Abstract
Background
Platelet factor 4 (PF4) is an abundant protein stored in platelet α-granules. Several patients have been described with platelet PF4 deficiency, including the Gray platelet syndrome (GPS), characterized by deficiency of α-granule proteins. Defective granule formation and protein-targeting are considered the predominant mechanisms. We have reported on a patient with thrombocytopenia and impaired platelet aggregation, secretion, and protein phosphorylation, associated with a mutation in transcription factor RUNX1. Platelet expression profiling showed decreased transcript expression of PF4 and its nonallelic variant PF4V1.
Objectives
To understand the mechanism leading to PF4 deficiency associated with RUNX1 haplodeficiency, we addressed the hypothesis that PF4 is a transcriptional target of RUNX1.
Methods/Results
Chromatin immunoprecipitation and gel-shift assays using PMA-treated human erythroleukemia (HEL) cells revealed RUNX1 binding to RUNX1 consensus sites at -1774/-1769 and -157/-152 on PF4 promoter. In luciferase reporter studies in HEL cells mutation of each site markedly reduced activity. PF4 promoter activity and protein were decreased by siRNA RUNX1 knockdown and increased by RUNX1 overexpression.
Conclusions
Our results provide the first evidence that PF4 is regulated by RUNX1 and that impaired transcriptional regulation leads to the PF4 deficiency associated with RUNX1 haplodeficiency. Because our patient had decreased platelet albumin and IgG (not synthesized by megakaryocytes), we postulate additional defects in RUNX1-regulated genes involved in vesicular trafficking. These studies advance understanding of the mechanisms in α-granule deficiency.
Keywords: α-granule, Gray platelet syndrome, platelet factor-4 deficiency, platelet function defect, RUNX1 haplodeficiency, transcriptional regulation
Introduction
Platelet factor 4 (PF4) (CXCL4), a CXC chemokine, is an abundant protein stored in α-granules of megakaryocytes (MK) and platelets and serves as a lineage-specific marker of megakaryocytic differentiation [1,2]. It is secreted from platelets upon activation. It binds with high affinity to heparin and to negatively-charged cell surface glycosaminoglycans, heparan sulfate, dermatan sulfate and chondroitin sulfate [3]. It is a negative regulator of megakaryopoiesis [4,5] and has a negative effect on angiogenesis [6,7]. PF4 plays a role in inflammation [8], atherosclerosis [9] and thrombosis [10]. PF4V1 (CXCL4L1), a nonallelic variant of PF4, is also expressed in platelets but at a low level relative to PF4 [11-13]. Compared to PF4 it has a 4.3% amino acid divergence in the coding region of the mature protein and 38% amino acid divergence in the signal peptide region [12]. PF4V1 has a greater effect in blocking angiogenesis in vivo and inhibiting endothelial cell chemotaxis in vitro [14].
Several patients have been reported with a deficiency of platelet PF4 and α-granule contents [15-19]. In some of them dense granule contents have been normal, and this selective α-granule deficiency has been referred to as the gray platelet syndrome (GPS) [15]. In others, dense granule contents have also been decreased giving rise to a combined granule deficiency (αδ-storage pool deficiency, αδ-SPD) [17,19]. GPS is a markedly heterogeneous disorder characterized by thrombocytopenia, large platelet size, and deficiency of α-granules and their contents, which includes proteins synthesized by MK (eg. PF4, β-thromboglobulin) and those incorporated by endocytosis into the granules (eg. albumin, fibinogen, IgG) [15,16]. The molecular mechanisms leading to α-granule deficiency in GPS or in combined αδ-SPD remain unclear. A number of mechanisms are postulated [15,16], including defective granule membrane formation and targeting of proteins to the α-granule, although the possibility exits that there may be decreased synthesis of PF4 and one or more of the deficient proteins. It is likely that the mechanisms are different in individual patients; the inheritance pattern of α-granule deficiency has been autosomal recessive in most reports, and autosomal dominant or sex-linked in some [15,17-19].
We have previously reported on a patient with thrombocytopenia, impaired agonist-stimulated platelet aggregation, secretion, phosphorylation of pleckstrin and myosin light chain (MLC), and GPIIb-IIIa activation [20,21]. This patient has a heterozygous mutation (G>T) in transcription factor RUNX1 (also called Core binding factor A2, CBFA2, or AML1), in intron 3 at the splice acceptor site for exon 4, leading to a frame shift with premature termination in the conserved RUNT domain [21]. RUNX1 is a transcription factor that plays a major role in hematopoiesis and megakaryopoiesis [22-24]. It is composed of two subunits and the α-subunit (RUNX1) is the DNA binding element of the complex and recognizes the DNA sequence TGT/cGGT. CBFβ, the β subunit, stabilizes RUNX1 binding to DNA but without direct DNA contact. RUNX1 mutations are associated with familial, autosomal dominant thrombocytopenia, platelet dysfunction, and predisposition to acute leukemia [19,25].
Detailed studies in our patient, including platelet expression profiling [26], showed decreased expression of several genes including chemokine PF4 and its non-allelic variant PF4V1. Moreover, decreased platelet PF4 has also been observed in several members of another family who were subsequently shown to have a RUNX1 mutation [17,19], although these subjects had dense granule SPD as well. Based on the findings in our patient of the decreased platelet expression of PF4 at mRNA and protein level, we postulated that PF4 was a direct transcriptional target of RUNX1. This would imply that at least in some patients with platelet PF4 deficiency, diminished synthesis would be a mechanism. In the present studies, we provide the first evidence that PF4 is regulated by RUNX1 and propose that impaired synthesis contributes to the deficiency of PF4 in some patients with α-granule abnormality.
