Abstract
Caspase-6 is an apoptotic cysteine protease that also governs disease progression in Huntington’s and Alzheimer’s Diseases. Caspase-6 is of great interest as a target for treatment of these neurodegenerative diseases, however the molecular basis of caspase-6 function and regulation remains poorly understood. In the recently reported structure of caspase-6, the 60’s and 130’s helices at the base of the substrate-binding groove extend upward, in a conformation entirely different from that of any other caspase. Presently, the central question about caspase-6 structure and function is whether the extended conformation is the catalytically competent conformation or whether the extended helices must undergo a large conformational rearrangement in order to bind substrate. We have generated a series of caspase-6 cleavage variants including a novel constitutively two-chain form and determined crystal structures of caspase-6 with and without the intersubunit linker. This series allows evaluation of the role of the prodomain and intersubunit linker on caspase-6 structure and function before and after substrate binding. Caspase-6 is inherently more stable than closely related caspases. Cleaved caspase-6 with both the prodomain and linker present is the most stable indicating that these two regions act in concert to increase stability, but maintain the extended conformation in the unliganded state. Most importantly, these data suggest that caspase-6 undergoes a significant conformational change upon substrate binding, adopting a structure that is more like canonical caspases.
Keywords: crystallography, apoptosis, protease, neurodegeneration, ligand binding
Introduction
Caspase-6 (Mch2) [1] is a member of a family of cysteine proteases originally discovered for their role in apoptosis (programmed cell death). Members of this family have subsequently been found to play major roles in inflammation and neurodegeneration as well. During apoptosis caspases participate in cascades whereby the upstream (initiator) caspases; caspase-8, -9 activate the downstream (executioner) caspases; caspase-3, -7 which cleave a specific group of cellular targets (for review see [2]). Some caspases also undergo self-processing [3]. Caspase-6 has traditionally been considered an executioner due to its homology to caspase-3 and-7, but classification of caspase-6 is difficult due to its unique roles in a number of biological processes. Over-expression of caspase-6 in mammalian cells does induce cell death, confirming its role in apoptosis [4]. Working as an executioner, caspase-6 has been reported to be the only caspase to cleave nuclear lamellar proteins Lamin A/C during apoptosis [5-7]. Caspase-6 has also been reported to serve as an initiator, activating caspase-8 [8-9]. Caspase-6 is reported to be activated by caspase-3 [10], which would place it downstream of both the typical initiator and executioner caspases, and by caspase-1, placing caspase-6 in an inflammatory pathway [11]. Thus caspase-6 has been reported to play executioner, initiator and inflammatory roles.
The most important roles for caspase-6 may actually be in biological cascades that result in neurodegeneration. Recent studies have uncovered the crucial role of caspase-6 in the onset of neurodegenerative disorders like Alzheimer’s, Huntington’s and Parkinson’s Diseases. Cleavage by caspase-6 has been shown to be important in formation of plaques composed of Aβ (amyloid β) peptide and neurofibrillary tangles composed of Tau filaments in Alzheimer’s Disease [12-18]. In mouse models of Huntington’s Disease, only cleavage at the caspase-6 site, but not at the caspase-3 site, is critical for neuronal dysfunction, indicating that caspase-6 cleavage, but not caspase-3 cleavage, is the disease-relevant event. Other Huntington Disease studies using recombinant caspases have implicated caspase-6 and not caspase-3, -7 or -8 in induction of human neuronal cell death [19]. In Parkinson’s Disease DJ-1 protein plays a protective role when cleaved by caspase-6 [20] and prevention of caspase-6 cleavage might be responsible for a subset of Parkinson’s Disease cases. Thus, caspase-6 continues to be implicated in a number of important physiological processes in addition to apoptosis.
The structure of caspase-6 [21] is similar in overall fold to the six other human caspases for which structures are available [22-27] all of which are dimeric when active. The structure of ligand-free caspase-6 differs significantly from all other caspases because two novel extended helices are observed flanking the caspase-6 active site. Caspases-1 [28], -3, and -7 [29-30] have an allosteric site at the dimer interface, which is also predicted to be present in caspase-6. All caspases share a common active-site cysteine-histidine dyad [31], and derive their name, cysteine aspartate proteases, from the presence of the catalytic cysteine at the active site and from their exquisite specificity for cleaving substrate proteins after aspartate residues [32]. Caspases catalyze cleavage of amide bonds via nucleophilic attack of the cysteine thiolate (Cys 163 in caspase-6) at the substrate amide carbonyl. During catalysis the histidine (His 121 in caspase-6) activates the catalytic cysteine and a water molecule. Mutation of either of these residues results in loss of catalytic activity [33].
All known caspases undergo zymogen activation either autocatalytically or through cleavage by an upstream initiator caspase. Caspases are expressed as inactive zymogens of a single polypeptide chain that is capable of dimerization. Cleavage of the procaspase polypeptide occurs at two locations, the prodomain and the intersubunit linker, resulting in the mature form of the enzyme. Caspase prodomain sizes range from 23 amino acids for caspase-6 to 219 amino acids for caspase-10. The initiator caspases typically have large prodomains containing caspase activation and recruitment domains (CARD) whereas executioner caspases have short prodomains. The roles of prodomains range from acting as folding chaperones [34], to mediating protein-protein interactions [35-36], to protecting nuclear localization signals [37]. The prodomain is released from the caspase dimer after cleavage, so the active form of the enzyme consists of two large and two small subunits. The large subunits contain the active site catalytic dyad residues and the small subunits contain most of the dimer interface and the allosteric site. The linker between the large and small subunits includes one or two Asp-containing cleavable sites (two for caspase-6). Caspase-6 has three reported cleavage sites that appear to be cleaved differently by autoproteolysis than when cleaved by other caspases: D23 (following the prodomain), D179 and D193 in the intersubunit linker [38]. The functional effects of both the prodomain and intersubunit linker appear to be unique for each caspase. In executioner caspases, intersubunit cleavage is critical for activation, however a unifying mechanism for activation of all caspases has not been discovered [39-40].
To control each of the caspases uniquely and develop caspase-specific therapies, it is essential that the molecular details of activation and regulation of each caspase are well understood. The prodomain and intersubunit linkers are the most distinctive regions of the caspases and thus hold the greatest promise for unlocking the mysteries of caspase-specific regulation and conformational change. Here we report the first systematic study of the interplay of the intersubunit-linker and prodomain in caspase-6, which also addresses the important questions raised by the reported structure of caspase-6. In the crystal structure of ligand-free caspase-6 [21] a region that forms loops and strands in all other caspases exists as two helices flanking the front and back of the caspase-6 active site. The two most pressing questions stemming from this unusual structure are first, whether the elongated helices are biologically relevant and second, whether substrate binding will induce conformational changes in these helices. Our crystal structures of caspase-6 with and without intersubunit linker demonstrate that the extended helices in caspase-6 are not dependant on cleavage form or crystallization conditions. Second, our spectroscopic analysis suggests that a significant conformational change leading caspase-6 to adopt a more canonical caspase fold is required for all the cleaved forms of caspase-6 to bind substrate.
Results
Cleavage of the prodomain and intersubunit linker has been reported to play a key role in activation of caspases. Despite its importance, the molecular details of these cleavage events and the effect of removal of the prodomain and intersubunit linker on caspase-6 function were impossible to study before now. Cleavage sites for caspase-6 autoactivation have been reported [38], however previous studies used genetic constructs which did not yield uniform populations of caspase-6. This prevented measurement of standard kinetic parameters such as Km and kcat disallowing quantitative comparison amongst caspase-6 cleavage variants or between caspases. Inhomogeneity, particularly in the intersubunit linker, also prevented any analysis of the contributions of the linker to caspase-6 structure or function. To rigorously assess the influence of cleavage and the presence of the linker on caspase structure, stability and function it is essential that strictly homogeneous preparations be made. To assess kinetic and biophysical properties of caspase-6 in all relevant cleaved states it was essential to engineer a new series of expression constructs. Our series is the only method to date that allows purification of uniformly cleaved caspase-6 variants with tight control over production of both the prodomain and intersubunit linker.
