Abstract
We studied the effect of the presence of Coccidioides on the production of nitric oxide (NO) by primary macrophages previously activated by IFN-γ and LPS. The fungal cells were isolated from cultures of arthroconidia that had been incubated for 24h in a medium that supported parasitic phase growth and were co-cultured with the macrophages. These live, first-generation parasitic cells of Coccidioides, referred to as spherule initials, suppressed NO production as well as iNOS mRNA expression by activated macrophages. Phagocytosis was not required for suppression of NO. We also showed that the culture supernatant of the spherule initials was capable of suppressing NO production, and that this activity was mediated by an as yet unidentified, secreted fungal factor(s). Heat-, paraformaldehyde- or X ray-treated spherule initials did not show this inhibitory effect. To our surprise, macrophages obtained from iNOS-deficient mice revealed phagocytic activity and killing efficiency which were comparable to that of macrophages isolated from wild type C57BL/6 mice. Although the cultured fungal pathogen can suppress NO production, this oxidative product is apparently not essential for in vitro killing of Coccidioides by activated macrophages. Our results suggest that other unidentified fungicidal mechanisms exist against Coccidioides which are apparently independent of NO production.
Keywords: nitric oxide, macrophage suppression, Coccidioides
1. Introduction
Coccidioides is the causative agent of coccidioidomycosis or San Joaquin Valley fever, one of the endemic mycoses in the United States with an estimated incidence of respiratory infection of 100,000 people annually [1]. This mycosis is also endemic to other regions of the Western Hemisphere, including Northern Mexico, Guatemala, Honduras, Colombia, Venezuela, Paraguay, and Argentina [2]. Human infection occurs primarily after inhalation of the fungal spores (arthroconidia). The majority of cases of coccidioidomycosis are either asymptomatic or result in self-limited pneumonia. Although this mycosis is rarely life threatening, most patients who do not recover spontaneously develop extrapulmonary infections [1].
Coccidioides encompasses two species, C. immitis and C. posadasii based on their genetic and biogeographical differences [3–5]. However, laboratory animal studies of the experimental pulmonary disease caused by C. immitis or C. posadasii have shown no significant differences in the morphogenesis or virulence of the two pathogens [6]. Coccidioides interaction with host phagocytes during disease onset is characterized by both an extracellular and intracellular relationship, which is dependent at least in part on the size of the parasitic cells. Arthroconidia are barrel-shaped cells that measure 2.5 to 3 μm in width by 3 to 6 μm in length. They are small enough to reach the alveoli if inhaled and are readily internalized by phagocytic cells. If these fungal cells survive exposure to the host defenses, they germinate by isotropic growth and give rise to small, round cells (spherule initials; approx. 15–20 μm in diameter) that continue to enlarge and differentiate into coenocytes (spherules). The latter typically range from 40 to 120 μm in diameter [7,8] and are too large to be engulfed by phagocytes. These multinucleate, parasitic cells subsequently undergo a process of cytoplasmic segmentation, which culminates in the formation of a multitude of endospores each of which are approximately 10 μm in diameter at the time of their release from the mature spherules. Endospores are also susceptible to phagocytosis by host innate immune cells.
Nitric oxide (NO) is known to be an important component of the arsenal of host oxidative defenses directed against microbial pathogens. NO is produced by enzymatic activity of the inducible isoform of nitric oxide synthase 2 (iNOS or NOS2), which is expressed by phagocytic cells (primarily macrophages). Host cytokines, such as IFN-γ and tumor necrosis factor (TNF), as well as microbial products, especially LPS, have been shown to activate and induce the synthesis of NO by macrophages [9]. The role of NO in host defense against several fungal pathogens has been previously discussed [10–14]. Mice which were genetically engineered to produce high levels of human interleukin (IL)-10 in response to Coccidioides infection were shown to be more susceptible to acute coccidioidal peritonitis and pneumonia and expressed lower levels of gamma IFN-γ, IL-12p40 and iNOS mRNA in their lungs than untreated mice [1]. The authors proposed that the reduced level of iNOS expression and increased susceptibility to infection of the transgenic mice implicated NO synthesis as a mechanism of resistance against coccidioidomycosis. In addition, these same investigators revealed that disease-resistant DBA/2 mice treated with aminoguanidine, an inhibitor of NO, increased their susceptibility to coccidioidal infection which further supported their notion that nitric oxide is directly involved in defense against this pathogen. In the present study we report the ability of Coccidioides to significantly reduce the level of in vitro production of NO by IFN-γ + LPS-activated murine primary macrophages. Our results show that the pathogen secretes a soluble factor(s) which is responsible for this suppression of NO production, as well as a corresponding down-regulation of iNOS expression by both peritoneal and bone marrow-derived macrophages. This is the first evidence that Coccidioides can directly suppress the production of a key product of the host oxidative defense repertoire.
2. Results
2.1. Suppression of NO production by primary peritoneal macrophages in the presence of spherule initials
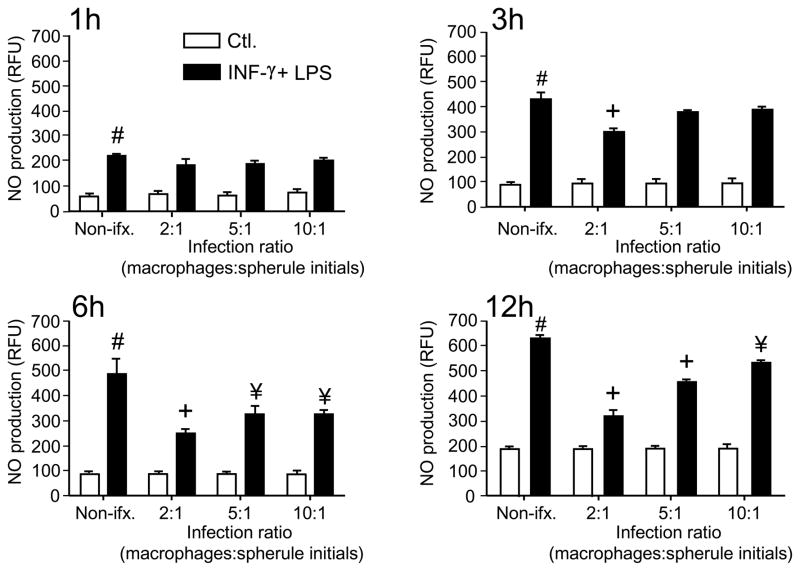
Primary peritoneal macrophages of C57BL/6 mice, which had been elicited with sodium thioglycolate, were isolated and either activated with IFN-γ plus LPS or untreated. Spherule initials from either C. posadasii or C. immitis were separately co-cultured with the phagocytic cells using a range of macrophage:fungus ratios. NO concentration was determined at different periods of co-culturing as shown in Fig. 1. We observed that production of NO by activated macrophages in the presence of spherule initials was suppressed compared to activated, non-infected phagocytes as early as 3 h after the addition of the fungal pathogen. Non-activated (control) macrophages showed minimal levels of production of NO in either the presence or absence of the fungal cells. The degree of suppression of NO production by activated macrophages varied according to the duration of exposure of the phagocytes to the pathogen and the MOI, with maximum suppression between 6 and 12 h at a MOI of 2:1 (P<0.001).
Fig. 1.
