Abstract
Objective
Large-scale genetic association studies have identified over 20 rheumatoid arthritis (RA) risk alleles among individuals of European ancestry. The influence of these risk alleles has not been comprehensively studied in African-Americans. We therefore sought to examine whether these validated RA risk alleles are associated with RA in an African-American population.
Methods
27 candidate SNPs were genotyped in 556 autoantibody-positive African-Americans with RA and 791 healthy African-American controls. Odds ratios (OR) and 95% confidence intervals (CI) for each SNP were compared to previously published ORs of RA patients of European ancestry. We then calculated a composite Genetic Risk Score (GRS) for each individual based on the sum of all risk alleles.
Results
There was overlap in the OR and 95% CI between the European and African-American populations in 24 of the 27 candidate SNPs. Conversely, 3 of the 27 SNPs (CCR6 rs3093023, TAGAP rs394581, TNFAIP3 rs6920220) demonstrated an OR in the opposite direction from those reported in RA patients of European ancestry. The GRS analysis indicated a small but highly significant probability that African-American cases were enriched for the European RA risk alleles relative to controls (p=0.00005).
Conclusion
The majority of RA risk alleles previously validated among European ancestry RA patients showed similar ORs in our population of African-Americans with RA. Furthermore, the aggregate GRS supports the hypothesis that these SNPs are risk alleles for RA in the African-American population. Future large-scale genetic studies are needed to validate these risk alleles and identify novel risk alleles for RA in African-Americans.
Rheumatoid arthritis (RA) is a phenotypically heterogeneous, systemic autoimmune disease characterized by chronic destructive inflammation in synovial joints. The disease can be subdivided into two groups (autoantibody-positive or autoantibody-negative) according to the presence or absence of either rheumatoid factor (RF) or autoantibodies to cyclic-citrullinated peptide (CCP). Genetic and environmental risk factors and their interaction are also known contributors to RA pathogenesis (1). Advances in human genetics have led to a dramatic increase in the number of disease risk alleles identified in persons of European ancestry with RA, with at least 20 common risk alleles discovered to date (2;3). However, because the vast majority of the large-scale genetic association studies have been conducted in autoantibody-positive persons of European ancestry, the question about whether these alleles are associated with RA risk in other ethnic groups remains unaddressed. We sought to study the association of these previously identified RA risk loci in a large group of well-characterized African-Americans with RA.
Specifically, we hypothesized that many of the risk loci identified in populations of European ancestry will also demonstrate risk for RA in African Americans. Most RA risk alleles outside of the MHC region have moderate effect sizes and have thus required large sample sizes for identification and replication. We anticipated that it would be difficult to demonstrate strong statistical support for individual risk alleles in our African American population of 556 autoantibody-positive RA cases and 791 healthy controls. To address this limitation we used two methodological approaches. We first tested whether individual risk allele OR are consistent (have overlapping confidence intervals) or inconsistent (non-overlapping confidence intervals) between the European and African-American populations. As a second step we derived an aggregate genetic risk score (GRS) in our population of African-American RA and controls and analyzed for differences in the composite effect of all European risk alleles with RA risk (4).
PATIENTS AND METHODS
Study subjects
We analyzed 27 SNPs in 556 African-Americans with autoantibody-positive RA, defined as a positive serum rheumatoid factor (RF) or a positive serum anti-CCP antibody. The analysis was limited to autoantibody-positive subjects because the risk alleles tested here were those previously validated in autoantibody-positive patients of European ancestry (2,5,6,7). All RA subjects were participants in the Consortium for the Longitudinal Evaluation of African-Americans with Early Rheumatoid Arthritis (CLEAR) Registry. The CLEAR Registry enrolls self-identified African-Americans who meet the American College of Rheumatology (ACR) 1987 diagnostic criteria for RA (8). CLEAR participants were recruited from the University of Alabama at Birmingham (UAB) (coordinating site); Emory University / Grady Hospital (Atlanta, GA); University of North Carolina at Chapel Hill; Medical University of South Carolina (Charleston, SC); and Washington University (St. Louis, MO). There are two phases of the CLEAR Registry. The longitudinal arm (CLEAR I) enrolled patients with early RA (<2 year disease duration) from 2000 to 2007 and follows them longitudinally until 5 years disease duration. CLEAR II started in 2007 and is an ongoing one-time evaluation of patients with any disease duration. In this study, we analyzed only the autoantibody-positive participants: 228 RA patients from CLEAR I and 328 RA patients from CLEAR II. Detailed demographic and clinical data and DNA samples are available on all CLEAR participants (see (http://medicine.uab.edu/rheum/70918/ for details).