Order of intersubunit linker cleavage
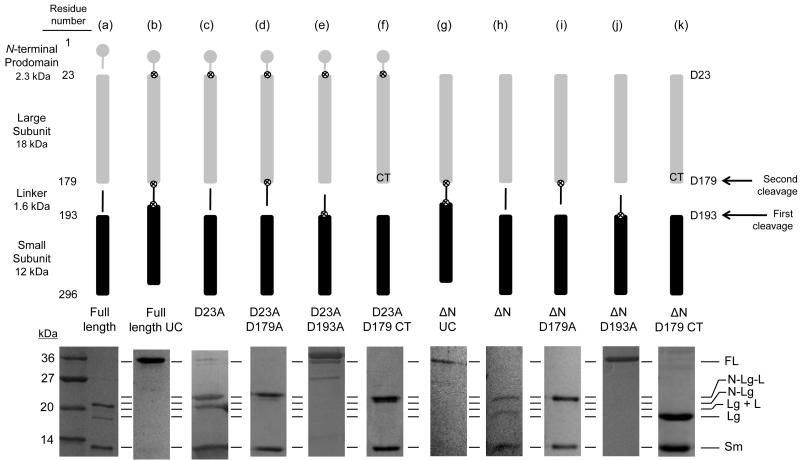
Expression of the full-length caspase-6 gene in bacteria leads to a heterogeneous population of caspase-6, consisting of three polypeptide chains (Fig. 1a). Substitution of the three previously reported cleavage sites [7] with alanine (D23A, D179A and D193A) to generate a full-length uncleavable (FLUC) construct prevented cleavage, producing full-length caspase-6 zymogen (Fig. 1b), confirming the importance of these sites for zymogen activation. Substitution of D23A led to formation of two prodomain plus large subunit fragments, with or without the linker (Fig. 1c), which are both larger than any of the fragments from full-length caspase-6. This suggests that the two large fragments from full-length caspase-6 are the large subunit with or without the intersubunit linker attached (Fig. 1a). In the D23A variant, substitution of D179A resulted in production of just one large subunit fragment with the intersubunit linker attached (Fig. 1d). On the other hand the D193A variant resulted in no zymogen processing whatsoever (Fig. 1e), suggesting that cleavage at D193 is necessary for subsequent cleavage at D179 (Fig. 1 arrows). To produce a homogenous caspase-6 without the linker, we generated constitutively two-chain (CT) caspase-6 in which the large and small subunits are translated independently (Fig. 1f). This novel constitutive two-chain method can be applied to other caspases to generate homogenously two-chain proteins. This is valuable as it does not rely on auto activation and can thus prevent the multiple self-proteolysis products.
Fig. 1.
Caspase-6 cleavage site-blocking variants. Cartoon representation of expression constructs showing cleavable (gap) or blocked ⊗ caspase cleavage sites (upper panel). Symbols for protein domains include prodomain (gray circle), large subunit (gray bar), intersubunit linker (black line), and small subunit (black bar). Aspartic acid residues mutated to alanine are depicted as ⊗. Residue numbers for each domain are indicated at the left of the upper panel. SDS-PAGE analysis of expressed constructs matured in E. coli cells and the observed cleavage patterns (lower panel). (a) Full-length caspase-6. (b) Full-length uncleavable caspase-6 variant (FLUC) D23A/D179A/D193A completely blocks zymogen processing. (c) D23A. (d) D23A/D179A. (e) D23A/D193A. (f) D23A/D179 Constitutive Two-chain (CT) variant expresses the prodomain-large subunit protein inclusive of residue 179 independently of the small subunit, which is expressed from an introduced ribosome-binding site and start codon at residue 193. (g-k) Expression constructs similar to b-f but lack the coding sequence for the N-terminal prodomain. FL= Full Length; N = N-terminal prodomain; Lg = Large subunit; Sm = Small subunit, L = Intersubunit linker. The order of cleavage events leading to activation is indicated by arrows on the right.
To study the interplay of the prodomain, a similar series of constructs were prepared wherein the N-terminal prodomain was deleted (ΔN). The pattern of intersubunit linker cleavage in the absence of the prodomain (Fig. 1g-j) was the same as in its presence. This suggests that the the prodomain has no influence on cleavage of the intersubunit linker. From this expression pattern it is clear that cleavage at D193 necessarily precedes cleavage at D179 by caspase-6, either in cis or in trans. In D193A variants, cleavage at D179 is not observed, suggesting that this site is not accessible to caspase-6 for self-processing, even in the absence of the prodomain. In contrast, in another study in HEK 293T cells, maturation of caspase-6 appeared to occur first at D179 and second at D193 [38]. Thus, the order of caspase-6 zymogen maturation may differ when mediated by another protease in vivo.
Role of prodomain and intersubunit linker on function
From our collection of caspase-6 cleavage variants, five representative constructs were selected to probe the role of the intersubunit linker and prodomain on caspase-6 structure and function. To confirm that the five caspase-6 variants were constituted as described, the mass spectra of each variant was analyzed (Table 1). Each of the variants exhibited precisely the expected molecular weight for the large and small subunits. Caspases require interaction between the two chains of the dimer to function. Removal of interactions such as these could decrease the dimeric propensity of caspases. Thus one possible role for the prodomain or intersubunit linker could be dimer stabilization. Using size exclusion chromatography full-length uncleavable caspase-6 zymogen (FLUC) migrated at the expected molecular weight of a dimer (Table 1, Supplementary Fig. S1). When cleaved at D193, in the presence or absence of the prodomain (D23A D179A and ΔN D179A) the expected molecular weight was observed. Conversely, both caspase-6 variants lacking the intersubunit linker, which also have higher pIs than the caspase-6 variants with the linker present, migrated more slowly than expected, probably due to interactions with the slightly negatively charged column material [41]. Sedimentation velocity analysis by analytical ultracentrifugation confirmed that both linker-lacking variants D23A D179 CT and ΔN D179 CT were indeed dimeric. Interestingly, the molecular weights measured by analytical ultracentrifugation of both linker-lacking variants were smaller than that observed for the zymogen FLUC, suggesting real differences in the shape and hydrodynamic radius of caspase-6 before and after cleavage. Nevertheless, neither the prodomain nor the intersubunit linker appear to change the dimeric state of the protein.
Table 1.
Influence of N-terminal prodomain and intersubunit linker on caspase-6 activity
| Caspase-6 Construct | Pro domain |
Linker | Expected MW |
Observed MW |
Km (μM) | kcat (s−1) | Observed MW SEC (kDa) |
Observed MW AUC (kDa) |
|---|---|---|---|---|---|---|---|---|
|
|
Yes | Yes | 34180 | 34179 | 43 ± 0.51 | 0.04 ± 0.001 | 66 | 85 |
|
|
Yes | Yes | 21892 12350 |
21894 12351 |
67 ± 3.2 | 0.84 ± 0.07 | 78 | NA |
|
|
Yes | No | 20265 12109 |
20264 12109 |
51 ± 3.5 | 0.82 ± 0.03 | 34 | 64 |
|
|
No | Yes | 19624 12350 |
19624 12351 |
66 ± 7 | 0.81 ± 0.01 | 56 | NA |
|
|
No | No | 18066 12350 |
18068 12351 |
45 ± 13 | 0.92 ± 0.14 | 35 | 64 |
UC - uncleavable. CT - constitutively two-chain. ΔN lacks the N-terminal prodomain. Expected molecular weight (MW) is calculated based on the gene sequence in the expression construct. Observed MW is based on mass spectrometric analysis. Kinetic parameters are based on substrate titrations measured from independent duplicate dilutions of substrate on three days. SEC Size Exclusion Chromatography. AUC Analytical Ultracentrifugation. NA Not Attempted.
Since all caspase-6 variants were dimeric, we expected they could be active. As is the case for other caspases [42], cleavage of caspase-6 is the critical activating event. Cleavage at the intersubunit linker was essential to activity, as uncleaved enzyme was inactive (Table 1). On the other hand, removal of the prodomain or intersubunit linker had essentially no impact on either Km or kcat, suggesting that the role of these regions of caspase-6 appears not to be in catalysis.
Prodomain and intersubunit linker affect stability
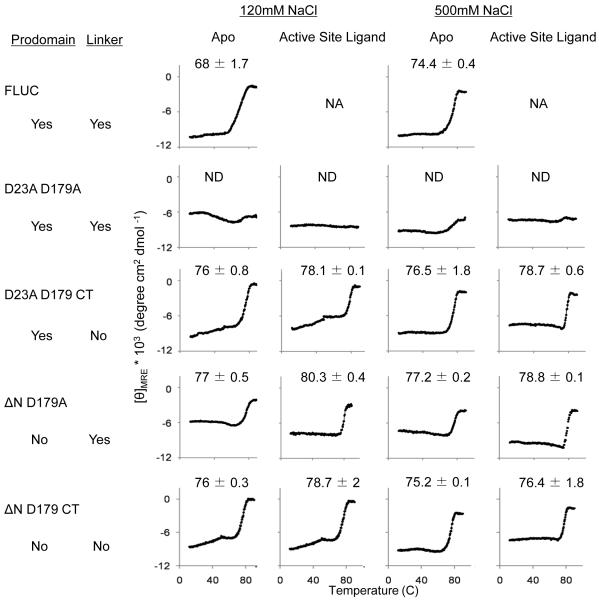
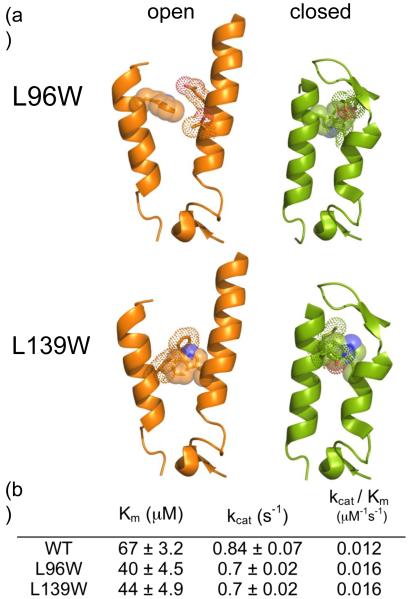
Whereas the linker or the prodomain had no effect on dimerization or catalytic efficiency, the presence of the linker and the prodomain did influence the stability of caspase-6. The thermal stability (Tm) was measured as the midpoint of the thermal denaturation transition (12 to 90°C) monitored by circular dichroism (CD) spectroscopy. At physiological salt concentrations (120 mM NaCl) the zymogen thermal stability was significantly lower than all cleaved forms of caspase-6 (Fig. 2a). Caspase-6 is most stable after a single cleavage with both the prodomain and the linker still attached (D23A D179A); no unfolding transition is observed up to 90°C (Fig. 2a, b). This effect seems to be dependent on the presence of both the intersubunit linker and the prodomain. When the intersubunit linker or prodomain were removed the protein was less stable and unfolded with a Tm of 76-77°C. In addition to monitoring Tm, CD spectra were collected before and after thermal denaturation (Fig. 2b). When most proteins fully unfold, the molar ellipticity (θ) approaches zero [43]. This full unfolding was observed for the zymogen FLUC and both of the linker-lacking variants. However, for both variants containing the intersubunit linker, full unfolding was not observed even at 90°C. This analysis further suggests that cleavage of the linker at D193 increases the stability of caspase-6 and loss of the linker (full detachment by subsequent cleavage at D179) contributes to loss of stability.