Spherule initials of Coccidioides suppress NO production by activated, primary macrophages isolated from C57BL/6 mice. Peritoneal macrophages were activated with IFN-γ + LPS, and NO concentration was measured by fluorescence using DAF-2DA and expressed as relative fluorescence units (RFU). Spherule initials of Coccidioides were co-cultured with macrophages to yield a multiplicity of infection (MOI) of 2:1, 5:1 or 10:1 (macrophages:fungal cells), and incubated 1, 3, 6 or 12 h in a 5% CO2 incubator at 37° C. All experiments were performed at least three times. Results of a representative experiment are shown. Error bars indicate standard errors of the mean (SEM). Statistically significant differences in NO production between test groups was determined using the Student t-test for the following comparisons: # vs non-activated macrophages = P<0.001, and + or ¥ vs activated, non-infected macrophages = P<0.001 & P<0.05, respectively. Non-ifx., non-infected macrophages; Ctl., non-activated, control macrophages.
2.2. Fungal suppression of NO production by macrophages requires live spherule initials but not engulfment of the pathogen
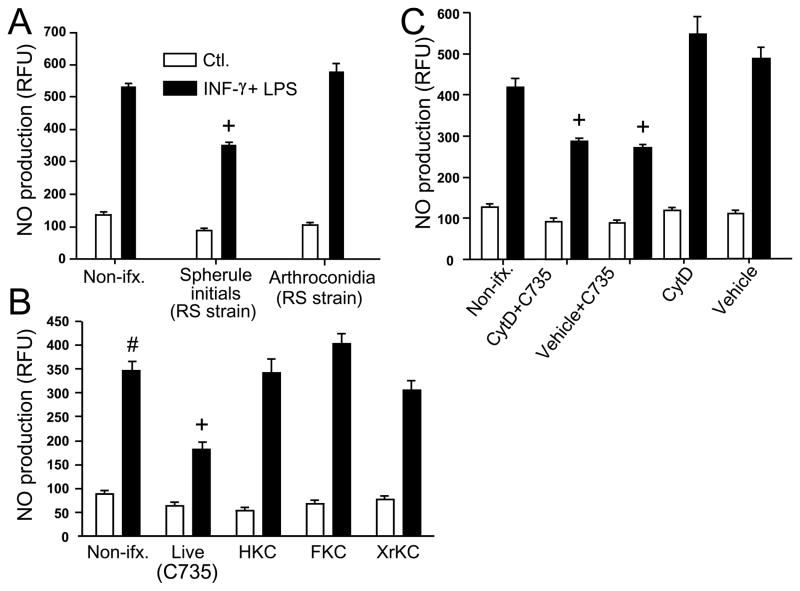
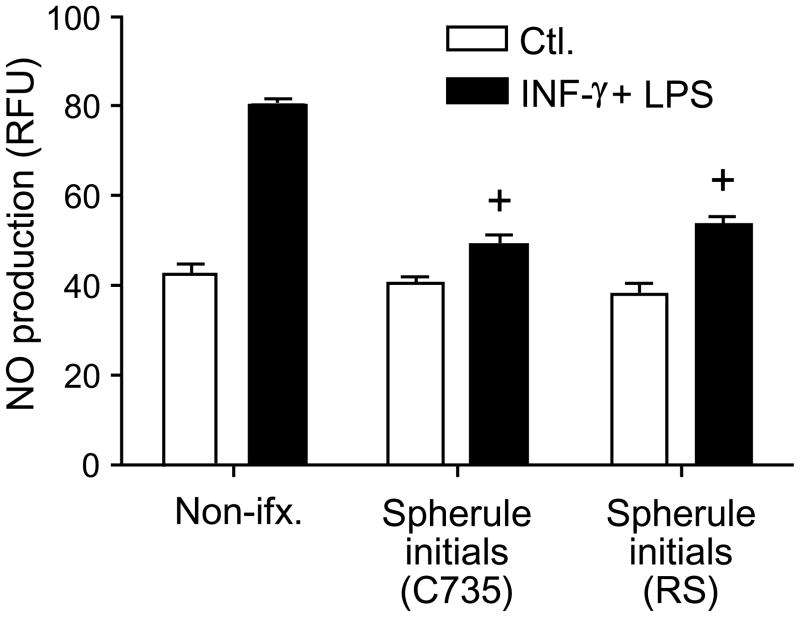
In contrast to the spherule initials, live arthroconida were not able to suppress NO production (Fig. 2A) using the same range of MOI reported in Fig. 1. In order to determine if live spherule initials were necessary for the observed suppression of NO, we subjected the fungal cells isolated from each species of Coccidioides (C735 and RS strains) to lethal effects of heat (HKC), paraformaldehyde fixation (FKC) or X-rays (XrKC). As shown in Fig. 2B, only the live spherule initials were able to suppress NO production. No statistically significant differences were observed between the two species of the pathogen (not shown).
Fig. 2.
A-C Spherule initials but not arthroconidia suppress NO production by activated peritoneal macrophages (A), live but not killed spherule initials are capable of the suppressive activity (B), and phagocytosis is not required for NO suppression (C). Macrophages were co-cultured with arthroconidia or spherule initials at a MOI of 2:1 (macrophages:fungal cells) for 12 h at 37° C (A), or incubated with killed spherule initials using the same MOI (B). Activated, peritoneal macrophages in (C) were first incubated with cytochalasin D (CytD) solubilized in DMSO, or DMSO alone (vehicle), and then co-cultured with spherule initials at the same MOI and incubation conditions as above. Experiments shown in A, B and C were performed at least three times by triplicate, with the same trend observed each time. A representative experiment is shown. A statistically significant difference in NO production was shown between + vs activated, non-infected macrophages or activated, macrophages infected with arthroconidia (A), + vs activated macrophages incubated with killed spherules (B), + and vs cytochalasin D- or DMSO-treated, activated macrophages, each of which = P<0.001. CytD, DMSO-solubilized cytochalasin D; HKC, heat-killed spherule initials; FKC, paraformaldehyde-fixed and killed spherule initials; XrKC, X ray-irradiated and killed spherule initials.
Harvested and activated peritoneal macrophages were exposed to cytochalasin D (CytD) at a concentration which inhibits actin polymerization and blocks phagocytic activity of the cells. We observed that the CytD-treated macrophages were still sensitive to the presence of spherule initials, suggesting that NO suppression is mediated either by fungal-macrophage contact, or by exposure of the phagocytes to a secreted factor(s) of the spherule initials (Fig. 2C).