Healthy African-American controls with similar sex, age, and geographic location were recruited through the CLEAR study. This included 132 controls from the CLEAR I study and 171 controls from the CLEAR II (total = 303). The remaining 501 African-American controls were recruited from the Birmingham, Alabama area. All participants were recruited with informed consent under the approval of each respective Institutional Review Board. Anti-CCP antibodies and RF were determined using methods as previously described (9).
Genotyping
SNP selection was based on previously identified and validated risk alleles in autoantibody-positive RA subjects of European ancestry (5,6,7,10–22), as well as those identified in a recent GWAS meta-analysis by Stahl et al., which included over 5,000 autoantibody-positive European RA cases and 20,000 controls (Table 2) (7).
Table 2. Comparison of ORs for Risk Alleles in the European (7) and African-American Populations.
A. Odds Ratios for 24 risk alleles (SNPs) for which ORs were consistent (overlapping confidence intervals) between African-American and European populations. B. Odds Ratios for 3 risk alleles (SNPs) for which ORs were inconsistent between African-American and European populations.
| SNP | Chr | Gene | Min/Maj Allele* |
MAF (cases) |
MAF (controls) |
P value | African-American OR |
European OR | MAF European Cases |
Statistical Power |
|---|---|---|---|---|---|---|---|---|---|---|
| rs3087243 | 2 | CTLA4 | A/G | 0.19 | 0.24 | 0.003 | 0.75 (0.62–0.91) | 0.87 (0.83–0.91) | 0.44 | 0.340 |
| rs11889341** | 2 | STAT4 | T/C | 0.15 | 0.13 | 0.071 | 1.22 (0.98–1.53) | 1.16 (1.10–1.23) | 0.22 | 0.231 |
| rs1980422 | 2 | CD28 | C/T | 0.22 | 0.19 | 0.073 | 1.19 (0.98–1.44) | 1.12 (1.06–1.18) | 0.24 | 0.216 |
| rs3761847 | 9 | TRAF1-C5 | A/G | 0.41 | 0.38 | 0.128 | 1.13 (0.97–1.32) | 1.13 (1.08–1.18) | 0.43 | 0.326 |
| rs6859219 | 5 | ANKRD55 | T/C | 0.03 | 0.04 | 0.144 | 0.73 (0.48–1.12) | 0.85 (0.78–0.93) | 0.18 | 0.139 |
| rs10499194† | 6 | TNFAIP3 | T/C | 0.18 | 0.16 | 0.164 | 1.16 (0.94–1.42) | 0.91 (0.87–0.96) | 0.27 | 0.148 |
| rs13031237 | 2 | REL | T/G | 0.10 | 0.08 | 0.185 | 1.20 (0.92–1.56) | 1.13 (1.07–1.18) | 0.37 | 0.146 |
| rs934734 | 2 | SPRED2 | G/A | 0.46 | 0.49 | 0.231 | 1.10 (0.94–1.28) | 1.13 (1.06–1.21) | 0.52 | 0.346 |
| rs2812378 | 9 | CCL21 | G/A | 0.45 | 0.42 | 0.232 | 1.10 (0.94–1.28) | 1.10 (1.05–1.16) | 0.34 | 0.228 |
| rs3890745 | 1 | TNFRSF14 | C/T*** | 0.52 | 0.54 | 0.270 | 0.92 (0.79–1.07) | 0.89 (0.85–0.94) | 0.32 | 0.318 |
| rs11586238† | 1 | CD58 | G/C | 0.09 | 0.10 | 0.281 | 0.87 (0.67–1.13) | 1.13 (1.07–1.19) | 0.24 | 0.164 |
| rs10919563† | 1 | PTPRC | A/G | 0.40 | 0.38 | 0.342 | 1.08 (0.92–1.26) | 0.88 (0.82–0.94) | 0.13 | 0.362 |