Fig. 2.
Circular dichroism (CD) of caspase-6 cleavage variants. (a) Thermal denaturation profiles of caspase-6 cleavage variants unliganded (apo) or with the active-site ligand VEID in physiological (120 mM) or high (500 mM) NaCl concentrations. The measured Tm did not vary between runs, however run-to-run variation in the shape and slope of the pre-melt transition precluded meaningful interpretation of this region of the melting curve. (b) CD spectra of caspase-6 cleavage variants at 12°C (black) or 90°C (gray). The [θ]208/[θ]222 values at 12°C are indicated above each spectra. Each variant was measured using two independently prepared and concentrated samples on two different days. The spectral features were observed in duplicate spectra and a representative spectrum for each variant is shown. NA Not applicable due to inability to functionally verify binding of VEID to the active site. ND Not Detectable.
To probe the basis of stabilization of caspase-6 in the presence of the intersubunit linker, we monitored unfolding at high salt concentrations (0.5 M NaCl). Salt-induced changes in melting temperature are likely to reflect an increased weight of the non-polar interactions relative to polar interactions, which are more strongly screened at high salt concentrations. The only caspase-6 cleavage variant that showed a change in Tm at high salt is the zymogen FLUC form, which increased by 6°C.
Together the prodomain and singly-cleaved linker increase caspase-6 stability. For caspase-7, binding of active-site ligand is similarly stabilizing as the Tm increases from 59°C for ligand-free caspase-7 to 76°C for active-site liganded caspase-7 [44]. To investigate whether significant stabilization was also an effect of substrate binding to caspase-6, we monitored stability in the presence of an active-site ligand. For all variants a 2-3°C increase in Tm was observed upon binding of the active-site ligand VEID-aldehyde, which is a peptide-based, covalently-binding substrate mimic.
Structural features of caspase-6
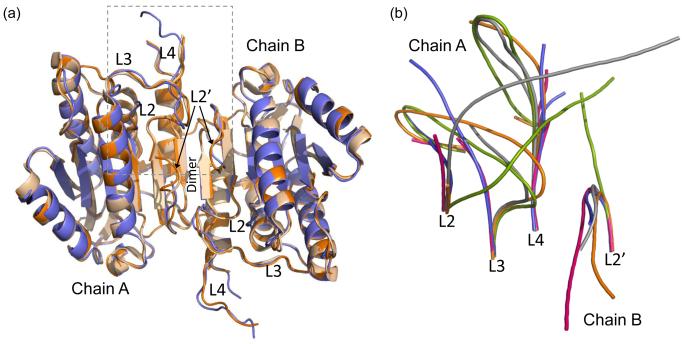
Knowing that the intersubunit linker stabilizes caspase-6, we sought to determine the structure of caspase-6 with and without the linker to note unique structural characteristics resulting from the linker. We report here the structures of two caspase-6 variants, ΔN D179 CT and ΔN D179A (Table 2). The two structures we determined are virtually identical to each other (Fig. 3a) and to the previously reported structure [21] in all of the structural details. Caspase-6 has a canonical dimeric caspase fold comprising a central β-sheet that spans the two halves of the dimer and constitutes the most important dimeric interactions. In the structure of ΔN D179 CT the L2 loop is disordered. In ΔN D179A the L2 loop and intersubunit linker are disordered. The disorder starts at approximately the same residue (R164). This suggests that any contacts made between the linker and the core of the protein may be transitory, or may require the presence of the prodomain. To date we have not succeeded in crystallizing any caspase-6 construct with the prodomain intact. Moreover, though various groups have performed diffraction experiments on several caspases with the prodomain present in the protein construct, no groups have ever observed the prodomain crystallographically [45-46].
Table 2.
Crystallographic Data Collection and Refinement Statistics
|
|
||
| PDB ID | 3K7E | 3NKF |
| Data collection statistics | ||
| Wavelength (Å) | 1.00 | 0.98 |
| Diffraction resolution (Å) | 3.0-64.8 (3-3.16) | 2.9-40 (2.90-2.96) |
| Measured reflections (n) | 57985 | 73593 |
| Unique reflections | 20696 | 21645 |
| Completeness (%) | 96.4 (97.3) | 99.6 (99.9) |
| Redundancy | 2.8 (2.8) | 3.4 (3.3) |
| I/σ(I) | 7.1 (2.0) | 20.6 (2.9) |
| Rmerge | 0.139 (0.653) | 0.085 (0.54) |
| Space group | P21 | P21 |
| a (Å) | 63.4 | 63.3 |
| b (Å) | 90.9 | 90.8 |
| c (Å) | 86.2 | 85.9 |
| α=γ (°) | 90 | 90 |
| β (°) | 91.5 | 91 |
| Refinement statistics | ||
| Atoms (n) | 6527 | 6665 |
| Water (n) | 61 | 27 |
| Rwork / Rfree (%) | 0.218/0.259 | 0.216/0.27 |
| RMSD bond length (Å) | 0.008 | 0.01 |
| RMSD bond angle (°) | 1.097 | 1.23 |
| Average B-factor (Å2) | 46.6 | 83.6 |
| Ramachandran plot | ||
| Core (%) | 89.7 | 89.4 |
| Allowed (%) | 8.9 | 10.5 |
| Generously allowed (%) | 0.5 | 0.1 |
| Disallowed (%) | 0.8 | 0 |
The numbers in parenthesis represent the data in the highest resolution bin.
Fig. 3.
Structures of apo-caspase-6 ΔN D179A and ΔN D179 CT. (a) Superposition of caspase-6 ΔN D179A (orange) and ΔN D179 CT (tan) and an independently determined caspase-6 structure (2WDP, blue). The boxed region indicates the substrate binding groove region that is shown in (b). (b) Superposition of unliganded mature caspase-6 ΔN D179A (orange) with liganded mature caspase-7 (1F1J, green) allosterically inhibited caspase-7 (1SHJ, pink), caspase-7 zymogen (1GQF, blue), and unliganded caspase-7 (1K86, gray) focusing on the substrate-binding loop region including loops L2, L3 and L4 from chain A of the dimer and L2’ from chain B of the dimer. Disordered regions not observed in the crystal structure account for the discontinuity in the middle of loops L3 and L4 in some of the structures.
The structures reported here were determined entirely independently from the previously reported mature ligand-free caspase-6 structure [21] and were deposited in the Protein Data Bank before that structure was released. Baumgartner et. al. used caspase-7 (PDB ID 1F1J) as an initial model and we used caspase-3 (PDB ID 1CP3) with all substrate-binding loops (L1, L2, L2’, L3 and L4) removed as a search model for molecular replacement and starting point for model building. These two independent structure determinations yield models (PDB ID 3K7E, 3NKF and 2WDP), which superimpose with an RMSD for backbone atoms of 0.73 Å, mutually underscoring the validity of this conformation of caspase-6 (Fig. 3a).
One central question about the structures of mature ligand-free caspase-6 is whether this structure represents the catalytically active conformation. In caspase-7, structures of the zymogen, mature ligand-free, mature active-site liganded and mature allosterically-inhibited are available. In caspase-7 only when ligand binds the active site do L2 and L2’ interact, holding L3 in the proper conformation for substrate binding [29, 45-49]. In caspase-6, L2, L2’ and L4 are in the inactive conformation, but even without being pinned down by L2, the L3 loop is in the active, down conformation (Fig. 3b). Like caspase-7, a structural reorganization appears to be required to convert caspase-6 to the catalytically competent conformation.