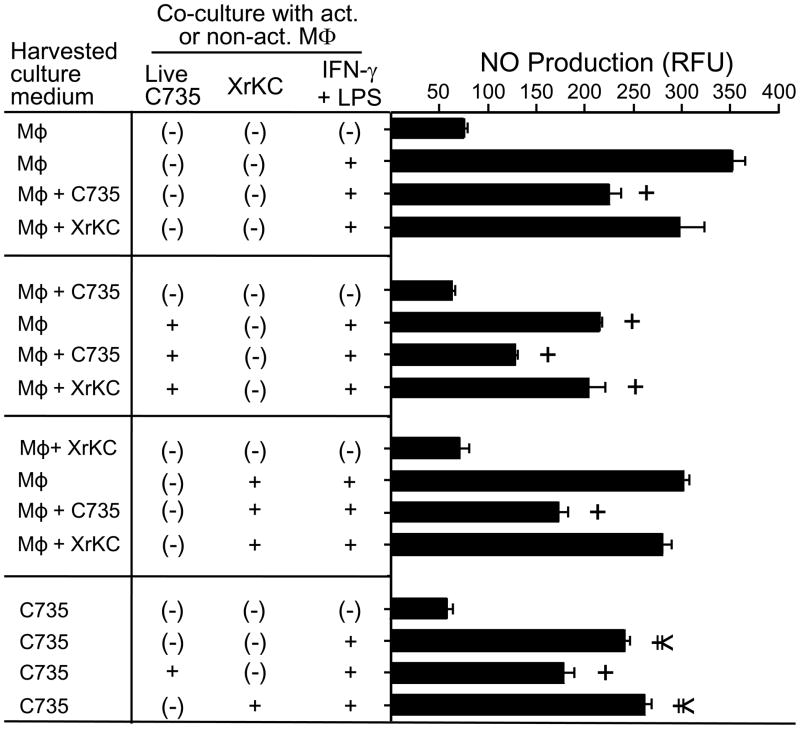
2.3. Suppression of NO production by primary macrophages is mediated by a secreted factor(s) derived from the spherule initials
To determine whether a secreted factor produced by the spherule initials was responsible for NO suppression, we isolated the supernatants of macrophage cultures which had been exposed to either live or dead fungal cells. We also separately isolated and evaluated supernatants from axenic cultures of macrophages and spherule initials. In each case the filtered supernatants were used to pre-treat fresh cultures of non-activated or activated macrophages in the presence/absence of either live or dead spherule initials. We observed that treatment of activated macrophages with supernatants from co-cultures of macrophages and live spherule initials, as well as exposure of these phagocytes to supernatants of axenic cultures of spherule initials suppressed NO production in a significant manner compared to cultures of activated macrophages alone (P<0.001 and P<0.05, respectively; Fig. 3). When we tested the supernatants of macrophage cultures which had been exposed to dead spherule initials by the same procedure, we did not observe a significant suppression of NO production by the phagocytic cells. An additive suppressive effect was apparent when the culture supernatant of live but not killed spherule initials plus activated macrophages were mixed with the test combination of macrophages and fungal cells. These results indicate that NO suppression by activated macrophages in vitro was mediated by an as yet unidentified soluble, secreted factor(s) derived from the live spherule initials.
Fig. 3.
The filtered culture supernatants of spherule initials produced by C. posadasii (strain C735) and C. immitis (strain RS; not shown) contain a biologically-active component(s) that can suppress NO production by activated peritoneal macrophages. The MOI and incubation conditions were the same as described in Fig.2. Fresh cultures of activated or non-activated macrophages were exposed to filtered supernatants of co-cultures of macrophage and live spherule initials (MΦ + C735), macrophage cultured with killed spherule initials (MΦ + XrKC), the filtered supernatant of macrophage cultures alone (MΦ), or the filtered supernatant of spherule initials alone (C735). A representative experiment is presented; the experiment was repeated three times with similar results. A statistically significant difference in NO production was shown between + and ¥ vs activated, non-infected macrophages, which = P<0.001 and P<0.05, respectively.
As a preliminary attempt to characterize the secreted component responsible for the partial inhibition of NO production, we fractioned the 24 h spherule initial culture supernatant by selective filtration to yield subfractions with molecular sizes that were <100 kDa, <50 kD, and <10 kDa. We observed that all fractions had a comparable degree of suppression of NO production by activated macrophages (not shown).
2.4. Peritoneal and bone marrow derived macrophages reveal comparable in vitro suppression of NO production in the presence of spherule initials
Our initial in vitro studies of NO suppression were conducted using peritoneal macrophages (PM), and we wondered if NO production by bone marrow-derived macrophages (BMDM) would be equally susceptible to the secreted products of spherule initials. BMDM were isolated from C57BL/6 mice using a standard procedure, activated as previously described for peritoneal macrophages, and co-cultured with live spherule initials at a MOI of 2:1 for 12 h. The degree of suppression of NO production by the activated BMDM was comparable to that observed in co-cultures of PM and spherule initials (Fig. 4).
Fig. 4.
Live spherule initials of Coccidioides can suppress NO production by bone marrow derived-macrophages (BMDM). The in vitro suppressive activities of C. posadasii (C735) and C. immitis (RS) were comparable. The MOI and incubation conditions were the same as described in Fig. 2. These experiments were performed at least three times by triplicate, with the same trend observed each time. A representative experiment is shown. A statistically significant difference in NO production was shown between + vs activated, non-infected BMDM, which = P<0.001.
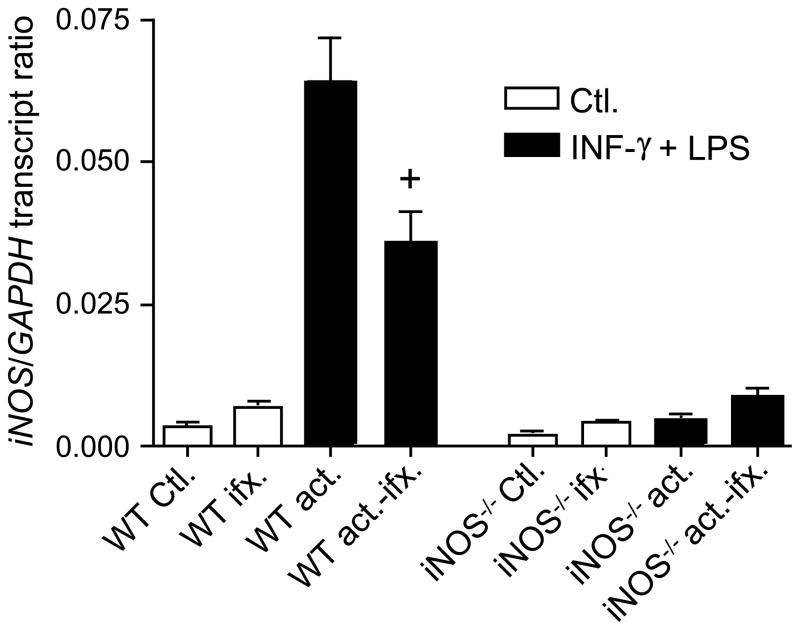
2.5. Spherule initials suppress iNOS expression in activated macrophages
Based upon our above observations of suppression of NO production, we expected that iNOS expression in macrophages co-cultured with live spherule initials would be down-regulated. Total mRNA was isolated from activated macrophages which had been cultured in the presence or absence of the fungal cells, and iNOS transcription levels were determined by quantitative real time-PCR normalized to the expression of a Coccidioides housekeeping gene that coded for glyceraldehyde 3-phosphate dehydrogenase (GAPDH). As shown in Fig. 5, iNOS mRNA was marginally detectable in non-activated macrophages, while non-infected WT macrophages previously activated with IFN-γ + LPS showed a marked induction of iNOS expression. However, in the presence of live spherule initials iNOS transcription by the activated peritoneal macrophages was reduced to about 0.5-fold of the expression level of non-infected phagocytes. This reduction in gene expression was comparable to the degree of suppression of NO production by the same infected and activated macrophages. These results suggest that the suppressive effect of spherule initials in vitro is at least partly manifested at the level of transcription.
Fig. 5.
Live spherule initials of Coccidioides can down-regulate iNOS expression by activated peritoneal macrophages. Expression of iNOS by activated (act.) and non-activated (Ctl.) macrophages of WT and iNOS−/− mice in the presence (ifx.) or absence of spherule initials was determined by quantitative real time-PCR, normalized to the expression of the constitutively-expressed GAPDH gene of C. posadasii. mRNA isolated from macrophages of iNOS−/− mice co-cultured with spherule initials as above served as controls. Error bars represent the standard error of the mean (SEM) of triplicate samples from one experiment representative of two. A statistically significant difference in NO production was shown between + vs activated, non-infected macrophages, which = P<0.01.