Materials and Methods
Patient Information
We have previously described [20,21] the clinical presentation and detailed studies in this 24 year old white male, documenting decreased platelet aggregation, secretion, activation of GPIIb-IIIa, pleckstrin and myosin light chain (MLC) phosphorylation, and PKC-θ level. Other findings in this patient include decreased platelet 12-lipoxygenase and 12-hydroxyeicosatetraenoic acid production [27]. This patient has a single point mutation in the conserved Runt homology domain of RUNX1 [21]. Platelet expression profiling studies showed [26] decreased expression of PF4, PF4V1 and several other genes. Platelet PF4 protein levels are decreased while β-thromboglobulin and dense granule ATP and ADP have been normal [20,21]. Control subjects used in current studies were healthy individuals, not on any medications. These studies were performed after approval by the Institutional Review Board.
Materials
All chemicals including phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma (St. Louis, MO, USA) and Fisher Scientific (Pittsburgh, PA, USA). The PCR enzymes, restriction endonucleases, and modifying enzymes were from Promega (Madison, WI) and New England Biolabs (Beverly, MA). JM109 competent cells, luciferase reporter pGL4-Basic and pRL-TK containing renilla luciferase gene, and Dual Luciferase Assay kit were from Promega. PCR primers and IRDye™700 probes for EMSA were synthesized by Integrated DNA Technologies (IDT, Coralville, IA). The DNA samples were sequenced at GENEWIZ (South Plainfield, NJ).
Cell lines and Cell culture
Human erythroleukemia (HEL) cells (from American Type Cell Culture, Rockville, MD) were cultured in RPMI-1640 medium containing 10% fetal bovine serum and antibiotics (penicillin, streptomycin, 1% each from Cellgro, Manassas, VA). HEL cells were grown in 10-30 nM PMA to induce megakaryocytic transformation [28,29].
Chromatin Immunoprecipitation (ChIP) Assay
ChIP analysis on PMA (10 nM, 24 hours)-treated HEL cells (1 × 108) was performed using ChIP-It kit (Active motif, Carlsbad, CA) as previously described [27]. HEL cell chromatin was sheared enzymatically for 15 min and immunoprecipitated with anti-RUNX1 antibody (Sc-8564x, Santa Cruz). PF4 and PF4V1 promoter regions were amplified from immunoprecipitated and input samples by PCR using specific primers (Supplemental Table 1). GAPDH was simultaneously amplified. Amplified products were analyzed by agarose gel electrophoresis. The observed bands were quantified by ImageJ software (1.43 version) developed by National Institute of Health. Fold enrichment was calculated by dividing the PCR signal with anti-RUNX1 antibody by that with control IgG.
Plasmid Construction and Mutagenesis
PF4 or PF4V1 promoter plasmids and their mutants were prepared using firefly luciferase reporter vector, pGL4-Basic. Promoter region of PF4 (-1936/-27 bp), PF4V1 (-1812/+25 bp) and PF4V1 (-359/+25) from ATG were amplified from HEL cell genomic DNA. Mutations were incorporated into RUNX1 sites -1774/-1769 and -157/-152 of PF4 promoter using megaprimer PCR method [30]. Primers used in these amplifications are shown in Supplemental Table 2. The PCR products were purified, blunt ended using end conversion mix kit (Novagen/EMD, San Diego, CA) and cloned into EcoRV site of pGL4-Basic vector. The recombinants were confirmed by sequencing.
Transfections and Luciferase Reporter Assays
In a 24-well plate HEL cells (1.5 × 105 per well) were co-transfected with promoter-reporter plasmid (500 ng) and an internal control plasmid pRL-TK (10 ng) at a ratio of 50:1 using Lipofectamine 2000 and PLUS™ reagent (Invitrogen). After one hour of transfection PMA (10-30 nM, as indicated) was added to the medium. Luciferase activity was measured at 48 hours using Dual-Luciferase Assay System. Promoter activity was calculated as a ratio of firefly luciferase activity to renilla luciferase activity and normalized relative to that of the empty vector. All transfection experiments were performed three to four times in triplicate.
Electrophoretic Mobility Shift Assay
Nuclear extracts were prepared from PMA-treated HEL cells using nuclear extraction kit (Active Motif, Carlsbad, CA). PF4 DNA probes -1782/-1761 and -166/-143 labeled with IR Dye™ 700 and containing intact or mutated RUNX1 sites (-1774/-1769 and -157/-152) (Supplemental Table 3) were used in the binding reactions. Nuclear protein (10-20 μg) was incubated on ice for 30 min with labeled probes. For competition assays, unlabelled wild type (Wt) or mutant probes were pre-incubated with protein for at least 30 min. For the supershift experiments, anti-RUNX1 antibody was incubated with protein before the addition of probe. Binding reactions were further incubated on ice for one hour and electrophoresed on 5% polyacrylamide gel. The gels were scanned on Odyssey Infrared Imaging System (Li-COR Biosciences). In addition, we studied the binding of recombinant RUNX1 protein (200 ng) (ORIGENE, Rockville, MD) to PF4 probes using buffer containing HEPES 0.3 mM, dithiothreital DTT 0.5 mM, Triton X-100 0.005%, NaCl 100 mM, bovine serum albumin 0.5 ug/ul, glycerol 10% and 5 mM MgCl2. For supershift assay anti-RUNX1 antibody (sc-8563X) or nonspecific IgG (both from Santa Cruz) was pre-incubated with protein as described above.
Analyses of Protein and mRNA Levels
Whole cell lysates (40 μg) from co-transfected HEL cells were subjected to SDS-PAGE, transferred to nitrocellulose membranes (Millipore, Bedford, MA), and probed with the anti-RUNX1 and anti-actin antibodies. Specific protein expressions were detected with IR-labeled secondary antibodies using Odyssey Infrared imaging system (Li-COR Biosciences). PF4 protein was measured by ELISA kit (RayBiotech, Inc, Norcross, GA).
