Abstract
Background
The long-term goal of the GKDZI (Genetics of Kidney Disease in Zuni Indians) Study is to identify genes, environmental factors, and genetic-environmental interactions that modulate susceptibility to renal disease and intermediate phenotypes.
Study Design
A community-based participatory research approach was used to recruit family members of individuals with kidney disease.
Setting & Participants
The study was conducted in the Zuni Indians, a small endogamous tribe located in rural New Mexico. We recruited members of extended families, ascertained through a proband with kidney disease and at least 1 sibling with kidney disease. 821 participants were recruited, comprising 7,702 relative pairs.
Predictor Outcomes & Measurements
Urine albumin-creatinine ratio (UACR) and hematuria were determined in 3 urine samples and expressed as a true ratio. Glomerular filtration rate (GFR) was estimated using the Modification of Diet in Renal Disease (MDRD) Study equation modified for American Indians. Probands were considered to have kidney disease if UACR was ≥0.2 in 2 or more of 3 spot urine samples or estimated GFR was decreased according to the CRIC (Chronic Renal Insufficiency Cohort) Study criteria.
Results
Kidney disease was identified in 192 participants (23.4%). There were significant heritabilities for estimated GFR, UACR, serum creatinine, serum urea nitrogen, and uric acid and a variety of phenotypes related to obesity, diabetes, and cardiovascular disease. There were significant genetic correlations of some kidney-related phenotypes with these other phenotypes.
Limitations
Limitations include absence of renal biopsy, possible misclassification bias, lack of direct GFR measurements, and failure to include all possible environmental interactions.
Conclusions
Many phenotypes related to kidney disease showed significant heritabilities in Zuni Indians, and there were significant genetic correlations with phenotypes related to obesity, diabetes, and cardiovascular disease. The study design serves as a paradigm for the conduct of research in relatively isolated, endogamous, underserved populations.
INDEX WORDS: Genetics, heritability, American Indians, kidney diseases, risk factors, glomerular filtration rate, urine albumin-creatinine ratio (UACR), creatinine, serum urea nitrogen (SUN), uric acid
The Zuni Indians are experiencing an epidemic of chronic kidney disease (CKD). The prevalence of end-stage renal disease is 20.0-, 4.4-, and 5.6-fold higher than in European and African Americans and the composite estimate for American Indians, respectively.1,2 Earlier studies, which were not population based, attributed most kidney disease to mesangiopathic glomerulonephritis.3–6 Presently, >95% of end-stage renal disease is attributable to diabetic nephropathy.
To decrease the burden of CKD, the Zuni Pueblo established the Zuni Kidney Project in partnership with the Indian Health Service, University of New Mexico, Southwest Foundation for Biomedical Research, and Dialysis Clinic Inc.2 We conducted a population-based cross-sectional survey that showed high prevalence estimates, age-and sex-adjusted to the Zuni population, for decreased estimated glomerular filtration rate (eGFR),7 albuminuria, and hematuria.8 Prevalence estimates for albuminuria and hematuria were higher for diabetic than nondiabetic participants.8,9
The GKDZI (Genetics of Kidney Disease in Zuni Indians) Study was initiated to identify genes, environmental factors, and genetic-environmental interactions that modulate susceptibility to CKD and intermediate phenotypes. This report presents heritability estimates and genetic correlations for CKD, diabetes, and cardiovascular disease phenotypes.
METHODS
Study Design
GKDZI is a community-based participatory research project. Institutional review boards from each institution approved the study, and informed consent was obtained. We recruited 821 members of extended families ascertained through probands with CKD and 1 or more affected sibling(s).
Setting
The Zuni Pueblo in rural New Mexico is relatively endogamous. The tribe has approximately 10,000 members, and 80% live in the pueblo. Median age is 26 years.10 Most adult tribal members work as artisans making jewelry, pottery, and fetishes, which are a contemporary art form that represents animals and icons important to the Zuni.
Participants
Probands were identified from Zuni Kidney Project survey participants.7–9,11,12 Eligibility criteria for probands and affected siblings included age 18 years or older and evidence of CKD, for example, urine albumin-creatinine ratio (UACR) ≥0.2 in at least 2 of 3 urine samples or decreased eGFR.13 We used parental identities to construct family trees and determine the relatedness of individual pairs. We recruited first-, second-, and third-degree relatives of probands and their spouses. First-degree relatives are parents, siblings, and offspring; second-degree relatives are aunts and uncles, nieces and nephews, grandparents, and grandchildren; and third-degree relatives are first cousins, great aunts, great uncles, etc. All family members with CKD were eligible. We used PEDSYS (Southwest Foundation for Biomedical Research, http://pedsys.sfbrgenetics.org)14 for data entry, quality control, report generation, and preparation of data files for statistical genetic analysis, and PedigreeDraw,15 a family tree drawing program (Jurek Software, www.pedigree-draw.com).
Variables
Participants were considered to have diabetes if they met at least 1 of the following conditions: (1) history of diabetes, (2) plasma glucose level ≥200 mg/dL, (3) hemoglobin A1c (HbA1c) level >7.0%,16,17 or (4) receiving diabetes medication(s). Diabetes status in participants with HbA1c level of 6.0%–7.0%, plasma glucose level <200 mg/dL, and no history of diabetes was considered “indeterminate.”7 Participants were classified as hypertensive if they met at least 1 of the following conditions: (1) history of hypertension; (2) systolic or diastolic blood pressure ≥140 and ≥90 mm Hg, respectively;18 or (3) using antihypertensive medication(s). Blood was drawn for chemistry profile, HbA1c,17 serum creatinine (SCr),19 and, in a subset, serum cystatin C (SCysC) measurement.20 Buffy coats were obtained by centrifugation for DNA isolation. We assessed phenotypes related to CKD (eGFR, UACR, SCr, and serum urea nitrogen [SUN]) or diabetes and cardiovascular disease (weight, body mass index [BMI], HbA1c, diabetes status, hypertension status, serum triglycerides, high-density lipoprotein [HDL] cholesterol, low-density lipoprotein [LDL] cholesterol, and total cholesterol) in artisans and nonartisans.
Data Sources and Measurement
Questionnaire Data and Biological Measurements Made in the Home
We administered a questionnaire2 that ascertained birth dates, parents’ identities, education, occupation, tribal affiliation, language spoken, and medical history. Height and weight were measured.2 We identified overweight (BMI 25–29 kg/m2) and obese (BMI ≥30 kg/m2) participants. We measured systolic and diastolic blood pressure 3 times separated by 1-minute intervals and used the respective average values to classify hypertension status.
Reducing Bias in Biological Samples
To minimize classification bias, we attempted to obtain 3 urine samples from each participant. The median interval between urine collections was 2 days. We compared classifications of albuminuria and hematuria using the first versus the mode of 3 urine samples. UACR was classified as normal (<0.03), incipient (0.03–0.19), or overt (≥0.20). If all 3 samples were discordant, we used the median value. Urine albumin was measured using nephelometry.21–23 The presence of 3 or more red blood cells per high-power field was considered evidence of hematuria.
Reducing Bias in eGFR
We used the 4-variable Modification of Diet in Renal Disease (MDRD) Study equation, modified for use in American Indians,7,8,24 and a SCysC-based equation (both formulas are given in the notes to the third table in this article) to estimate GFR based on a single serum sample.25–28 Limitations of these equations include need for race-specific coefficients and lack of widespread calibration in SCr and SCysC assays.25 SCr level is influenced by muscle mass. eGFR may underestimate GFR in people with near-normal kidney function.25–28 We categorized eGFR using the National Kidney Foundation’s Kidney Disease Outcomes Quality Initiative (KDOQI)29 and the CRIC (Chronic Renal Insufficiency Cohort) Study age-specific criteria.13
Study Size
Study size was derived from previous studies of genetic effects on complex diseases in Mexican American,30,31 American Indian,32,33 and Alaskan Eskimo34,35 extended families. The GKDZI sample size was similar to these studies, in which we obtained significant heritabilities.
Statistical Methods
Questionnaire data were entered into a Microsoft Access database using double data entry and computerized range checks. Discrepancies were resolved by direct comparison with the questionnaire. Initial data entry error rates were low (2.1%) and decreased to <0.5%. Laboratory data were transferred electronically to the database.
Statistical analyses were conducted using SOLAR (Southwest Foundation for Biomedical Research, http://solar.sfbrgenetics.org).36,37 Data were expressed as mean ± standard error. For tests of differences between groups and calculation of heritabilities and correlations, all traits were inverse normalized. Observations were ranked and replaced by the expected value for that rank from a standard normal distribution. Multiple UACR measurements from an individual were evaluated using the Friedman nonparametric test for repeated measures. Phenotypic correlations of eGFR calculated using the MDRD Study equation modified for American Indians (eGFRMDRD-AI) versus eGFRSCysC were computed in SOLAR. Categorical classifications by eGFRMDRD-AI and eGFRSCysC were compared using a weighted κ, 95% confidence interval (CI), and symmetry tests. Differences were considered statistically significant for P < 0.05.