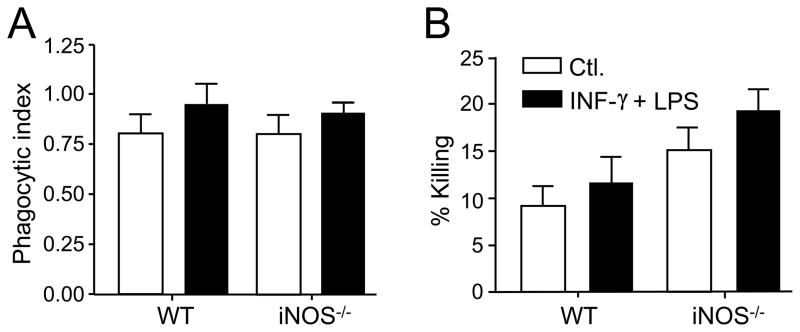
2.6. Macrophages derived from WT and iNOS−/− C57BL/6 mice show comparable levels of phagocytosis and killing efficiency of spherule initials in vitro
The phagocytic index was determined by microscopic examination of fluorescently-stained spherule initials co-cultured with activated or non-activated peritoneal macrophages isolated from either WT or iNOS gene knock-out mice. A near-identical phagocytic index was determined for the interaction of the fungal pathogen with either activated or non-activated macrophages derived from these two strains of mice (Fig. 6A).
Fig. 6.
A,B Assays of phagocytosis and killing efficiency of spherule initials by peritoneal macrophages isolated from WT and NOS−/− mice suggest that NO production is not essential for fungicidal activity against the parasitic cells of Coccidioides. (A) Determination of the phagocytic indices for activated and non-activated peritoneal macrophages isolated from either WT or iNOS−/− mice and co-cultured with spherule initials using a MOI of 2:1 (macrophages:fungal cells). (B) Estimate of killing efficiency of activated and non-activated peritioneal macrophages derived from WT or iNOS−/− mice based upon recovered numbers of viable spherule initials which had interacted with (adhered to or engulfed by) the phagocytes. No statistically significant difference existed between the two sets of conditions, although a trend was evident that the macrophages isolated from iNOS−/− mice showed a slightly higher killing efficiency in vitro. Error bars represent the standard error of the mean (SEM) of three independent determinations (n=6).
We also performed fungicidal assays using peritoneal macrophages isolated from the WT or iNOS−/− mice and co-cultured with viable spherule initials. The total number of viable fungal cells recovered after phagocytosis (non-engulfed or adherent spherule initials) was determined and used to estimate the per cent killing by macrophages isolated from the WT and iNOS−/− mice. No statistically significant difference in fungal cell killing efficiency was observed between the activated and non-activated macrophages derived from the two strains of mice. However, there was an apparent trend in the case of iNOS−/−-derived macrophages which showed consistently higher killing efficiency under the incubation conditions used in our study. These data suggest that nitric oxide production is not essential for fungicidal activity of macrophages against the spherule initials of Coccidioides.
3. Discussion
Using a macrophage-free system and a NO generator (3-morpholinosydnonimine [SIN-1]), we previously reported that arthroconidia of C. posadasii were more sensitive to NO exposure than either spherule initials or endosporulating spherules, suggesting that the latter two parasitic morphotypes of Coccidioides may be capable of detoxification of nitric oxide in vitro [7]. The ability of Coccidioides to influence host macrophage production of NO had not been further explored. On the other hand, several fungal pathogens which also cause human systemic infections have been reported to be capable of direct or indirect suppression of macrophage production of NO in vitro, including Cryptoccocus neoformans, Candida albicans and Paracoccidioides brasiliensis [15–19]. However, the mechanisms by which these fungi influence nitric oxide production have not been determined.
In the present study, we demonstrated that live spherule initials of Coccidioides were able to suppress both NO production and iNOS expression by activated peritoneal and bone marrow-derived macrophages grown in vitro. We revealed that this inhibitory effect was not dependent on phagocytosis, and could be abolished if the parasitic cells were killed by heat (70° C), chemical-fixation, or exposure to X-ray irradiation. Contrary to studies reported above for C. neoformans [15, 19] and C. albicans [16, 17], we found that direct contact between the parasitic cells of Coccidioides and peritoneal macrophages was not required to suppress NO production. We demonstrated that the filtered culture supernatants of spherule initials grown in the presence or absence of activated phagocytes were able to suppress NO production by activated peritoneal macrophages grown in freshly prepared media. In a preliminary attempt to characterize the molecular size of the secreted factor(s) responsible for this inhibition, our results were inconclusive. Biological activity was observed over a wide range of molecular size, from 100 kDa to <10 kDa. Experiments are underway using high pressure liquid chromatographic (HPLC) methods to identify the suppressive components secreted by the spherule initials.
We demonstrated that the inhibitory effect on NO production is fungal morphotype-specific. Arthroconidia were unable to suppress the production of nitric oxide by macrophages in vitro. We have reported that the outer cell wall layer of arthroconidia contains abundant cysteine-rich proteins (hydrophobins) that function as a hydration barrier [7]. When arthrocondia were exposed to SIN-1 in solution at a concentration of 1.25 mM, the cells remained in colloidal suspension due to their hydrophobicity and showed about 40% growth inhibition. Presumably, NO was able to penetrate the physical barrier of the conidial wall. As arthroconidia undergo isotropic growth in vitro or in vivo, their outer wall layer is shed and the inner layers of the spherule initials are hydrated. However, spherule initials exposed to SIN-1 at the same concentration as above showed only 20% growth inhibition. At this stage of parasitic cell development soluble products are secreted by the parasitic cells into the growth medium and/or remain associated with the surface of the microbe [20]. In fact, we have shown that spherule initials are coated with an immunodominant, proline/aspartate-rich glycoprotein [21], which prompted us to suspect on the basis of the reported function of the cell surface-associated glycoprotein (GP43) in P. brasiliensis which was shown to inhibit the release of NO by zymosan stimulated-macrophages [18], that this cell surface antigen may be involved in NO suppression. Spherule initials of a mutant strain of C. posadasii lacking the gene that encodes this spherule outer wall glycoprotein (ΔSOWgp), together with the wild type parental strain, were tested for their ability to suppress NO production by activated peritoneal macrophages. We observed that the two fungal strains were equally capable of the inhibitory process (data not shown), indicating that SOWgp is not a factor directly involved in the suppression of NO production in vitro.