PF4 and PF4V1 mRNA levels in platelets and PMA (30 nM)-treated HEL cells were measured by real-time PCR using the Mastercycler ep realplex (Eppendorf). Real-time PCR was performed using the SYBR Green mix (Applied Biosystems). GAPDH was used as the housekeeping gene. The forward primer used for PF4 was 5’-TTCTGCGCCTCACGCCC-3’; for PF4V1 was 5’-CGCCACCCGCCAGGAGAT-3’; the common reverse primer was 5’- TGGGACGGACCTGG GAG-3’ [13]. Relative quantification method (2-ΔΔCt) was used to analyze the changes in expression.
siRNA knockdown of RUNX1
RUNX1 siRNA pool (sc-37677) and control siRNA (sc-44233) were from Santa Cruz Biotechnologies, Inc. HEL cells (1.5× 105) were cotransfected with PF4 promoter construct (500 ng) and RUNX1 siRNA (200 nM, added twice 48 hours apart) or mock siRNA, transferred to 2X medium containing PMA (30 nM) and incubated at 37°C with 5% CO2. Plasmid pRL-TK containing renilla luciferase gene was used as an internal control. Luciferase activity and protein levels were determined at 96 hours.
RUNX1 Overexpression Studies
HEL cells (1.5×105 per well) were cotransfected with 2 μg each of reporter plasmid (Wt or mutant) and RUNX1 expression plasmid RUNX1-pCMV6-XL4 or empty vector pCMV6-XL4 (ORIGENE, Rockville, MD). After transfections PMA (30 nM) was added to cells and incubated at 37°C with 5% CO2. Luciferase activity and protein levels were assessed at 48 hours.
Bioinformatics
Potential RUNX1 binding sites on PF4 and PF4V1 promoter regions were analyzed by TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html) and TESS (http://www.cbil.upenn.edu/tess).
Results
Binding of RUNX1 to PF4 promoter by Chromatin Immunoprecipitation
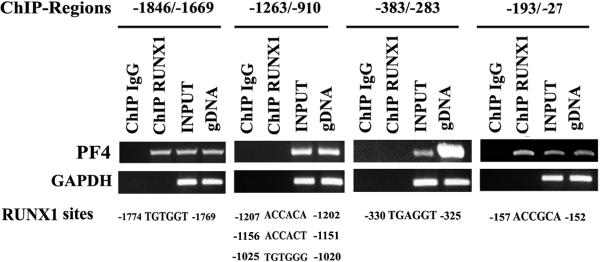
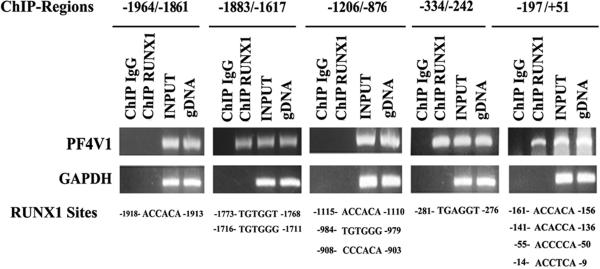
Computer analysis revealed 6 consensus RUNX1 sites in ~2 kb upstream region (Fig. 1). PCR primers were designed to amplify 4 regions of PF4 promoter encompassing these RUNX1 sites. The regions -1846/-1669 and -193/-27 showed amplification of DNA precipitated by anti-RUNX1-antibody but not by the IgG control (Fig. 1). DNA regions -1263/-910 and -383/-283 showed no amplification from immunoprecipitated DNA. These studies indicate that RUNX1 binds to the regions -1846/-1669 bp and -193/-27 bp with each containing one RUNX1 site.
Fig. 1. Chromatin immunoprecipitation (ChIP) analysis of PF4 promoter region.
Immunoprecipitation was performed using PMA-treated HEL cells and control IgG (column 1) of each panel or anti-RUNX1 antibody (column 2). Samples were analyzed by PCR using primers specific to PF4 promoter regions and GAPDH. Columns 3 and 4 of each panel show PCR amplification of total input DNA and genomic DNA, respectively. Also shown are the RUNX1 consensus sites in each region. Regions -1846/-1669 and -193/-27 revealed RUNX1 binding; the fold enrichment was 30.0 ± 9.6 (mean ± SE, n=3) and 11.9 ± 0.9 (n=3) fold, respectively.
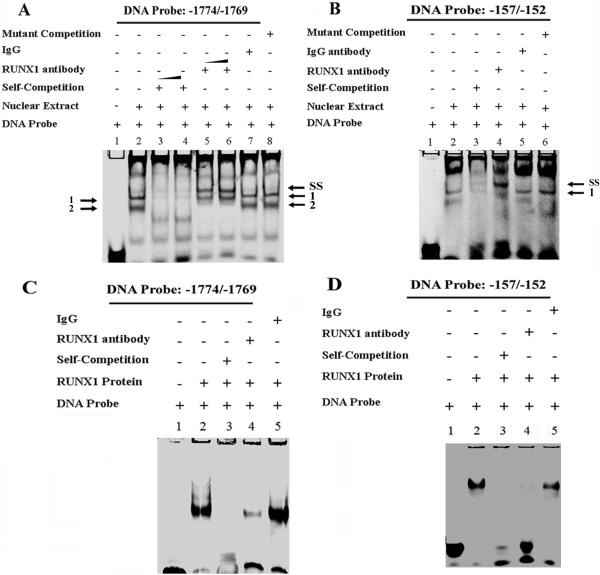
EMSA on RUNX1 sites -1774/-1769 and -157/-152 in PF4 Promoter
EMSA was performed using probes encompassing the RUNX1 sites amplified in ChIP studies (Fig. 1). With the probe containing -1774/-1769 RUNX1 site protein-binding was noted (two bands) (Fig. 2A, Lane 2), which was competed by increasing amounts of unlabeled excess (150-200 fold) probe (lanes 3-4). A supershift (arrow SS) was observed along with loss of band 2 with RUNX1 antibody (lanes 5-6) but not with IgG (lane 7). Unlabeled probe with RUNX1 site mutated did not compete with the specific complexes (lane 8). With the probe encompassing the RUNX1 site -157/-152 (Fig. 2B), protein binding was observed (lane 2), which was competed by excess unlabeled probe (lane 3). Anti-RUNX1 antibody supershifted the complex (lane 4; arrow SS). IgG (lane 5) and unlabeled mutant probe (lane 6) did not alter the protein-binding. These studies indicate that RUNX1 binds to the consensus sites at -1774/ -1769 and -157/-152. Additional studies were performed using recombinant RUNX1 (Figs. 2 C and D). As shown in figure 2C the probe with RUNX1 site -1774/-1769 showed binding with recombinant RUNX1 protein (lane 2), which was abolished by self-competition (lane 3) and RUNX1 antibody (lane 4) but not by nonspecific IgG (Lane 5). Similarly, the probe with RUNX1 site -157/-152 showed binding (fig. 2D, lane 2), which was competed by self-competition (lane 3) and abolished by RUNX1 antibody (lane 4) but not IgG (lane 5). Thus, RUNX1 binds to the consensus sites -1774/-1769 and -157/-152.