| rs548234 | 6 | PRDM1 | C/T | 0.08 | 0.07 | 0.436 | 1.12 (0.84–1.49) | 1.10 (1.05–1.16) | 0.33 | 0.100 |
| rs2476601 | 1 | PTPN22 | A/G | 0.02 | 0.01 | 0.507 | 1.23 (0.66–2.29) | 1.94 (1.81–2.08) | 0.10 | 0.665 |
| rs10488631 | 7 | IRF5 | T/C | 0.03 | 0.03 | 0.530 | 1.16 (0.72–1.87) | 1.25 (1.14–1.37) | 0.13 | 0.156 |
| rs3218253 | 22 | IL2RB | A/G | 0.15 | 0.13 | 0.575 | 1.07 (0.85–1.33) | 1.09 (1.03–1.15) | 0.26 | 0.121 |
| rs706778 | 10 | IL2RA | C/T | 0.50 | 0.49 | 0.656 | 1.04 (0.89–1.21) | 1.11 (1.06–1.17) | 0.44 | 0.268 |
| rs26232 | 5 | C5orf13, | T/C | 0.29 | 0.30 | 0.687 | 0.97 (0.82–1.14) | 0.93 (0.88–0.98) | 0.29 | 0.138 |
| rs540386 | 11 | TRAF6 | T/C | 0.25 | 0.25 | 0.756 | 0.97 (0.81–1.16) | 0.88 (0.83–0.94) | 0.14 | 0.307 |
| rs874040 | 4 | RBPJ | C/G | 0.34 | 0.34 | 0.771 | 1.02 (0.87–1.21) | 1.18 (1.12–1.24) | 0.33 | 0.530 |
| rs13315591† | 3 | PXK | C/T | 0.34 | 0.35 | 0.799 | 0.98 (0.83–1.15) | 1.13 (1.04–1.23) | 0.10 | 0.326 |
| rs4750316† | 10 | PRKCQ | C/G | 0.39 | 0.38 | 0.813 | 1.02 (0.87–1.19) | 0.87 (0.82–0.92) | 0.19 | 0.417 |
| rs6822844† | 4 | IL2, IL21 | T/G | 0.02 | 0.02 | 0.829 | 1.06 (0.63–1.77) | 0.90 (0.84–0.95) | 0.18 | 0.069 |
| rs4810485† | 20 | CD40 | T/G | 0.07 | 0.07 | 0.908 | 1.02 (0.75–1.38) | 0.85 (0.80–0.90) | 0.25 | 0.196 |
| A | ||||||||||
| SNP | Chr | Gene |
Min/Maj Allele |
MAF (cases) |
MAF (controls) |
P value |
African- American OR |
European OR |
MAF European Cases |
Statistical Power |
| rs3093023 | 6 | CCR6 | A/G | 0.14 | 0.17 | 0.035 | 0.79 (0.64–0.98) | 1.11 (1.06–1.16) | 0.47 | 0.175 |
| rs394581 | 6 | TAGAP | C/T | 0.47 | 0.43 | 0.080 | 1.15 (0.98–1.34) | 0.91 (0.87–0.96) | 0.30 | 0.225 |
| rs6920220 | 6 | TNFAIP3 | A/G | 0.10 | 0.11 | 0.321 | 0.88 (0.68–1.13) | 1.22 (1.16–1.29) | 0.22 | 0.383 |
| B | ||||||||||
Note that four SNPs had OR in opposite directions.
Minor and major allele frequencies refer to the African-American population.
Genotyping results have previously been reported on this SNP which is a proxy SNP for rs7574865 (see Discussion).
Minor allele for SNP rs3890745 (TNFRSF14) was switched for computing the African American OR to reflect the minor allele found in the European population.
Demonstrate OR in the opposite direction compared to European RA but the CIs overlap between the two populations indicating no statistically significant difference.
Note that all three OR were in opposite directions and the confidence limits did not overlap.
SNP genotyping was performed at the Broad Institute using Sequenom iPlex, as previously described (15). For quality control we required that each SNP pass the following criteria: (1) genotype missing rate < 10%; (2) minor allele frequency > 1%; and (3) Hardy-Weinberg equilibrium with p>0.001. We then excluded individuals with data missing for > 10% of SNPs passing quality control.
