Abstract
Dyslexia is a complex learning disability with evidence for a genetic basis. Strategies that may be useful for dissecting its genetic basis include the study of component phenotypes, which may simplify the underlying genetic complexity, and use of an analytic approach that accounts for the multilocus nature of the trait to guide the investigation and increase power to detect individual loci. Here we present results of a genetic analysis of spelling disability as a component phenotype. Spelling disability is informative in analysis of extended pedigrees because it persists into adulthood. We show that a small number of hypothesized loci are sufficient to explain the inheritance of the trait in our sample, and that each of these loci maps to one of four genomic regions. Individual trait models and locations are a function of whether a verbal IQ adjustment is included, suggesting mediation through both IQ-related and unrelated pathways.
Keywords: Reading disability, Linkage analysis, Complex disorder, MCMC, Learning disability
Introduction
Dyslexia is a complex disorder that affects 5–12% of school-aged children (Katusic et al. 2001; Shaywitz et al. 1990). It is characterized by unexpected difficulty in learning to read, despite adequate educational opportunities, with such difficulties not being attributable to other identifiable factors such as cognitive delay, psychiatric or neurological disorders, or sensory impairment. Deficits involve several cognitive processes that are important for normal development of reading skills, including storage of phonologically coded words in short-term memory, maintaining correspondence between letters and sounds, and conscious awareness of the sounds in auditory words (Wagner and Torgesen 1987). The phenotypic presentation changes throughout development from childhood to adulthood. In younger children, deficits typically involve difficulties in learning to name letters and associate sounds with letters with subsequent difficulties with real and pseudo-word reading, while deficits in older children manifest as problems with fluent reading, spelling, and written expression (Berninger et al. 2001; Bruck 1993a). Deficits in fluent oral reading and spelling persist into adulthood for about half of individuals with a childhood diagnosis (Schulte-Korne et al. 2007) even after other manifestations of this learning disability appear to have been corrected (Berninger et al. 2006; Berninger et al. 2001; Bruck 1990, 1992, 1993b; Felton et al. 1990; Pennington et al. 1990; Ramus et al. 2003; Wilson and Lesaux 2001). Spelling has been found to be a reliable phenotype for dyslexia not only in English, for which the spelling-sound correspondences are variable (Berninger et al. 2008a), but also in languages such as German with invariant spelling-sound correspondences (Roeske et al. 2009). Spelling deficits are therefore a key aspect of the phenotype, particularly in older children (Shaywitz et al. 1999) and adults (Grossglenn et al. 1985; Hatcher et al. 2002; Pennington et al. 1986).
There is ample evidence that dyslexia has a complex genetic basis. Both twin studies (DeFries et al. 1991; Pennington et al. 1991; Willcutt et al. 2000) and family studies (Chapman et al. 2003a; Gilger et al. 1994; Igo et al. 2006a; Naples et al. 2009; Raskind et al. 2005; Schulte-Korne et al. 2007; Wijsman et al. 2000; Wolff and Melngailis 1994; Ziegler et al. 2005) have consistently provided evidence for a genetic etiology for a discrete diagnosis of dyslexia, and for a genetic basis to many component phenotypes associated with dyslexia. However, there are multiple correlated phenotypes involved, and, as for most complex traits, almost certainly multiple underlying etiological factors. Such heterogeneity complicates identification of underlying genes, regardless of the study design. Given the available data, two strategies can mitigate the effects of this complexity. The first involves efficient use of available data, including all available pedigree and marker data, combined with efficient use of available phenotypic data. This includes use of larger pedigrees when available, full use of multipoint marker information, and use of quantitative traits rather than artificially dichotomized traits (Wijsman and Amos 1997). By using the data efficiently, it is easier to attain the higher power necessary to overcome the effects of heterogeneity, in the context of an existing sample. The second strategy focuses on single component phenotypes rather than joint or summarized phenotypes (Grigorenko et al. 1997). The use of component phenotypes is a natural consequence of the use of continuous traits. Also, under the assumption that the use of component phenotypes simplifies the mode of inheritance, it may reduce the problem of heterogeneity. It may therefore be an advantageous strategy if there are different underlying genetic architectures acting on different aspects of the phenotype.
Choice of the analytic approach used is another key issue in the search of the genetic basis of complex traits. Two approaches that are useful in this context include a variety of methods that correlate phenotypic similarity and estimates of marker identity-by-descent proportions, and linkage and segregation analysis methods that can accommodate multilocus modes of inheritance. The former approach includes variance components (VC) approaches (Almasy and Blangero 1998; Amos 1994), which capture the trait model through a simplified genetic variance parameterization based on joint marker and trait similarity between pairs of individuals and can be used on a variety of pedigree structures. VC methods are widely used, but can be sensitive to issues including sample ascertainment, leading to both excess false-positive and false-negative results (Forrest and Feingold 2000; Williams et al. 1997). It is also difficult to predict the outcome of a genome scan from simple measures of heritability, which sometimes are used as an initial filter to determine the trait(s) to carry forward to a genome scan (Huang et al. 2007; Kan et al. 2007). In contrast, joint oligogenic segregation and linkage analysis (OSA) allows for a multilocus trait model and is a fully model-based approach for the trait, thus improving power and resolution in some situations. The approach can be used to investigate the genetic model space prior to a genome scan, with demonstration of clear models in the oligogenic model space typically leading to successful trait genome scans (Daw et al. 2007; Igo et al. 2006a; Raskind et al. 2005). Complex models and full use of available marker data on extended pedigrees depend on availability of computationally sophisticated and demanding implementations, but these are now available (Heath 1997; Marchani et al. 2009; Rosenthal and Wijsman 2010; Wijsman et al. 2006).