Macrophages and neutrophils are considered to be the first members of the innate immune repertoire to be directed in defense against Coccidioides infection [6,7]. This innate immune response against intracellular pathogens is activated by Toll-like receptors (TLRs), which trigger direct antimicrobial activity that is typically mediated primarily by nitric oxide [22,23]. Production of reactive nitrogen intermediates by activated macrophages has been shown to have a fungicidal effect on a broad spectrum of causative agents of mycoses, including P. brasiliensis, C. neoformans, C. albicans and Sporothrix schenckii among others [10–14]. However, a comparable in vitro study of phagocyte interaction with Coccidioides revealed that a low percentage of the parasitic cells are killed by the activated host cells [24], and fungicidal activity against the respiratory pathogen declines during transformation from arthroconidia to spherule initials [25]. In general spherules have been reported to exhibit resistance to the fungicidal activity of macrophages, with <10% killed in vitro [26]. We have shown that the killing efficiency against spherule initials elicited by macrophages isolated from an iNOS knock-out strain of mice was comparable to slightly greater than the killing efficiency of activated macrophages obtained from the wild-type strain. In addition, we did not observe any difference in the phagocytic index between macrophages obtained from these two sources. These results argue that NO production by activated macrophages during phagocytosis does not have a direct fungicidal effect on Coccidioides. Similar results were reported by Farah and coworkers [27], who observed no statistically significant difference in the ability of iNOS−/−- and WT murine strain-derived BMDM to kill C. albicans yeast in vitro. In contrast to our findings, however, they reported that macrophages from iNOS−/− mice phagocytosed a significantly greater percentage of yeast than BMDM isolated from wild type, control mice. In another study, it was proposed that NO is indirectly involved in macrophage candidacidal activity [28]. The authors suggested that nitric oxide may serve as a candidastatic molecule, and other candidacidal oxidative and non-oxidative mechanisms are directly involved in the pathogen killing process. Perhaps our report of the apparent increase in killing efficiency of Coccidioides by macrophages isolated from the iNOS−/− mice is the result of a compensatory effect on expression of other fungicidal products. The report by Jimenez and coworkers [1] cited in our introductory remarks which reported a correlation between an elevated level of murine IL-10 production, reduced expression of iNOS and increased susceptibility to coccidioidal disease could be interpreted as a consequence of the down-regulation of the cellular immune response to infection, and not the direct effect of a decrease in NO synthesis. In their examination of the influence of aminoguanadine treatment of mice on susceptibility to Coccidioides infection, the same authors have pointed out that this compound is not a specific inhibitor of iNOS expression and the resulting increase in susceptibility to disease, therefore, may not be due directly to reduced NO production as suggested. With these results in mind and trying to find a possible explanation for our findings, we decided to measure the IL-10 levels in culture supernatants of activated or non-activated macrophages in the presence or absence of spherule initials of Coccidioides at 24h of co-culture. We did not observe any difference in the IL-10 levels of the different co-cultures (data not shown), indicating that IL-10 production is not involved in the suppression of NO production by this fungal pathogen. The functions of nitric oxide produced by murine phagocytes extend beyond its direct microbicidal activity. For instance, NO has been implicated in the regulation of expression of IL-6, IL-12 and IFN-γ and, thereby, has been demonstrated to influence both innate and adaptive immune responses to infection [29,30]. It is possible that the suppression of NO production by macrophages in the presence of Coccidioides may have a greater influence on regulatory signals related to cellular immunity than direct microbicidal activity of the phagocytes. Immunological investigations of the in vivo response of WT and iNOS−/− mice to pulmonary infection with Coccidioides are in progress, and the results of these studies will be presented in a separate report.
Destruction of NO has been attributed to numerous bacterial and fungal gene products, including globins, cytochrome redutases, peroxiredoxins, flavohaemoglobins, flavorubredoxins, and nitrite reductases [31–37]. Flavohemoglobin denitrosylase and S-nitrosoglutathione reductase have been suggested to be involved in the consumption and metabolism of NO which contribute to the protection of C. neoformans yeast against nitrosative activity in vitro [38]. Coccidioides genes that encode flavohemoglobins I, II and III, S-nitrosoglutathione, thiol reductase, thioredoxin reductase and nitrate reductase have been identified by analysis of the genome sequences of C. posadasii and C. immitis using the Basic Local Alignment Search Tool (BLAST) [4,5]. Additional work is necessary to determine whether specific gene products and/or transcriptional regulators of Coccidioides are responsible for the suppression of NO production by activated macrophages, and to explore the role of this suppressive activity on the course of disease in our murine model of coccidioidomycosis.
4. Materials and methods
4.1. Reagents
Phosphate-buffered saline (PBS), Hanks balanced salt solution (HBSS), 100X penicillin-streptomycin solution, sodium pyruvate, nonessential amino acids, HEPES buffer, Dulbecco’s modified Eagle’s medium (DMEM), and RPMI-1640 were obtained from Mediatech Inc. (Manassas, VA). Fetal bovine serum (FBS) was obtained from HyClone/Fisher Scientific (Logan, UT). Trypsin and ACK lysis buffer (0.15 M ammonium chloride, 1 mM potassium bicarbonate, 0.1 mM ethylenediaminetetraacetic acid, pH 7.4) were obtained from Gibco/Invitrogen (Carlsbad, CA). Cytochalasin D and ultrapure water (LAL reagent grade) was obtained from MP Biomedicals, LLC (Solon, OH). Murine IFN-γ was purchased from BD Biosciences/Pharmingen (San Jose, CA), and ultrapure E. coli-derived lipopolysaccharide (LPS) was obtained from InvivoGen (San Diego, CA). Nutrients used in the fungal culture media were obtained from Difco Laboratories (Detroit, MI). All other chemicals and solutions were purchased from Sigma Chemical Company (St. Louis, MO), unless otherwise specified.
4.2. Fungal strains, media and growth conditions
Coccidioides posadasii (isolate C735) and C. immitis (isolate RS) were used for all experimental procedures reported in this study. The saprobic phase of Coccidioides was grown on glucose-yeast-extract agar (GYE; 1% glucose, 0.5% yeast extract, 2% agar) at 30° C for 3 to 4 weeks. Arthroconidia (asexual reproductive products of the saprobic phase) were isolated from these plate cultures by washing with PBS, and the cell suspensions were used to inoculate a defined glucose-salts medium for growth of the parasitic phase at 39° C in the presence of 10% CO2 [39]. The first generation of the parasitic cycle of Coccidioides is fairly well synchronized, which has permitted the isolation of different cell types (e.g., spherule initials, pre-segmented spherules, segmented spherules, or endosporulating spherules) produced after different times of incubation [21]. Spherule initials are first generation parasitic cells which were isolated from the glucose-salts medium after 24 h of incubation, and are isotropically germinated arthroconidia that are distinguished from pre-segmented spherules by their smaller size (15–20 μm diam.). Spherule initials are encountered by the host innate defense cells during the primary stage of infection [7].
For preparation of killed spherule initials, a suspension containing 107 fungal cells was incubated either at 70° C for 2 h (heat-killed [HK] cells), fixed with 1% paraformaldehyde for 1 h at room temperature with agitation (FK cells), or inactivated by exposure of cell suspensions to X-ray irradiation (XrKC) using 160 kV and 6.3 mA for 30 min while the spherule initials were contained in a 100 × 15 mm Petri plate that was placed in a Faxitron X-ray cabinet (Faxitron X-ray Corp., Lincolnshire, IL). After the different treatments fungal cells were washed three times with HBSS, counted in a hemocytometer and adjusted to the desired organism concentration prior to use. Control plating experiments confirmed that these three separate treatments resulted in nonviable cells.
4.3. Tissue culture medium
A conditioned culture medium (CCM) was obtained from cultures of a murine fibroblast cell line L929 (ATCC, Manassas, VA) and was used to provide a source of macrophage colony-stimulating factor (M-CSF) to foster the differentiation of bone marrow cell precursors into macrophages [40]. Briefly, fibroblasts were cultured in DMEM supplemented with 10% heat-inactivated FBS, 10 mM HEPES buffer (pH 7.4), 1 μM β-mercaptoethanol, 0.1 mM nonessential amino acids, 4 mM of L-glutamine and 1 mM sodium pyruvate. After the cells reached confluence plus 1 day, the medium supernatant (CCM L929) was harvested and passed through a sterile 0.2 μm pore filter.