Heritability Estimation and Quantitative Genetic Analysis
We estimated the heritability (h2) of eGFRMDRD-AI, UACR, and other intermediate phenotypes and genetic correlations using maximum likelihood variance decomposition methods36,38 in SOLAR. Heritability is the ratio of the additive genetic variance to the total phenotypic variance.39,40 We calculated heritabilities using residual variances after accounting for effects of covariates. We constructed 2 sets of models, the first including age, sex, and their higher order terms and interactions as covariates and the second including these covariates plus diabetes, hypertension, and artisan status. We estimated the proportion of variance attributable to covariates by comparing models with and without covariates. Covariates were included on the basis of their biological, rather than statistical, significance. We computed Kullback-Leibler R2 values for dichotomous traits. We used family relationships and phenotypic measures in family members to infer the proportion of phenotypic variance attributable to the additive effects of genes and the genetic correlation between pairs of phenotypes. The calculations do not require information for specific genotypes. If other nonadditive sources of genetic variation exist, for example, dominance or epistasis, the reported heritabilities and correlations would represent lower bounds. We did not include household membership, which may fluctuate. Only if the exposure pattern for an environmental variable or of shared household membership mimicked Mendelian transmission would the heritability and correlation estimates be inflated by nongenetic factors.
Using the variance component model, P values for heritabilities were obtained by comparing the likelihood of a model in which h2 is estimated to the likelihood of a model in which h2 is constrained to zero. Twice the difference in the natural log likelihoods is asymptotically distributed as a ½:½ mixture of a χ2 distribution with 1 df and a point mass at zero. This approach allows an explicit test of whether correlations among family members are caused in part by additive genetic effects.
Bivariate analyses yield genetic and environmental correlations among phenotypes. Genetic correlations indicate the extent to which the additive effects of the same set of genes influence more than 1 phenotype. Environmental correlations include environmental effects and nonadditive genetic effects.
RESULTS
Participants
We recruited 821 participants from 30 families, of which 19 contained 3 or more generations and 11 contained 2 generations. After linking participants through offspring and marriage, 805 merged into 1 family. Among 821 participants, there were 7,702 relative pairs (Table 1). Median age was 36.7 years, and interquartile range was 21.1 years. Of the participants, 405 (49.3%) were female, 446 (54.3%) reported 12 or more years of education, 463 (56.4%) were artisans, and 686 (83.6%) spoke Shiwi, the Zuni language.
Table 1.
Distribution of Relative Pairs Among GKDZI Study Participants
| Relationship | Zuni Relative Pairs |
|---|---|
| Parent-offspring | 530 |
| Siblings | 448 |
| Half siblings | 352 |
| Avuncular (eg, uncle-niece) | 966 |
| Grand avuncular | 370 |
| Grandparent-grandchild | 158 |
| 1st cousin | 987 |
| 1st cousin once removed | 1,244 |
| 2nd cousin | 790 |
| Other relationship | 1,857 |
| Total | 7,702 |
Note: N = 821.
Abbreviation: GKDZI, Genetics of Kidney Disease in Zuni Indians.
Evidence of Kidney Disease in Diabetic and Nondiabetic Participants
Participants were considered to have CKD if they had a UACR >0.03 on 2 or more urine samples or decreased eGFR according to age-specific criteria used in CRIC. Hematuria was not used in the classification of CKD because we did not ascertain its cause. CKD was present in 192 participants (23.4%). Of these, 83 had diabetes, 108 did not have diabetes, and 1 had indeterminate diabetes status. Albuminuria was present in 105 (97.2%) nondiabetic and 73 (98.7%) diabetic participants with CKD stages 1–4 (P < 0.001). eGFR was decreased in 24 (28.9%) diabetic and 7 nondiabetic (6.5%) participants with CKD stages 1–5. Many nondiabetic participants with albuminuria and/or decreased eGFR (n = 108) had features of metabolic syndrome, for example, hypertension (52.8%), overweight (37.0%), and obesity (45.4%). All 12 participants with end-stage renal disease had diabetes. Inclusion of hematuria as a diagnostic criterion would have yielded an additional 32 participants with CKD.
Prevalence of Kidney Disease in Multigenerational Families
CKD was present in 2 or more generations in 23 families. In nondiabetic participants, 4, 3, and 2 generations were present in 1, 3, and 10 families, respectively. Two generations of diabetic participants with CKD were present in 10 families, and 3 generations, in 1 family.
UACR, Hematuria, and eGFR
Urine Albumin-Creatinine Ratio
Three urine samples or 2 concordant samples were obtained for 798 participants. Classifications using the initial versus the mode of 3 urine samples were similar (Table 2; weighted κ, 0.89 [95% CI, 0.85–0.92]).41 The symmetry test (s = 6.0; P = 0.1) showed no directional bias. However, the initial UACR tended to overestimate the prevalence of albuminuria. In participants with incipient albuminuria on the initial urine sample, 18 (15%) subsequently had 2 normal UACR determinations. Three participants (5%) with overt albuminuria on the initial urine sample subsequently had 2 normal UACR determinations. In participants with an initial normal UACR, 9 (1.5%) subsequently were reclassified as incipient, and 1 (0.2%), as overt albuminuria.
Table 2.
Comparison of UACR Classified by Initial Versus Mode of 3 Urine Samples
| Initial Sample | Mode of 3 Samples |
||
|---|---|---|---|
| Normal | Incipient | Overt | |
| Normal | 607 | 9 | 1 |
| Incipient | 18 | 97 | 6 |
| Overt | 3 | 2 | 55 |
Abbreviation: UACR, urinary albumin-creatinine ratio.
Hematuria
We determined the mode for the presence of red blood cells in 3 urine samples in 739 participants. The initial urine sample had 3 or more red blood cells per high-power field in 86 (11.6%) participants. However, using the mode of 3 urine samples, only 44 (51.2%) of these participants had 3 or more red blood cells per high-power field. In 653 (88.4%) participants without hematuria on the initial urine sample, only 12 (1.8%) had a mode of 3 or more red blood cells per high-power field (McNemar test, s = 16.7; P < 0.001). Despite modest agreement (κ = 0.58 [95% CI, 0.48–0.68]), the lack of symmetry indicated a bias toward a false-positive result of a single test for hematuria. Of 192 participants classified as having kidney disease using UACR or decreased eGFRMDRD-AI, only 24 (12.5%) had 3 or more red blood cells per high-power field.
Estimated GFR
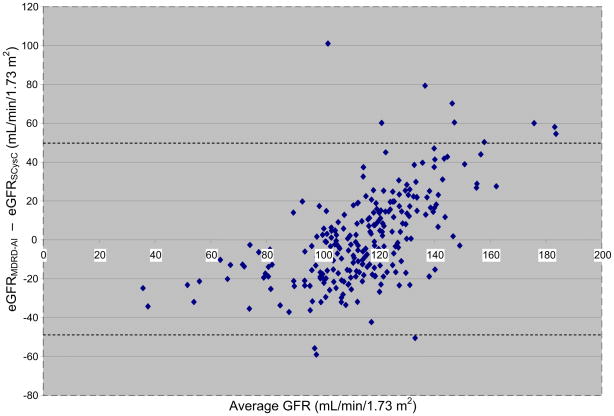
We compared the classification of kidney disease obtained using the MDRD Study equation modified for use in American Indians26,42 and the SCysC eGFR equation43 in 245 participants. There was moderately good agreement (κ = 0.50 [95% CI, 0.37–0.63]) using KDOQI stages.29 Agreement for the age-specific CRIC criteria13 was modest (κ = 0.39 [95% CI, 0.00–0.78]). Using KDOQI criteria, Tidman et al44 also reported good agreement between eGFRMDRD and eGFRSCysC. Although the MDRD Study equation modified for American Indians tended to classify low GFR more often than the SCysC eGFR equation, the difference was not significant (P = 0.1). Phenotypic correlations between the eGFRMDRD-AI and eGFRSCysC were moderately strong in the study sample as a whole and in groups stratified by age, sex, and diabetes status (Table 3). The Bland-Altman plot showed a slight bias because at lower values, the eGFRMDRD-AI tended to be lower than the eGFRSCysC. The opposite was true at higher values (Fig 1). Overall, differences between the eGFRMDRD-AI and eGFRSCysC were small. Therefore, we used eGFRMDRD-AI, which was less expensive to measure, in subsequent analyses. Hemodialysis patients were not included in the comparison of GFR-estimating equations because neither SCr nor SCysC concentrations are in steady state.
Table 3.