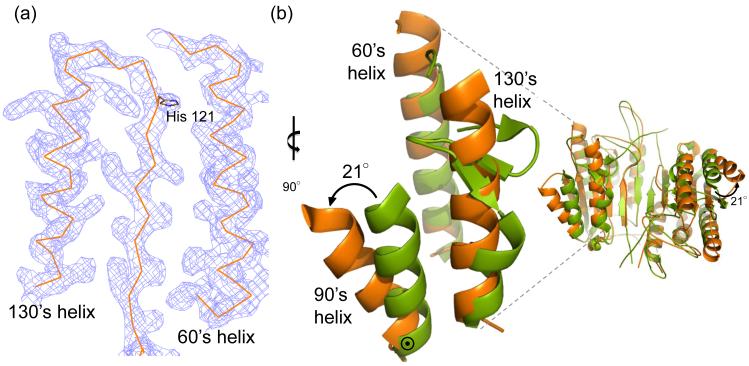
The conformation of mature ligand-free caspase-6 differs in several dramatic details from the structures of any of the previously reported caspases. Two helices are extended in caspase-6 (Fig. 4a) relative to canonical caspases. Superposition of caspase-6 with active caspase-7 reveals the 60’s helix (amino acids 57 – 70) and 130’s helix (amino acids 128 – 142) are one and two helical turns longer, respectively, in caspase-6 (Fig. 4b). In all other caspases the 130’s region shows a strand-turn-strand conformation like that seen in caspase-7. Inspection of the structure revealed a packing network linking the extended portion of the 130’s helix to the extended portion of the 60’s helix (Fig. 4c). This network comprises Tyr 128 from the 130’s helix interacting with the His 121 backbone amide. The His 121 side chain, which is part of the catalytic dyad is rotated away from the catalytic site and held in place by Glu 53 and Glu 63. These interactions are not present in caspase-7 (Fig. 4d). Moreover, key residues in the 60’s to 130’s network, such as Glu 63, are not conserved in any other caspases, and Glu 53 is only present in caspase-1 (Fig. 4e). In other caspases residues unfavorable for helix formation, including Gly and Pro potentially define why extended helices may not form in other caspases. Interestingly His 121, the catalytic base in the cysteine-histidine dyad, mediates these interactions. This His 121 is rotated away from the catalytic nucleophile Cys 163 and is held in a position that is 9.0 Å away. For catalysis these residues should be approximately 2.5 Å apart and interacting. Thus this conformation seems to lock caspase-6 into an inactive state.
Fig. 4.
Unique structural features of caspase-6. (a) 2Fo-Fc electron density map contoured at 1 σ in the region of the 60’sand 130’s helices. Cα trace is shown in orange. (b) Comparison of mature ligand-free caspase-6 60’s, 90’s and 130’s helices (orange) to the homologous region of mature ligand-free caspase-7 (1K86, green). ⊙ denotes the hinge around which the 90’s helix pivots by 21°. (c) Interactions (dashes) holding the 60’s and 130’s network of helices together are mediated by the inactive conformation of catalytic-dyad residue, His 121. (d) The homologous region to (c) in the active-site liganded caspase-7 structure with caspase-6 numbering shown. Indicated residues (drawn in sticks) have the same amino acid identity in both caspase-6 and caspase-7, although the numbering is different. R64, T67 and H121 in caspase-6 numbering are R87, T90 and H144 in caspase-7 numbering. (e) Sequence alignment for the 60’s, 90’s and 130’s helices for all caspases. Amino acid numbering for each caspase is indicated. Strictly conserved residues (orange letters), important network residues (gray highlight) and the catalytic histidine (yellow highlight) in apoptotic initiators, executioners (Exec) and inflammatory (Inflam) caspases are shown. (f) Surface of caspase-6 near the 90’s helix (drawn with 2WDP coordinates, in which the 90’s helix side chains are better resolved) showing the pocket between the 90’s and 130’s helices
The 90’s helix (amino acids 90 – 104) also adopts a conformation not observed in any other caspase. In caspase-6 the 90’s helix pivots outward by 21° around a hinge at the base (C-terminus) of the 90’s helix, resulting in a 10 Å movement of the helix N-terminus relative to caspase-7 (Fig. 4b). Four monomers were observed in the asymmetric unit of this crystal structure. The 90’s helix in two monomers was in one crystal contact, while the 90’s helices in the other two monomers were packed completely differently. Thus, the open conformation of the 90’s helix appears not to result from crystal packing and may reflect a biologically relevant conformation. The outward movement of the 90’s helix relative to the 130’s helix produces a cavity (Fig. 4f), which is not observed in caspase-7.
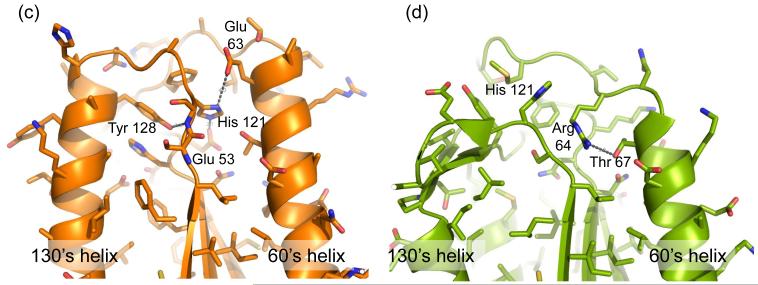
The outward conformation of 90’s helix raises several questions about its physiological role. One possibility is that the 90’s helix acts as a gate, changing conformations between the unliganded and substrate-bound states. Upon binding of a substrate the 90’s helix might move closer to the 130’s helix, thus converting the caspase-6 90’s helix to a canonical, closed structure. To investigate whether the relative position of the 90’s helix with respect to the 130’s helix would affect substrate binding, we sought residues that when mutated to a bulky amino acid would lock the 90’s helix in an open conformation. Every residue at the interface of 90’s and 130’s helices was screened by visualization and modeling on both caspase-6 and closed, substrate-bound caspase-7 (1F1J). Two residues, L96 (90’s helix) and L139 (130’s helix), were chosen to mutate to Trp as the Trp modeled at these positions could be accommodated in the open conformation but no side chain rotamers of Trp were compatible with the closed 90’s helix (Fig. 5a). The L96W and L139W variants did not show much change in activity as assessed by Km or kcat when compared to wild-type (Fig. 5b) suggesting that the relative orientation of the 90’s helix does not affect substrate binding.
Fig. 5.
Mutants to probe the unique 90’s helix conformation. (a) Models of the 90’s and 130’s helices from caspase-6 (orange) in the open conformation and caspase-7 (green) in closed conformation. Caspase-6 positions 96 and 139 are shown mutated from Leu to Trp. Caspase-6 L96 and L139 correspond to residues M116 and L162 respectively in caspase-7. L96 and L139 can accommodate at least one rotomer of Trp in the open conformation, but no rotomers of Trp in the closed conformation, as evidenced by the observed clashes between the introduced Trp and adjacent side chains. (b) The kinetic parameters for caspase-6 ΔN D179A (WT), ΔN L96W D179A (L96W) and ΔN L139W D179A (L139W).
Influence of active-site ligand on structure
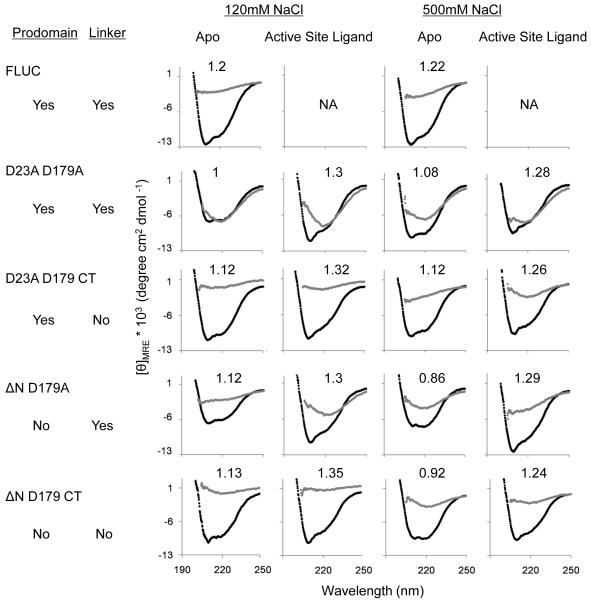
A comparison of the CD spectra of apo and active-site liganded caspase-6 for all cleavage variants showed a consistent increase in the ratio [θ]208/[θ]222 (Fig. 2b) indicating a loss in helical content (signal at 222 nm) upon substrate binding. In ΔN D179 CT, for example, this change resulted in the loss of 18% of CD signal at 222 nm (Fig. 6a,c). Upon binding of active-site ligand, caspase-7 does not show a similar change, losing only 2.3% of the 222 nm CD signal (Fig. 6b, c). This suggests that caspase-6 undergoes a conformational change upon binding active-site ligand that is more substantial than in other caspases. The overall CD spectral shape for caspase-6 and -7 with ligand bound are similar to one another perhaps suggesting similarity in the final bound conformations. When caspase-7 with a DEVD peptide ligand bound in the active site is superimposed with caspase-6 (Fig. 6d), it is clear that some conformational changes must occur in order to accommodate substrate. At a minimum one would predict that the loop above the 130’s helix, which clashes with the caspase-7 substrate, should reorient. Our data go further suggesting that the network between the 60’s and 130’s helices may also reorganize, losing helical structure in order to bind substrate.