Statistical Analysis
We performed SNP associations with RA risk using the software package Plink v1.06 (http://pngu.mgh.harvard.edu/purcell/plink) (23). ORs reflected the differences between cases and controls of minor allele frequencies in the African-American population, and they were tested with Pearson’s Chi square. The analysis included 556 autoantibody-positive RA cases and 791 controls after filtering was done for quality control. We did not perform a Bonferroni correction because each of the SNPs tested is a validated risk allele for RA in the European population. Had we used a Bonferroni correction for multiple comparisons, we would not have declared statistical significance unless the p-value was less than the nominal alpha level of 0.002 (0.05 divided by 27).
We compared ORs in European ancestry RA to those in our African-American RA population. The OR and 95% confidence intervals from European subjects were derived from a meta-analysis of 5,539 autoantibody positive RA cases and 20,169 controls of European descent (7). Differences in ORs were declared significant between the European and African-American populations if the OR 95% confidence limits did not overlap. The statistical power to detect a significant association of SNPs with RA in African-Americans was calculated using the Genetic Power Calculator (24). These power calculations were based on: the ORs for association with RA in the European population; the sample size of African-Americans; and the minor allele frequencies in the African-American population.
As a final step, we derived an aggregate genetic risk score (GRS) in our population of autoantibody positive African-American RA and controls (4). For each individual the sum (or count) of the number of risk alleles across the 27 SNPs was calculated (possible range of values from 0 to 54). We chose a GRS count approach rather than a weighted GRS (where each SNP in the GRS is weighted by the OR), as we did not want to assume that the OR in African-Americans was the same as the OR in individuals of European ancestry. The total number of risk vs. non-risk alleles between cases and controls was then assessed using contingency table analysis, and the hypothesis that the type of allele was independent of case-control status was assessed with Pearson's chi square statistic. Equivalently, the GRS score (probability that the individual had risk alleles) was modeled as a binomial response and case-control status as a dichotomous predictor using generalized linear models. Both approaches were implemented in the statistical package R (25).
RESULTS
The characteristics of the RA participants in CLEAR I and CLEAR II are shown in Table 1. Demographic characteristics were similar with the exception of the mean age of RA onset and disease duration, which were significantly higher in CLEAR II than in CLEAR I because of differences in inclusion criteria for the two CLEAR arms.
Table 1. Characteristics of 556 autoantibody RA patient participants in CLEAR I (longitudinal evaluation of early RA) and CLEAR II (cross-sectional evaluation of established RA).
Significant differences in demographic parameters between study groups were tested with T-tests for independent samples (Age at RA, Disease duration) and normal approximations for testing differences between large sample proportions (remaining variables). P-values for these tests are listed in the right column. Where data was unavailable for some of the subjects, sample sizes showing the numbers of subjects available for analysis are shown within the table
| CLEAR I (n=228) | CLEAR II (n=328) | P value | |
|---|---|---|---|
| Age at RA onset, mean (SD) | 48.8 (13.4) | 43.4 (14.0) | <0.0001 |
| Disease duration at enrollment, months (SD) | 12.8 (7.3) | 133.8 (112.7) | <0.0001 |
| Gender (female), % | 84.2 | 83.5 (N = 324) | 0.83 |
| Anti-CCP antibodies, % positive | 85.1 | 87.1 (N = 271) | 0.52 |
| Rheumatoid factor, % positive | 95.7 (N = 211) | 96.6 (N = 322) | 0.62 |
| Both CCP and RF positive (%) | 80.6 (N = 211) | 82.6 (N = 265) | 0.56 |
| HLA SE positive (%) | 45.9 (N = 218) | 39.5 (N = 258) | 0.16 |
| Smoker (current) (%) | 29.8 | 25.3 | 0.24 |
| Smoker (ever) (%) | 52.2 | 54.6 | 0.58 |
Association between genetic risk alleles and RA in African-Americans
The allele frequencies of each of the candidate SNPs tested for association with RA risk in our African-American cohort are shown in Table 2. Notably, some RA risk alleles were present at a substantially lower frequency in African-American than European populations (e.g., ANKRD55, PTPN22, IRF5, IL2-IL21). We have previously reported allele frequencies and ORs of 5 SNPs in the STAT4 region (rs11889341; rs10931481; rs7574865; rs8179673; and rs10181656) in 723 of African-American RA patients (including the 556 autoantibody-positive patients reported here, plus autoantibody-negative patients) and 660 of these same controls (26). The MAFs for African-American RA patients and controls for the STAT4 rs11889341 T allele were 0.141 and 0.133, respectively using ABI TaqMan assays. These values are consistent with the results obtained on the Sequenom platform used in the current study.