Correlations between eGFRMDRD-AI and eGFRSCysC
| No. of Participants | Correlation ± SEa | |
|---|---|---|
| All participants | 245 | 0.449 ± 0.05 |
| Age | ||
| Younger | 123 | 0.479 ± 0.07 |
| Older | 122 | 0.405 ± 0.08 |
| Sex | ||
| Female | 135 | 0.431 ± 0.05 |
| Male | 110 | 0.494 ± 0.05 |
| Diabetes status | ||
| Diabetic | 52 | Not computableb |
| Nondiabetic | 193 | 0.407 ± 0.06 |
Note: eGFRMDRD-AI was calculated in milliliters per minute per 1.73 m2 using the 4-variable MDRD (Modification of Diet and Renal Disease) Study equation modified for American Indians42: 186 × ([SCr]−1.154) × (age−0.203) × 0.742 [if female] × 1.106. eGFRSCysC was calculated in milliliters per minute per 1.73 m2 as: 33.7 + (−0.047 × age) + (68.4/SCysC).
Abbreviations: eGFR, estimated glomerular filtration rate; SCr, serum creatinine; SCysC, serum cystatin C; SE, standard error.
All correlations, P < 0.05.
Correlations between eGFRMDRD-AI and eGFRSCysC in diabetic participants could not be computed due to small sample size (n = 52).
Figure 1.
Bland-Altman plot shows the difference between glomerular filtration rate estimated using the Modification of Diet and Renal Disease Study equation modified for American Indians (eGFRMDRD-AI) and serum cystatin C–based formula (eGFRSCysC) plotted against the average of the eGFRMDRD-AI and eGFRSCysC.
Intermediate Phenotypes
Of the participants, 277 (33.7%) were classified as overweight (BMI, 25–29 kg/m2), and 325 (39.6%), as obese (BMI ≥ 30 kg/m2). There were 140 diabetic (17.1%) participants, of whom 17 (12.1%) were newly diagnosed. Hypertension was present in 286 (34.8%) participants. Of these, 105 (36.7%) were using an angiotensin-converting enzyme inhibitor or angiotensin receptor blocker and 93 (32.5%) were not previously aware of their hypertension.
Intermediate phenotypes related to CKD, obesity, diabetes, and cardiovascular disease stratified by sex (Table 4), diabetes (Table 5), and hypertension status (Table 6) are shown. There were significant differences by sex for eGFRMDRD-AI, SCr, uric acid, hematuria, weight, BMI, HbA1c, systolic blood pressure, diastolic blood pressure, LDL cholesterol, and total cholesterol (Table 4). eGFRMDRD-AI, UACR, SUN, uric acid, weight, BMI, HbA1c, systolic blood pressure, HDL cholesterol, LDL cholesterol, and triglycerides differed among diabetic versus nondiabetic participants (Table 5). HDL and LDL cholesterol were similar in hypertensive and nonhypertensive participants; however, all other phenotypes differed by hypertension status (Table 6).
Table 4.
Traits Related to Kidney Disease, Diabetes, and CVD in GKDZI Study Participants, by Sex
| Trait | All Participants | No. (women/men) | Women | Men | Pa |
|---|---|---|---|---|---|
| eGFRMDRD-AI (mL/min/1.73m2) | 116.14 ± 28.7 | 396/411 | 113.53 ± 31.2 | 118.65 ± 25.9 | 0.002 |
| UACR | 129.28 ± 566.1 | 397/413 | 100.68 ± 417.5 | 156.77 ± 678.3 | 0.3 |
| SCr (mg/dL) | 0.916 ± 0.88 | 404/416 | 0.864 ± 0.99 | 0.968 ± 0.77 | <0.001 |
| SUN (mg/dL) | 12.29 ± 6.4 | 404/415 | 12.52 ± 7.6 | 12.06 ± 5.1 | 0.7 |
| Uric acid (mg/dL) | 5.92 ± 1.7 | 403/415 | 5.17 ± 1.4 | 6.64 ± 1.6 | <0.001 |
| Hematuriab | 2.18 ± 10.6 | 397/412 | 3.04 ± 14.6 | 1.34 ± 3.3 | <0.001 |
| Weight (lb) | 166.99 ± 40.6 | 405/416 | 161.81 ± 38.8 | 172.04 ± 41.8 | <0.001 |
| BMI (kg/m2) | 29.44 ± 6.8 | 405/416 | 30.99 ± 7.4 | 27.93 ± 5.9 | <0.001 |
| HbA1c (%) | 5.81 ± 1.4 | 405/416 | 5.91 ± 1.4 | 5.72 ± 1.3 | 0.02 |
| SBP (mm Hg) | 122.54 ± 16.6 | 405/416 | 117.78 ± 15.0 | 127.17 ± 16.8 | <0.001 |
| DBP (mm Hg) | 77.70 ± 11.5 | 405/416 | 75.07 ± 10.0 | 80.27 ± 12.3 | <0.001 |
| HDL-C (mg/dL) | 49.47 ± 16.1 | 385/387 | 48.30 ± 14.5 | 50.64 ± 17.4 | 0.07 |
| LDL-C (mg/dL) | 98.29 ± 31.2 | 350/361 | 94.54 ± 27.5 | 101.93 ± 34.0 | 0.01 |
| Triglycerides (mg/dL) | 171.50 ± 131.4 | 402/414 | 169.35 ± 108.8 | 173.59 ± 150.3 | 0.5 |
| Total cholesterol (mg/dL) | 180.15 ± 38.3 | 402/414 | 175.16 ± 35.2 | 184.99 ± 40.5 | <0.001 |
Note: Unless otherwise indicated, values shown are mean ± standard error.
Abbreviations and definitions: BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated using the Modification of Diet in Renal Disease Study equation modified for American Indians; GKDZI, Genetics of Kidney Disease in Zuni Indians; HbA1c, glycated hemoglobin; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure; SCr, serum creatinine; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
All traits were inverse normalized to test differences between the 2 groups.
Hematuria is defined as 3 or more red blood cells per high-power field.
Table 5.
Traits Related to Kidney Disease, Diabetes, and CVD in GKDZI Study Participants, by Diabetes Status
| Trait | No. (no DM/DM)a | Participants Without DM | Participants With DM | Pb |
|---|---|---|---|---|
| eGFRMDRD-AI (mL/min/1.73 m2) | 679/128 | 117.89 ± 26.2 | 106.79 ± 38.5 | <0.001 |
| UACR | 678/131 | 59.23 ± 242.5 | 492.57 ± 1,236.6 | <0.001 |
| SCr (mg/dL) | 680/140 | 0.82 ± 0.30 | 1.39 ± 2.0 | 0.08 |
| SUN (mg/dL) | 679/140 | 11.23 ± 3.7 | 17.40 ± 12.0 | <0.001 |
| Uric acid (mg/dL) | 678/140 | 5.98 ± 1.7 | 5.59 ± 1.6 | 0.001 |
| Hematuriac | 677/131 | 2.16 ± 11.3 | 2.09 ± 5.2 | 0.4 |
| Weight (lb) | 680/140 | 164.62 ± 38.5 | 179.01 ± 47.6 | <0.001 |
| BMI (kg/m2) | 680/140 | 28.91 ± 6.5 | 32.08 ± 7.5 | <0.001 |
| HbA1c (%) | 680/140 | 5.37 ± 0.41 | 7.96 ± 2.2 | <0.001 |
| SBP (mm Hg) | 680/140 | 120.87 ± 15.9 | 130.77 ± 17.8 | <0.001 |
| DBP (mm Hg) | 680/140 | 77.49 ± 11.4 | 78.77 ± 12.0 | 0.9 |
| HDL-C (mg/dL) | 639/133 | 50.29 ± 16.3 | 45.55 ± 14.4 | 0.004 |
| LDL-C (mg/dL) | 594/117 | 99.31 ± 30.8 | 93.14 ± 32.6 | 0.03 |
| Triglycerides (mg/dL) | 676/140 | 158.95 ± 106.4 | 232.11 ± 204.6 | <0.001 |
| Total cholesterol (mg/dL) | 676/140 | 180.35 ± 37.3 | 179.16 ± 42.9 | 0.2 |
Note: Unless otherwise indicated, values shown are mean ± standard error.
Abbreviations and definitions: BMI, body mass index; DBP, diastolic blood pressure; CVD, cardiovascular disease; DM, diabetes mellitus; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated using the Modification of Diet and Renal Disease Study equation modified for American Indians; GKDZI, Genetics of Kidney Disease in Zuni Indians; HbA1c, glycated hemoglobin (based on American Diabetes Association criteria); HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure; SCr, serum creatinine; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
Diabetes status could not be determined in 1 participant.
All traits were inverse normalized to test differences between the 2 groups.
Hematuria is defined as 3 or more red blood cells per high-power field.
Table 6.