Fig. 2. EMSA using nuclear extracts from PMA-treated HEL cells or recombinant RUNX1 and DNA probes with RUNX1 sites -1774/-1769 and -157/-152.
(A) EMSA using DNA probe with -1774/-1769 RUNX1 site. Lane 1, DNA probe alone; lane 2, probe with nuclear extract showing bands 1 and 2; lanes 3 and 4, competition with 150 and 200-fold excess unlabeled probe; lanes 5 and 6, supershift with 4 and 6 ug RUNX1 antibody (arrow SS shows supershifted complex) along with loss of band 2; lane 7, effect of nonspecific IgG; lane 8, competition with unlabeled probe containing mutated RUNX1 site. (B) EMSA using DNA probe with RUNX1 site -157/-152. Lane 1, probe alone; lane 2, probe with nuclear extract; lane 3, competition with 300-fold excess unlabeled probe; lane 4, with RUNX1 antibody showing supershifted band, arrow SS; lane 5, effect of nonspecific IgG; lane 6, competition with unlabeled probe with mutated RUNX1 site. (C) EMSA using probe -1774/-1769 and recombinant protein RUNX1 (200 ng). Lane 1, probe alone; lane 2, probe with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, inhibition in the binding by RUNX1 antibody; and lane 5, effect of nonspecific IgG. (D) EMSA using recombinant protein RUNX1 and oligonucleotide probe -157/-152. Lane 1, probe alone; lane 2, probe with recombinant RUNX1; lane 3, competition with excess unlabeled probe; lane 4, inhibition of binding with RUNX1 antibody; and lane 5, effect of nonspecific IgG.
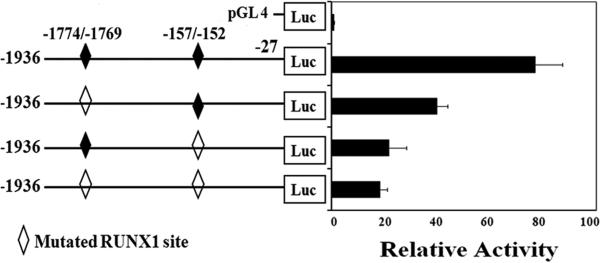
Luciferase Reporter Studies on PF4 Promoter Activity
PF4 RUNX1 sites -1774/-1769 and -157/-152 were individually mutated (Supplemental Table 2). Mutation of each RUNX1 site markedly decreased reporter activity, with the greatest decrease when both sites were mutated (Fig. 3) indicating that each site contributes to PF4 promoter activity.
Fig. 3. Luciferase reporter studies on PF4 promoter in PMA-treated HEL cells.
Reporter constructs with PF4 promoter regions with wild type (closed symbol) and mutated (open symbols) RUNX1 sites were transfected into HEL cells and activity was measured at 48 hours. Shown are mean and SEM of 3 experiments in triplicate.
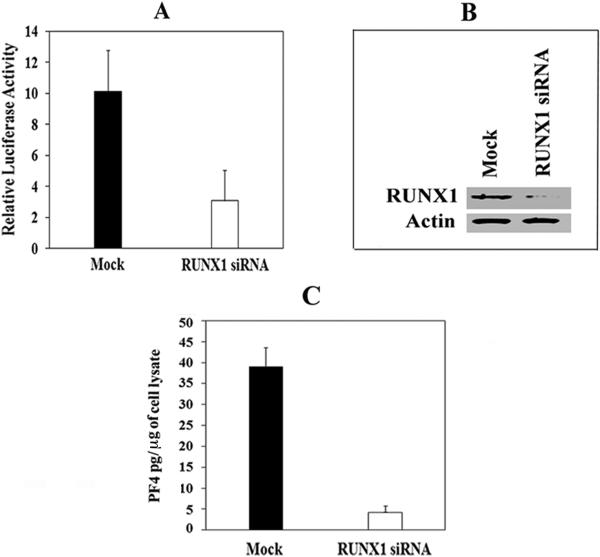
Effect of RUNX1 siRNA on PF4 Promoter Activity and PF4 Protein
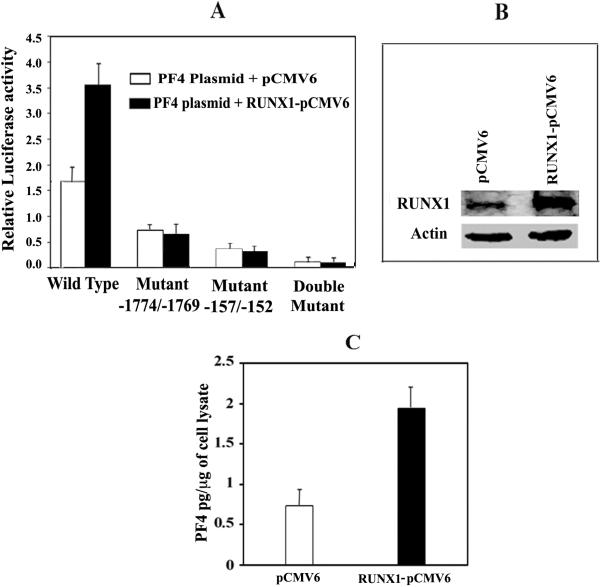
RUNX1 siRNA caused a ~70% reduction in PF4 promoter activity compared to that with mock siRNA (Fig. 4A). There was a marked reduction in endogenous RUNX1 (Fig. 4B) and PF4 proteins (Fig. 4C). These studies provide evidence for in vivo regulation of PF4 by RUNX1.