Using a variety of ascertainment schemes, subject populations, and analytic approaches, at least 20 potential susceptibility loci for dyslexia and dyslexia-related traits have been identified, as reviewed in (Schumacher et al. 2007). The first two of these loci, DYX1 on chromosome 15q21, and DYX2 on chromosome 6p21-p22, were selected for linkage analysis on the basis of a cytogenetic polymorphism (Smith et al. 1983) or a hypothesis about a relationship between autoimmunity and dyslexia (Cardon et al. 1994; Cardon et al. 1995), respectively. Subsequently, many studies focused on these two loci and found evidence for their involvement in a variety of dyslexia endophenotypes (Schumacher et al. 2007). Many of the other dyslexia candidate loci were identified in individual families or by targeting specific chromosome locations (Schumacher et al. 2007). To date, nine genome-wide scans for dyslexia (Brkanac et al. 2008; de Kovel et al. 2004; Fagerheim et al. 1999; Fisher et al. 2002; Igo et al. 2006a; Kaminen et al. 2003; Nopola-Hemmi et al. 2001; Raskind et al. 2005) and one genome-wide association scan for an electrophysiologic measure related to speech processing (Roeske et al. 2009) have been reported. For many studies that analyzed components, the bulk of the linkage evidence was provided by one or a few endophenotypes. Although spelling impairment is a robust and persistent feature of dyslexia, only the German group has concentrated on this phenotype (Nöthen et al. 1999; Schulte-Korne et al. 1998; Schumacher et al. 2006); therefore, information about the specific genetic contributions to spelling impairment is limited. Their targeted analyses provided support for the locus on chromosome 15 (Nöthen et al. 1999), but not for those on chromosomes 6 or 18 (Schulte-Korne et al. 1998; Schumacher et al. 2006). Evidence for linkage of spelling impairment to chromosome 1p34-p36 and for association with a chromosome 6p haplotype were among the most robust findings in targeted studies of these loci in other ethnic populations (Turic et al. 2003; Tzenova et al. 2004). However, linkage results for spelling phenotypes were weak in the two genome-wide scans performed by Fisher et al. (Fisher et al. 2002).
Here we present a genetic analysis of measures of spelling ability in families ascertained through a proband with reading difficulties. Our general strategy in the study of the genetic basis of dyslexia is to focus on component measures, with preliminary aggregation and segregation analyses used to prioritize measures for further genome scans (Chapman et al. 2003b; Hsu et al. 2002; Igo et al. 2006a; Raskind et al. 2000; Raskind et al. 2005). We focus here on spelling deficits because they are a key aspect of the phenotype and because they were previously shown to have high correlation with other measures of reading difficulty in our sample (Hsu et al. 2002), as well as a correlation pattern between relatives that is consistent with a genetic basis (Raskind et al. 2000). The persistence of spelling deficits in adults with earlier reading difficulties makes it a particularly useful trait for study in extended pedigrees, as characterizes our sample. Our goals were to determine whether there is evidence for a pattern of inheritance for performance on the spelling measures, and if so, to identify regions of the genome with evidence for linkage to these measures.
Methods
Sample
The sample used in this study has been described in detail elsewhere (Berninger et al. 2001; Igo et al. 2006a; Raskind et al. 2000). In brief, individuals between ages 6 and 16 years were ascertained initially based on low reading performance. Ascertainment to the study was further restricted to subjects with a minimum verbal IQ (VIQ) of at least 90, and low reading abilities relative to VIQ, as assessed by ten measures of reading or spelling, including the two spelling measures used here (Berninger et al. 2001). Subjects who were impaired on any of the ten measures were selected as probands; in fact, most probands were impaired on most or all of the measures (average 6.7 measures/proband) (Berninger et al. 2001). Probands were primarily of European descent, and were not excluded if they had a diagnosis of ADHD, but were excluded for other conditions associated with learning disabilities. Family members of each proband who agreed to participate were then added to the study, using a sequential sampling strategy (Cannings et al. 1978) to extend pedigrees through family members with the most extreme, and therefore informative, values for the same qualifying phenotypes (Boehnke and Moll 1989). This study obtained informed consent from all participants and was approved by the University of Washington institutional review board. A total of 2152 individuals in 307 families were included in the study, with family sizes of 3–23 individuals in 2–4 generations. For the current analysis, 1783 of the individuals were included in covariate adjustments, with 369 subjects omitted due to missing phenotype data. 1951 individuals in 260 families were included in the segregation analysis, and 1957 individuals in 260 families were included in the joint segregation-linkage analysis, with omissions due to one or more of lack of phenotype information, lack of genotype information, or uninformative pedigree structure.