Traits Related to Kidney Disease, Diabetes, and CVD in GKDZI Study Participants, by HTN Status
| Trait | No. (no HTN/HTN) | Participants Without HTN | Participants With HTN | Pa |
|---|---|---|---|---|
| eGFRMDRD-AI (mL/min/1.73 m2) | 533/274 | 119.15 ± 26.6 | 110.27 ± 31.6 | <0.001 |
| UACR | 533/277 | 36.75 ± 148.4 | 307.32 ± 921.1 | <0.001 |
| SCr (mg/dL) | 534/286 | 0.799 ± 0.18 | 1.13 ± 1.5 | <0.001 |
| SUN (mg/dL) | 533/286 | 11.26 ± 3.5 | 14.20 ± 9.5 | <0.001 |
| Uric acid (mg/dL) | 532/286 | 5.74 ± 1.6 | 6.24 ± 1.7 | <0.001 |
| Hematuriab | 532/277 | 2.45 ± 12.7 | 1.66 ± 4.2 | 0.05 |
| Weight (lb) | 535/286 | 159.73 ± 35.9 | 180.57 ± 45.2 | <0.001 |
| BMI (kg/m2) | 535/286 | 28.35 ± 6.4 | 31.48 ± 7.0 | <0.001 |
| HbA1c (%) | 535/286 | 5.48 ± 0.88 | 6.43 ± 1.9 | <0.001 |
| SBP (mm Hg) | 535/286 | 115.34 ± 11.2 | 136.00 ± 16.8 | <0.001 |
| DBP (mm Hg) | 535/286 | 73.65 ± 8.3 | 85.29 ± 12.8 | <0.001 |
| HDL-C (mg/dL) | 501/271 | 49.46 ± 15.6 | 49.49 ± 16.9 | 0.6 |
| LDL-C (mg/dL) | 464/247 | 96.37 ± 28.3 | 101.90 ± 35.7 | 0.1 |
| Triglycerides (mg/dL) | 530/286 | 152.92 ± 105.0 | 205.94 ± 164.6 | <0.001 |
| Total cholesterol (mg/dL) | 530/286 | 175.7 ± 35.4 | 188.39 ± 41.9 | <0.001 |
Note: Unless otherwise indicated, values shown are mean ± standard error.
Abbreviations and definitions: BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated using the Modification of Diet and Renal Disease Study equation modified for American Indians; GKDZI, Genetics of Kidney Disease in Zuni Indians; HbA1c, glycated hemoglobin (based on American Diabetes Association criteria); HDL-C, high-density lipoprotein cholesterol; HTN, hypertension; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure; SCr, serum creatinine; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
All traits were inverse normalized to test differences between the 2 groups.
Hematuria is defined as 3 or more red blood cells per high-power field.
Approximately half the participants were artisans. Distributions of UACR, SUN, uric acid, systolic blood pressure, diastolic blood pressure, HDL cholesterol, LDL cholesterol, and total cholesterol were significantly higher in artisans (Table 7).
Table 7.
Traits Related to Kidney Disease, Diabetes, and CVD in GKDZI Study Participants, by Heavy Metal Exposure
| Trait | No. (nonartisans/artisans) | Participants Who Were Not Artisans | Participants Who Were Artisans | Pa |
|---|---|---|---|---|
| eGFRMDRD-AI (mL/min/1.73m2) | 348/459 | 118.15 ± 29.8 | 114.61 ± 27.8 | 0.2 |
| UACR | 349/461 | 78.26 ± 315.2 | 167.91 ± 696.4 | 0.009 |
| SCr (mg/dL) | 358/462 | 0.94 ± 1.1 | 0.90 ± 0.68 | 0.2 |
| SUN (mg/dL) | 358/461 | 12.87 ± 7.3 | 11.83 ± 5.7 | 0.002 |
| Uric acid (mg/dL) | 357/461 | 5.69 ± 1.7 | 6.09 ± 1.6 | 0.005 |
| Hematuriab | 349/460 | 2.29 ± 14.8 | 2.09 ± 5.4 | 0.9 |
| Weight (lb) | 358/463 | 166.30 ± 41.7 | 167.53 ± 39.8 | 0.1 |
| BMI (kg/m2) | 358/463 | 29.81 ± 7.2 | 29.15 ± 6.5 | 0.7 |
| HbA1c (%) | 358/463 | 5.79 ± 1.5 | 5.83 ± 1.3 | 0.3 |
| SBP (mm Hg) | 358/463 | 119.43 ± 15.9 | 124.94 ± 16.8 | <0.001 |
| DBP (mm Hg) | 358/463 | 75.17 ± 10.9 | 79.67 ± 11.6 | 0.003 |
| HDL-C (mg/dL) | 335/437 | 48.05 ± 15.6 | 50.57 ± 16.3 | 0.02 |
| LDL-C (mg/dL) | 311/400 | 93.25 ± 26.9 | 102.21 ± 33.6 | 0.001 |
| Triglycerides (mg/dL) | 356/460 | 172.05 ± 141.4 | 171.08 ± 123.3 | 0.9 |
| Total cholesterol (mg/dL) | 356/460 | 174.16 ± 35.6 | 184.78 ± 39.6 | <0.001 |
Note: Unless otherwise indicated, values shown are mean ± standard error.
Abbreviations and definitions: BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated by the Modification of Diet and Renal Disease Study equation modified for American Indians; GKDZI, Genetics of Kidney Disease in Zuni Indians; HbA1c, glycated hemoglobin; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; SBP, systolic blood pressure; SCr, serum creatinine; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
All traits were inverse normalized to test differences between the 2 groups.
Hematuria is defined as 3 or more red blood cells per high-power field.
Heritability and Genetic Correlations of Intermediate Phenotypes
Heritability estimates and proportions of variance attributable to covariates for phenotypes related to CKD and cardiovascular disease are listed (Table 8). Heritabilities for eGFRMDRD-AI, UACR, SCr, SUN, uric acid, SCysC, weight, BMI, HbA1c, systolic blood pressure, diastolic blood pressure, hypertension status, HDL cholesterol, LDL cholesterol, triglycerides, and total cholesterol were significantly different from zero. Heritabilities for hematuria and diabetes status failed to reach statistical significance. Inclusion of additional covariates in model 2 did not substantially change the estimated heritabilities. The proportion of total phenotypic variance attributable to covariates increased as the number of covariates increased. Proportions of variance attributable to covariates for model 2 ranged from 0.044 for LDL cholesterol to 0.437 for systolic blood pressure.
Table 8.
Heritabilities of Traits Related to Kidney Disease and to Obesity, Diabetes, and CVD
| Trait | Model 1 |
Model 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| No. | h2 ± SE | P | PVC | No. | h2 ± SE | P | PVC | |
| eGFRMDRD-AI | 818 | 0.32 ± 0.07 | <0.001 | 0.233 | 818 | 0.33 ± 0.07 | <0.001 | 0.233 |
| UACR | 797 | 0.28 ± 0.08 | <0.001 | 0.074 | 796 | 0.25 ± 0.08 | <0.001 | 0.161 |
| SCr | 809 | 0.23 ± 0.07 | <0.001 | 0.274 | 809 | 0.24 ± 0.07 | <0.001 | 0.274 |
| SUN | 807 | 0.22 ± 0.07 | <0.001 | 0.133 | 807 | 0.20 ± 0.08 | 0.001 | 0.159 |
| Uric acid | 816 | 0.29 ± 0.07 | <0.001 | 0.223 | 816 | 0.32 ± 0.07 | <0.001 | 0.244 |
| Hematuria | 761 | 0.07 ± 0.06 | 0.1 | 0.113 | 715 | 0.06 ± 0.06 | 0.2 | 0.114 |
| SCysC | 245a | 0.50 ± 0.13 | <0.001 | 0.235 | 245a | 0.46 ± 0.13 | <0.001 | 0.260 |
| Weight | 816 | 0.58 ± 0.08 | <0.001 | 0.077 | 815 | 0.53 ± 0.08 | <0.001 | 0.137 |
| BMI | 817 | 0.51 ± 0.08 | <0.001 | 0.108 | 816 | 0.44 ± 0.08 | <0.001 | 0.174 |
| HbA1c | 807 | 0.25 ± 0.07 | <0.001 | 0.133 | 806 | 0.28 ± 0.07 | <0.001 | 0.322 |
| Diabetes status | 819 | 0.07 ± 0.15 | 0.3 | 0.213b | 819 | 0.16 ± 0.16 | 0.1 | 0.304b |
| SBP | 818 | 0.31 ± 0.07 | <0.001 | 0.272 | 817 | 0.15 ± 0.06 | 0.003 | 0.437 |
| DBP | 818 | 0.24 ± 0.07 | <0.001 | 0.201 | 817 | 0.31 ± 0.06 | 0.007 | 0.363 |
| HTN status | 820 | 0.58 ± 0.15 | <0.001 | 0.168b | 819 | 0.60 ± 0.15 | <0.001 | 0.236b |
| Triglycerides | 809 | 0.27 ± 0.07 | <0.001 | 0.029 | 809 | 0.27 ± 0.07 | <0.001 | 0.089 |
| HDL-C | 770 | 0.39 ± 0.07 | <0.001 | 0.033 | 770 | 0.37 ± 0.07 | <0.001 | 0.063 |
| LDL-C | 667 | 0.31 ± 0.09 | <0.001 | 0.023 | 667 | 0.30 ± 0.09 | <0.001 | 0.044 |
| Total cholesterol | 813 | 0.37 ± 0.07 | <0.001 | 0.070 | 813 | 0.36 ± 0.07 | <0.001 | 0.085 |
Note: Glucose excluded because many participants were not fasting. Model 1: adjusted for age, sex, age2, age × sex, and age2 × sex. Model 2: adjusted for age, sex, age2, age × sex, age2 × sex, plus being an artisan and diabetic and HTN status. For diabetes, diabetes status was not used as a covariate; for HTN, SBP and DBP, hypertension status was not used as a covariate.