Fig. 4. Effect of RUNX1 siRNA on PF4 promoter activity and PF4 protein.
(A) PF4 promoter activity: HEL cells were cotransfected with mock or RUNX1 siRNA and PF4 luciferase-reporter construct (-1936/-27 bp). Shown is the mean and SEM of 2 experiments in triplicate. (B) Immunoblotting of RUNX1 and actin in HEL cells. (C) PF4 by ELISA (pg/μg HEL cell protein).
Effect of Overexpression of RUNX1 on PF4 Promoter Activity and PF4 protein
RUNX1 expression plasmid doubled the PF4 promoter activity as compared to an empty vector (Fig. 5A). With mutation of the RUNX1 sites -1774/-1769 and -157/-152, individually or together, there was decreased activity as expected, and there was no increase in activity with RUNX1 overexpression. RUNX1 (Fig. 5B) and PF4 (Fig. 5C) protein were upregulated by RUNX1 expression plasmid. These results provide further evidence that RUNX1 regulates PF4 expression.
Fig. 5. Effect of transient overexpression of RUNX1 on PF4 promoter activity and PF4 protein.
(A) PF4 promoter activity at 48 hours. HEL cells were cotransfected with RUNX1-pCMV6 expression vector (filled bars) or empty pCMV6 vector (open bars), along with PF4 luciferase-reporter construct (-1936/-27), wild-type or with mutations in site -1774/-1769, -157/-152 or both. Shown is mean ± SEM of 3 experiments in triplicate. (B) Western blotting analysis showing RUNX1 overexpression in HEL cells. Actin is a loading control. (C) Effect of RUNX1 overexpression on PF4 by ELISA (pg/μg HEL cell protein).
Studies on PF4V1
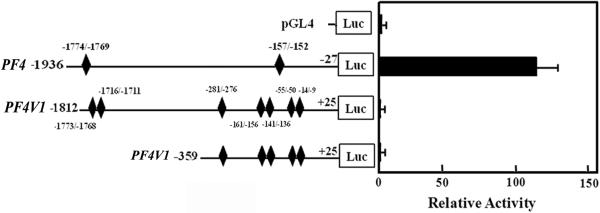
Measurements by real-time PCR showed that PF4 is expressed ~40 fold higher than PF4V1 (not shown), in line with previous studies [13]. ChIP analysis of PF4V1 upstream region (Fig. 6) showed 3 regions, -1883/-1617, -334/-242, and -197/+51, which were amplified from DNA suggesting that RUNX1 binds to these regions. Luciferase reporter studies using constructs of ~ 1.8 kb or 359 bp size of PF4V1 promoter showed barely detectable activity compared to PF4 promoter (Fig. 7). In PMA-treated HEL cells PF4 message was detected at 0 hours and increased by 11, 33 and 40 fold at 48, 72 and 96 hours, respectively (not shown). PF4V1 was undetectable at 0 hours and present at 96 hours, but 4 orders of magnitude lower than PF4. Together, these studies indicate a differential regulation of PF4 compared to PF4V1.
Fig. 6. Chromatin immunoprecipitation (ChIP) analysis of PFV1 promoter.
Immunoprecipitation was performed in PMA-treated HEL cells using control IgG (column 1 of each panel) or anti-RUNX1 antibody (column 2). PCR was performed using primers specific to PF4V1 promoter regions and GAPDH. Columns 3 and 4 show PCR amplification of total input DNA and genomic DNA, respectively. Regions -1883/-1617, -334/-242 and -187/+51 revealed enrichment with anti-RUNX1 antibody. The fold enrichment was 6.1 ± 0.9, 6.8 ± 0.5 and 5.5 ± 1.2 fold, respectively (n=3 for each region).
Fig. 7. Comparison of PF4 and PF4V1 promoter activity in PMA-treated HEL cells.
RUNX1 consensus sites detected by ChIP analysis are shown for both PF4 and PF4V1. Luciferase activity was measured at 48 hours.
Discussion
Our studies provide evidence that transcription factor RUNX1 regulates the expression of PF4 in MK and platelets. ChIP assay revealed in vivo RUNX1 binding to the PF4 promoter (Fig. 1). EMSA using nuclear extracts and recombinant RUNX1 showed RUNX1 binding to two sites (Fig. 2), and mutations of these sites reduced PF4 promoter activity (Fig. 3). siRNA downregulation of RUNX1 reduced (Fig. 4) and RUNX1 overexpression enhanced (Fig. 5) PF4 promoter activity and PF4 protein. Together with the decreased PF4 in our patient platelets [21], these findings provide, to our knowledge, the first evidence that PF4 promoter is regulated by RUNX1 and that this is altered in human RUNX1 haplodeficiency leading to decreased platelet PF4.