We compared the ORs and 95% CI in our African-American population to those in a recently published meta-analysis of European ancestry RA patients (7). Of the 27 SNPs tested, 24 demonstrated OR and 95% CI that overlap between the European and African-American populations (Table 2A, Figure 1). Only 1 of the 24 SNPs, CTLA4 rs3087243, showed a statistically significant association with RA in African-Americans (OR 0.75, [95% CI 0.62–0.91] p=0.003). Seven of the 24 SNPs (TNFAIP3 rs10499194, CD58 rs11586238, PTPRC rs10919563, PXK rs13315591, PRKCQ rs4750316, IL2,IL21 rs6822844, CD40 4810485) demonstrated OR in the opposite direction to that seen in European ancestry RA, but the 95% CIs overlapped between the two populations, and there was no statistically significant difference between African-American RA and controls.
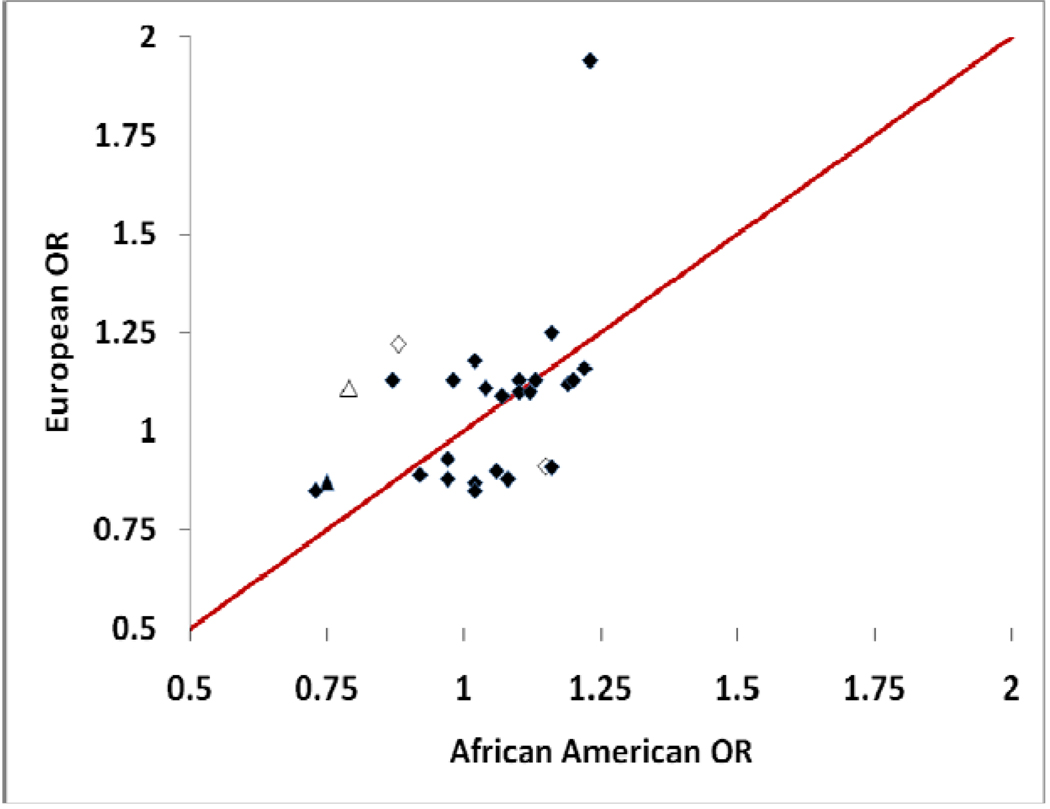
Figure 1. Scatter plot of the odds ratios of SNPs with replicated association in European populations on the ordinate and OR of the same SNPs tested in the African-American (A-A) population on the abscissa.