Phenotypes
Two tests were used as measures of spelling: the Wide Range Achievement Test-3 (WRAT3SP) (Wilkinson 1993) and the Wechsler Individual Achievement Test-II (WI-ATSP) (Wechsler 2002). WRAT3SP and WIATSP are both standardized to have mean 100 (SD = 15). VIQ was used for some analyses as a covariate, as provided by prorated VIQ scores from the Wechsler Intelligence Scale for Children—Third Edition (Wechsler 1992) for subjects <17 years old, or the Wechsler Adult Intelligence Scale—Revised (Wechsler 1981) for subjects >16 years old. For each test, standard protocols were used to provide standardized scores. Throughout, we present all spelling scores in units of these standardized scores. As these spelling tests measure the same trait on the same scale (they have an estimated correlation in this sample of 0.86), the mean of these two measures was used to construct the measure Spelling Average (SpAvg) for further analysis. Analysis was also conducted on the individual measures with similar results (not shown).
Genotypes
The genotyping for the sample was obtained in two batches from the Marshfield Genotyping Service. Because the genotyping batches were obtained at different times, two somewhat different marker panels were used: Marshfield panels 10 and 13. Of the 477 total markers used in the analyses, 296 were present in both genome scan panels. Every individual within a pedigree was typed on the same panel, so differences in the marker panels used occurred only between families. The marker data were therefore merged by treating the genotypes at all markers on the panel for which the individual was not typed as missing for analysis purposes. Marker positions were then combined, using the Rutgers map as a reference (Matise et al. 2007) and the Haldane map function (Haldane 1919). All references to map positions henceforth are based on the Haldane map function. The genotype data were also used for quality control of both pedigree structure and the genotypes, as described earlier for this sample (Chapman et al. 2004; Raskind et al. 2005). In brief, this included identifying relationship inconsistencies, Mendelian inconsistencies, highly unlikely genotypes, and individual markers with excessive errors, and making appropriate data corrections or eliminations.
Statistical analysis
Covariate adjustments
It is important to adjust the model for covariates to eliminate as many non-genetic factors that may affect our analysis as possible, reduce within-genotype variance, and increase power to detect genetic effects (Wijsman and Amos 1997). Therefore, a predictive linear model for SpAvg was constructed to account for external factors that appeared to be important in our sample. We used a piecewise linear model to adjust for age because of evidence, in this sample, of age effects (Hsu et al. 2002), even for these standardized spelling measures that already are based on an age-adjustment, using the standard test protocols. Age was further subdivided into a dichotomous variable (cohort) indicating whether an individual is a youth or adult, with the “youth” group (<300 months or 25 years) serving as the baseline. This accommodates effects of development in the youths, which are expected to result in greater age-effects than would be seen in adults. This age division was chosen as it was a nadir in the age distribution of our sample, making it a logical point at which to divide the proband and parental generations. Due to the small number of individuals near age 300 months, the exact point of division does not strongly influence the results. Sex was also included as a covariate (with the males serving as the baseline) because of previous observations in this sample (Hsu et al. 2002), male bias in the probands in this sample (Raskind et al. 2000), and the observation of gender differences in many (Berninger et al. 2008b; Flannery et al. 2000; Hsu et al. 2002; Katusic et al. 2001; Rutter et al. 2004; Wadsworth et al. 1992), although not all (Shaywitz et al. 1990; Wadsworth et al. 1992), studies of learning disabilities.
Several models were considered without explicitly accounting for relationships in the sample. Covariates considered included age, cohort, sex, interactions between age and cohort, and the three-way interaction between age, cohort and sex. The best-fitting model, as judged by the significance of the coefficient of each covariate (p < 0.05) and overall fit of the model, had linear terms for age, with different slopes in the two cohorts, and an offset for sex (i.e., SpAvg = β0 + β1(age) + β2(cohort) + β3(sex) + β4(age)(cohort)). Further oligogenic segregation and joint segregation-linkage analyses were then conducted using residuals from this model (SpAvg(Adj)), but also incorporating the pedigree structure.
It remains a point of contention whether or not one should account for VIQ in studies of the genetic basis of dyslexia. Many samples have been collected on the basis of an IQ-discrepancy between actual and predicted reading disabilities (Fisher et al. 1999; Raskind et al. 2000), while other studies do not impose this restriction (Gayan et al. 1999). However, some studies have reported higher estimates of heritability (Knopik et al. 2002; Remschmidt et al. 1999; Stevenson et al. 1987; Wadsworth et al. 2000) when a discrepancy measure is used. We therefore carried out all analyses both with and without adjustment for VIQ under the hypothesis that a genome scan might shed light on this issue. Thus, once a best-fit model was chosen based on the covariates, above, an additional model was created that includes VIQ as a covariate (SpAvg = β0 + β1(age) + β2(cohort) + β3(sex) + β4(age)(cohort) + β5(VIQ)). As for the previous adjustment, we used the residuals from this model (SpAvg(VIQ)) for further oligogenic segregation and joint segregation-linkage analyses.