Abbreviations and definitions: BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated using the Modification of Diet and Renal Disease Study equation modified for American Indians; GKDZI, Genetics of Kidney Disease in Zuni Indians; h2, heritability; HbA1c, glycated hemoglobin; HDL-C, high-density lipoprotein cholesterol; HTN, hypertension; LDL-C, low-density lipoprotein cholesterol; PVC, proportion of variance due to covariates; SBP, systolic blood pressure; SE, standard error of variance; SCr, serum creatinine; SCysC, serum cystatin C; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
Pilot study only.
For dichotomous traits, Kullback-Leibler R2 replaces PVC.
Genetic correlations indicate the extent of pleiotropy, or shared genetic effects, among phenotypes. There were no significant genetic correlations, indicating no detectable shared genetic effects, among kidney-related phenotypes. Genetic and environmental correlations of CKD phenotypes with obesity, diabetes, and cardiovascular disease phenotypes are listed (Table 9). Information about specific genotypes is not required to estimate genetic correlations. Genetic and environmental correlations correspond to partitioning of the covariance into the additive genetic component and a component subsuming environment and nonadditive genetic effects.
Table 9.
Genetic and Environmental Correlations Between Intermediate Phenotypes Related to Kidney Disease and Traits Related to Obesity, Diabetes, and CVD
| Trait 1 | Trait 2 | ρg ± SE | P | ρe ± SE | P |
|---|---|---|---|---|---|
| eGFRMDRD-AI | BMI | 0.155 ± 0.16 | 0.3 | −0.099 ± 0.09 | 0.3 |
| Weight | 0.086 ± 0.15 | 0.6 | −0.084 ± 0.09 | 0.4 | |
| HbA1c | −0.194 ± 0.19 | 0.3 | 0.089 ± 0.07 | 0.2 | |
| SBP | 0.166 ± 0.17 | 0.3 | −0.101 ± 0.07 | 0.2 | |
| DBP | 0.161 ± 0.19 | 0.4 | −0.012 ± 0.07 | 0.9 | |
| HTN status (yes/no) | 0.305 ± 0.19 | 0.1 | −0.135 ± 0.07 | 0.07 | |
| HDL-C | 0.094 ± 0.16 | 0.6 | 0.227 ± 0.07 | 0.004 | |
| Total cholesterol | −0.029 ± 0.17 | 0.9 | 0.076 ± 0.08 | 0.3 | |
| Triglycerides | 0.036 ± 0.19 | 0.9 | −0.182 ± 0.07 | 0.01 | |
| UACR | BMI | 0.212 ± 0.16 | 0.2 | 0.004 ± 0.09 | 0.9 |
| Weight | 0.110 ± 0.15 | 0.5 | 0.045 ± 0.09 | 0.6 | |
| HbA1c | 0.204 ± 0.20 | 0.3 | 0.173 ± 0.07 | 0.02 | |
| SBP | 0.421 ± 0.15 | 0.02 | 0.177 ± 0.07 | 0.02 | |
| DBP | 0.478 ± 0.18 | 0.02 | 0.107 ± 0.07 | 0.1 | |
| HTN status (yes/no) | 0.563 ± 0.16 | 0.003 | 0.264 ± 0.06 | <0.001 | |
| HDL-C | 0.063 ± 0.17 | 0.7 | 0.082 ± 0.08 | 0.3 | |
| Total cholesterol | 0.185 ± 0.17 | 0.3 | 0.095 ± 0.08 | 0.2 | |
| Triglycerides | 0.070 ± 0.20 | 0.7 | 0.111 ± 0.07 | 0.1 | |
| SCr | BMI | −0.187 ± 0.18 | 0.3 | 0.077 ± 0.08 | 0.3 |
| Weight | −0.182 ± 0.18 | 0.3 | 0.093 ± 0.09 | 0.3 | |
| HbA1c | 0.231 ± 0.20 | 0.3 | −0.083 ± 0.07 | 0.2 | |
| SBP | −0.103 ± 0.19 | 0.6 | 0.013 ± 0.07 | 0.8 | |
| DBP | −0.193 ± 0.20 | 0.4 | −0.020 ± 0.06 | 0.8 | |
| HTN status (yes/no) | −0.261 ± 0.21 | 0.2 | 0.076 ± 0.07 | 0.3 | |
| HDL-C | −0.011 ± 0.18 | 0.9 | −0.220 ± 0.07 | 0.003 | |
| Total cholesterol | 0.080 ± 0.19 | 0.7 | −0.085 ± 0.07 | 0.2 | |
| Triglycerides | −0.011 ± 0.21 | 0.9 | 0.148 ± 0.07 | 0.03 | |
| SUN | BMI | 0.044 ± 0.18 | 0.8 | 0.008 ± 0.09 | 0.9 |
| Weight | 0.135 ± 0.17 | 0.4 | −0.027 ± 0.09 | 0.8 | |
| HbA1c | 0.664 ± 0.23 | 0.005 | −0.022 ± 0.07 | 0.7 | |
| SBP | −0.379 ± 0.20 | 0.06 | −0.048 ± 0.07 | 0.5 | |
| DBP | −0.273 ± 0.21 | 0.2 | −0.072 ± 0.07 | 0.3 | |
| HTN status (yes/no) | −0.372 ± 0.21 | 0.08 | 0.171 ± 0.08 | 0.02 | |
| HDL-C | −0.227 ± 0.18 | 0.2 | −0.289 ± 0.07 | <0.001 | |
| Total cholesterol | 0.059 ± 0.19 | 0.8 | −0.159 ± 0.07 | 0.03 | |
| Triglycerides | −0.169 ± 0.23 | 0.5 | 0.145 ± 0.07 | 0.04 | |
| Uric acid | BMI | 0.296 ± 0.14 | 0.06 | 0.198 ± 0.08 | 0.02 |
| Weight | 0.267 ± 0.14 | 0.07 | 0.186 ± 0.09 | 0.05 | |
| HbA1c | 0.223 ± 0.19 | 0.3 | −0.052 ± 0.07 | 0.5 | |
| SBP | 0.290 ± 0.17 | 0.1 | 0.123 ± 0.07 | 0.09 | |
| DBP | 0.124 ± 0.19 | 0.5 | 0.220 ± 0.07 | 0.001 | |
| HTN status (yes/no) | 0.188 ± 0.19 | 0.3 | 0.092 ± 0.07 | 0.2 | |
| HDL-C | −0.303 ± 0.15 | 0.06 | −0.094 ± 0.08 | 0.2 | |
| Total cholesterol | 0.134 ± 0.17 | 0.4 | 0.138 ± 0.07 | 0.7 | |
| Triglycerides | 0.325 ± 0.17 | 0.09 | 0.190 ± 0.07 | 0.009 |
Note: Glucose excluded because many participants were not fasting. No significant genetic or environmental correlation with LDL-C.
Abbreviations and definitions: BMI, body mass index; CVD, cardiovascular disease; DBP, diastolic blood pressure; eGFR, estimated glomerular filtration rate; eGFRMDRD-AI, estimated glomerular filtration rate calculated by the Modification of Diet and Renal Disease Study equation modified for American Indians; HbA1c, glycated hemoglobin; HDL-C, high-density lipoprotein cholesterol; HTN, hypertension; ρg, genetic correlation coefficient; ρe, environmental correlation coefficient; SBP, systolic blood pressure; SCr, serum creatinine; SE, standard error of variance; SUN, serum urea nitrogen; UACR, urinary albumin-creatinine ratio.
Genetic correlations of UACR with systolic blood pressure, diastolic blood pressure, and hypertension status indicate that some genes of the hypertension axis influence UACR. The genetic correlation between SUN and HbA1c reflects the pleiotropic effects of genes involved in ambient glucose levels and/or glycosylation. There were no significant genetic correlations and no evidence for pleiotropy for eGFRMDRD-AI, SCr, or uric acid with any phenotypes related to cardiovascular disease, diabetes, or obesity. There was no evidence of pleiotropy for BMI, weight, HDL cholesterol, LDL cholesterol, total cholesterol, or triglycerides with any intermediate phenotypes for CKD; thus, these may represent genetically independent phenotypes.
Environmental correlations with traits related to obesity, diabetes, and blood pressure tended to be lower than the genetic correlations, but were more likely to attain statistical significance. Environmental and genetic correlations with eGFRMDRD-AI and SCr tended to be opposite in sign. For UACR, all genetic and environmental correlations were positive, and correlations with HbA1c, systolic blood pressure, and hypertension status were statistically significant.