Fig. 6.
Comparison of caspase-6 and caspase-7 structures and CD spectra in the presence or absence of the active-site ligands VEID or DEVD. (a) CD spectra of caspase-6 bound to active-site ligand VEID (gray) or in the apo state with no ligand bound (black). (b) CD spectra of caspase-7 bound to active-site ligand DEVD (gray) or in the apo state with no ligand bound (black). (c) Comparision of [θ]208/[θ]222 for apo and substrate-bound states of caspase-6 and caspase-7. (d) Superposition of mature ligand-free caspase-6 ΔN D179CT (orange) with caspase-7 (1F1J, green) bound to the substrate-like ligand DEVD (purple dots).
Discussion
Our studies reveal three major findings, which address the most compelling questions about the structure of caspase-6. First, our data suggest stabilizing interactions between the prodomain and intersubunit linker. Second, we have observed a dramatically different conformation of caspase-6 compared to all other caspases, in the 60’s, 90’s and 130’s region in all of the cleaved forms of caspase-6, reinforcing the fact that the structure of caspase-6 differs from the existing caspase crystal structures; the non-canonical conformation is not an artifact of the construct nor crystallization or structure determination methods. We have shown that movement of the 90’s helix is not essential for caspase function. Finally, our data suggest that binding of active-site ligands induces a significant conformational change comprising a loss in caspase-6 helicity, consistent with conversion to a canonical caspase structure.
Prodomain and intersubunit linker interactions
Caspase-6 with the prodomain and singly-cleaved linker is the most stable form of the protein. Loss of the prodomain makes caspase-6 only slightly less stable. When caspase-6 looses the linker it is destabilized whether the prodomain is present or not. Together these data suggest that the intersubunit linker itself makes important contacts with the core that stabilize caspase-6. When the prodomain and intersubunit linker are present, caspase-6 is so stable that no unfolding is observed even at temperatures as high as 90°C. This suggests that critical contacts for stabilizing caspase-6 are formed between the intersubunit linker and the prodomain, either directly or indirectly. These stabilizing interactions also exist at high salt concentrations, suggesting a hydrophobic basis for the interaction, which is somewhat surprising given the four negative charges present in the intersubunit linker.
Neither the intersubunit linker nor the prodomain has ever been visualized in any caspase structure and our structures are no exception. Given our observation that the intersubunit linker and prodomain function together to stabilize caspase-6, it is perhaps not surprising that we were unable to visualize the intersubunit linker in ΔN D179A as it lacked the prodomain as an interacting partner. Nevertheless, the role of the prodomain in other executioner caspases has been studied. The small prodomain in caspase-6 is typical of the executioner caspases. Removal of the procaspase-6 prodomain has been reported to be essential for caspase-6 mediated activation of caspase-8 [9]. Differences in the pattern of caspase-6 cleavage in that study, (e.g. the observation of the intersubunit linker attached to the small subunit, which we have never observed), make it difficult to confirm these findings. In our studies the presence of the prodomain had no effect on the activity of caspase-6 against peptide substrates. One possible role of the prodomain could be to block access of large substrates to caspase-6. The prodomain of caspase-3 assists in assembly of the proper conformation of caspase-3 [34], but its removal does not impact dimerization [50]. Removal of the caspase-7 prodomain results in a more apoptotically active form than the full-length protein (with the prodomain intact), suggesting that the prodomain plays a protective role in cell survival [51]. In caspase-9 removal of the CARD-containing prodomain also results in increased catalytic activity [52-53]. Thus a stabilizing role of the prodomain may be unique to caspase-6.
The roles of intersubunit linkers have also been studied. In caspase-3 intersubunit cleavage precedes removal of the prodomain [54], but in caspase-7 the prodomain is removed before intersubunit cleavage [55]. In caspase-3, mutation of the intersubunit linker also had a very minor effect on activity, but did seem to improve stability of caspase-3 as the linker mutants were more prone to aggregation [34]. This suggests that perhaps the intersubunit linker in caspase-3 plays a similar stabilizing role as in caspase-6.
The intersubunit linker composition and length are not conserved amongst caspases. Caspase-3 is cleaved between the large and small subunits, but no part of the intersubunit linker is enzymatically removed. The caspase-3 intersubunit linker contains a DDD “safety catch” that keeps caspase-3 from being activated inappropriately by upstream caspases [56]. Overall, intersubunit linkers in various caspases appear to play unique roles and regulate function by different mechanisms in each caspase, implicating this region as one of the critical regions for defining unique caspase specificities. Our findings for caspase-6 are the first to suggest that there are interactions between the prodomain and intersubunit linker that stabilize a caspase.
Function of caspases at the appropriate biological time points and in chemically stressful conditions like apoptosis is essential for organismal survival. Perhaps for this reason, caspase-6 has evolved as a very stable protein, in all forms having melting temperatures greater than 70°C, far above physiological temperatures. In cells, caspase-6 exists as a full-length zymogen in the cytosol while cleaved caspase-6 accumulates in the nucleus, allowing cleavage of targets [57]. The molecular details of how caspase-6 is recognized, transported and subsequently regulated are not known. Given the decrease in the stability upon removal of the intersubunit linker, it is possible that the prodomain and linker function as a timer for destruction of caspase-6, perhaps by the proteosome, as caspases have been reported to be proteosome substrates.
Unique conformation of 60’s, 90’s and 130’s helices
In the structures of unliganded caspase-6 the conformation of the 60’s, 90’s and 130’s helices are different than in any other caspase. Our independently solved structures (3K7E, 3NKF) and the existing unliganded caspase-6 structure (2WDP) reinforce the fact that the structure of caspase-6 does indeed differ from the existing caspase crystal structures and the non-canonical conformation is not an artifact of the construct used or of the crystallization or structure determination methods.
The open conformation of the 90’s helix with respect to 130’s helix is intriguing in that it appears visually to be a potential serendipitous allosteric site. To investigate whether the open form of the 90’s helix was coupled to the extended conformation of the 60’s and 130’s helix we introduced the L96W and L139W mutations. Neither of these mutations, predicted to be incompatible with closure of the 90’s helix, led to decreases in the catalytic efficiency. This suggests that the conformation of the 90’s helix is not directly coupled to changes in substrate binding or activity.
The extended 60’s and 130’s helices are unique to mature ligand-free caspase-6. These striking features raise several questions about the role of caspase-6 in apoptosis and beyond. We consider two potential roles of the extended helical conformation. When the 60’s and 130’s helices are extended they form a network spanning the base of the substrate-binding region (Fig. 4c). This conformation is likely not compatible with binding substrate (Fig. 6d) and holds the catalytic dyad in an inactive conformation (Fig. 4c). During apoptosis the extended 130’s and 60’s helices conformation could potentially restrict the recognition of apoptotic substrates, which could explain why caspase-6 is less apoptotic than other executioner caspases [4]. For example, the 60’s and 130’s helical complex might be compatible with recruitment or recognition of Lamin A/C proteins making caspase-6 the only caspase to cleave these nuclear membrane proteins. It is unlikely that other caspases can adopt this extended helical conformation. Most caspases have residues such as Gly or Pro in the middle of the 130’s region, lowering the helical propensity and likely preventing formation of the extended helix. Caspase-6 lacks Gly or Pro residues in this region and is the only caspase whose sequence is compatible with the Glu 53-His 121-Glu 63 interaction, which aids in maintaining the extended 60’s and 130’s helices.
In addition to changing the shape of the substrate-binding groove, it seems significant that the extended 60’s-130’s conformation locks the catalytic cysteine residue into an inactive conformation. Caspase-6 has also evolved to be more stable than caspase-7. This structural stabilization can be rationalized in terms of the 60’s to 130’s network and the down conformation of the L3 loop. The low gain of stability in caspase-6, upon active-site ligand binding may be explained by the down conformation of L3, making the L2-L2’ interactions more accessible. Though these data might structurally explain the anomalous stability of caspase-6, the question still remains as to why caspase-6 needs to be more stable than other caspases. Based on our data, it is tempting to speculate about a biological role for a cleaved (mature), very stable but inactive form of caspase-6. Given that caspase-3 appears to act cytosolically, it could be the case that caspase-6 undergoes zymogen maturation in the cytosol and then must remain inactive until after transport to the nucleus.
Substrate induced conformational change
Two lines of evidence speak to a change in the structure of caspase-6 upon substrate binding. First, upon active-site binding there is a modest but measurable increase in the thermal stability. Secondly and more importantly, we observe a significant change in the circular dichroism spectra upon substrate binding, which is consistent with a loss in helical structure. This change in CD spectra upon substrate binding, which has also been suggested by others [58], is similar in all cleavage-site variants, indicating that these conformational changes are independent of the prodomain and linker. Thus it appears that upon binding of substrate at the active site, caspase-6 undergoes a significant conformational change in the region of the 60’s and 130’s helices, liberating the catalytic H121 and forming the canonical caspase fold.
