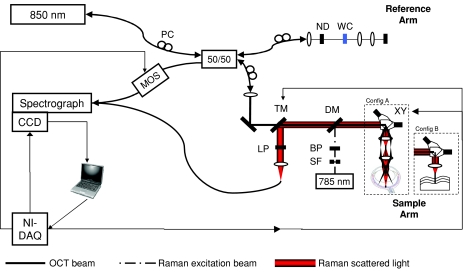
The overall RS-SDOCT system schematic is shown in Fig. 2. The Raman illumination consists of free space optics integrated into the OCT sample arm. The multimode-shaped source passes through a spatial filter and a 785-nm bandpass filter to produce a spectrally and spatially clean collimated beam. A standard 785-nm Raman dichroic mirror (Chroma Technologies, Bellows Falls, Vermont) allows this beam to be coaligned with the OCT beam and directed toward the sample. During Raman acquisition, the mirrors in the xy galvanometer pair are stationary. Because the galvanometer pair and the objective lens are arranged in a telecentric manner, RS can thus be performed along any A-scan in the OCT image. Acquisition from the central A-scan is preferred for maximum Raman signal, but is not essential. An achromatic doublet lens (f = 30 mm, NA = 0.42) serves as the objective and focuses the light to a 15-μm spot. An achromat lens is important to achieve relatively consistent collection efficiency of the Raman scattered light across the entire fingerprint region. The axial response of the RS collection was measured by translating a thin (12.5-μm) polyethylene sheet through the focus and measuring the intensity of the 1308-cm−1 peak. The full width at half maximum (FWHM) of the axial response function was then calculated to be 425 μm. The power of the Raman beam at the sample is 40 mW. The Raman scattered light then returns back through the sampling optics and dichroic mirror, and is redirected toward the Raman collection fiber by a mirror mounted to a computer-controlled translation stage. Before fiber coupling, an 818-nm long pass filter further rejects elastic scatter. To characterize the performance of the RS subsystem, we acquired spectra from a silicon wafer (Tacq = 1 sec), and measured the signal-to-noise ratio to 22.3 dB. During OCT imaging, the translation stage is shifted aside to allow the OCT beam to freely pass. As such, the system does not allow for simultaneous RS and OCT. The two datasets are acquired sequentially. Registration of the Raman collection within the OCT image is performed as described in a previously reported RS-OCT system with independent detection arms.13
Figure 2.
Overall RS-SDOCT system schematic. Configurations of sampling optics used for collection from retina (config A), and from standard samples (config B). PC – polarization control paddles, ND – neutral density filter, WC – water-filled cuvette, TM – translatable mirror, LP – long pass filter, DM – dichroic mirror, BP – band pass filter, SF – spatial filter, xy – xy galvanometer pair, MOS – MEMS optical switch, and NI-DAQ – National Instruments multifunction DAQ.
The OCT source is coupled to a 50∕50 fiber splitter, which directs light to the sample and reference arms. Polarization control paddles are placed in the source, reference, and sample arms to fine tune the splitting ratio and optimize the detected interference signal. The sample arm includes an xy galvanometer pair for generating the transverse scan dimension in OCT and the achromatic doublet objective lens, which focuses the OCT beam to a spot size of 18 μm. The lens was selected to achieve nearly isotropic spatial resolution, which is important for high quality imaging. The reference arm consists of a neutral density filter and an achromatic objective lens matching that of the sample. Insertion of the neutral density filter allows the appropriate level of reference attenuation to optimize imaging sensitivity while preventing saturation of the detector.19 The optimized sensitivity of the system is measured to be -86 dB, at which point the corresponding power of the OCT beam at the sample is 14 μW. OCT sensitivity can be improved by averaging multiple images after tomographic reconstruction. When 25 frames are averaged, the system can achieve a sensitivity of -100 dB. Care was taken to match the dispersion of the reference and sample arms to minimize degradation of the axial resolution. The measured axial resolution is 11 μm in air (8 μm in tissue). An optical MEMS switch (Thorlabs, Newton, New Jersey) is placed in the detection arm of the fiber interferometer. During acquisition of OCT images, the interference signal passes through to the spectrograph; however, during Raman acquisition, the switch directs the OCT signal returning from the reference arm away from the spectrograph so as not to overwhelm the Raman signal.
The entire system is run off a single notebook computer with a LabVIEW software interface. A multifunction DAQ device (National Instruments, Austin, Texas) allows software control of an optical MEMS switch, the translation stage mounted mirror in the sample arm, and the xy galvanometer pair. As previously noted, the instrument operates in either Raman or OCT mode. During OCT mode, the xy mirrors scan the beam across the sample, the MEMS optical switch directs the OCT interference signal to the spectrograph, and RS is disabled by positioning the mirror mounted to the translation stage, such that the Raman scatter is not collected. Furthermore, the detector reads out a cropped portion of CCD array that consists of only the bottom 20 rows of the chip (Fig. 1). This allows spectra to be collected at the maximum possible line rate of 2.1 kHz. In Raman mode, the OCT illumination beam is blocked by the translatable mirror, the xy mirrors are fixed, and the MEMS optical switch directs the light from the OCT reference arm away from the spectrograph. The detector then bins the set of rows on the top half of the chip that are illuminated by the Raman signal. During RS, the last acquired OCT image is retained and the position of the Raman collection is graphically overlaid onto the display to guide user positioning of sample and∕or xy mirrors.