DISCUSSION
This study shows the value of studying extended families to assess the heritability of CKD and intermediate phenotypes in an endogamous population. The significant heritability estimates obtained and the proportion of phenotypic variance contributed by covariates support our hypothesis that genetic and environmental factors modulate susceptibility to kidney disease in the Zuni.
Heritability measures the proportion of phenotypic variance attributed to the additive effects of genes within a population, and the sources, nature, and magnitude of phenotypic variance differ between populations. Therefore, comparisons of heritabilities between populations must be interpreted with caution. Familial clustering and heritability estimates for kidney function and related phenotypes in the GKDZI were similar to those in other studies.32,45–51 Significant heritabilities for eGFR have been observed in European Americans (0.25–0.31)49 and African Americans (0.17).47 In the Framingham Heart Study, there were significant heritabilities for eGFR (0.36)48 and log-transformed systolic blood pressure in unadjusted models (0.38) and models adjusted for sex, BMI, and alcohol consumption (0.47).46 In hypertensive families of African descent, heritability estimates adjusted for age and sex for measured and Cockcroft-Gault–estimated creatinine clearance were 0.52 and 0.82, respectively.45 In American Indians in the Strong Heart Study, heritability estimates for diastolic blood pressure and BMI were 0.34 and 0.44, respectively.32
Heritabilities have been reported for traits related to diabetic nephropathy.50,52–57 The heritability of UACR was 0.21 in Pima Indians50 and as reported in Fogarty et al52 and Krolewski et al54, 0.27 and 0.23, respectively, in non-Hispanic white families. In Finnish families, heritability for albuminuria was 0.30.53 Fogarty et al52 found significant genetic correlations of UACR with systolic and diastolic blood pressure. Studies in non-Hispanic whites with type 2 diabetes yielded high heritabilities for eGFR (0.75) and UACR (0.46).55 However, these traits were not genetically correlated.56 The high prevalence of albuminuria in nondiabetic Zuni Indians with hypertension and obesity is in concert with observations in Australian Aborigines.58
Genetic factors contribute to the variance in renal function and intermediate phenotypes in many populations. Genetic signals for traits related to diabetic nephropathy have been detected in non-Hispanic whites, African Americans, Mexican Americans, and American Indians. Imperatore et al,50 using segregation analysis, reported evidence for a major gene influencing susceptibility to diabetic nephropathy in Pima Indians. Other analyses have focused on candidate chromosomal regions,59–61 genome-wide microsatellite markers,54,57,62–69 and single-nucleotide polymorphism markers.70
In the present study, there were no significant genetic correlations between diabetes status and CKD-related intermediate phenotypes. There was a significant genetic correlation between HbA1c and SUN. There were no significant genetic correlations of HbA1c with SCr, eGFR, or UACR. Given the increased risk of CKD in diabetic patients, the significant heritability of HbA1c and the strong genetic correlation between HbA1c and SUN, genes that influence glycemic control may influence kidney function.71 The genetic correlations of UACR with systolic blood pressure, diastolic blood pressure, and hypertension status indicate the pleiotropic effects of genes that influence these traits. This may explain in part why albuminuria is a strong predictor of cardiovascular disease.72 There were no significant genetic correlations among the kidney-related phenotypes.
Although some differences in intermediate phenotypes among participants stratified by sex, diabetes, hypertension, and artisan vocation achieved statistical significance, they may not be clinically significant. Heavy metals used by artisans may have contributed to the observed differences in several intermediate phenotypes, for example, uric acid, systolic blood pressure, diastolic blood pressure, LDL cholesterol, and total cholesterol. We previously showed high levels of cadmium and lead in household dust of Zuni artisans.73 Environmental lead exposure is a risk factor for the onset70,74–76 and progression of CKD.77 Environmental cadmium exposure is associated with diabetes78 and CKD.79 Jewelry, pottery, and fetish making occurs primarily in the home. We cannot determine what differences in selected variables reflect genetic versus nongenetic factors until genetic analyses have been completed.
The present study has several limitations. First, given the high prevalence of diabetes and absence of kidney biopsies, there are 2 potential sources of misclassification bias: (1) nondiabetic kidney disease in diabetic participants and (2) diabetic kidney disease in participants who have not yet met diagnostic criteria for diabetes. No participants with diabetes and kidney disease were classified as having nondiabetic kidney disease. We may have underestimated the prevalence of nondiabetic kidney disease and the number of generations with nondiabetic kidney disease in a given family. Second, despite the agreement between eGFRMDRD-AI and eGFRSCysC, absence of a direct measure of GFR represents a significant limitation, especially because eGFR was based on a single SCr or SCysC determination. Third, the high prevalence of CKD increased anxiety, thus limiting the willingness of many to participate. Fourth, not all genetic-environmental interactions were assessed.
The study also has several strengths: (1) a community-based participatory research study design, (2) large multigenerational extended families, (3) 3 urine samples to classify UACR and hematuria, (4) comparison of eGFRMDRD-AI and eGFRSCysC, and (5) collection of vocational data.
In summary, many phenotypes related to kidney disease show significant heritabilities in Zuni Indians, and there are significant genetic correlations with phenotypes related to obesity, diabetes, and cardiovascular disease. The study design serves as a paradigm for the conduct of research in relatively isolated, endogamous, and under-served populations.
Acknowledgments
We acknowledge the contribution and support of the Zuni Governors and Tribal Council and sincerely thank the Zuni people for welcoming us in their homes. We thank Drs Narva and Whitfield and the Strong Heart Study investigators for their support. Excellent technical support was provided by Amuche Ezeilo, Meg Lamey, Serena Cumber, and Kelly Utterback. Finally, the authors thank the National Institute of Diabetes and Digestive and Kidney Diseases (Dr Rebekah Rasooly) and the Advisory Committee for their guidance.
Support: This study was supported in part by grants DK066660-03 and DK57300-05 from the National Institutes of Health (NIH); 5M01RR00997 from the University of New Mexico Clinical Research Center; and P30 ES-012072 from the National Institute of Environmental Health Sciences, and Dialysis Clinic Inc. At the Southwest Foundation for Biomedical Research, these studies were conducted in facilities constructed with support from the Research Facilities Improvement Program grant C06 RR013556 from the National Center for Research Resources, NIH. The AT&T Genomics Computing Center supercomputing facilities used for statistical genetic analyses were supported in part by a gift from the SBC Foundation. The statistical genetics computer package, SOLAR, is supported by grant R01 MH059490 from the National Institute of Mental Health.
Footnotes
Financial Disclosure: The authors declare that they have no relevant financial interests.