The medium used for experiments with the bone marrow-derived macrophages (BMDM) consisted of DMEM supplemented with 10% FBS as above, 10 mM HEPES buffer (pH 7.4) and 25% CCM L929. Where indicated below, penicillin-streptomycin was added at final concentrations of 100 IU/ml penicillin and 100 μg/ml streptomycin to prevent bacterial contamination.
4.4. Isolation and culturing of murine primary macrophages
C57BL/6 (wild type [WT]) mice were purchased from Charles River Laboratory Inc. (Wilmington, MA). B6.129P2-Nos2tm1Lau/J (iNOS−/−) mice were obtained from Jackson Laboratory (Bar Harbor, ME) and bred in the Small Animal Facility at the University of Texas at San Antonio. Mice were kept under pathogen-free conditions in enclosed filter top cages and provided autoclaved standard animal chow and chlorinated tap water ad libitum. Both strains of mice (females, 8–12 weeks-old) were injected intraperitoneally (i.p.) with 2 ml of sterile 3% thioglycolate medium to stimulate in vivo recruitment of peritoneal macrophages. After 72 h the animals were sacrificed, cells in the peritioneal fluid were recovered by peritoneal lavage and transferred to 10 ml of ice-cold-RPMI 1640 supplemented with 1% penicillin–streptomycin, 10 mM HEPES and 1 mM sodium pyruvate. The peritoneal macrophages (PM) were washed once with the same medium and aliquots were transferred either to 96-well tissue culture plates (OptipluxTM black or clear bottom; Falcon/Becton Dickinson; Franklin Lakes, NJ) at a concentration of 0.2 × 106 cells per well for nitric oxide (NO) determinations, or to 48-well tissue culture plates (Corning Inc., Corning, NY) at 0.3 × 106 cells per well for phagocytosis and killing assays. Peritoneal macrophages were allowed to adhere to the surface of the plates for 2 h. After non-adherent cells were removed by washing, the adherent cells were incubated in the presence of 5% CO2 at 37° C in RPMI supplemented as above with the addition of 10% heat-inactivated FBS for 48 h.
To cultivate BMDM, tibias and femurs were aseptically removed from euthanized C57BL/6 or iNOS−/− mice and dissected free of adherent tissue. After removal of the muscle tissue, the bones were washed with PBS and disinfected with 70% alcohol for 3 min, washed twice with PBS, and transferred to a fresh Petri plate containing DMEM. Using sterile instruments, both ends of the bones were cut, and the marrow flushed out with approx. 5 ml of DMEM using a syringe and a 25-gauge, hollow-bore needle. The tissue was suspended by pipetting, passed through a sterile 70 μm pore nylon mesh to remove bone and other debris, and centrifuged at 650 X g for 5 min at room temperature (RT). The pellet from each mouse was resuspended in 15 ml of ACK lysis buffer for 10 min at RT to lyse erythrocyte precursors, followed by centrifugation as above. The cell pellets were then resuspended in 15 ml of DMEM and centrifuged again. Finally, each cell pellet was resuspended in CCM L929 with antibiotics as previously described, adjusted to 4 × 106 cells/ml, and added to separate 250-ml culture flasks. Fresh CCM L929 with antibiotics was added to each flask after 3 days of incubation, and the cells were then allowed to expand for an additional 3 days. Cells were harvested on the sixth day, washed twice with HBSS followed by addition 10 ml 0.2 mg/ml EDTA per flask, and incubated for an additional 20 min. The surface of the flasks were gently scraped using a sterile rubber policeman to remove tightly adherent macrophages. The suspension was centrifuged at 650 X g for 5 min, resuspended in DMEM, and transferred to CCM L929 at the desired cell concentration (0.5 × 106/ml). Aliquots (0.2 ml) of the cell suspensions were transferred to the wells of 96- or 48-well plates as above 36 h before initiation of the assays of NO production and phagocytosis, respectively. The BMDM cell concentration approximately doubled from the time of seeding [41].
4.5. Infection of primary macrophages
PM and BMDM were cultured in the presence or absence of 10 ng/ml recombinant murine IFN-γ plus 100 ng/ml LPS for 16–18 h to yield activated and non-activated macrophages, respectively. The macrophage growth medium was replaced with 0.2 ml or 0.3 ml of RPMI when using 96- or 48-well culture plates, respectively, and 0.01 ml of a suspension of Coccidioides spherule initials was added to yield a multiplicity of infection (MOI) of 10:1, 5:1 or 2:1 (macrophages:spherule initials). The macrophages and fungal cells were co-cultured for 1, 3, 6 or 12 h in a 5% CO2 incubator at 37° C. Based on early results of the NO production assays, a MOI of 2:1 and 12 h of incubation was chosen for subsequent experiments. In certain experiments, macrophage monolayers were lysed to determine the fungal burden based on counts of fungal colony-forming units (CFU), or the cells were fixed to determine the phagocytosis index as described below.
4.6. Measurement of nitric oxide (NO)
Nitric oxide (NO) production was determined using a nitric oxide synthase (NOS) detection reagent (diaminofluorescein-2 diacetate [DAF-2DA]; Cell Technology, Inc., Mountain View, CA) by following the manufacturer’s instructions. DAF-2DA is a non-fluorescent cell permeable reagent that can measure free NO and iNOS activity in living cells under physiological conditions. Upon entering the macrophage, intracellular esterases hydrolyze the diacetate moiety, trapping free DAF-2 within the cell that is covalently modified by NO; thus, production of NO converts the non-fluorescent dye to its fluorescent derivate [42].
Briefly, after macrophage activation with IFN-g plus LPS, the cells were washed with warm HBSS. A working solution of DAF-2DA in HBSS (pH 7.0 – 7.5, in the absence of serum/BSA or phenol red) was added to wells of plates containing adherent macrophages and incubated for 1 h to pre-load the dye into the cells. Macrophages were then washed with HBSS to remove excess dye, followed by addition of non-supplemented RPMI (without phenol red) containing an appropriate concentration of spherule initials and incubated for different time periods as described above. NO concentration was determined by fluorescence intensity using a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA) with excitation at 488 nm and emission at 515 nm. Estimated levels of NO production were expressed as relative fluorescence units (RFU). Assays of NO production were performed under conditions designed to minimize endotoxin contamination. Endotoxin-free water was used for buffer and medium preparation in all experiments.
4.7. Phagocytic assays
Spherule initials (5 × 106) were suspended in HBSS and stained with a fluorescent probe (FUN® 1; Molecular Probes Inc, Eugene, OR) at a concentration of 10 μM according to the manufacturer’s instructions. The stained fungal cells were added to activated or non-activated macrophages at the selected MOI described above and incubated for 3 h. The co-cultured cells were then washed twice with HBSS and incubated with trypan blue (1 mg/ml in HBSS buffer) for 15 min at 25° C to quench the fluorescence of non-ingested fungal cells. Macrophages were washed again as above and fixed with 1% paraformaldehyde for 1h at 4° C. Phagocytosis was quantified by phase-contrast and fluorescence microscopy. The phagocytic index was calculated on the basis of the total number of ingested fungal cells per 200 macrophages observed microscopically.
To determine whether phagocytosis was necessary for suppression of NO production, peritoneal macrophages were treated with cytochalasin D. Macrophages were washed in HBSS and adjusted to the desired cell number. Cytochalasin D prepared at a final concentration of 5 mg/ml dimethylsulfoxide (DMSO; ATCC, Manassas, VA), or DMSO alone, was added to the macrophage cultures and incubated for 1 hour at 37ºC. Macrophages were then co-cultured with live spherule initials and NO production was measured as described above.