Data processing to produce OCT images and Raman spectra is performed in realtime. To create an OCT image, the reference arm signal is stored before each image acquisition, and then subtracted from subsequent spectra to remove the dc component of the interference signal. After dc subtraction, the spectrum is inflated and remapped from wavelength space to linear k-space to ensure appropriate mapping from the spectral domain to the spatial domain.20 Fast Fourier transform then produces the depth scan. At this stage, multiple OCT images can be averaged to improve imaging sensitivity. After factoring in data transfer and processing time, the display rate of processed OCT images (512×512) is 2 frames∕sec. A number of additional system calibrations are necessary prior to processing the Raman spectra. Variations in system throughput are corrected for using a National Institute of Standards and Technology (NIST) calibrated quartz-tungsten-halogen lamp. To account for minor day-to-day variations in the spectral position of the Raman excitation laser line, the relative wavenumber axis is calibrated with standards such as acetaminophen and naphthalene. Laser-induced fluorescence from the system optics and general background signal is removed by subtracting background spectra. After the sample spectrum is white light corrected and background subtracted, the tissue autofluorescence is removed using a modified polynomial fitting algorithm.21
To validate the ability of the common-detector RS-OCT system to acquire data from tissues, ex-vivo images and spectra were collected from dissected rodent calvaria (skull cap) and the rodent retina. In-vivo data of human skin were collected from the hand of a volunteer. Data collection from the rodent retina was performed through the intact eye, and therefore required a modified instrument configuration (Fig. 2, sample arm configuration A) to account for the focusing properties of the eye itself. Because the physical optics of the eye focus distant light onto the retina, the instrument configuration was modified to illuminate the sample with collimated light. In the sample arm, a second objective lens was inserted to create a 2-f telecentric scan geometry that illuminated the pupil of the eye with a collimated beam (dOCT = 2.1 mm, dRaman = 2.4 mm). The additional dispersion introduced in the sample arm by the second lens and the eye itself was compensated for in the OCT reference arm by adding a matching second objective lens and a 5-mm water-filled cuvette. All data were collected under protocols approved by the Vanderbilt University institutional review board and institutional animal care and use committee.
Results
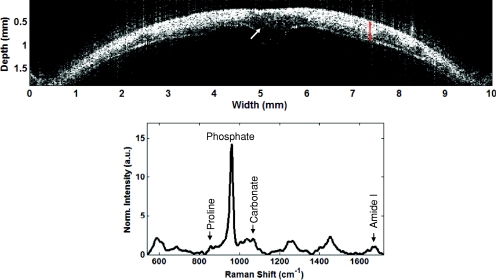
To demonstrate the capability of the common detector RS-OCT instrument in highly scattering tissues, data was acquired from the dissected calvaria of a mouse (Fig. 3). The OCT image [Fig. 3a] is the average of three acquisitions. The image allows clear visualization of the inner and outer surfaces of the calvaria, along with fair contrast between the hyper-reflective mineralized bone tissue and the less reflective collagenous tissue associated with the sutures that hold the plates of the skull together.23 This is most evident in the center of the image at the suture connecting the left and right parietal plates. The corresponding Raman spectra [Fig. 3b, tacq = 30 sec] acquired from the sample is representative of a typical Raman signature of mineralized tissue. The features typically used for analysis22 include the prominent phosphate peak at 960 cm−1, the carbonate peak at 1072 cm−1, the amide-I peak at 1667 cm−1, and the subtle proline peak at 857 cm−1.
Figure 3.
RS-OCT of a dissected murine calvaria. (a) OCT image. Arrow indicates dark hyporeflective region likely associated with the suture of the left and right parietal skull plates. Red area indicates the region where the corresponding Raman spectrum was acquired. Axial scale assumes ncalvaria = 1.55.23 (b) Raman spectrum, normalized to mean spectral intensity. Arrows indicate positions of peaks typically utilized in Raman spectral analysis of mineralized tissues, including the proline peak at 857 cm−1, the phosphate peak at 960 cm−1, the carbonate peak at 1072 cm−1, the amide I peak at 1667 cm−1.
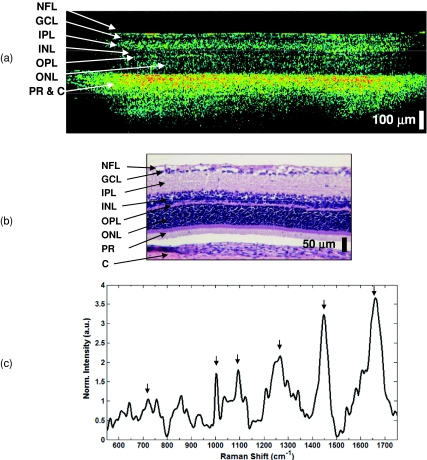
As previously mentioned, the OCT performed in the band from 800 to 900 nm is best suited for imaging the retina. To demonstrate the ability of the common-detector RS-OCT instrument to characterize both the morphology and biochemistry of the retina, data were collected from the retina of a 4-week-old rat pup through the intact eye (Fig. 4). The OCT image [Fig. 4a] is an average of 20 OCT frames and is depicted in the false color scale typically associated with OCT of the retina.24 The image demonstrates the sufficient resolution and imaging range of the system to depict the layers of the retina. A set of five Raman spectra (tacq = 90 sec) was acquired from multiple axes across the image to characterize the general Raman signature. To achieve sufficient signal-to-noise, the Raman power at the sample was increased from 40 to 100 mW, which was necessary due to the gradual loss of clarity in the eye after the animal was sacrificed and the lack of pupil dilation. The mean spectrum is shown in Fig. 4c. The spectral features show similarities to those reported in the literature,25, 26 including peaks at 1265 cm−1 (amide III), 1440 cm−1 (CHx), and 1660 cm−1 (amide I), which are commonly seen in most soft tissues. Protein signatures present in Raman spectra include those arising from DNA and RNA molecules present in the nuclear layers, the most intense of which occur at 723, 1003, and 1094 cm−1.