References
- 1.US Renal Data System. USRDS 2000 Annual Data Report: Atlas of End-Stage Renal Disease in the United States. Bethesda, MD: National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Disease; 2000. [Google Scholar]
- 2.Stidley CA, Shah VO, Narva AS, et al. A population-based, cross-sectional survey of the Zuni Pueblo: a collaborative approach to an epidemic of kidney disease. Am J Kidney Dis. 2002;39:358–368. doi: 10.1053/ajkd.2002.30557. [DOI] [PubMed] [Google Scholar]
- 3.Hoy WE, Megill DM. End-stage renal disease in southwestern Native Americans, with special focus on the Zuni and Navajo Indians. Transplant Proc. 1989;21:3906–3908. [PubMed] [Google Scholar]
- 4.Hoy WE, Smith SM, Hughson MD, Megill DM. Mesangial proliferative glomerulonephritis in southwestern American Indians. Transplant Proc. 1989;21:3909–3912. [PubMed] [Google Scholar]
- 5.Hoy WE, Hughson MD, Smith SM, Megill DM. Mesangial proliferative glomerulonephritis in southwestern American Indians. Am J Kidney Dis. 1993;21:486–496. doi: 10.1016/s0272-6386(12)80394-5. [DOI] [PubMed] [Google Scholar]
- 6.Smith SM, Tung KS. Incidence of IgA-related nephritides in American Indians in New Mexico. Hum Pathol. 1985;16:181–184. doi: 10.1016/s0046-8177(85)80067-8. [DOI] [PubMed] [Google Scholar]
- 7.Scavini M, Stidley CA, Paine SS, et al. The burden of chronic kidney disease among the Zuni Indians: the Zuni Kidney Project. Clin J Am Soc Nephrol. 2007;2:509–516. doi: 10.2215/CJN.02780806. [DOI] [PubMed] [Google Scholar]
- 8.Shah VO, Scavini M, Stidley CA, et al. Epidemic of diabetic and nondiabetic renal disease among the Zuni Indians: the Zuni Kidney Project. J Am Soc Nephrol. 2003;14:1320–1329. doi: 10.1097/01.asn.0000059920.00228.a0. [DOI] [PubMed] [Google Scholar]
- 9.Tentori F, Stidley CA, Scavini M, et al. Prevalence of hematuria among Zuni Indians with and without diabetes: the Zuni Kidney Project. Am J Kidney Dis. 2003;41:1195–1204. doi: 10.1016/s0272-6386(03)00351-2. [DOI] [PubMed] [Google Scholar]
- 10.Office of the Zuni Tribal Census. Tribal Census 2000. 2000. [Google Scholar]
- 11.Scavini M, Stidley CA, Shah VO, et al. Prevalence of diabetes is higher among female than male Zuni indians. Diabetes Care. 2003;26:55–60. doi: 10.2337/diacare.26.1.55. [DOI] [PubMed] [Google Scholar]
- 12.Stidley CA, Shah VO, Scavini M, et al. The Zuni Kidney Project: a collaborative approach to an epidemic of kidney disease. J Am Soc Nephrol. 2003;14(suppl 2):S139–143. doi: 10.1097/01.asn.0000070151.95421.87. [DOI] [PubMed] [Google Scholar]
- 13.Feldman HI, Appel LJ, Chertow GM, et al. The Chronic Renal Insufficiency Cohort (CRIC) Study: design and methods. J Am Soc Nephrol. 2003;14(suppl 2):S148–153. doi: 10.1097/01.asn.0000070149.78399.ce. [DOI] [PubMed] [Google Scholar]
- 14.Dyke B. Population Genetics Laboratory Technical Report. Vol. 2. San Antonio, TX: Southwest Foundation for Biomedical Research; 1994. A Pedigree Data Management System. User’s Manual. [Google Scholar]
- 15.Mamelka PM, Dyke B, MacCluer JW. Population Genetics Laboratory Technical Report. Vol. 1. San Antonio, TX: Southwest Foundation for Biomedical Research; 1993. Pedigree/Draw for the Apple Macintosh. [Google Scholar]
- 16.American Diabetes Association. Standards of medical care for patients with diabetes mellitus. Diabetes Care. 2006;29(suppl 1):S4–42. [PubMed] [Google Scholar]
- 17.Little RR. Recent progress in glycohemoglobin (HbA1c) testing. Diabetes Care. 2000;23:265–266. doi: 10.2337/diacare.23.3.265. [DOI] [PubMed] [Google Scholar]
- 18.The Sixth Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure. Arch Intern Med. 1997;157:2413–2446. doi: 10.1001/archinte.157.21.2413. No authors listed. [DOI] [PubMed] [Google Scholar]
- 19.Murray RL. Creatinine. In: Pesce AJ, Kaplan LA, editors. Methods in Clinical Chemistry. St Louis, MO: Mosby; 1987. [Google Scholar]
- 20.Knight EL, Verhave JC, Spiegelman D, et al. Factors influencing serum cystatin C levels other than renal function and the impact on renal function measurement. Kidney Int. 2004;65:1416–1421. doi: 10.1111/j.1523-1755.2004.00517.x. [DOI] [PubMed] [Google Scholar]
- 21.Schwab SJ, Christensen RL, Dougherty K, Klahr S. Quantitation of proteinuria by the use of protein-to-creatinine ratios in single urine samples. Arch Intern Med. 1987;147:943–944. [PubMed] [Google Scholar]
- 22.Sternberg JC. A rate nephelometer for measuring specific proteins by immunoprecipitin reactions. Clin Chem. 1977;23:1456–1464. [PubMed] [Google Scholar]
- 23.Watts GF, Bennett JE, Rowe DJ, et al. Assessment of immunochemical methods for determining low concentrations of albumin in urine. Clin Chem. 1986;32:1544–1548. [PubMed] [Google Scholar]
- 24.Nelson RG, Greene T, Beck GJ, Van Lente F, Wong X, Knowler WC. Estimating GFR by the MDRD and Cockcroft-Gault equations in Pima Indians [abstract] J Am Soc Nephrol. 2003;14:134A. [Google Scholar]
- 25.Hunsicker LG, Adler S, Caggiula A, et al. Predictors of the progression of renal disease in the Modification of Diet in Renal Disease Study. Kidney Int. 1997;51:1908–1919. doi: 10.1038/ki.1997.260. [DOI] [PubMed] [Google Scholar]
- 26.Poggio ED, Wang X, Greene T, Van Lente F, Hall PM. Performance of the Modification of Diet in Renal Disease and Cockcroft-Gault equations in the estimation of GFR in health and in chronic kidney disease. J Am Soc Nephrol. 2005;16:459–466. doi: 10.1681/ASN.2004060447. [DOI] [PubMed] [Google Scholar]
- 27.Rule AD, Larson TS, Bergstralh EJ, Slezak JM, Jacobsen SJ, Cosio FG. Using serum creatinine to estimate glomerular filtration rate: accuracy in good health and in chronic kidney disease. Ann Intern Med. 2004;141:929–937. doi: 10.7326/0003-4819-141-12-200412210-00009. [DOI] [PubMed] [Google Scholar]
- 28.Verhave JC, Gansevoort RT, Hillege HL, de Zeeuw D, Curhan GC, de Jong PE. Drawbacks of the use of indirect estimates of renal function to evaluate the effect of risk factors on renal function. J Am Soc Nephrol. 2004;15:1316–1322. [PubMed] [Google Scholar]
- 29.National Kidney Foundation. K/DOQI Clinical Practice Guidelines for Chronic Kidney Disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(suppl 1):S1–266. [PubMed] [Google Scholar]
- 30.MacCluer JW, Stern MP, Almasy L, et al. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr Rev. 1999;57(suppl 5):S59–65. doi: 10.1111/j.1753-4887.1999.tb01790.x. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell BD, Kammerer CM, Blangero J, et al. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 32.North KE, Howard BV, Welty TK, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the Strong Heart Family Study. Am J Epidemiol. 2003;157:303–314. doi: 10.1093/aje/kwf208. [DOI] [PubMed] [Google Scholar]
- 33.North KE, Williams K, Williams JT, et al. Evidence for genetic factors underlying the insulin resistance syndrome in American Indians. Obes Res. 2003;11:1444–1448. doi: 10.1038/oby.2003.193. [DOI] [PubMed] [Google Scholar]
- 34.Tejero ME, Voruganti VS, Cai G, et al. Pleiotropic effects on subclasses of HDL, adiposity, and glucose metabolism in adult Alaskan Eskimos. Am J Hum Biol. doi: 10.1002/ajhb.21015. [published online ahead of print November 30, 2009] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voruganti VS, Cai G, Cole SA, et al. Common set of genes regulates low-density lipoprotein size and obesity-related factors in Alaskan Eskimos: results from the GOCADAN Study. Am J Hum Biol. 2006;18:525–531. doi: 10.1002/ajhb.20527. [DOI] [PubMed] [Google Scholar]
- 36.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blangero J, Almasy L. Multipoint oligogenic linkage analysis of quantitative traits. Genet Epidemiol. 1997;14:959–964. doi: 10.1002/(SICI)1098-2272(1997)14:6<959::AID-GEPI66>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 38.Amos C, de Andrade M, Zhu D. Comparison of multivariate tests for genetic linkage. Hum Hered. 2001;51:133–144. doi: 10.1159/000053334. [DOI] [PubMed] [Google Scholar]
- 39.Falconer DS. Introduction to Quantitative Genetics. 2. London, UK: Longman; 1981. p. 112. [Google Scholar]
- 40.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. p. 170. [Google Scholar]
- 41.Altman DG. Practical Statistics for Medical Research. London, UK: Chapman & Hall; 1991. [Google Scholar]
- 42.Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: a new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470. doi: 10.7326/0003-4819-130-6-199903160-00002. [DOI] [PubMed] [Google Scholar]