The hallmark of the GPS, a markedly heterogeneous disorder, is deficiency of PF4 and other α-granule proteins that are synthesized by MK as well as of proteins incorporated into the granules by endocytosis but not synthesized in MK [15,16]. The molecular mechanisms leading to α–granule deficiency in GPS and in αδ–SPD are unknown; they have been attributed to failure of α-granule maturation during MK differentiation, transport of proteins to α-granules, and synthesis of granule membranes [15,16]. Proteomic studies in a GPS patient suggest the basic defect is a failure to incorporate endogenously-synthesized MK proteins into α-granules, which is more severe than for endocytosed proteins [16]. Some GPS patients have elevated plasma PF4 [15] suggesting that PF4 synthesis was normal and the primary defect was impaired granule biogenesis with leakage of PF4. Our findings suggest that platelet PF4 level in RUNX1 haplodeficiency is low due to a defect in transcriptional regulation and synthesis of PF4. This is relevant to other reported patients with α-granule deficiency, especially those with RUNX1 mutations [17,19]. GPS has been reported [18] in a patient with a X-linked thrombocytopenia, thalassemia, and Arg216Gln mutation in GATA-1, a major regulator of megakaryopoiesis. GATA-1 knockdown mice have granule abnormalities with a decrease in PF4 mRNA in primary MK [31]. Moreover, a mutation in VPS33B protein (a member of the Sec1/Munc18 protein family) involved in vesicle trafficking has been associated with human α-granule deficiency in the arthrogryposis multiplex congenital, renal dysfunction and cholestasis (ARC) syndrome [32]. As of now, these are the only reported human mutations associated with α-granule deficiency. Our results extend this to RUNX1, an important transcription factor regulating hematopoiesis. There is evidence that RUNX1 and GATA-1 interact to regulate αIIb promoter [33], which may be relevant for PF4 as well. Overall, the molecular mechanisms in α-granule deficiency are heterogenous and distinct, and include defects in transcriptional regulation of proteins, and defects in vesicle trafficking and granule packaging.
In addition to PF4 deficiency, our patient platelets have decreased albumin and IgG [21], two proteins not synthesized by MK but incorporated into the α-granule by endocytosis and granule targeting. However, platelet fibrinogen was normal [21], possibly because its uptake occurs by a different mechanism, mediated by GPIIb-IIIa [34,35]. The mechanisms regulating platelet/MK incorporation of albumin and IgG are unclear. In endothelial cells albumin uptake is protein-kinase C-dependent [36]; our patient's platelets have decreased PKC-θ [21], providing a potential mechanism. The decreased albumin and IgG suggest that RUNX1 haplodeficiency is associated with additional alterations, in granule membrane proteins or vesicle trafficking. Indeed, there is some direct evidence for this: platelet transcript levels of RAB31 and RAB1B are decreased in our patient [26]. Small GTP-binding RAB proteins, such as RAB31 and RAB1B, play essential roles in vesicle trafficking and granule targeting [37,38]. RAB31 is present in platelet membrane and granule fractions [38]. RAB1B plays a role in vesicle transport between endoplasmic reticulum and golgi [39,40]. Mutations in other GTPases, RAB27A and RAB27B, and in RAB geranylgeranyltransferase are implicated in animal models of platelet granule defects and defective vesicular transport [15,41-44]. Moreover, DNM3 (dynamin 3) transcript was decreased in our patient platelets [26]. This microtubule-associated force-producing protein can hydrolyze GTP and participates in MK growth and development [45]. Dynamins as a group are implicated in vesicular trafficking, in particular endocytosis [46-48]. Further studies in RUNX1 haplodeficiency on these proteins are likely to yield new insights.
Our studies are beginning to unravel the complex nature of the platelet defects in RUNX1 haplodeficiency and the genes regulated in platelets/MK by RUNX1. We have shown that RUNX1 regulates 12-lipoxygenase (ALOX12)[27], myosin light chain (MYL9) [49], protein kinase C-θ (PRKCQ) [50] and PF4 (PF4), and each appears relevant to platelet production or function. Together with previously reported other gene alterations [26] the paradigm that emerges is that RUNX1 mutations induce alterations in diverse platelet and MK processes.
Because PF4V1 transcript level was also decreased in our patient [26] we studied its regulation. In normal platelets PF4V1 expression was markedly less than of PF4. Although there was RUNX1 binding to the promoter region (Fig. 6) luciferase reporter studies using ~2 kb 5’-upstream promoter region showed little activity compared to PF4 construct (Fig. 7). In HEL cells there was minimal increase in PF4V1 mRNA PMA-treatment compared to that in PF4 mRNA. Thus, PF4V1 is regulated distinctly differently from PF4.
In summary, our studies demonstrate that PF4 is regulated at the transcriptional level by RUNX1, and provide a mechanism for PF4 deficiency associated with RUNX1 haplodeficiency in our patient and others [19]. While defective protein targeting has been considered the main mechanism leading to PF4 deficiency in GPS, our studies provide evidence that, in some patients, PF4 deficiency results from impaired PF4 synthesis. These studies advance understanding of the impaired platelet mechanisms in RUNX1 haplodeficiency and GPS, and emphasize the complex nature of the associated platelet/megakaryocytic defects.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the assistance of Denise Tierney in manuscript preparation.
This work was supported by grants from the National Institutes of Health (R01HL85422 and R01HL56724) (AKR). KA was supported by National Institutes of Health T32 training grant HL007777.