The conformational changes that occur at the caspase-6 active site appear to be very different in caspase-6 than in all other caspases. Although IAPs (Inhibitor of Apoptosis Proteins) are known to bind and inhibit caspase-3 and -7 active sites [59], they have not been reported to bind to caspase-6. This ability to discriminate may well be linked to the highly stable conformation of caspase-6 we observe. This may be related to the fact that binding active-site ligand to caspase-6 does increase stability, but the extent of stabilization is not nearly as great as in caspase-7. In wild-type caspase-7 a 17°C increase in the Tm is observed upon binding of an active-site ligand [44] coincident with a large conformational change in the active site loops. These changes in stability in caspase-7 can be rationalized by an understanding of the interactions of the substrate-binding loops with each other and with substrate. Caspase-6 is intrinsically much more stable in the mature, ligand-free state (Tm > 76°C for caspase-6 depending on cleavage state versus 59°C for caspase-7), so the increase in thermal stability upon substrate binding is small for caspase-6 (approximately 2°C). This suggests that losses in stability in reorganizing the 60’s and 130’s bundle to form the canonical caspase fold can be compensated by cross dimer interactions of the substrate binding loops. Thus the mechanism of caspase-6 regulation and substrate binding appears to be very different than in other caspases. Understanding the conformational transition from the stable, catalytically restricted form observed in our crystal structures to the active conformation may be useful for building caspase-6 specific regulators. One might envision that small molecules that could bind to the stable inactive form of caspase-6 seen in our crystal structures could provide a new mechanism for pharmacological inhibition of caspase-6.
Materials and methods
Production of caspase-6 cleavage-site variant expression constructs
The human caspase-6 cDNA cloned into pET23b (generous gift from Guy Salvesen) was used for early experiments. This construct codes for three amino acids that are remnants of the cloning procedure: one extra alanine codon at the 3’ end of the caspase-6 gene and codons for LE at the 5’ end of the gene just before the his-tag coding region. For most of the work described here a synthetic E. coli codon-optimized full-length His-tagged casp-6 gene coding for amino acids 1-293 (Celtek Bioscience) was ligated into the NdeI/BamHI sites of pET11a vector (Stratagene). This codon-optimized caspase-6 gene dramatically improved expression levels for wild-type caspase-6 protein, so this gene was used to make all of the caspase-6 variants. The cleavage-site variants used in these studies were generated by substitution of the codons for amino acids D23, D179 and D193 by the alanine codon GCG. These mutations were introduced by the Quikchange site-directed mutagenesis method (Stratagene). To generate the ΔN constructs the DNA encoding the prodomain (amino acids 1-24, MSSASGLRRGHPAGGEENMTETD) was truncated by PCR amplification of the coding region for amino acids 24-293 from the full-length synthetic variant constructs and ligated into the NdeI/BamHI sites of pET11a.
When protein expressed from the full-length gene was incubated at room temperature it undergoes further maturation at D179 leading to the loss of the intersubunit linker, but room temperature incubation also leads to time-dependant loss of caspase-6 activity (data not shown). In order to obtain homogenous preparations of caspase-6 lacking the intersubunit linker, we generated an expression construct in which a stop codon TAA-Ribosome binding site-TATACATATG was inserted after position 179 by Quikchange mutagenesis (Stratagene). These constructs, which are programmed to produce the caspase-6 large and small subunits as two independent polypeptides, are referred to as constitutive two-chain (CT). The CT constructs omit the intersubunit linker (amino acids 180-193, NTEKLDNITEVD). The mutations L96W and L139W were introduced in the caspase-6 ΔN D179A expression plasmid by Quikchange site-directed mutagenesis method (Stratgene).
Caspase-6 expression and purification
The gene constructs in pET23b (human sequence) or pET11a vectors (E. coli optimized sequence) were transformed into the BL21(DE3) T7 express strain of E. coli (NEB). The cultures were grown in 2xYT media with Amp (100 mg/L, Sigma-Aldrich) at 37°C until they reached OD600 = 0.6. The temperature was reduced to 20°C and cells were induced with 1 mM IPTG (Anatrace) to express soluble His-tagged protein. Cells were harvested after 4 hr for the FLUC or ΔN UC variants and after 18 hr for all other variants to ensure complete processing. Cell pellets stored at −20°C were freeze-thawed and lysed in a microfluidizer (Microfluidics, Inc.) in 300 mM NaCl, 2 mM imidazole, 50 mM phosphate pH 8.5. Lysed cells were centrifuged at 17K rpm to remove cellular debris. The filtered supernatant was loaded onto a 5 ml HiTrap Ni-affinity column (GE Healthcare). The column was washed with 300 mM NaCl, 50 mM imidazole, 50 mM phosphate pH 8.5 and the protein was eluted with 300 mM NaCl, 250 mM imidazole, 50 mM phosphate pH 8.5. The eluted fraction was diluted by 5-fold into 2 mM DTT, 20 mM Tris pH 8.5 buffer to reduce the salt concentration. This protein sample was loaded onto a 5 ml Macro-Prep High Q column (Bio-Rad Laboratories, Inc.). The column was developed with a linear NaCl gradient and eluted in 120 mM NaCl, 2 mM DTT, and 20 mM Tris pH 8.5 buffer. The eluted protein was stored in −80°C in the above buffer conditions. The E. coli optimized gene construct improved the yield from 2-3 mg/L to 7-8 mg/L. The identity of the purified caspase-6 variants was analyzed by SDS-PAGE and ESI-MS to confirm mass and purity.
Activity Assays
For kinetic measurements of caspase activity, 100 nM freshly purified protein (within hours of purification and without ever being frozen, to prevent changes in cleavage pattern or activity) was assayed over the course of 7 minutes in a caspase-6 activity buffer containing 100 mM HEPES pH 7.5, 10% sucrose, 0.1% CHAPS, 5 mM DTT and 30 mM NaCl [60]. For substrate titrations, a range of 0-500 μM fluorogenic substrate, VEID-AMC, (N-acetyl-Val-Glu-Ile-Asp-AMC (7-amino-4-methylcoumarin), Enzo Lifesciences) Ex365/Em495, was added to initiate the reaction. Assays were performed in duplicates at 37°C in 100 μL volumes in 96-well microplate format using a Molecular Devices Spectramax M5 spectrophotometer. Initial velocities versus substrate concentration were fit to a rectangular hyperbola using GraphPad Prism (Graphpad Software) to determine kinetic parameters Km and kcat. Enzyme concentrations were determined by active site titration with quantitative, inhibitor VEID-CHO (N-Acetyl-Val-Glu-Ile-Asp-Aldehyde, Enzo Lifesciences). Active site titration setups were incubated over a period of 2 hours in 120 mM NaCl, 2mM DTT, 20 mM Tris pH 8.5 at nanomolar concentrations. Optimal labeling was observed when protein was added to VEID-CHO solvated in DMSO in 96-well V-bottom plates, sealed with tape, and incubated at room temperature in a final volume of 200 μL. 90 μL aliquots were transferred to black-well plates in duplicate, and assayed with 50 fold molar excess of substrate. The protein concentration was determined to be the lowest concentration at which full inhibition was observed.
Mass spectrometry
Caspase-6 variants in 120 mM NaCl, 2 mM DTT, and 20 mM Tris pH 8.5 were diluted in 0.1% formic acid to a final concentration of 8-10 μM in a100 μl volume and analyzed on an Esquire-LC electrospray ion trap mass spectrometer (Bruker Daltonics, Inc.) set up with an ESI source and positive ion polarity. A PROTO 300 C4 5 μM column (Higgins Analytical, Inc.) was used to desalt the protein on the LC prior to ionization in the mass spectrometer. The MS instrument system was equipped with an HP1100 HPLC system (Hewlett-Packard). Scanning was carried out between 600-1400 m/z and the final spectra obtained were an average of 10 individual spectra.
Size Exclusion Chromatography
The oligomeric state of the caspase-6 variants was determined by running protein samples on a Superdex 200 10/300 GL (GE Healthcare) gel-filtration column. Protein samples were buffer exchanged into 100 mM NaCl, 10 mM Phosphate buffer pH 7.5 using Vivaspin 500, 3K molecular weight cutoff membrane concentrators (Sartorium Stedim Biotech) and 100 μl of 2 mg/ml protein was loaded onto the column. Protein was eluted with 100 mM NaCl, 10 mM Phosphate buffer pH 7.5. The identity of the peak fractions and the loaded protein sample were analyzed by SDS-PAGE. Four different molecular weight standards from the Gel-filtration calibration kit LMW (GE Healthcare) were run in the same conditions and a standard plot was generated to calculate the molecular weights of the caspase-6 variants.