Oligogenic segregation analysis
We conducted oligogenic segregation analysis to determine the genetic model required to explain the observed data. The goal of this analysis was to identify how many diallelic quantitative trait loci (QTLs) are needed to explain the data, as well as the effect sizes of the models, their dominance structures, and their parameter values. Past experience has shown that the number of QTLs thus revealed is a useful indicator of whether QTLs are likely to be localized in a genome scan (Igo et al. 2006a; Igo et al. 2006b). This analysis was conducted using Loki 2.4.7 and provides estimates of the posterior distributions of model parameters using a Bayesian reversible-jump MCMC approach (Heath, 1997). Analysis is carried out conditional on user-specified prior distributions on the QTL parameters, pedigree data, and phenotype data. Reversible-jump MCMC allows the number of parameters in the model to vary; thus we can estimate the number of QTLs in addition to their parameter values. We used the adjusted SpAvg scores (as described in the previous section) for input to the analysis, with the prior distribution on the number of QTLs in the model a truncated Poisson distribution with mean 2, a maximum number of QTLs of 17, and a value for the variance, τβ, on the genotype effects that was four times the adjusted phenotypic variance. Details about the number of QTLs assumed in the prior distribution are relatively unimportant, as long as they allow a sufficient range of values for the MCMC sampler in evaluating the model space (Wijsman and Yu 2004). Use of τβ in this range is generally a good choice of parameter value (Wijsman and Yu 2004), and was chosen by evaluating a range of values from ¼ to 8 times the phenotypic variance. Insensitivity of conclusions was determined by comparing runs of the segregation analysis using the various values of this parameter. Analysis runs consisted of 100,000 iterations, with a burn-in of 1000 iterations, and a thinning interval of 10.
Statistical inference was based on the posterior distribution of model parameters, summarized through marginal distributions of the modal classes of models. Individual QTLs were further summarized as follows. The alleles for each diallelic QTL were labeled as ‘A’ and ‘a’ for the allele contributing to high and low scores, respectively, with pA referring to the allele frequency of the A allele. The AA (or homozygote) genotype effect was defined as the difference between the mean genotype values of the AA and aa genotypes, or μAA − μaa. A similar definition for the Aa (heterozygote) effect was defined as the difference between the mean genotype values of the Aa and aa genotypes, or μAa − μaa.
Joint linkage-segregation analysis
Joint linkage and segregation analysis was conducted to estimate the position of QTLs in the genome, and to improve precision of the estimates found in the segregation analysis through the additional constraints provided by the linked markers. The joint linkage and segregation analysis was conducted using Loki 2.4.7, as described above. This analysis conditions on the marker data as well as the prior distributions of the parameters, pedigree data, and phenotype data. The model, prior on the number of QTLs and their genotype effects, and number of iterations are the same as in the segregation analysis, but with a smaller thinning interval of 5. We used the genome-scan markers and map described above, with allele frequencies estimated from the data, based on a Dirichlet (1, 1, …, 1) prior distribution. As for the segregation analysis, inference was based on posterior model distributions. For posterior QTL model distributions, we summarized all QTLs with localization to a chromosome. We used Bayes Factors (Kass and Raftery 1995) to evaluate the strength of evidence for linkage, where the Bayes Factor for linkage is the ratio of the posterior to prior odds of the location of a QTL in a particular interval, with the prior odds based on the prior distributions used for analysis. For computation of Bayes Factors, we accumulated the information in 2 cM non-overlapping bins across the genome. For the strongest linkage signals, we also obtained estimated frequentist p-values by simulating 1000 replicates of marker genotypes in a 40 cM region around the strongest signals under the null hypothesis of no linkage, followed by re-analysis with the real trait data (Igo and Wijsman 2008).
Results
Spelling scores
Summaries of the averaged spelling scores are shown in Table 1. The mean across the full sample (97.7) is slightly below the expected mean, given the use of standardized scores in the current study. Males have lower scores than females overall (94.1 vs. 101.6 in males and females, respectively) as well as separately in both youths (89.7 vs. 96.5 for males and females, respectively) and adults (98.1 vs. 105 for males and females, respectively). The variance in scores is higher in adult males than in adult females, but is similar for the two groups in the youths. Within the sample, probands had lower scores than their relatives, with means of 83.1 and 84.5 for WIATSP and WRAT3SP, respectively. As expected by the ascertainment, these spelling scores are low, relative to what would be expected in the context of the average proband VIQ of 110.8 and standard deviation 12.72 in this sample (Berninger et al. 2006; Brkanac et al. 2008).
Table 1.