Solid markers indicate OR that are consistent between populations (Table 2A); open markers are those SNPs in which the OR are inconsistent between populations (Table 2B). The two triangle markers are those SNPs (rs3087243, CTLA4 and rs3093023, CCR6) with statistically significant association with RA in this African-American sample. The diamond at the top of the figure is PTPN22, which has an OR=1.94 and OR=1.23 in European ancestry RA patients and African-American RA patients, respectively.
As shown in Table 2B and Figure 1, we found evidence for differences in ORs between the two ethnic groups in 3 of the 27 SNPs (CCR6 rs3093023, TAGAP rs394581, and TNFAIP3 rs6920220). Of these three SNPs, only CCR6 rs3093023 showed a statistically significant association with RA in African-Americans (OR 0.79, [95% CI 0.64–0.98] p=0.035). All three of these risk alleles had OR that were in the opposite directions between populations.
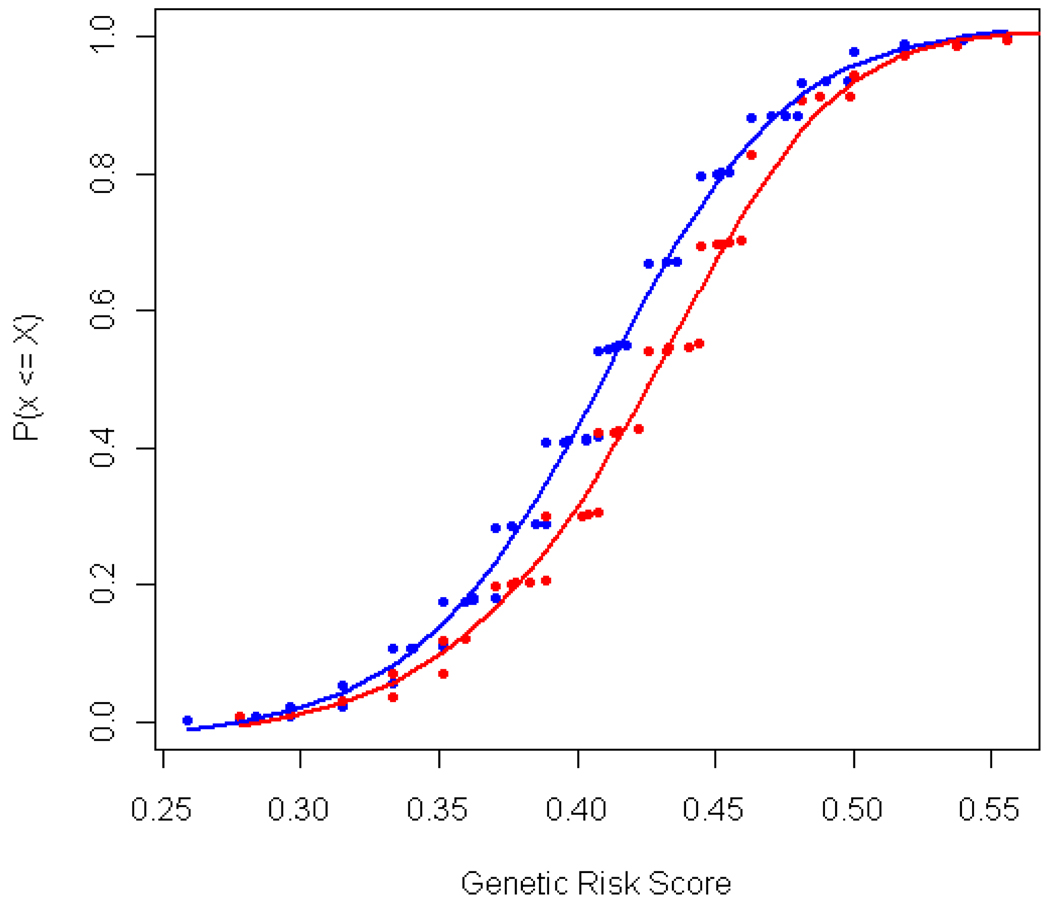
To overcome the limitation in statistical power due to the relatively small size of our African-American RA cohort, we calculated a genetic risk score (GRS) based on all RA risk alleles for each person in the study. This approach combines the effect of each risk allele into a single aggregate score, which can then be used to test whether the GRS differentiates between RA cases and controls. The GRS analysis demonstrated that the probability of individuals having risk alleles was higher in cases (mean GRS = 0.43) than in controls (0.41) (p=0.00005, Figure 2). To exclude the possibility that the observed difference was due to a single SNP with a large effect size, we repeated the GRS analysis without the CTLA4 rs3087243 SNP; the result remains significant (0.41 cases, 0.40 controls, p=0.0003). This observation supports the hypothesis that many risk alleles found in European RA patients are also found in African-Americans with RA.