Figure 4.
RS-OCT of the rodent retina. (a) OCT image. The layers of the retina visible, from inner to outer layers, include the thin, bright nerve fiber layer (NFL), hyporeflective ganglion cell layer (GCL), thicker hyper-reflective inner plexiform layer (IPL), the hyporeflective inner nuclear layer (INL), the thin, bright outer plexiform layer (OPL), the hyporeflective outer nuclear layer (ONL), and the photoreceptor layer and choroid (PR and C), which are difficult to distinguish and are labeled as a single layer. (b) Corresponding histology. (c) Mean Raman spectrum acquired from five axes equally spread across the retina. The set of peaks most prominent in the retina are identified (arrows), and include amide III (1265 cm−1), CHx (1440 cm−1), and amide I (1660 cm−1), as well as those from DNA∕RNA at 723, 1003, and 1094 cm−1.
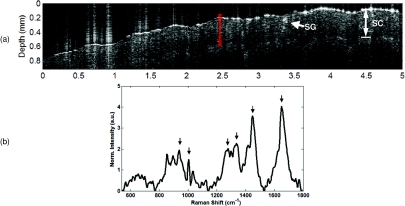
Figure 5 demonstrates the ability of the integrated RS-OCT instrument to acquire in-vivo data from human skin. The OCT image shown in Fig. 5a is a single unaveraged image acquired from the palm of the hand of a volunteer. Because the image is not averaged, it depicts the minimum sensitivity of the system, which still allows visualization of distinct morphological features in the skin.27 The boundary between the stratum corneum and the underlying layers of the epidermis can be seen as a dark band across the length of the image. The image also captures the presence of a hyper-reflective structure in the outer region of the epidermis that is likely a sweat gland. The position where the corresponding Raman spectrum was acquired is indicated by the red overlay. The Raman spectrum shown in Fig. 5b (also a single unaveraged acquisition tacq = 30 sec) is representative of typical skin signatures,9 with peaks at 936 cm−1 (C-C backbone of collagen), 1003 cm−1 (phenylalanine), 1280 cm−1 (amide III), 1335 cm−1 (C-H), 1440 cm−1 (CHx), and 1660 cm−1 (amide I).
Figure 5.
RS-OCT of in-vivo human skin on the palm of the hand. (a) OCT image. A hyper-reflective feature that is likely a sweat gland (SG) is seen within the stratum corneum (SC). Red area indicates location of Raman spectrum. Axial scale assumes nskin = 1.38.27 (b) Raman spectrum of skin. The prominent skin peaks are identified (arrows) at 936 cm−1 (C-C backbone of collagen protein), 1003 cm−1 (phenylalanine), 1280 cm−1 (amide III), 1335 cm−1 (C-H), 1440 cm−1 (CHx), and 1660 cm−1 (amide I). (Color online only.)
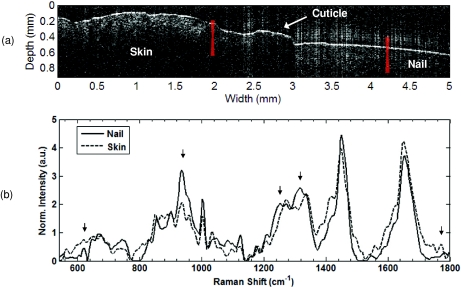
Finally, RS-OCT data were acquired from the finger of a volunteer, where the base of the fingernail inserts into the skin (Fig. 6). As with Fig. 5a, the OCT image [Fig. 6a] shown is unaveraged. The nail is seen on the right side of the image. The root of the nail inserts into the skin below the cuticle, which can be seen as the raised region in the center of the image. To the left of the cuticle is the skin on the back of the finger. We acquired Raman spectra [Fig. 6b] from the nail and the skin with an acquisition time of 30 sec for each spectrum. The location the spectra were acquired from is indicated by the red overlays in Fig. 6a. The nail spectrum is characterized by features that correspond to that of keratin proteins in a β-sheet configuration. This includes the sharp, distinct nature of the 620-cm−1 peak involved in the disulfide bonds that give keratin arrangements their strength, the relative intensity of the 936-cm−1 C-C peak from the keratin protein backbone, the relative intensity and position of the amide III peak at 1251 cm−1 (indicative of the β-sheet confirmation), and the C-H peak at 1317 cm−1.28 These features are distinct from those of the skin mentioned earlier. For example, the spectral difference between the skin and nail spectra at the 1770-cm−1 lipid peak indicates the lack of contribution from soft tissue components in the nail.