Analytical Ultracentrifugation
Sedimentation velocity experiments were run on Beckman optima XL-1 analytical ultracentrifuge equipped with an An-60 Ti analytical rotor. 400 μl of 1mg/ml protein in 100 mM NaCl, 10 mM Phosphate buffer pH 7.5 was spun at 20 °C in a quartz cell at 30K rpm while monitoring the absorbance at 280 nm. The density of the buffer and the specific volume of the protein were calculated using SEDNTERP [61]. Data was analyzed by SEDFIT [62] using continuous c(M) distribution model.
Stability measurements by circular dichroism
Thermal denaturation of caspase-6 variants was monitored by loss of circular dichroism signal at 222 nm over a range of 12-90°C and CD spectra (250-190 nm) were measured on a J-720 circular dichroism spectrometer (Jasco) with a peltier controller. Active-site liganded caspase-6 variants were prepared by 2-hour room temperature incubation with 3 molar equivalents of VEID-CHO (N-Acetyl-Val-Glu-Ile-Asp-aldehyde, Enzo Lifesciences) at a concentration of 6-12 μM in 120 mM NaCl, 2mM DTT, 20 mM Tris, pH 8.5 buffer. Caspase-7 samples were prepared in a similar fashion by incubating with DEVD-CHO (N-Acetyl-Asp-Glu-Val-Asp-aldehyde, Enzo Lifesciences). To ensure complete binding of active-site ligand to the protein, the protein was assayed with 50 molar excess of substrate VEID-AMC for caspase-6 and DEVD-AMC (N-acetyl-Asp-Glu-Val-Asp -AMC (7-amino-4-methylcoumarin) for caspase-7. A 95-97 % inhibition was observed indicating that the protein was fully occupied with the substrate mimic. Liganded and apo proteins were then buffer exchanged 6 times (7-fold dilution each time) into 10 mM Phosphate buffer, pH 7.5 with 120 mM NaCl or 500 mM NaCl using Vivaspin 500, 3K MWCO membrane concentrators (Sartorium Stedim Biotech) for repeated dilution and buffer exchange. After buffer exchange the final concentration of DTT was ~ 10 nM. A protein concentration of ~6 μM, assessed by absorbance at 280 nm (Nanodrop 2000C spectrophotometer) was used for analysis. Data collected in duplicates on separate days from different batches of purification was fit using Origin Software (OriginLab) using sigmoid fit to determine the melting temperature.
Crystallization and data collection
Purified ΔN D179 CT caspase-6 (construct lacking both the prodomain and the intersubunit linker) in 20mM Tris pH 8.0, 2mM DTT, 120mM NaCl was concentrated using Amicon Ultrafree 5K NMWL membrane concentrators (Millipore Inc.) to ~ 7 mg/ml and crystal trays were set up using Hampton crystal screen solutions. Initial crystal hits were further optimized using different temperatures, pH, salt, protein to precipitant ratios, additives to obtain larger crystals. The best diffracting crystals grew to 30 × 100 × 150 μm in 0.1 M CH3COONa·3H2O pH 4.6, 2M NaCl (Hampton Research, HR2-112), and 3% ethanol in 2-3 days at 25°C in a sitting drop set up.
Purified ΔN D179A (construct lacking prodomain with the intersubunit linker) was concentrated to ~10 mg/ml and the final salt concentration was adjusted to 500 mM. Crystals of approximate dimensions 40 × 100 × 200 μm grew in 0.1 M CH3COONa·3H2O pH 4.6, 2M NaCl, 3% ethanol in 48 hours at 25°C in a hanging drop crystal tray set up.
The ΔN D179 CT caspase crystals cryo-protected in mother liquor with 30% glycerol diffracted to 3 Å and ΔN D179A crystals cryo-protected in 20% glycerol diffracted to 2.9 Å at 100K. Complete data sets of diffraction images were collected for both these crystals at X6A beamline at the Brookhaven National laboratories-National Synchrotron Light Source, Upton, NY.
Structure determination
Diffraction data for ΔN D179 CT were processed in the primitive monoclinic space group P21 with MOSFLM [63] and scaled with Scala [64] in CCP4i [65] (Table 1). Initially we used the highest resolution caspase structure (PDB ID 2DKO, 1.06 Å resolution) but were not able to solve the structure with this search model. The search model successfully used for molecular replacement using Phaser [66] was a monomer of Caspase-3 (PDB ID: 1CP3). To avoid phase bias we omitted the substrate binding loops comprising residues 53-66 (L1 loop), 164-174 (L2), 184-195 (L2’), 203-211(L3), 247-260 (L4). The solution from Phaser had an increasing LLG of 44, 155, 225, 344 and TFZ of 5.0, 15.2, 8.4, 12.5 for each of the monomers. This solution comprising four monomers in the asymmetric unit was built into electron density by iterative rounds of model building in Coot [67] and TLS, NCS restrained refinement [68] using CCP4i. Initially, poor, ambiguous density was seen in the 116-132 region. After several rounds of extensive building and density modification in CCP4i, composite omit maps were generated in CNS [69] and a helical density was seen for the ambiguous 130 region. Structure was refined using automated refinement in Phenix [70] to a final R/Rfree of 0.22/0.26 and was validated by Procheck [71] using CCP4i. The final model is PDB ID 3K7E.
Diffraction data for ΔN D179A was processed and scaled in the primitive monoclinic space group P21 using HKL2000 [72] (Table 2). Since the unit cell parameters were identical to the ΔN D179 CT Caspase-6 (PDB ID: 3K7E) the Rfree set was retained and was refined against 3K7E structure without loops to reduce model bias. This solution comprising four monomers in the asymmetric unit was built into electron density by iterative rounds of model building in Coot and NCS restrained refinement using CCP4i. TLS refinement was added only during the final refinement cycle. Water molecules were placed and the structure was refined to a final R/Rfree of 0.216/0.27. The final structure was validated by Procheck. The final model is PDB ID 3NKF.
Supplementary Material
Acknowledgements
The authors thank Witold A. Witkowski for the caspase-7 protein used in the circular dichroism studies and Prof. Scott C. Garman and Prof. Karsten Theis for helpful discussions regarding crystallographic structure determination. The authors also acknowledge Ama Ruth Boadu for growing the first crystals of mature ligand-free caspase-6. This work was supported by the National Institutes of Health (GM80532), the American Cancer Society (IRG 93-033) and a Young Investigator Grant from the Beckman Foundation. EV was supported by the National Science Foundation (S21000025700000 and DGE-0504485). We thank the staff at X6A beam line, which is funded by the National Institute of Health (GM-0080), and the National Synchrotron Light Source at Brookhaven National Laboratory, which is supported by the U.S. Department of Energy (DE-AC02-98CH10886).
Abbreviations
- CT
constitutive two-chain
- CD
circular dichroism spectroscopy
- FLUC
full-length uncleavable
- VEID
valine-glutamate-isolucine-aspartate (caspase-6 recognition sequence)
- DEVD
Aspartate-glutamate-valine-aspartate (caspase-7 recognition sequence)
- FMK
fluoromethyl ketone
- CHO
aldehyde
- AMC
amino methyl coumarin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein Data Bank Accession Number
Coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 3K7E (ΔN D179 CT) and 3NKF (ΔN D179A).