4.8. Evaluation of the effects of secreted fungal products on NO production
We determined whether products secreted by the spherule initials suppressed NO production. Supernatants from 12 h co-cultures of peritoneal macrophages with either live or killed spherule initials, as well as supernatants of spherule initials cultured alone in RPMI medium were passed through 0.22-μm pore filters. The filtered supernatants were then added to wells containing fresh, activated or non-activated macrophages, either in the presence or absence of live or killed spherule initials, and assayed for NO production as described above.
4.9. Fractionation of culture supernatants of spherule initials
Culture supernatants from spherule initials of C. posadasii (strain C735) were fractionated using sterile centrifugal filter units (Millipore) with different molecular size cutoffs (10-, 50-, and 100-kDa) as recommended by the manufacturer. After centrifugation, the concentrated supernatants were reconstituted back to their original volume and tested as above for their ability to suppress production of NO by activated macrophages.
4.10. Determination of levels of iNOS mRNA expression by QRT-PCR
Total RNA was extracted from the activated or non-activated macrophage in the presence or absence of spherule initials at 6h of co-culture, and using a RNeasy Mini Kit (Qiagen, Valencia, CA) as described in the manufacturer’s instructions. Reverse transcription was performed using 0.1 μg total macrophage RNA in a 20 μl reaction mixture (Promega, Madison, WI) which contained oligo-dT and SuperScript III reverse transcriptase (Invitrogen). To determine the relative quantity of iNOS gene transcripts, the amplification of the cDNA sample (1 μl) was monitored using the fluorescent DNA-binding dye, SYBR Green, in a 7900HT Fast Quantitative Real Time-PCR (QRT-PCR) System (Applied Biosystem; Foster City, CA). The primers used for determination of the relative levels of iNOS transcription (GenBank accession number NM_010927) were as follows: 5’-GCT GGC CAA TGA GGT ACT CA-3’ (forward) and 5’-ACC TGC TCC TCG CTC AAG TT-3’ (reverse). Measurement of the expression of the Coccidioides constitutive gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH, GenBank accession number XM_001473623)) was conducted for normalization of the data using the following PCR primers: 5’-CTT CCG TGT TCC TAC CC-3’ (forward) 5’-CAA CCT GGT CCT CAG T-3’ (reverse). Levels of iNOS mRNA expression are reported as the ratios of iNOS to GAPDH transcript numbers.
4.11. Killing assays
Quantitation of macrophage fungicidal activity was conducted as previously described [43]. Activated or non-activated peritoneal macrophages were cultured and infected with Coccicioides spherule initials at a MOI of 2:1. A total of 0.15 × 106 fungal cells were added to each well containing 0.30 × 106 macrophages. After 12 h of incubation, the adherent macrophages were washed twice (0.25 ml each) with culture media without FBS, and the total wash was incubated on GYE agar for 72 h at 37° C to determine the number of fungal colony-forming units (CFUs). These counts were an estimate of the number of non-phagocytosed fungal cells. The macrophages were lysed following the washes by incubation with 0.5 ml of sterile deionized water for 30 min at 37° C, and the suspension was mixed vigorously. The process of lysis was repeated, but without incubation to obtain a final volume of 1 ml. A 0.1 ml aliquot of this suspension containing released fungal cells were streaked on GYE plates as above and the number of viable spherule initials was determined as described above. These counts represent the number of viable fungal cells after phagocytosis by the peritoneal macrophages, and permitted us to estimate the percentage of engulfed spherule initials that were killed by the phagocytic cells.
4.12. Statistical analysis
All data were expressed as mean ± standard error of the mean (SEM). ANOVA followed by either Tukey’s test or the Student’s t test were applied. Statistical calculations were performed using GraphPad Prism version 4.0 for Macintosh (GraphPad software, San Diego, CA). Values of P<0.05 were considered statistically significant.
Acknowledgments
Support for this study was provided by Public Health Service grants AI071118 and AI070891 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health, awarded to GTC. Additional support was provided by the Margaret Batts Tobin Foundation, San Antonio, TX. The authors are grateful for the technical assistance of Ms. Natalia Castro-Lopez.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jimenez M, del P, Wall L, Fierer J. High levels of interleukin-10 impair resistance to pulmonary coccidioidomycosis in mice in part through control of nitric oxide synthase 2 expression. Infect Immun. 2006;74:3387–95. doi: 10.1128/IAI.01985-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pappagianis D. Epidemiology of coccidioidomycosis. In: Stevens DA, editor. Coccidioidomycosis: A Text. New York: Plenum Press; 1980. pp. 68–85. [Google Scholar]
- 3.Fisher MC, Koening GL, White TJ, Taylor JW. Molecular and phenotypic description of Coccidioides posadasii sp. nov. previously recognized as the non-California population of Coccidioides immitis. Mycologia. 2002;94:73–84. [PubMed] [Google Scholar]
- 4.Neafsey DE, Barker BM, Sharpton TJ, Stajich JE, Park DJ, Whiston E, et al. Population genomic sequencing of Coccidioides fungi reveals recent hybridization and transposon control. Genome Res. 2010;20:938–46. doi: 10.1101/gr.103911.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009;19:1722–31. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue J, Chen X, Selby D, Hung CY, Yu JJ, Cole GT. A genetically engineered live attenuated vaccine of Coccidioides posadasii protects BALB/c mice against coccidioidomycosis. Infect Immun. 2009;77:3196–208. doi: 10.1128/IAI.00459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole GT, Xue J, Seshan K, Borra P, Borra R, Tarcha E, et al. Virulence mechanisms of Coccidioides. In: Heitman J, Filler SG, Mitchell AP, editors. Molecular Principles of Fungal Pathogenesis. Washington, D. C: American Society for Microbiology Press; 2006. pp. 363–91. [Google Scholar]
- 8.Sun SH, Cole GT, Drutz DJ, Harrison JL. Electron-microscopic observations of the Coccidioides immitis parasitic cycle in vivo. J Med Vet Mycol. 1986;24:183–92. [PubMed] [Google Scholar]
- 9.Lyons CR, Orloff GJ, Cunningham JM. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J Biol Chem. 1992;267:6370–4. [PubMed] [Google Scholar]
- 10.Aguirre KM, Gibson GW. Differing requirement for inducible nitric oxide synthase activity in clearance of primary and secondary Cryptococcus neoformans infection. Med Mycol. 2000;38:343–53. doi: 10.1080/mmy.38.5.343.353. [DOI] [PubMed] [Google Scholar]
- 11.Diez-Orejas R, Molero G, Moro MA, Gil C, Nombela C, Sanchez-Perez M. Two different NO-dependent mechanisms account for the low virulence of a non-mycelial morphological mutant of Candida albicans. Med Microbiol Immunol. 2001;189:153–60. doi: 10.1007/s430-001-8022-6. [DOI] [PubMed] [Google Scholar]