Figure 6.
RS-OCT of the region surrounding the proximal nail fold. (a) OCT image. Nail is labeled on the right side of the image, and inserts into the nail bed beneath the cuticle, which is located in the transverse dimension from 2.5 to 3.0 mm. To the left of the cuticle is the skin on the back of the finger. Raman spectra acquired from the regions are indicated from the red overlays. (b) Raman spectra of the skin and nail. Peaks of interest are indicated with arrows, and include the sharp 620-cm−1 C-S peak, the 936-cm−1 C-C protein backbone peak, the relative intensity and position of the amide III (1251 cm−1), and C-H (1317 cm−1) peaks, all of which are indicative of the β-sheet keratin proteins that make up the nail. In contrast, the 1770-cm−1 lipid peak seen in soft tissue and skin is not seen in the nail.(Color online only.)
Discussion
We report the development of a combined RS-OCT instrument employing common detection hardware, and demonstrate applications of the device for morphological and biochemical characterization of ex-vivo rodent calvaria and retina, along with in-vivo analysis of human skin. Previously reported implementations of RS-OCT consisted of two independent systems combined at the sample arm level.13, 14 In the case of both RS and OCT, the extensive hardware required for independent detection is both costly and cumbersome. However, spectral domain configurations of OCT utilize a detection platform similar to RS. The novel design we demonstrate here takes advantage of this fact and integrates the detection arms of both modalities into a single spectrograph and CCD. The result is a fully integrated system that demonstrates for the first time, to our knowledge, in-vivo characterization of both the biochemical composition and microstructure of tissues with a common-detector RS-OCT system.
The results shown here demonstrate the instrument’s capabilities, and provide a set of examples describing potential applications. The images and spectra from the calvaria (Fig. 3) demonstrate the ability of the device to characterize highly scattering tissue. In the field of bone biology, histomorphometric measurement of calvarial thickness in mouse models is a time-consuming and destructive assay used to characterize bone formation. In addition to structural measures, a number of other destructive or time-consuming assays are performed to characterize compositional properties related to mineralization and bone formation.29 The clear definition of the inner and outer surfaces of the calvaria in OCT suggests that it would be possible to perform nondestructive measurement of calvaria thickness, while compositional properties of the bone such as mineral-to-collagen ratio, mineral crystallinity, and carbonate substitution can be calculated from analysis of the Raman spectrum.22 The ability to perform nondestructive high resolution imaging and biochemical characterization of murine calvaria in realtime represents one example of the benefits a common-detector RS-OCT system can provide in the context of ex-vivo analysis of dissected tissues.
Biochemical and morphological analysis of retinas in animal models is another promising potential application of the common-detector RS-OCT device. Retinal imaging is the most prominent clinical application of OCT; however, recent studies have demonstrated that the utility of OCT can be extended to the evaluation of rodent models of retinal pathology as well. For example, OCT was found to serve as a useful tool for monitoring tumor progression in a rat model of retinoblastoma30 and retinal degeneration in a transgenic mouse model.31 The ex-vivo images acquired with the common-detector RS-OCT instrument [Fig. 4a] allow visualization of most layers of the retina, as can be confirmed by comparison to the corresponding histology cross section ]Fig. 4b]. These images certainly allow evaluation of retinal architecture, and could also be used to perform quantitative measurements of the thickness of specific retinal layers. Such analysis would be well complemented by RS, which is capable of identifying the molecular progression of disease. For example, recent work has demonstrated the sensitivity of RS to advanced glycation end products, which are molecules widely believed to play an important role in the ill-defined pathogenesis of age-related macular degeneration.32 The expanding role of OCT in animal studies, coupled with the sensitivity of RS to biochemical features invisible to OCT, suggests that common-detector RS-OCT has the potential to become a valuable research tool for the study of retinal disease in animal models. The application of RS-OCT to human retinal analysis is not possible at this time, however, due to the fact that the laser exposure levels required to perform RS are well in excess of the Americal National Standards Institute (ANSI) standards.
The ability of common-detector RS-OCT to perform in-vivo characterization of tissue morphology and biochemical composition is demonstrated through evaluation of human skin (Fig. 5). The OCT images depict the stratified nature of the skin, and can even visualize the presence of microstructural features within the stratum corneum. Based on a previously published report of OCT in the skin that details the appearance of sweat glands in the stratum corneum,33 and the fact that the subject’s skin was healthy, clean, and debris-free, we believe this feature is likely a sweat gland. It is unlikely the feature is simply an artifact. In spectral domain OCT, artifacts can be seen as a result of strong reflections from the primary surface. Such artifacts can be observed near 0.9 mm in the transverse dimension of Fig. 5a, and are seen both above and below the primary surface. The feature we believe to be a sweat gland has a much different appearance and is only seen below the skin surface. The Raman spectrum depicts signal in all the primary molecular moieties associated with the skin, including peaks associated with the collagen backbone, phenylalanine, and amide I and III.34 The ultimate strength of RS-OCT arises from its ability to utilize OCT imaging to guide positioning of the RS probe beam, as well as the instruments ability to use the RS subsystem to interrogate the biochemical composition of morphological features in an OCT image. The fundamental concept that RS-OCT is capable of characterizing both the morphological and biochemical properties of tissue is thus demonstrated through the analysis of tissues surrounding the proximal nail fold on the finger of a human volunteer.