Supported by grants NIH R01 HL85422 (AKR) and NIH R01 HL56724 (AKR). KA was supported by NIH T32 Training Grant HL007777.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Holt JC, Niewiarowski S. Biochemistry of alpha granule proteins. Semin Hematol. 1985;22:151–63. [PubMed] [Google Scholar]
- 2.Kowalska MA, Rauova L, Poncz M. Role of the platelet chemokine platelet factor 4 (PF4) in hemostasis and thrombosis. Thromb Res. 2010;125:292–6. doi: 10.1016/j.thromres.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 3.Sachais BS, Higazi AA, Cines DB, Poncz M, Kowalska MA. Interactions of platelet factor 4 with the vessel wall. Semin Thromb Hemost. 2004;30:351–8. doi: 10.1055/s-2004-831048. [DOI] [PubMed] [Google Scholar]
- 4.Gewirtz AM, Zhang J, Ratajczak J, Ratajczak M, Park KS, Li C, Yan Z, Poncz M. Chemokine regulation of human megakaryocytopoiesis. Blood. 1995;86:2559–67. [PubMed] [Google Scholar]
- 5.Lambert MP, Rauova L, Bailey M, Sola-Visner MC, Kowalska MA, Poncz M. Platelet factor 4 is a negative autocrine in vivo regulator of megakaryopoiesis: clinical and therapeutic implications. Blood. 2007;110:1153–60. doi: 10.1182/blood-2007-01-067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maione TE, Gray GS, Petro J, Hunt AJ, Donner AL, Bauer SI, Carson HF, Sharpe RJ. Inhibition of angiogenesis by recombinant human platelet factor-4 and related peptides. Science. 1990;247:77–9. doi: 10.1126/science.1688470. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi K, Ogawa K, Katsube T, Shimao K, Konno S, Shimakawa T, Yoshimatsu K, Naritaka Y, Yagawa H, Hirose K. Platelet factor 4 gene transfection into tumor cells inhibits angiogenesis, tumor growth and metastasis. Anticancer Res. 2005;25:847–51. [PubMed] [Google Scholar]
- 8.Ferroni P, Basili S, Davi G. Platelet activation, inflammatory mediators and hypercholesterolemia. Curr Vasc Pharmacol. 2003;1:157–69. doi: 10.2174/1570161033476772. [DOI] [PubMed] [Google Scholar]
- 9.Yu G, Rux AH, Ma P, Bdeir K, Sachais BS. Endothelial expression of E-selectin is induced by the platelet-specific chemokine platelet factor 4 through LRP in an NF-kappaB-dependent manner. Blood. 2005;105:3545–51. doi: 10.1182/blood-2004-07-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eslin DE, Zhang C, Samuels KJ, Rauova L, Zhai L, Niewiarowski S, Cines DB, Poncz M, Kowalska MA. Transgenic mice studies demonstrate a role for platelet factor 4 in thrombosis: dissociation between anticoagulant and antithrombotic effect of heparin. Blood. 2004;104:3173–80. doi: 10.1182/blood-2003-11-3994. [DOI] [PubMed] [Google Scholar]
- 11.Green CJ, Charles RS, Edwards BF, Johnson PH. Identification and characterization of PF4varl, a human gene variant of platelet factor 4. Mol Cell Biol. 1989;9:1445–51. doi: 10.1128/mcb.9.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisman R, Surrey S, Ramachandran B, Schwartz E, Poncz M. Structural and functional comparison of the genes for human platelet factor 4 and PF4alt. Blood. 1990;76:336–44. [PubMed] [Google Scholar]
- 13.Lasagni L, Grepin R, Mazzinghi B, Lazzeri E, Meini C, Sagrinati C, Liotta F, Frosali F, Ronconi E, Alain-Courtois N, Ballerini L, Netti GS, Maggi E, Annunziato F, Serio M, Romagnani S, Bikfalvi A, Romagnani P. PF-4/CXCL4 and CXCL4L1 exhibit distinct subcellular localization and a differentially regulated mechanism of secretion. Blood. 2007;109:4127–34. doi: 10.1182/blood-2006-10-052035. [DOI] [PubMed] [Google Scholar]
- 14.Struyf S, Burdick MD, Proost P, Van Damme J, Strieter RM. Platelets release CXCL4L1, a nonallelic variant of the chemokine platelet factor-4/CXCL4 and potent inhibitor of angiogenesis. Circ Res. 2004;95:855–7. doi: 10.1161/01.RES.0000146674.38319.07. [DOI] [PubMed] [Google Scholar]
- 15.Nurden AT, Nurden P. The gray platelet syndrome: clinical spectrum of the disease. Blood Rev. 2007;21:21–36. doi: 10.1016/j.blre.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Maynard DM, Heijnen HF, Gahl WA, Gunay-Aygun M. The alpha granule proteome: novel proteins in normal and ghost granules in gray platelet syndrome. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.03932.x. doi: 10.1111/j.538-7836.2010.03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss HJ, Witte LD, Kaplan KL, Lages BA, Chernoff A, Nossel HL, Goodman DS, Baumgartner HR. Heterogeneity in storage pool deficiency: studies on granule-bound substances in 18 patients including variants deficient in alpha-granules, platelet factor 4, beta-thromboglobulin, and platelet-derived growth factor. Blood. 1979;54:1296–319. [PubMed] [Google Scholar]
- 18.Tubman VN, Levine JE, Campagna DR, Monahan-Earley R, Dvorak AM, Neufeld EJ, Fleming MD. X-linked gray platelet syndrome due to a GATA1 Arg216Gln mutation. Blood. 2007;109:3297–9. doi: 10.1182/blood-2006-02-004101. [DOI] [PubMed] [Google Scholar]
- 19.Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, Rossier C, Antonarakis SE, Israels S, McNicol A, Weiss H, Horwitz M, Scott HS. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–72. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- 20.Gabbeta J, Yang X, Sun L, McLane MA, Niewiarowski S, Rao AK. Abnormal inside-out signal transduction-dependent activation of glycoprotein IIb-IIIa in a patient with impaired pleckstrin phosphorylation. Blood. 1996;87:1368–76. [PubMed] [Google Scholar]
- 21.Sun L, Mao G, Rao AK. Association of CBFA2 mutation with decreased platelet PKC-θ and impaired receptor-mediated activation of GPIIb-IIIa and pleckstrin phosphorylation: proteins regulated by CBFA2 play a role in GPIIb-IIIa activation. Blood. 2004;103:948–54. doi: 10.1182/blood-2003-07-2299. [DOI] [PubMed] [Google Scholar]
- 22.de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–48. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 23.Mikhail FM, Sinha KK, Saunthararajah Y, Nucifora G. Normal and transforming functions of RUNX1: a perspective. J Cell Physiol. 2006;207:582–93. doi: 10.1002/jcp.20538. [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa M, Asai T, Saito T, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Ogawa S, Kurokawa M, Hirai H. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 25.Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–75. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 26.Sun L, Gorospe JR, Hoffman EP, Rao AK. Decreased platelet expression of myosin regulatory light chain polypeptide (MYL9) and other genes with platelet dysfunction and CBFA2/RUNX1 mutation: insights from platelet expression profiling. J Thromb Haemost. 2007;5:146–54. doi: 10.1111/j.1538-7836.2006.02271.x. [DOI] [PubMed] [Google Scholar]