References Cited
- 1.Fernandes-Alnemri T, Litwack G, Alnemri E. Mch2, a new member of the apoptotic Ced-3/Ice cysteine protease gene family. Cancer research. 1995;55(13):2737. [PubMed] [Google Scholar]
- 2.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384(Pt 2):201–32. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita E, et al. Caspase-9 processing by caspase-3 via a feedback amplification loop in vivo. Cell Death Differ. 2001;8(4):335–44. doi: 10.1038/sj.cdd.4400824. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki A, et al. Regulation of caspase-6 and FLIP by the AMPK family member ARK5. Oncogene. 2004;23(42):7067–75. doi: 10.1038/sj.onc.1207963. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi A, et al. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci U S A. 1996;93(16):8395–400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orth K, et al. The CED-3/ICE-like protease Mch2 is activated during apoptosis and cleaves the death substrate lamin A. J Biol Chem. 1996;271(28):16443–6. [PubMed] [Google Scholar]
- 7.Srinivasula SM, et al. The Ced-3/Interleukin 1 beta Converting Enzyme-like Homolog Mch6 and the Lamin-cleaving Enzyme Mch2 alpha Are Substrates for the Apoptotic Mediator CPP32. Journal of Biological Chemistry. 1996;271(43):27099–27106. doi: 10.1074/jbc.271.43.27099. [DOI] [PubMed] [Google Scholar]
- 8.Cowling V, Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death and Differentiation. 2002;9(10):1046–1056. doi: 10.1038/sj.cdd.4401065. [DOI] [PubMed] [Google Scholar]
- 9.Cowling V, Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 2002;9(10):1046–56. doi: 10.1038/sj.cdd.4401065. [DOI] [PubMed] [Google Scholar]
- 10.Srinivasula SM, et al. Generation of constitutively active recombinant caspases-3 and -6 by rearrangement of their subunits. J Biol Chem. 1998;273(17):10107–11. doi: 10.1074/jbc.273.17.10107. [DOI] [PubMed] [Google Scholar]
- 11.Guo H, et al. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13(2):285–92. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- 12.Wellington C, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. Journal of Biological Chemistry. 1998;273(15):9158. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 13.Lu D, et al. Caspase cleavage of the amyloid precursor protein modulates amyloid - protein toxicity. Journal of neurochemistry. 2003;87(3):733–741. doi: 10.1046/j.1471-4159.2003.02059.x. [DOI] [PubMed] [Google Scholar]
- 14.Graham R, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125(6):1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 15.Wellington C, et al. Inhibiting caspase cleavage of huntingtin reduces toxicity and aggregate formation in neuronal and nonneuronal cells. Journal of Biological Chemistry. 2000;275(26):19831. doi: 10.1074/jbc.M001475200. [DOI] [PubMed] [Google Scholar]
- 16.Weidemann A, et al. Proteolytic processing of the Alzheimer’s disease amyloid precursor protein within its cytoplasmic domain by caspase-like proteases. Journal of Biological Chemistry. 1999;274(9):5823. doi: 10.1074/jbc.274.9.5823. [DOI] [PubMed] [Google Scholar]
- 17.Galvan V, et al. Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. National Acad Sciences; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu D, et al. A second cytotoxic proteolytic peptide derived from amyloid -protein precursor. Nature Medicine. 2000;6(4):397–404. doi: 10.1038/74656. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Goodyer C, LeBlanc A. Selective and protracted apoptosis in human primary neurons microinjected with active caspase-3,-6,-7, and-8. Journal of Neuroscience. 2000;20(22):8384. doi: 10.1523/JNEUROSCI.20-22-08384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giaime E, et al. Loss of function of DJ-1 triggered by Parkinson’s disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death & Differentiation. 2009;17(1):158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumgartner R, et al. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423(3):429–39. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- 22.Wilson KP, et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370(6487):270–5. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 23.Schweizer A, Briand C, Grütter M. Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. Journal of Biological Chemistry. 2003;278(43):42441. doi: 10.1074/jbc.M304895200. [DOI] [PubMed] [Google Scholar]
- 24.Rotonda J, et al. The three-dimensional structure of apopain/CPP32, a key mediator of apoptosis. Nature Structural & Molecular Biology. 1996;3(7):619–625. doi: 10.1038/nsb0796-619. [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7(6):423–32. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard H, et al. The three-dimensional structure of caspase-8: an initiator enzyme in apoptosis. Structure. 1999;7(9):1125–1133. doi: 10.1016/s0969-2126(99)80179-8. [DOI] [PubMed] [Google Scholar]
- 27.Renatus M, et al. Dimer formation drives the activation of the cell death protease caspase 9. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14250. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc Natl Acad Sci U S A. 2006;103(20):7595–600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy JA, et al. Discovery of an allosteric site in the caspases. Proc Natl Acad Sci U S A. 2004;101(34):12461–6. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy JA, Wells JA. Dissecting an allosteric switch in caspase-7 using chemical and mutational probes. J Biol Chem. 2009;284(38):26063–9. doi: 10.1074/jbc.M109.001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denault JB, Salvesen GS. Caspases: keys in the ignition of cell death. Chem Rev. 2002;102(12):4489–500. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 32.Chereau D, et al. Structural and functional analysis of caspase active sites. Biochemistry. 2003;42:4151–4160. doi: 10.1021/bi020593l. [DOI] [PubMed] [Google Scholar]
- 33.Wilson KP, et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature. 1994;370(6487):270–5. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 34.Feeney B, Clark AC. Reassembly of active caspase-3 is facilitated by the propeptide. Journal of Biological Chemistry. 2005;280(48):39772–39785. doi: 10.1074/jbc.M505834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin H, et al. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399(6736):549–57. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- 36.Zhou P, et al. Solution structure of Apaf-1 CARD and its interaction with caspase-9 CARD: a structural basis for specific adaptor/caspase interaction. Proc Natl Acad Sci U S A. 1999;96(20):11265–70. doi: 10.1073/pnas.96.20.11265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaoita Y. Inhibition of nuclear transport of caspase-7 by its prodomain. Biochem Biophys Res Commun. 2002;291(1):79–84. doi: 10.1006/bbrc.2002.6408. [DOI] [PubMed] [Google Scholar]
- 38.Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793(3):592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 39.Boatright KM, et al. A unified model for apical caspase activation. Mol Cell. 2003;11(2):529–41. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 40.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9(3):459–70. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 41.Baker OJ, et al. Size Exclusion Chromatography of Proteins. In: Wu CS, editor. Handbook of Size Exclusion Chromatorgraphy and Related Techniques. 2nd Edition. Marcel Dekker, Inc.; New York, NY: 2004. pp. 439–462. Revised and Expanded. [Google Scholar]
- 42.Karki P, Dahal GR, Park IS. Both dimerization and interdomain processing are essential for caspase-4 activation. Biochem Biophys Res Commun. 2007;356(4):1056–61. doi: 10.1016/j.bbrc.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 43.Scholtz JM, et al. Urea unfolding of peptide helices as a model for interpreting protein unfolding. Proc Natl Acad Sci U S A. 1995;92(1):185–9. doi: 10.1073/pnas.92.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Witkowski WA, Hardy JA. L2’ loop is critical for caspase-7 active site formation. Protein Sci. 2009;18(7):1459–68. doi: 10.1002/pro.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai J, et al. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107(3):399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 46.Riedl SJ, et al. Structural basis for the activation of human procaspase-7. Proc Natl Acad Sci U S A. 2001;98(26):14790–5. doi: 10.1073/pnas.221580098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agniswamy J, Fang B, Weber IT. Plasticity of S2-S4 specificity pockets of executioner caspase-7 revealed by structural and kinetic analysis. Febs J. 2007;274(18):4752–65. doi: 10.1111/j.1742-4658.2007.05994.x. [DOI] [PubMed] [Google Scholar]
- 48.Chai J, et al. Structural basis of caspase-7 inhibition by XIAP. Cell. 2001;104(5):769–80. doi: 10.1016/s0092-8674(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 49.Wei Y, et al. The structures of caspases-1, -3, -7 and -8 reveal the basis for substrate and inhibitor selectivity. Chem Biol. 2000;7(6):423–32. doi: 10.1016/s1074-5521(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 50.Pop C, et al. Removal of the pro-domain does not affect the conformation of the procaspase-3 dimer. Biochemistry. 2001;40(47):14224–35. doi: 10.1021/bi011037e. [DOI] [PubMed] [Google Scholar]
- 51.Denault JB, Salvesen GS. Human caspase-7 activity and regulation by its N-terminal peptide. J Biol Chem. 2003;278(36):34042–50. doi: 10.1074/jbc.M305110200. [DOI] [PubMed] [Google Scholar]
- 52.Shiozaki EN, et al. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11(2):519–27. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 53.Stennicke HR, et al. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274(13):8359–62. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- 54.Martin SJ, et al. The cytotoxic cell protease granzyme B initiates apoptosis in a cell-free system by proteolytic processing and activation of the ICE/CED-3 family protease, CPP32, via a novel two-step mechanism. EMBO J. 1996;15(10):2407–16. [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X, et al. Granzyme B mimics apical caspases. Description of a unified pathway for trans-activation of executioner caspase-3 and -7. J Biol Chem. 1998;273(51):34278–83. doi: 10.1074/jbc.273.51.34278. [DOI] [PubMed] [Google Scholar]
- 56.Roy S, et al. Maintenance of caspase-3 proenzyme dormancy by an intrinsic “safety catch” regulatory tripeptide. Proc Natl Acad Sci U S A. 2001;98(11):6132–7. doi: 10.1073/pnas.111085198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warby SC, et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17(15):2390–404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 58.Kang BH, et al. The structure of procaspase 6 is similar to that of active mature caspase 6. Biochem J. 2002;364(Pt 3):629–34. doi: 10.1042/BJ20011787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suzuki Y, et al. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J Biol Chem. 2001;276(29):27058–63. doi: 10.1074/jbc.M102415200. [DOI] [PubMed] [Google Scholar]
- 60.Stennicke HR, Salvesen GS. Biochemical Characteristics of Caspases-3,-6,-7, and-8. Journal of Biological Chemistry. 1997;272(41):25719–25723. doi: 10.1074/jbc.272.41.25719. [DOI] [PubMed] [Google Scholar]
- 61.Hayes D, Laue T, Philo J. Sednterp: Sedimentation Interpretation Program. 1995 Version. [Google Scholar]
- 62.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical journal. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leslie A. Jnt CCP4/ESF-EACBM Newsl. Protein Crystallogr. 1992;26:27–33. [Google Scholar]
- 64.Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D: Biological Crystallography. 2005;62(1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 65.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 66.McCoy A, et al. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40(4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D: Biological Crystallography. 2004;60(12):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 68.Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D: Biological Crystallography. 1997;53(3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 69.Brunger A, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallographica Section D: Biological Crystallography. 1998;54(5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 70.Adams P, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallographica Section D: Biological Crystallography. 2002;58(11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 71.Laskowski R, et al. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26(2):283–291. [Google Scholar]
- 72.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. Macromolecular Crystallography, Part A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.