There were a few important experimental considerations that enabled acquisition of consistent datasets from the RS-OCT instrument, most importantly during in-vivo measurements. Namely, the speed of both the RS and OCT subsystems required that the sample remain fairly stable over the course of the data acquisition times. In the case of RS, spectral quality degraded if motion artifact was not minimized by restraining the palm or finger to a sample stage prior to measurement. Stabilization of the sample was also important to minimize the effect of fringe washout, which occurs due to sample motion over the acquisition time of a single A-scan.35 The result is noticeable image fading, which can also arise from movements and vibrations transferred to the fiber optic components of the OCT system. Thus, all the fibers were affixed to the table, and the instrument was built on a vibration dampening pneumatic table. To minimize image distortion in OCT due to these motion-related artifacts, the spectra were collected at the fastest possible frame rate and are also unaveraged. Therefore, the images shown in Figs. 5a, 6a represent the performance limit of the OCT subsystem with respect to sensitivity and speed. These limitations are a function of the deep-depletion, back-illuminated detector array chosen to ensure feasibility of RS. To further advance potential in-vivo applications of common-detector RS-OCT, it will certainly be necessary to incorporate an improved detector capable of improving the speed of both RS and OCT.
Commercially available deep-depletion, back-illuminated detectors have limitations in readout rate and dynamic range, which in turn limit OCT imaging speed and sensitivity. These detectors are typically operated at slow readout rates with a premium placed on sensitivity. The corresponding detection electronics therefore have a limited bandwidth to ensure low read noise. To increase the imaging speed, it would be necessary to both decrease the exposure time of the detector and increase the bandwidth of the detection electronics to improve readout rates. The detected spectra in RS are typically shot-noise limited by the relatively intense background tissue autofluorescence, which indicates that the detector speed could be increased to some degree without adversely affecting the integrity of RS. Frame transfer cameras with shorter exposure times and a second buffer array are available, and offer the potential to moderately improve the imaging performance when taking OCT “snapshots.” Decreasing exposure time would reduce the susceptibility of OCT to fringe washout; however such technology would only allow rapid acquisition of at most a few image frames and would not facilitate a faster continuous OCT frame rate. To further increase OCT sensitivity based on the current system design, the sample must simply be illuminated with increased laser power without saturating the detector. Currently, the detector is illuminated very near the saturation level, which is typically proportional to the full well capacity of the detector elements. A detector with increased pixel full well capacity and thus increased saturation levels would alleviate this constraint. To our knowledge, such a detector is not currently available from standard commercial manufacturers. Nevertheless, the realization of a detector with the properties described could improve OCT imaging performance and improve the in-vivo imaging potential of the device.
In addition to improving the sensitivity of OCT imaging, increasing the achievable imaging depth would also benefit the system’s ease of use and imaging performance. As previously described, the OCT spectral resolution limits the achievable imaging depth range to 1.27 mm. Although this imaging range exceeds the optical penetration depth of the 855-nm source in the samples we imaged earlier, an expanded range would simplify localization of the sample as well as mitigate the depth-dependent sensitivity roll-off that results from the finite spectrograph spectral resolution.36 The OCT spectral resolution in the system we describe is limited by the horizontal size of the individual CCD pixels. Decreasing the horizontal pixel size and increasing the number of horizontal pixels from 1024 to 2048 would be beneficial. In addition, as seen in Fig. 1, the detectable spectral range begins at 780 nm, despite the fact that the OCT source intensity falls below the noise floor near 800 nm and the RS filter cutoff occurs at 818 nm. If a CCD with increased pixel density was obtained, limiting illumination of CCD to the 800- to 920-nm spectral band with a custom grating would further improve the spectral resolution, thereby expanding the achievable imaging range, and increasing imaging performance as a function of depth.
Despite the fact that the camera used in this work can benefit from further optimization, we have demonstrated the ability of a commercially available detector commonly used in tissue RS to acquire OCT images of microstructural details in tissue. The significance of this demonstration lies in the implication that a few important modifications to an existing RS instrument can enable OCT imaging at a fraction of the cost of adapting an existing OCT instrument to perform RS. The essential components to perform this modification include the OCT light source, a 2×2 fiber coupler, and an input adapter for the spectrograph, along with the appropriate optics for the reference and sample arms. 2-D OCT images can be generated by translating the sample with a motorized stage in place scanning the sampling beam with a galvanometer. The instrument can be further simplified by replacing the components that automate the transition between OCT and RS (MEMS optical switch and the translation stage mounted mirror in the sample arm) with manual flip mirrors. Although these simplifications would come at the expense of the system’s ease of use, they provide basic design guidelines that would enable current users of RS to complement biochemically specific spectra with OCT images of tissue microstructure. Another common wavelength source for tissue RS is 830 nm; however, as previously stated, there are no OCT sources that overlap the 830-nm RS fingerprint region known to the authors. If an OCT source with a spectral range was fully detected within the range covered by typical 830-nm RS spectrographs (830 to 1000 nm), it would certainly be well suited for incorporation into a RS-OCT system similar to that described here. Alternatively, adaptation of the spectrograph dispersion to cover the appropriate spectral ranges of an existing OCT source and the 830-nm RS fingerprint region is also possible, granted a sufficient spectral resolution is achieved.