Distributions of Average WRAT3SP and WIATSP spelling scores (SpAvg) and verbal IQ (VIQ) scores by category of subject
| Group | SpAvg |
VIQ |
||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Female youths | 96.52 | 13.67 | 109.73 | 12.90 |
| Male youths | 89.70 | 13.97 | 109.03 | 13.14 |
| Female adults | 104.95 | 10.53 | 108.23 | 11.69 |
| Male adults | 98.14 | 15.14 | 109.47 | 12.45 |
| All females | 101.56 | 12.59 | 108.83 | 12.20 |
| All males | 94.07 | 15.18 | 109.26 | 12.78 |
| All youths | 92.73 | 14.24 | 109.34 | 13.03 |
| All adults | 101.72 | 13.36 | 108.82 | 12.07 |
| All subjects | 97.74 | 14.46 | 109.05 | 12.50 |
Covariate adjustments
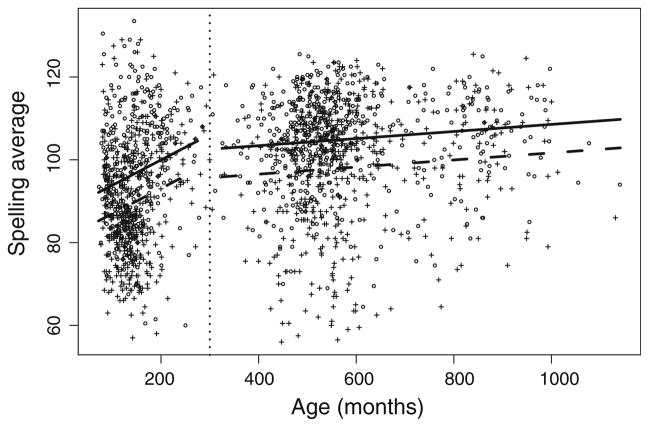
Age, sex and cohort all were significant covariates (p < 0.05) as predictors of SpAvg, with or without inclusion of VIQ as a covariate (Table 2). The best-fitting predictive model in the absence of inclusion of VIQ is: SpAv-g = 87.71 + 0.06(age) + 12.28(cohort) − 6.88(sex) − 0.05 (age)(cohort), with R2 = 0.17. This model (Fig. 1) suggests that females have SpAvg which exceeds that in males by 6.88, and that the intercept for adults exceeds that of youths by 12.28. Furthermore, youth spelling scores improve with age at a rate of 0.06 per month, but this rate does not continue in adults, whose scores change negligibly with age (average rate of improvement <0.01 per month). The best-fitting model when we include VIQ, which is also a significant predictor of SpAvg (p < 0.05), is: SpAvg = 26.73 + 0.08(age) + 19.92 (cohort) − 7.04(sex) − 0.08(age)(cohort) + 0.53(VIQ), with R2 = 0.38. The coefficients have similar interpretations to those in the model without VIQ, with a slightly higher separation in the intercept between adults and youths (19.92 after VIQ adjustment compared to 12.28 without such adjustment), a slightly faster improvement with age in youths (0.08 per month with VIQ adjustment, compared to 0.06 without such adjustment), and negligible change in adults with age. In addition, every 10 points of increase in VIQ corresponds to an average increase in SpAvg(VIQ) of 5.3.
Table 2.
Coefficients of the covariate adjustment models
| Model | Predictor | Estimated coefficient | 95%a confidence interval |
|---|---|---|---|
| SpAvg | Intercept | 87.71 | (84.47, 90.95) |
| Age (months) | 0.06 | (0.04, 0.08) | |
| Cohort | 12.28 | (7.62, 16.93) | |
| Sex | −6.88 | (−8.11, −5.65) | |
| (Age)(Cohort) | −0.05 | (−0.07, −0.03) | |
| SpAvg(VIQ) | Intercept | 26.73 | (21.08, 32.39) |
| Age (months) | 0.08 | (0.06, 0.10) | |
| Cohort | 19.92 | (15.84, 24.00) | |
| Sex | −7.04 | (−8.11, −5.98) | |
| (Age)(Cohort) | −0.08 | (−0.10, −0.06) | |
| VIQ | 0.53 | (0.49, 0.58) |
Because we do not account for relatedness in the linear adjustment model, the variance of the regression coefficients is underestimated, and the true coverage of the confidence intervals is likely to be less than 95%. The estimates of the coefficients themselves are not affected
Fig. 1.
Scatterplot of age (in months) vs. Spelling Average (SpAvg), with best-fitting covariate adjustment indicated. The lines show the predicted average SpAvg as a function of sex, with a break at age 300 months (vertical dotted line) between the youth and adult cohorts, and with the difference in intercept between males and females the value of the estimated coefficient for the sex effect. Females circles, solid lines; Males: plus symbols, dashed lines
Oligogenic segregation analysis
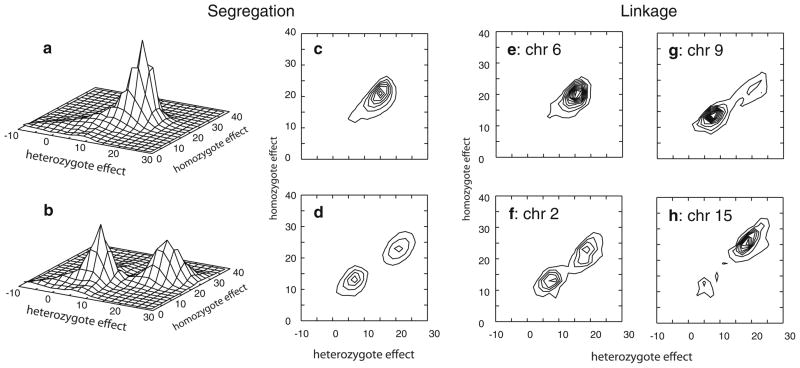
Oligogenic segregation analysis revealed a simple model structure for SpAvg(Adj). Only one notable QTL (model 1) was identified in the posterior oligogenic distribution (Fig. 2a, c). The model identified had a homozygote effect of 19.5, indicating that individuals with the AA genotype are estimated to have SpAvg(Adj) approximately 19.5 points higher than the aa genotype. The heterozygote genotype effect was about 72% of the homozygote effect, such that the mean phenotype among individuals with the putative Aa genotype was ~14 points higher than those with the aa genotype. Therefore this QTL model describes a mode of inheritance which can be described as a co-dominant model for the A allele, with the mean phenotype among heterozygotes closer to the high than the low genotype mean.