Figure 2. Cumulative distribution of the GRS scores between cases (red) and controls (blue).
The y axis, P (x <= X), is the cumulative probability that individual cases or controls have less than the given GRS score. The GRS score is defined here as the individual proportion of risk alleles carried for the 27 SNPs which have validated association with RA in populations of European ancestry. The mean is 0.43 for cases and 0.41 for controls.
Discussion
To date, the majority of GWAS and subsequent meta-analyses of GWAS data in RA have focused on subjects of European and East Asian ancestry. It has become clear from these and other large scale genetic studies of complex diseases such as RA that genetic risk loci can differ among these different ethnic groups (18;22;27). There is a paucity of data from well-characterized, large groups of African-Americans with RA. In this study we sought to test the hypothesis that RA risk alleles validated in populations of European ancestry would also be associated with RA in African-Americans. The power to test individual risk alleles in our African American sample is limited compared to studies in European RA. The results of this study, however, demonstrated that cumulatively (via the GRS analysis), risk alleles for RA in Europeans also confer risk for African Americans. Therefore, we conclude that the two populations are best characterized as genetically homogeneous with respect to validated risk alleles for RA.
We found that the ORs between European and African-American populations were consistent for 24 of 27 European RA risk loci. One interpretation of this finding is that the genetic etiology of RA risk in the two populations is very similar. However, only one European RA risk allele achieved statistically significant association with risk of RA in the African-American population (CTLA4 rs3087243). Although the CLEAR cohort is the largest group of African-American RA patients currently available for analysis, we had limited power to detect genetic associations. This limited power, as shown in Table 2, can be attributed to the small effect size of many individual risk alleles and the low frequency of some of the risk alleles in the African-American population. The limited power to detect association signals also affected our ability to demonstrate between population inconsistencies in OR. For example, the low frequency PTNP22 SNP rs2476601 (MAF = 0.01) showed the largest between-population difference (Figure 1), with OR of 1.23 in African-Americans and 1.94 in Europeans. However, due to the large confidence interval limit on the African-American OR, we were unable conclude that these OR were different between populations (Table 2).
We observed differences in direction of OR between European and African American populations for three loci (CCR6 rs3093023, TAGAP rs394581, and TNFAIP3 rs6920220). There are at least three explanations for these differences. First, inconsistent ORs might be due to weakened correlations between tag SNPs and the causal allele (not yet identified) with actual risk with RA. It is known from the International Haplotype Mapping Project (HapMap) that LD structure between European and African American populations may be different for common SNPs in any given locus. Such an effect may be particularly striking if the causal allele is rare in the population (e.g., frequency <5%)(28). The second possible explanation is that these differences may be explained by genetic heterogeneity; the European risk allele may operate differently in its effect on RA risk for African-Americans. Such population specific differences may reflect gene by gene or gene by environmental interactions. The third possible explanation is that the inconsistency may simply be due to chance, given the number of hypotheses tested. Further investigation of these alleles among individuals of African ancestry is needed in explore these possibilities.
Although we had limited statistical power because of our small sample size, when the SNP risk alleles are viewed together in the GRS analysis, there is evidence that the European risk alleles are also risk alleles in the African American population. The GRS was found to be significantly different between RA cases and controls, even after excluding the CTLA4 SNP with the largest effect size. However this approach indicates that even though most of the individual markers were not statistically associated with RA in African-Americans, they may contribute to a panel of alleles that collectively confer risk.
Future studies of large, well-characterized cohorts of African-American RA patients and controls are needed to definitively determine whether the European RA risk alleles are associated with RA risk in African-Americans. Large scale GWAS in African-American RA patients are also needed in order to explore novel risk alleles among this genetically admixed ethnic group. We believe that detailed genetic studies of African-Americans with RA will lead to important insights into the pathogenesis of this disease.