- 27.Kaur G, Jalagadugula G, Mao G, Rao AK. RUNX1/core binding factor A2 regulates platelet 12-lipoxygenase gene (ALOX12): studies in human RUNX1 haplodeficiency. Blood. 2010;115:3128–35. doi: 10.1182/blood-2009-04-214601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cupit LD, Schmidt VA, Gnatenko DV, Bahou WF. Expression of protease activated receptor 3 (PAR3) is upregulated by induction of megakaryocyte phenotype in human erythroleukemia (HEL) cells. Exp Hematol. 2004;32:991–9. doi: 10.1016/j.exphem.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Long MW, Heffner CH, Williams JL, Peters C, Prochownik EV. Regulation of megakaryocyte phenotype in human erythroleukemia cells. J Clin Invest. 1990;85:1072–84. doi: 10.1172/JCI114538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta AK. Efficient amplification using ‘megaprimer’ by asymmetric polymerase chain reaction. Nucleic Acids Res. 1995;23:4530–1. doi: 10.1093/nar/23.21.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vyas P, Ault K, Jackson CW, Orkin SH, Shivdasani RA. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 1999;93:2867–75. [PubMed] [Google Scholar]
- 32.Lo B, Li L, Gissen P, Christensen H, McKiernan PJ, Ye C, Abdelhaleem M, Hayes JA, Williams MD, Chitayat D, Kahr WH. Requirement of VPS33B, a member of the Sec1/Munc18 protein family, in megakaryocyte and platelet alpha-granule biogenesis. Blood. 2005;106:4159–66. doi: 10.1182/blood-2005-04-1356. [DOI] [PubMed] [Google Scholar]
- 33.Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–41. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 34.Handagama PJ, Amrani DL, Shuman MA. Endocytosis of fibrinogen into hamster megakaryocyte alpha granules is dependent on a dimeric gamma A configuration. Blood. 1995;85:1790–5. [PubMed] [Google Scholar]
- 35.Klinger MH, Kluter H. Immunocytochemical colocalization of adhesive proteins with clathrin in human blood platelets: further evidence for coated vesicle-mediated transport of von Willebrand factor, fibrinogen and fibronectin. Cell Tissue Res. 1995;279:453–7. doi: 10.1007/BF00318157. [DOI] [PubMed] [Google Scholar]
- 36.Lynch JJ, Ferro TJ, Blumenstock FA, Brockenauer AM, Malik AB. Increased endothelial albumin permeability mediated by protein kinase C activation. J Clin Invest. 1990;85:1991–8. doi: 10.1172/JCI114663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–25. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 38.Bao X, Faris AE, Jang EK, Haslam RJ. Molecular cloning, bacterial expression and properties of Rab31 and Rab32. Eur J Biochem. 2002;269:259–71. doi: 10.1046/j.0014-2956.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez C, Garcia-Mata R, Brandon E, Sztul E. COPI recruitment is modulated by a Rab1b-dependent mechanism. Mol Biol Cell. 2003;14:2116–27. doi: 10.1091/mbc.E02-09-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plutner H, Cox AD, Pind S, Khosravi-Far R, Bourne JR, Schwaninger R, Der CJ, Balch WE. Rab1b regulates vesicular transport between the endoplasmic reticulum and successive Golgi compartments. J Cell Biol. 1991;115:31–43. doi: 10.1083/jcb.115.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Detter JC, Zhang Q, Mules EH, Novak EK, Mishra VS, Li W, McMurtrie EB, Tchernev VT, Wallace MR, Seabra MC, Swank RT, Kingsmore SF. Rab geranylgeranyl transferase alpha mutation in the gunmetal mouse reduces Rab prenylation and platelet synthesis. Proc Natl Acad Sci U S A. 2000;97:4144–9. doi: 10.1073/pnas.080517697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak EK, Gautam R, Reddington M, Collinson LM, Copeland NG, Jenkins NA, McGarry MP, Swank RT. The regulation of platelet-dense granules by Rab27a in the ashen mouse, a model of Hermansky-Pudlak and Griscelli syndromes, is granule-specific and dependent on genetic background. Blood. 2002;100:128–35. doi: 10.1182/blood.v100.1.128. [DOI] [PubMed] [Google Scholar]
- 43.Swank RT, Jiang SY, Reddington M, Conway J, Stephenson D, McGarry MP, Novak EK. Inherited abnormalities in platelet organelles and platelet formation and associated altered expression of low molecular weight guanosine triphosphate-binding proteins in the mouse pigment mutant gunmetal. Blood. 1993;81:2626–35. [PubMed] [Google Scholar]
- 44.Wilson SM, Yip R, Swing DA, O'Sullivan TN, Zhang Y, Novak EK, Swank RT, Russell LB, Copeland NG, Jenkins NA. A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc Natl Acad Sci U S A. 2000;97:7933–8. doi: 10.1073/pnas.140212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reems JA, Wang W, Tsubata K, Abdurrahman N, Sundell B, Tijssen MR, van der Schoot E, Di Summa F, Patel-Hett S, Italiano J, Jr., Gilligan DM. Dynamin 3 participates in the growth and development of megakaryocytes. Exp Hematol. 2008;36:1714–27. doi: 10.1016/j.exphem.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gold ES, Underhill DM, Morrissette NS, Guo J, McNiven MA, Aderem A. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med. 1999;190:1849–56. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–8. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 48.Pelkmans L, Puntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–9. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- 49.Jalagadugula G, Mao G, Kaur G, Goldfinger LE, Dhanasekaran DN, Rao AK. Regulation of platelet myosin light chain (MYL9) by RUNX1: implications for thrombocytopenia and platelet dysfunction in RUNX1 haplodeficiency. Blood. 2010 doi: 10.1182/blood-2010-06-289850. DOI 10.1182/blood-2010-06-289850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jalagadugula GS, Kaur G, Mao G, Dhanasekaran DN, Rao AK. Platelet/megakaryocyte PKC-θ is a transcriptional target of RUNX1/CBFA2: studies in human RUNX1 haplodeficiency. Blood. 2008;112:649. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.