Fig. 2.
QTL models obtained in oligogenic segregation analysis (a–d), and in joint oligogenic segregation and linkage analysis (e–h). a, b Oligogenic segregation analysis with SpAvg(Adj) and SpAvg(VIQ), respectively, with scale of vertical axis denoting relative frequency of model parameters for a particular set of values for pairs of genotype effects; c, d contour plots of the same results shown in a, b; e QTL model obtained from chromosome 6 in joint segregation and linkage analysis on model SpAvg(Adj); and QTL models obtained for SpAvg(VIQ) on f chromosome 2, g chromosome 9, and h chromosome 15. Individual contour lines in all panels depict increasing relative frequency of QTL models, with individual lines incrementing at 0.75% of total QTLs in the analysis run
When we included VIQ as a covariate, the posterior model structure for the resulting adjusted trait, SpAvg(VIQ), was slightly more complicated. In this case, segregation analysis identified two notable QTLs in the posterior oligogenic model distribution. The first model (model 1) is essentially a dominant model with respect to the A allele with both homozygote and heterozygote effects of ~21–23 (Fig. 2b, d; Table 3). This model is similar to model 1 identified for SpAvg(Adj) in the difference between the AA and aa genotype means. However, the dominance structure is different than model 1 for SpAvg(Adj), as is the allele frequency (Table 3). The second model (model 2) is essentially an additive model, with a heterozygote effect (5.7) that is 48% of the homozygote effect (11.9). In addition, this model yielded an intermediate allele frequency for the A allele (Table 3).
Table 3.
Mean posterior model parameters for individual QTLs
| Analysis | Trait | Model | Chr | Location (cM) | pA (95% CI) | μAa − μaa (95% CI) | μAA − μaa (95% CI) |
|---|---|---|---|---|---|---|---|
| SA | SpAvg(Adj) | 1 | NA | NA | 0.40 (0.13, 0.67) | 14.1 (8.3, 19.9) | 19.5 (13.3, 25.7) |
| SpAvg(VIQ) | 1 | NA | NA | 0.21 (0.09, 0.32) | 20.8 (15.4, 26.1) | 22.9 (17.0, 28.8) | |
| 2 | NA | NA | 0.54 (0.08, 1.0) | 5.7 (0.9, 12.3) | 11.9 (5.6, 18.3) | ||
| SA + LA | SpAvg(Adj) | 1 | 6 | 167 | 0.43 (0.23, 0.64) | 14.9 (10.3, 19.5) | 19.3 (14.4, 24.2) |
| SpAvg(VIQ) | 1 | 2 | 137 | 0.24 (0.12, 0.36) | 18.6 (13.8, 23.5) | 21.8 (16.1, 27.5) | |
| 2 | 2 | 128 | 0.55 (0.18, 0.93) | 7.5 (3.2, 11.8) | 12.9 (8.9, 17.0) | ||
| 2 | 9 | 155 | 0.47 (0.16, 0.78) | 7.8 (3.7, 12.0) | 13.4 (9.7, 17.1) | ||
| 1 | 15 | 23 | 0.19 (0.10, 0.27) | 19.1 (14.3, 24.0) | 25.9 (20.6, 31.2) |
SA oligogenic segregation analysis, SA + LA joint oligogenic segregation and linkage analysis
Joint segregation and linkage analysis
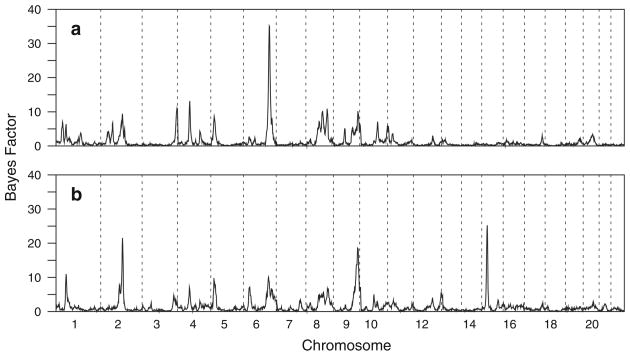
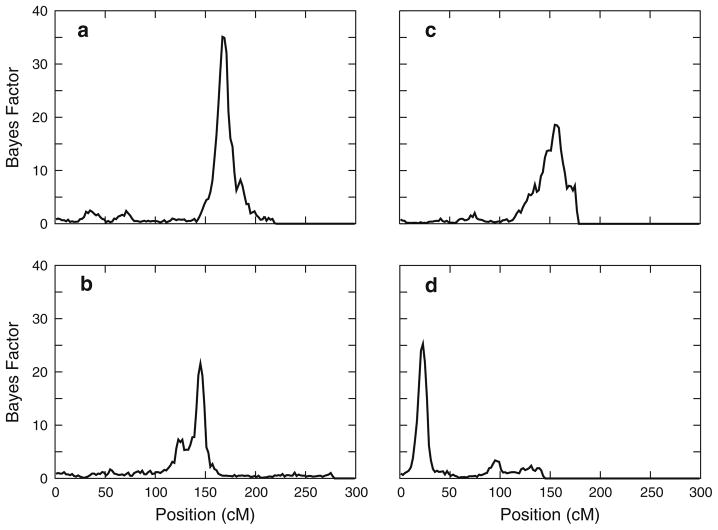
A genome scan using joint segregation and linkage analysis identified strong evidence for linkage of SpAvg(Adj) to a single region of the genome, on chromosome 6q (Fig. 3a). A strong maximum Bayes Factor of 35.1 was obtained at 167 cM (Fig. 4a), with the closest flanking markers D6S1277 and D6S1027. Consistent with previous simulations on this sample under the null hypothesis of no linkage (Igo and Wijsman 2008), a region-wide p-value for this Bayes Factor was found to be about 0.005. The genome scan also provided modest evidence for linkage of SpAvg(Adj) to chromosomes 3, 4, 8, and 9 (Bayes factors between 10 and 15), but these signals were not pursued.