Acknowledgements
The authors gratefully acknowledge all the study patients for their contribution to this work. We also thank Drs. David Allison, Hemant Tiwari, Maria Danila, Monica Crawford, and Jeffrey Faggard for helpful discussions and review of the manuscript. This research was supported by NIH grants T32 HL072757. The CLEAR Registry is supported by NIH contract N01-AR-6-2278. RMP is supported by grants from NIAMS-NIH (R01-AR056768 and R01-AR057108) and holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund.
Reference List
- 1.Mahdi H, Fisher BA, Kallberg H, Plant D, Malmstrom V, Ronnelid J, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009;41(12):1319–1324. doi: 10.1038/ng.480. [DOI] [PubMed] [Google Scholar]
- 2.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, Guiducci C, et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat Genet. 2009;41(12):1313–1318. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plenge RM. Recent progress in rheumatoid arthritis genetics: one step towards improved patient care. Curr Opin Rheumatol. 2009;21(3):262–271. doi: 10.1097/BOR.0b013e32832a2e2d. [DOI] [PubMed] [Google Scholar]
- 4.Karlson EW, Chibnik LB, Kraj P, Cui J, Keenan BT, Ding B, et al. Cumulative Association of Twenty-Two Genetic Variants with Seropositive Rheumatoid Arthritis Risk. Ann Rheum Dis. 2010 doi: 10.1136/ard.2009.120170. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, Burtt NP, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet. 2009;41(7):820–823. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson B, et al. Genome-wide association study meta-analysis identifies 7 novel rheumatoid arthritis risk loci. Nat Genet. 2010;42(6):508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 9.Mikuls TR, Holers VM, Parrish L, Kuhn KA, Alarcón GS, Conn DL, et al. Anti-cyclic citrullinated peptide antibody and rheumatoid factor isotypes in African Americans with early rheumatoid arthritis. Arthritis Rheum. 2006;54(9):3057–3059. doi: 10.1002/art.22200. [DOI] [PubMed] [Google Scholar]
- 10.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40(10):1156–1159. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barton A, Thomson W, Ke X, Eyre S, Hinks A, Bowes J, et al. Re-evaluation of putative rheumatoid arthritis susceptibility genes in the post-genome wide association study era and hypothesis of a key pathway underlying susceptibility. Hum Mol Genet. 2008;17(15):2274–2279. doi: 10.1093/hmg/ddn128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, et al. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet. 2004;75(2):330–337. doi: 10.1086/422827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang M, Rowland CM, Garcia VE, Schrodi SJ, Catanese JJ, van der Helm-van Mil AH, et al. A large-scale rheumatoid arthritis genetic study identifies association at chromosome 9q33.2. PLoS Genet. 2008;4(6):e1000107. doi: 10.1371/journal.pgen.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurreeman FA, Padyukov L, Marques RB, Schrodi SJ, Seddighzadeh M, Stoeken-Rijsbergen G, et al. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLoS Med. 2007;4(9):e278. doi: 10.1371/journal.pmed.0040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39(12):1477–1482. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34(4):395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 19.Thomson W, Barton A, Ke X, Eyre S, Hinks A, Bowes J, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39(12):1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plenge RM, Padyukov L, Remmers EF, Purcell S, Lee AT, Karlson EW, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77(6):1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, et al. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81(6):1284–1288. doi: 10.1086/522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki A, Yamada R, Kochi Y, Sawada T, Okada Y, Matsuda K, et al. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat Genet. 2008;40(10):1224–1229. doi: 10.1038/ng.205. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 25.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. Ref Type: Online Source. [Google Scholar]
- 26.Kelley JM, Hughes LB, Malik A, Danila MI, Edberg Y, Alarcón GS, et al. Genetic variants of STAT4 associated with rheumatoid arthritis in persons of Asian and European ancestry do not replicate in African-Americans. Ann Rheum Dis. 2010 doi: 10.1136/ard.2009.113183. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HS, Korman BD, Le JM, Kastner DL, Remmers EF, Gregersen PK, et al. Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum. 2009;60(2):364–371. doi: 10.1002/art.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome- wide associations. PLoS Biol. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]