- 43.Perkins BA, Nelson RG, Ostrander BE, et al. Detection of renal function decline in patients with diabetes and normal or elevated GFR by serial measurements of serum cystatin C concentration: results of a 4-year follow-up study. J Am Soc Nephrol. 2005;16:1404–1412. doi: 10.1681/ASN.2004100854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tidman M, Sjostrom P, Jones I. A comparison of GFR estimating formulae based upon s-cystatin C and s-creatinine and a combination of the two. Nephrol Dial Transplant. 2008;23:154–160. doi: 10.1093/ndt/gfm661. [DOI] [PubMed] [Google Scholar]
- 45.Bochud M, Elston RC, Maillard M, et al. Heritability of renal function in hypertensive families of African descent in the Seychelles (Indian Ocean) Kidney Int. 2005;67:61–69. doi: 10.1111/j.1523-1755.2005.00055.x. [DOI] [PubMed] [Google Scholar]
- 46.Brown WM, Beck SR, Lange EM, et al. Age-stratified heritability estimation in the Framingham Heart Study families. BMC Genet. 2003;4(suppl 1):S32. doi: 10.1186/1471-2156-4-S1-S32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeWan AT, Arnett DK, Atwood LD, et al. A genome scan for renal function among hypertensives: the HyperGEN Study. Am J Hum Genet. 2001;68:136–144. doi: 10.1086/316927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fox CS, Yang Q, Cupples LA, et al. Genomewide linkage analysis to serum creatinine, GFR, and creatinine clearance in a community-based population: the Framing-ham Heart Study. J Am Soc Nephrol. 2004;15:2457–2461. doi: 10.1097/01.ASN.0000135972.13396.6F. [DOI] [PubMed] [Google Scholar]
- 49.Hunt SC, Hasstedt SJ, Coon H, et al. Linkage of creatinine clearance to chromosome 10 in Utah pedigrees replicates a locus for end-stage renal disease in humans and renal failure in the fawn-hooded rat. Kidney Int. 2002;62:1143–1148. doi: 10.1111/j.1523-1755.2002.kid557.x. [DOI] [PubMed] [Google Scholar]
- 50.Imperatore G, Knowler WC, Pettitt DJ, Kobes S, Bennett PH, Hanson RL. Segregation analysis of diabetic nephropathy in Pima Indians. Diabetes. 2000;49:1049–1056. doi: 10.2337/diabetes.49.6.1049. [DOI] [PubMed] [Google Scholar]
- 51.Paterson AD, Magistroni R, He N, et al. Progressive loss of renal function is an age-dependent heritable trait in type 1 autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2005;16:755–762. doi: 10.1681/ASN.2004090758. [DOI] [PubMed] [Google Scholar]
- 52.Fogarty DG, Rich SS, Hanna L, Warram JH, Krolewski AS. Urinary albumin excretion in families with type 2 diabetes is heritable and genetically correlated to blood pressure. Kidney Int. 2000;57:250–257. doi: 10.1046/j.1523-1755.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- 53.Forsblom CM, Kanninen T, Lehtovirta M, Saloranta C, Groop LC. Heritability of albumin excretion rate in families of patients with type II diabetes. Diabetologia. 1999;42:1359–1366. doi: 10.1007/s001250051450. [DOI] [PubMed] [Google Scholar]
- 54.Krolewski AS, Poznik GD, Placha G, et al. A genome-wide linkage scan for genes controlling variation in urinary albumin excretion in type II diabetes. Kidney Int. 2006;69:129–136. doi: 10.1038/sj.ki.5000023. [DOI] [PubMed] [Google Scholar]
- 55.Langefeld CD, Beck SR, Bowden DW, Rich SS, Wagenknecht LE, Freedman BI. Heritability of GFR and albuminuria in Caucasians with type 2 diabetes mellitus. Am J Kidney Dis. 2004;43:796–800. doi: 10.1053/j.ajkd.2003.12.043. [DOI] [PubMed] [Google Scholar]
- 56.Placha G, Canani LH, Warram JH, Krolewski AS. Evidence for different susceptibility genes for proteinuria and ESRD in type 2 diabetes. Adv Chronic Kidney Dis. 2005;12:155–169. doi: 10.1053/j.ackd.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Placha G, Poznik GD, Dunn J, et al. A genome-wide linkage scan for genes controlling variation in renal function estimated by serum cystatin C levels in extended families with type 2 diabetes. Diabetes. 2006;55:3358–3365. doi: 10.2337/db06-0781. [DOI] [PubMed] [Google Scholar]
- 58.Rowley KG, Iser DM, Best JD, O’Dea K, Leonard D, McDermott R. Albuminuria in Australian Aboriginal people: prevalence and associations with components of the metabolic syndrome. Diabetologia. 2000;43:1397–1403. doi: 10.1007/s001250051545. [DOI] [PubMed] [Google Scholar]
- 59.Freedman BI, Rich SS, Yu H, Roh BH, Bowden DW. Linkage heterogeneity of end-stage renal disease on human chromosome 10. Kidney Int. 2002;62:770–774. doi: 10.1046/j.1523-1755.2002.00534.x. [DOI] [PubMed] [Google Scholar]
- 60.Iyengar SK, Fox KA, Schachere M, et al. Linkage analysis of candidate loci for end-stage renal disease due to diabetic nephropathy. J Am Soc Nephrol. 2003;14(suppl 2):S195–201. doi: 10.1097/01.asn.0000070078.66465.55. [DOI] [PubMed] [Google Scholar]
- 61.Vardarli I, Baier LJ, Hanson RL, et al. Gene for susceptibility to diabetic nephropathy in type 2 diabetes maps to 18q22.3-23. Kidney Int. 2002;62:2176–2183. doi: 10.1046/j.1523-1755.2002.00663.x. [DOI] [PubMed] [Google Scholar]
- 62.Bowden DW, Colicigno CJ, Langefeld CD, et al. A genome scan for diabetic nephropathy in African Americans. Kidney Int. 2004;66:1517–1526. doi: 10.1111/j.1523-1755.2004.00915.x. [DOI] [PubMed] [Google Scholar]
- 63.Imperatore G, Hanson RL, Pettitt DJ, Kobes S, Bennett PH, Knowler WC. Sib-pair linkage analysis for susceptibility genes for microvascular complications among Pima Indians with type 2 diabetes. Pima Diabetes Genes Group. Diabetes. 1998;47:821–830. doi: 10.2337/diabetes.47.5.821. [DOI] [PubMed] [Google Scholar]
- 64.Iyengar SK, Abboud HE, Goddard KA, et al. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2007;56:1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- 65.McKnight AJ, Maxwell AP, Sawcer S, et al. A genome-wide DNA microsatellite association screen to identify chromosomal regions harboring candidate genes in diabetic nephropathy. J Am Soc Nephrol. 2006;17:831–836. doi: 10.1681/ASN.2005050493. [DOI] [PubMed] [Google Scholar]
- 66.Mottl AK, Vupputuri S, Cole SA, et al. Linkage analysis of glomerular filtration rate in American Indians. Kidney Int. 2008;74:1185–1191. doi: 10.1038/ki.2008.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schelling JR, Abboud HE, Nicholas SB, et al. Genome-wide scan for estimated glomerular filtration rate in multi-ethnic diabetic populations: the Family Investigation of Nephropathy and Diabetes (FIND) Diabetes. 2008;57:235–243. doi: 10.2337/db07-0313. [DOI] [PubMed] [Google Scholar]
- 68.Voruganti VS, Nath SD, Cole SA, et al. Genetics of variation in serum uric acid and cardiovascular risk factors in Mexican Americans. J Clin Endocrinol Metab. 2009;94:632–638. doi: 10.1210/jc.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voruganti VS, Goring HH, Mottl A, et al. Genetic influence on variation in serum uric acid in American Indians: the Strong Heart Family Study. Hum Genet. 2009;126:667–676. doi: 10.1007/s00439-009-0716-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henderson DA. The aetiology of chronic nephritis in Queensland. Med J Aust. 1958;45:377–386. doi: 10.5694/j.1326-5377.1958.tb86396.x. [DOI] [PubMed] [Google Scholar]
- 71.Kovesdy CP, Sharma K, Kalantar-Zadeh K. Glycemic control in diabetic CKD patients: where do we stand? Am J Kidney Dis. 2008;52:766–777. doi: 10.1053/j.ajkd.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 72.Donnelly R, Yeung JM, Manning G. Microalbuminuria: a common, independent cardiovascular risk factor, especially but not exclusively in type 2 diabetes. J Hypertens Suppl. 2003;21:S7–12. [PubMed] [Google Scholar]
- 73.Gonzales M, Shah V, Bobelu A, et al. Concentrations of surface-dust metals in Native American jewelry-making homes in Zuni Pueblo, New Mexico. Arch Environ Health. 2004;59:245–249. doi: 10.3200/AEOH.59.5.245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nuyts GD, Daelemans RA, Jorens PG, Elseviers MM, Van de Vyver FL, De Broe ME. Does lead play a role in the development of chronic renal disease? Nephrol Dial Transplant. 1991;6:307–315. doi: 10.1093/ndt/6.5.307. [DOI] [PubMed] [Google Scholar]
- 75.Wedeen RP, Maesaka JK, Weiner B, et al. Occupational lead nephropathy. Am J Med. 1975;59:630–641. doi: 10.1016/0002-9343(75)90224-7. [DOI] [PubMed] [Google Scholar]
- 76.Wedeen RP, Malik DK, Batuman V. Detection and treatment of occupational lead nephropathy. Arch Intern Med. 1979;139:53–57. [PubMed] [Google Scholar]
- 77.Lin JL, Lin-Tan DT, Hsu KH, Yu CC. Environmental lead exposure and progression of chronic renal diseases in patients without diabetes. N Engl J Med. 2003;348:277–286. doi: 10.1056/NEJMoa021672. [DOI] [PubMed] [Google Scholar]
- 78.Schwartz GG, Il’yasova D, Ivanova A. Urinary cadmium, impaired fasting glucose, and diabetes in the NHANES III. Diabetes Care. 2003;26:468–470. doi: 10.2337/diacare.26.2.468. [DOI] [PubMed] [Google Scholar]
- 79.Satarug S, Haswell-Elkins MR, Moore MR. Safe levels of cadmium intake to prevent renal toxicity in human subjects. Br J Nutr. 2000;84:791–802. [PubMed] [Google Scholar]