Fig. 3.
Bayes Factors obtained across all autosomes. a Trait under analysis was SpAvg(Adj), b trait under analysis was SpAvg(VIQ). Vertical dotted lines delimit breaks between chromosomes
Fig. 4.
Bayes Factors on individual chromosomes with strongest evidence of linkage. a SpAvg(Adj) and chromosome 6, b SpAvg(VIQ) on chromosome 2, c SpAvg(VIQ) on chromosome 9, d SpAvg(VIQ) on chromosome 15
The genome scan for SpAvg(VIQ) identified three regions with notable evidence for linkage (Fig. 3b). The strongest evidence for linkage was found on chromosome 15q, with a maximum Bayes Factor of 25.3 (p = 0.005) at 23 cM (Fig. 4d), between flanking markers D15S822 and GATA50C03. There was also strong evidence for linkage to chromosome 2q, with a somewhat bimodal Bayes Factor distribution and a maximum Bayes Factor of 21.6 (p = 0.015), at 145 cM between flanking markers near D2S2972 and D2S1326 (Fig. 4b). Finally, there was evidence for linkage to chromosome 9, with a maximum Bayes Factor of 18.6 (p = 0.013) at 155 cM between flanking markers D9S934 and D9S1838 (Fig. 4c). Weaker evidence for linkage was also found on chromosomes 1 and 6 (Bayes Factors of 10–15), including the region on chromosome 6 with strong evidence for linkage for SpAvg(Adj), but these signals were not pursued.
The QTL model that explains the evidence for linkage of SpAvg(Adj) to chromosome 6 is essentially identical to QTL model 1 predicted in the oligogenic segregation analysis of SpAvg(Adj) (Fig. 2c vs. e; Table 3). Extraction of the model parameters from the posterior distribution of models with a location on chromosome 6 identified a single major QTL model. Similar to the model predicted by segregation analysis, the homozygote effect for this QTL is ~19.3, the heterozygote effect is 14.9 (or ~77% of the homozygote effect), and the allele frequency, pA = 0.43. This therefore provides evidence that the QTL model identified in the oligogenic segregation analysis in the absence of marker information maps to this region of chromosome 6 in this sample.
Extraction of model parameters from each of the individual chromosome analyses with strong evidence of linkage for SpAvg(VIQ) similarly identified models predicted by the oligogenic segregation analysis (Fig. 2). For chromosome 2, the pair of QTL models identified in oligogenic segregation analysis (Fig. 2b, d) is the same pair of QTL models as was identified by summarizing the parameters for the QTLs that localize to chromosome 2 (Fig. 2f; Table 3). There was evidence that this pair of models had different locations on chromosome 2: the mean location of QTLs that contribute to model 1 was at 137.4 cM, while the equivalent location for QTLs contributing to model 2 was at 127.9 cM, consistent with the bimodal Bayes Factor distribution (Fig. 4b). For chromosomes 9 and 15 only one of the two segregation analysis models explained the evidence for linkage to each chromosome. The model that explains the evidence for linkage to chromosome 9 is essentially the same as model 2 identified in oligogenic segregation analysis of SpAvg(VIQ) (Fig. 2d vs. g; Table 3), while QTLs with the parameter values for model 1 are essentially missing from chromosome 9. In contrast, for chromosome 15, QTLs with parameters similar to those of model 2 are missing, but there is a model that is similar to model 1 (Fig. 2d vs. h; Table 3). However, this strong model on chromosome 15 has slightly larger genotypic effects (25.9 and 19.1 for the homozygote and heterozygote effect) than does the segregation analysis model 1 (22.9 and 20.8, respectively), and also shows more evidence for a codominant model than does the segregation analysis model 1: the heterozygote effect in the context of linkage analysis was 74% of the homozygote effect, compared to 91% when obtained from segregation analysis alone. The allele frequency, pA = 0.19, remains close to that predicted by the segregation analysis, pA = 0.21.
Discussion
We have presented results of a genome scan for spelling ability in families collected through a proband with dyslexia. Our results suggest that the inheritance of spelling ability has a relatively simple underlying model structure in our sample, with evidence for four genomic regions that contain loci that contribute to its genetic basis. In addition, our results suggest that conclusions about the genetic basis depend on whether spelling scores are adjusted for VIQ, as we find evidence for linkage to chromosome 6q in the absence of VIQ adjustment, but we find evidence for linkage to chromosomes 2q, 9q and 15q only in the presence of VIQ adjustment, with only weak residual evidence of linkage to chromosome 6q. This suggests that VIQ could be a moderating factor for some, but not all, genetic contributions to spelling ability. For example, different mechanisms may be involved, such as differences in perception vs. retention of information. These results also suggest that strategies that both do and do not account for VIQ may have value in the search for genetic factors contributing to spelling ability and other measures related to dyslexia.