The system we describe represents development of the dual-modal RS-OCT instruments we have previously discussed;13, 14 however, alternative approaches have been reported to combine the biochemical specificity of vibrational spectroscopy with high resolution morphological imaging. Techniques utilizing coherent anti-Stokes Raman spectroscopy (CARS) have been reported for in-vivo video-rate microscopy,37 as well as CARS-based interferometric imaging,38 however, the cost of the ultrafast laser sources necessary for such approaches contrasts starkly with the simplicity of the stripped-down common-detector RS-OCT instrument proposed in the previous paragraph.
The benefit of complementing Raman spectral analysis with OCT imaging is based on the fact that point-wise RS provides no direct spatially relevant information. Accordingly, OCT images can be useful to guide positioning of the Raman probe beam as well as provide contextual morphological and microstructural information to benefit tissue characterization. Similarly, Raman spectra can provide information related to the biochemical content of features within an OCT image, as well as valuable data related to general compositional properties that are not directly accessible from images of tissue reflectivity. The underlying value of RS-OCT is based on the complementary nature of the RS and OCT datasets, and could be beneficial to a wide range of applications beyond those demonstrated here. Biochemical and morphological analysis can potentially assist in screening and diagnosis of diseases such as cancer, enhance the guidance and monitoring of medical therapies, and even benefit nonbiomedical applications where high-resolution cross sectional imaging and high specificity compositional analysis are critical, such as art preservation and restoration.
Conclusions
We report on the design and demonstration of a fully integrated RS-OCT system that employs a common detection arm to acquire both morphological and biochemical features of tissue. While previous implementations of RS-OCT simply provided coaligned sampling beams, the instrument we demonstrate uses a common-detection arm to perform both RS and OCT. The significance of the common-detector design lies in the fact that it reduces the hardware requirements for RS-OCT and simplifies implementation. The instrument uses a single spectrograph to perform 855-nm OCT in the spectral domain, and RS with a 785-nm source, whose biological fingerprint spectral range overlaps the OCT bandwidth. The two beams are coaligned in the sample arm, allowing the instrument to perform sequential, coregistered acquisition of the complementary datasets. OCT can be performed at 2 frames∕sec with a sensitivity of -86 dB and a spatial resolution of 8 μm (axial, in tissue) by 18 μm (transverse). Averaging consecutive frames allows the imaging sensitivity to improved to better than -100 dB. Tissue Raman spectra corresponding to specific A-scans in the OCT image are acquired in 30 sec with 40 mW of laser power. The capabilities of the instrument are demonstrated ex vivo in the calvaria and retina of rodents, and in vivo in human skin.
Acknowledgments
The authors would like to acknowledge assistance from Dean Paras and Jonathon Cayce in the Biomedical Optics Laboratory, and Professor John Penn in the Vanderbilt Eye Institute. This work was supported by the National Institutes of Health R21 CA133477–01∕02. Kalkman gratefully acknowledges the support of the Smart Mix Program of the Netherlands Ministry of Economic Affairs, and the Netherlands Ministry of Education, Culture, and Science. Faber is supported by a personal grant in the Vernieuwingsimpuls program (AGT07544) by the Netherlands Organization of Scientific Research (NWO) and the Technology Foundation STW.
References
- Fujimoto J. G., Brezinski M. E., Tearney G. J., Boppart S. A., Bouma B., Hee M. R., Southern J. F., and Swanson E. A., “Optical biopsy and imaging using optical coherence tomography,” Nat. Med. 1(9), 970–972 (1995). 10.1038/nm0995-970 [DOI] [PubMed] [Google Scholar]
- Mahadevan-Jansen A., “Raman spectroscopy: from benchtop to bedside,” in Biomedical Photonics Handbook, Vo Dinh T., Ed., pp. 30:31–27, CRC Press, Boca Raton, FL: (2003). [Google Scholar]
- Bowd C., Zangwill L. M., Berry C. C., Blumenthal E. Z., Vasile C., Sanchez-Galeana C., Bosworth C. F., and Sample P. A., “Detecting early glaucoma by assessment of retinal nerve fiber layer thickness and visual function,” Invest. Ophth. Vis. Sci. 42(9), 1993–2003 (2001). [PubMed] [Google Scholar]