- 12.Fernandes KS, Cohelo AL, Lopes-Bezerra LM, Barja-Fidalgo C. Virulence of Sporothrix schenckii conidia and yeast cells and their susceptibility to nitric oxide. Immunol. 2000;101:563–9. doi: 10.1046/j.1365-2567.2000.00125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez A, De Gregori W, Velez D, Restrepo A, Cano LE. Nitric oxide participation in the fungicidal mechanism of gamma-interferon-activated murine macrophages against Paracoccidioides brasiliensis conidia. Infect Immun. 2000;68:2546–52. doi: 10.1128/iai.68.5.2546-2552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera J, Mukherjee J, Weiss LM, Casadevall A. Antibody efficacy in murine pulmonary Cryptococcus neoformans infection: a role for nitric oxide. J Immunol. 2002;168:3419–27. doi: 10.4049/jimmunol.168.7.3419. [DOI] [PubMed] [Google Scholar]
- 15.Kawakami K, Zhang T, Qureshi MH, Saito A. Cryptococcus neoformans inhibits nitric oxide production by murine peritoneal macrophages stimulated with interferon-γ and lipopolysaccharide. Cell Immunol. 1997;180:47–54. doi: 10.1006/cimm.1997.1166. [DOI] [PubMed] [Google Scholar]
- 16.Chinen T, Qureshi MH, Koguchi Y, Kawakami K. Candida albicans suppresses nitric oxide (NO) production by interferon-gamma (IFN-γ )- and lipopolysaccharide (LPS)-stimulated murine peritoneal macrophages. Clin Exp Immunol. 1999;115:491–7. doi: 10.1046/j.1365-2249.1999.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schröppel K, Kryk M, Hermann M, Leberer E, Röllinghoff M, Bogdan C. Suppression of type 2 NO-synthase activity in macrophages by Candida albicans. Int J Med Microbiol. 2001;290:659–68. doi: 10.1016/S1438-4221(01)80003-5. [DOI] [PubMed] [Google Scholar]
- 18.Flavia-Popi AF, Lopes JD, Mariano M. GP43 from Paracoccidioides brasiliensis inhibits macrophage functions. An evasion mechanism of the fungus. Cell Immunol. 2002;218:87–94. doi: 10.1016/s0008-8749(02)00576-2. [DOI] [PubMed] [Google Scholar]
- 19.Xiao G, Miyazato A, Inden K, Nakamura K, Shiratori K, Nakagawa K, et al. Cryptococcus neoformans inhibits nitric oxide synthesis caused by CpG- oligodeoxynucleotide-stimulated macrophages in a fashion independent of capsular polysaccharides. Microbiol Immunol. 2008;52:171–9. doi: 10.1111/j.1348-0421.2008.00019.x. [DOI] [PubMed] [Google Scholar]
- 20.Cole GT, Samson RA. The conidia. In: Al-Doory Y, editor. Mould Allergy. Philadelphia: Lea & Febiger; 1984. pp. 66–103. [Google Scholar]
- 21.Hung CY, Yu JJ, Seshan KR, Reichard U, Cole GT. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect Immun. 2002;70:3443–56. doi: 10.1128/IAI.70.7.3443-3456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viriyakosol S, Fierer J, Brown GD, Kirkland TN. Innate immunity to the pathogenic fungus Coccidioides posadasii is dependent on Toll-like receptor 2 and Dectin-1. Infect Immun. 2005;73:1553–60. doi: 10.1128/IAI.73.3.1553-1560.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, et al. Toll-like receptor triggering of vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–3. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- 24.Drutz D, Huppert M. Coccidioidomycosis: factors affecting the host-parasite interaction. J Infect Dis. 1983;147:372–90. doi: 10.1093/infdis/147.3.372. [DOI] [PubMed] [Google Scholar]
- 25.Frey CL, Drutz DJ. Influence of fungal surface components on the interaction of Coccidioides immitis with polymorphonuclear neutrophils. J Infect Dis. 1986;153:933–43. doi: 10.1093/infdis/153.5.933. [DOI] [PubMed] [Google Scholar]
- 26.Ampel NM, Bejarano GC, Galgiani JN. Killing of Coccidioides immitis by human peripheral blood mononuclear cells. Infect mmun. 1992;60:4200–4. doi: 10.1128/iai.60.10.4200-4204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farah CS, Saunus JM, Hu Y, Kazoullis A, Ashman RB. Gene targeting demonstrates that inducible nitric oxide synthase is not essential for resistance to oral candidiasis in mice or for killing of Candida albicans by macrophages in vitro. Oral Microbiol Imunol. 2009;24:83–8. doi: 10.1111/j.1399-302X.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 28.Vazquez-Torres A, Jones-Carson J, Balish E. Nitric oxide production does not directly increase macrophage candidacidal activity. Infect Immun. 1995;63:1142–4. doi: 10.1128/iai.63.3.1142-1144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang FP, Niedbala W, Wei XQ, Xu D, Feng GJ, Robinson JH, et al. Nitric oxide regulates Th1 cell development through the inhibition of IL-12 synthesis by macrophages. Eur J Immunol. 1998;28:4062–70. doi: 10.1002/(SICI)1521-4141(199812)28:12<4062::AID-IMMU4062>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 30.Bogdan C. The function of nitric oxide in the immune system. In: Mayer B, editor. Handbook of Experimental Pharmacology. New York: Springer; 2000. pp. 443–92. [Google Scholar]
- 31.Hromatka BS, Noble SM, Johnson AD. Transcriptional response of C. albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell. 2005;16:4814–26. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nittler MP, Hocking-Murray D, Foo CK, Sill A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol Biol Cell. 2005;16:4792–813. doi: 10.1091/mbc.E05-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poole RK. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem Soc Trans. 2005;33:176–80. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 34.Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohaemoglobin hmp. J Biol Chem. 2006;281:28039–47. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- 35.Mills PC, Rowley G, Spiro S, Hinton JC, Richardson DJ. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiol. 2008;154:1218–28. doi: 10.1099/mic.0.2007/014290-0. [DOI] [PubMed] [Google Scholar]
- 36.Missall TA, Pusareti ME, Donlin MJ, Chambers KT, Corbett JA, Lodge JK. Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: implications for virulence. Eukaryot Cell. 2006;5:518–29. doi: 10.1128/EC.5.3.518-529.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das P, Lahiri A, Lahiri A, Chakravortty D. Novel role of the nitrite transporter NirC in Salmonella pathogenesis: SPI2-dependent suppression of inducible nitric oxide synthase in activated macrophages. Microbiol. 2009;155:2476–89. doi: 10.1099/mic.0.029611-0. [DOI] [PubMed] [Google Scholar]
- 38.De Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–8. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 39.Levine HB. Purification of the spherule-endospore phase of Coccidioides immitis. Sabouraudia. 1961;1:112–5. doi: 10.1080/00362176285190231. [DOI] [PubMed] [Google Scholar]
- 40.Stanley ER, Heard PM. Factors regulating macrophage production and growth. Purification and some properties of the colony stimulating factor from medium conditioned by mouse L cells. J Biol Chem. 1977;252:4305–12. [PubMed] [Google Scholar]
- 41.Iovine NM, Pursnani S, Voldman A, Wasserman G, Blaser MJ, Weinrauch Y. Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect Immun. 2008;76:986–93. doi: 10.1128/IAI.01063-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gumpricht E, Dahl R, Yerushalmi B, Devereaus MW, Sokol RJ. Nitric oxide ameliorates hydrophobic bile acid-induced apoptosis in isolated rat hepatocytes by non-mitochondrial pathways. J Biol Chem. 2002;277:25823–830. doi: 10.1074/jbc.M112305200. [DOI] [PubMed] [Google Scholar]
- 43.Hung CY, Seshan KR, Yu JJ, Schaller R, Xue J, Basrur V, Gardner MJ, Cole GT. A metalloproteinase of Coccidioides posadasii contributes to evasion of host detection. Infect Immun. 2005;73:6689–703. doi: 10.1128/IAI.73.10.6689-6703.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]