- Zuccaro G., Gladkova N., Vargo J., Feldchtein F., Zagaynova E., Conwell D., Falk G., Goldblum J., Dumot J., Ponsky J., Gelikonov G., Davros B., Donchenko E., and Richter J., “Optical coherence tomography of the esophagus and proximal stomach in health and disease,” Am. J. Gastroenterol. 96(9), 2633–2639 (2001). 10.1111/j.1572-0241.2001.04119.x [DOI] [PubMed] [Google Scholar]
- Mogensen M., Joergensen T. M., Nurnberg B. M., Morsy H. A., Thomsen J. B., Thrane L., and Jemec G. B., “Assessment of optical coherence tomography imaging in the diagnosis of non-melanoma skin cancer and benign lesions versus normal skin: observer-blinded evaluation by dermatologists and pathologists,” Dermatol. Surg. 35(6), 965–972 (2009). 10.1111/j.1524-4725.2009.01164.x [DOI] [PubMed] [Google Scholar]
- Van Der Meer F. J., Faber D. J., Sassoon D. M. B., Aalders M. C., Pasterkamp G., and van Leeuwen T. G., “Localized measurement of optical attenuation coefficients of atherosclerotic plaque constituents by quantitative optical coherence tomography,” IEEE T. Med. Imag. 24(10), 1369–1376 (2005). 10.1109/TMI.2005.854297 [DOI] [PubMed] [Google Scholar]
- Cilesiz I., Fockens P., Kerindongo R., Faber D., Tytgat G., ten Kate F., and van Leeuwen T., “Comparative optical coherence tomography imaging of human esophagus: How accurate is localization of the muscularis mucosae?” Gastrointest. Endosc. 56(6), 852–857 (2002). 10.1016/S0016-5107(02)70359-3 [DOI] [PubMed] [Google Scholar]
- Mahadevan-Jansen A., Mitchell M. F., Ramanujam N., Malpica A., Thomsen S., Utzinger U., and Richards-Kortum R., “Near-infrared Raman spectroscopy for in vitro detection of cervical precancers,” Photochem. Photobiol. 68(1), 123–132 (1998). 10.1111/j.1751-1097.1998.tb03262.x [DOI] [PubMed] [Google Scholar]
- Lieber C. A., Majumder S. K., Ellis D. L., Billheimer D. D., and Mahadevan-Jansen A., “In vivo nonmelanoma skin cancer diagnosis using Raman microspectroscopy,” Laser. Surg. Med. 40(7), 461–467 (2008). 10.1002/lsm.20653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haka A. S., Shafer-Peltier K. E., Fitzmaurice M., Crowe J., Dasari R. R., and Feld M. S., “Diagnosing breast cancer by using Raman spectroscopy,” Proc. Natl. Acad. Sci. USA 102(35), 12371–12376 (2005). 10.1073/pnas.0501390102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim M. G., Song L. M., Marcon N. E., and Wilson B. C., “In vivo near-infrared Raman spectroscopy: demonstration of feasibility during clinical gastrointestinal endoscopy,” Photochem. Photobiol. 72(1), 146–150 (2000). [DOI] [PubMed] [Google Scholar]
- Ko A. C., Choo-Smith L. P., Hewko M., Leonardi L., Sowa M. G., Dong C. C., Williams P., and Cleghorn B., “Ex vivo detection and characterization of early dental caries by optical coherence tomography and Raman spectroscopy,” J. Biomed. Opt. 10(3), 031118 (2005). 10.1117/1.1915488 [DOI] [PubMed] [Google Scholar]
- Patil C. A., Bosschaart N., Keller M. D., van Leeuwen T. G., and Mahadevan-Jansen A., “Combined Raman spectroscopy and optical coherence tomography device for tissue characterization,” Opt. Lett. 33(10), 1135–1137 (2008). 10.1364/OL.33.001135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J. W., Zawadzki R. J., Liu R., Chan J. W., Lane S. M., and Werner J. S., “Optical coherence tomography and Raman spectroscopy of the ex-vivo retina,” J. Biophoton. 2(6–7), 398–406 (2009). 10.1002/jbio.200910022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraga J. J., Feld M. S., and Rava R. P., “Rapid near-infrared Raman-spectroscopy of human tissue with a spectrograph and CCD detector,” Appl. Spectrosc. 46(2), 187–190 (1992). 10.1366/0003702924125555 [DOI] [Google Scholar]
- Hausler G. and Lindner M. W., “Coherence radar and spectral radar—new tools for dermatological diagnosis,” J. Biomed. Opt. 3(1), 21–31 (1998). 10.1117/1.429899 [DOI] [PubMed] [Google Scholar]
- Boustany N. N., Manoharan R., Dasari R. R., and Feld M. S., “Ultraviolet resonance Raman spectroscopy of bulk and microscopic human colon tissue,” Appl. Spectrosc. 54(1), 24–30 (2000). 10.1366/0003702001948330 [DOI] [Google Scholar]
- Kawabata T., Mizuno T., Okazaki S., Hiramatsu M., Setoguchil T., Kikuchi H., Yamamoto M., Hiramatsu Y., Kondo K., Baba M., Ohta M., Kamiya K., Tanaka T., Suzuki S., and Konno H., “Optical diagnosis of gastric cancer using near-infrared multichannel Raman spectroscopy with a 1064-nm excitation wavelength,” J. Gastroenterol. 43(4), 283–290 (2008). 10.1007/s00535-008-2160-2 [DOI] [PubMed] [Google Scholar]
- Leitgeb R., Hitzenberger C. K., and Fercher A. F., “Performance of fourier domain vs. time domain optical coherence tomography,” Opt. Express. 11(8), 889–894 (2003). 10.1364/OE.11.000889 [DOI] [PubMed] [Google Scholar]
- Wojtkowski M., Leitgeb R., Kowalczyk A., Bajraszewski T., and Fercher A. F., “In vivo human retinal imaging by Fourier domain optical coherence tomography,” J. Biomed. Opt. 7(3), 457–463 (2002). 10.1117/1.1482379 [DOI] [PubMed] [Google Scholar]
- Lieber C. A. and Mahadevan-Jansen A., “Automated method for subtraction of fluorescence from biological Raman spectra,” Appl. Spectrosc. 57(11), 1363–1367 (2003). 10.1366/000370203322554518 [DOI] [PubMed] [Google Scholar]
- Carden A. and Morris M. D., “Application of vibrational spectroscopy to the study of mineralized tissues (review),” J. Biomed. Opt. 5(3), 259–268 (2000). 10.1117/1.429994 [DOI] [PubMed] [Google Scholar]
- Ascenzi A. and Fabry C., “Technique for dissection and measurement of refractive index of osteones,” J. Biophys. Biochem. Cytol. 6(1), 139–142 (1959). 10.1083/jcb.6.1.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izatt J. A., Rollins A. M., Ung-arunyawee R., Yazdanfar S., and Kulkami M. D., “System integration and signal∕image processing,” in Handbook of Optical Coherence Tomography, Bouma B. and Tearney G., Eds., pp. 143–174, Marcel Dekker, New York: (2002). [Google Scholar]
- Beattie J. R., Brockbank S., McGarvey J. J., and Curry W. J., “Effect of excitation wavelength on the Raman spectroscopy of the porcine photoreceptor layer from the area centralis,” Mol. Vis. 11, 825–832 (2005). [PubMed] [Google Scholar]
- Beattie J. R., Brockbank S., McGarvey J. J., and Curry W. J., “Raman microscopy of porcine inner retinal layers from the area centralis,” Mol. Vis. 13, 1106–1113 (2007). [PMC free article] [PubMed] [Google Scholar]
- Ding H. F., Lu J. Q., Wooden W. A., Kragel P. J., and Hu X. H., “Refractive indices of human skin tissues at eight wavelengths and estimated dispersion relations between 300 and 1600 nm,” Phys. Med. Biol. 51(6), 1479–1489 (2006). 10.1088/0031-9155/51/6/008 [DOI] [PubMed] [Google Scholar]
- Edwards H. G. M., Hunt D. E., and Sibley M. G., “FT-Raman spectroscopic study of keratotic materials: horn, hoof and tortoiseshell,” Spectrochim. Acta. Part A: Molecular Biomolecular Spectrosc. 54(5), 745–757 (1998). 10.1016/S1386-1425(98)00013-4 [DOI] [PubMed] [Google Scholar]
- Arnett T. R. and Henderson B., Methods in Bone Biology, Chapman Hall, New York: (1998). [Google Scholar]
- Ruggeri M., Tsechpenakis G., Jiao S., Jockovich M. E., Cebulla C., Hernandez E., Murray T. G., and Puliafito C. A., “Retinal tumor imaging and volume quantification in mouse model using spectral-domain optical coherence tomography,” Opt. Express. 17(5), 4074–4083 (2009). 10.1364/OE.17.004074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. H., Puoris’haag M., Maguluri G. N., Umino Y., Cusato K., Barlow R. B., and de Boer J. F., “Monitoring mouse retinal degeneration with high-resolution spectral-domain optical coherence tomography,” J. Vis. 8(1), 17 11–11 (2008). 10.1167/8.1.17 [DOI] [PubMed] [Google Scholar]
- Glenn J. V., Beattie J. R., Barrett L., Frizzell N., Thorpe S. R., Boulton M. E., McGarvey J. J., and Stitt A. W., “Confocal Raman microscopy can quantify advanced glycation end product (AGE) modifications in Bruch’s membrane leading to accurate, nondestructive prediction of ocular aging,” Faseb. J. 21(13), 3542–3552 (2007). 10.1096/fj.06-7896com [DOI] [PubMed] [Google Scholar]
- Lademann J., Otberg N., Richter H., Meyer L., Audring H., Teichmann A., Thomas S., Knüttel A., and Sterry W., “Application of optical non-invasive methods in skin physiology: a comparison of laser scanning microscopy and optical coherent tomography with histological analysis,” Skin Res. Technol. 13(2), 119–132 (2007). 10.1111/j.1600-0846.2007.00208.x [DOI] [PubMed] [Google Scholar]
- Caspers P. J., Lucassen G. W., Wolthuis R., Bruining H. A., and Puppels G. J., “In vitro and in vivo Raman spectroscopy of human skin,” Biospectrosc. 4(5 Suppl), S31–39 (1998). 10.1002/(SICI)1520-6343(1998)4:5+3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- Yun S. H., Tearney G., de Boer J., and Bouma B., “Motion artifacts in optical coherence tomography with frequency-domain ranging,” Opt. Express 12(13), 2977–2998 (2004). 10.1364/OPEX.12.002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrer C., Belabas N., Likforman J. P., and Joffre M., “Spectral resolution and sampling issues in Fourier-transform spectral interferometry,” J. Opt. Soc. Am. B-Opt. Phys. 17(10), 1795–1802 (2000). 10.1364/JOSAB.17.001795 [DOI] [Google Scholar]
- Evans C. L., Potma E. O., Puoris’haag M., Cote D., Lin C. P., and Xie X. S., “Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy,” Proc Natl. Acad. Sci. USA 102(46), 16807–16812 (2005). 10.1073/pnas.0508282102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D. L. and Boppart S. A., “Nonlinear interferometric vibrational imaging,” Phys. Rev. Lett. 92(12), 123905-1 to 123905-4 (2004). 10.1103/PhysRevLett.92.123905 [DOI] [PubMed] [Google Scholar]