Our analyses identified simple QTL models that had clear locations, once the genome scan was completed. These results are similar to several others that have been obtained on this sample (Chapman et al. 2003b; Igo et al. 2006a; Raskind et al. 2005), as well as for other disorders (Gagnon et al. 2005; Marchani et al. 2010): in each case a small number of QTLs identified in segregation analysis lead to evidence of linkage of these QTLs, with typically a single QTL model mapping to a particular genomic region. These results illustrate an important advantage to using a computationally tractable approach for fitting oligogenic models in the absence of prior information about the model complexity. Identification of clear models in the oligogenic model space tends to result in genomic regions with evidence for linkage that can be explained by those models. Traits that do not have clear models in the initial oligogenic segregation analysis also tend to fail to yield positive evidence for linkage in a genome scan (results not shown). Use of oligogenic segregation analysis therefore provides a relatively fast way to screen potential traits for use in a genome scan in a particular sample. Also, for any complex multilocus trait, sampling variation is likely to yield different numbers of identifiable QTLs in different samples, and relative simplicity of the model space in a particular study should not be over-generalized to the general population without further investigation.
It is necessary to avoid over-interpreting the results of the models estimated here, however. The sample used here for analysis was selected through a proband with a reading disability. We were unable to apply an ascertainment correction both because of the complex ascertainment of the probands, and because the method of analysis does not yet have such an option. Therefore, certain parameters are likely to be highly biased. In particular, the allele frequency describing the genotypes attributable to low spelling scores are likely to be severely inflated. It is also possible that the apparent “recessivity” of some of the models may be in part a result of ascertainment through children. Also, the ability of a particular sample to resolve models in oligogenic segregation analysis is a function of the size of the sample and the sizes of the pedigrees in the sample. When the sample and constituent pedigrees are only of modest size, small differences in models may not be resolvable. This appeared to be the case here for the two similar models mapping to chromosomes 2 and 15; these were similar enough in the absence of marker data that they “merged” into a single model in the segregation analysis, only to be more differentiable once the markers provided additional information about the model parameters.
To the best of our knowledge, our results represent the first such genome scan of spelling ability that focused on a sample ascertained in part through spelling difficulties. The only other such genome scan for spelling ability was carried out in normal twins (Bates et al. 2007), and also highlighted regions with evidence for linkage on chromosomes 6q and 15q. The regions noted by Bates et al. do not appear to be identical to those identified in our scan, but both regions are sufficiently close that overlap cannot be ruled out. Notably, the region identified here on chromosome 15q is close to the region containing DYX1, which may have the best support for linkage across multiple linkage analyses involving dyslexia (Chapman et al. 2004; Fisher et al. 2002; Grigorenko et al. 1997; Smith et al. 1983), including positive results from one study that evaluated spelling ability (Schulte-Korne et al. 1998). This suggests that components of the genetic basis of spelling ability may be shared with other measures of dyslexia, which would not be at all surprising. However, we have also identified new candidate regions in our current scan, which now become candidates for investigation in other samples, particularly those that have spelling measures available.
Acknowledgments
We are grateful to the families for their willingness to devote the time necessary to participate in these studies. We appreciate the expert help of Department of Educational Psychology graduate students Sylvia Abbott, Allison Brooks, Ana Rueda Brown, Rebecca Brooksher, Julie Busse, Kristina Byrd, Belle Chenault, Gerry Curtin, Kate Eschen, Julie Gibson, Sarah Hellewege, Diana Hoffer, Renee Hartman, Stephanie King, Linelle Milatchov, Stacy Ogier, Tanya Prather, James Rodriguez and Jared Taylor, in administering the test battery. We also thank Jennifer Thomson for her help in working with families, supervising the testers, and entering data; John Wolff for providing technical help in processing all the blood and DNA samples; and Hiep Nguyen for providing computer support. This study was supported by R01 HD054562 from the National Institute of Child Health and Development, and T32 GM081062 from the National Institute of General Medical Sciences. Genome scan genotyping was provided by N01-HV-48141 from the National Institute of Heart, Lung, and Blood to the Marshfield Genotyping Service.
Contributor Information
Kevin Rubenstein, Department of Biostatistics, University of Washington, Box 357232, Seattle, WA, USA.
Mark Matsushita, Division of Medical Genetics, Department of Medicine, University of Washington, Box 357720, Seattle, WA 98195-7720, USA.
Virginia W. Berninger, Department of Educational Psychology, University of Washington, Box 353600, Seattle, WA, USA
Wendy H. Raskind, Division of Medical Genetics, Department of Medicine, University of Washington, Box 357720, Seattle, WA 98195-7720, USA, Department of Psychiatry and Behavioral Sciences, University of Washington, Seattle, WA, USA
Ellen M. Wijsman, Email: wijsman@u.washington.edu, Division of Medical Genetics, Department of Medicine, University of Washington, Box 357720, Seattle, WA 98195-7720, USA, 4333 Brooklyn Ave, NE, Box 989460, Seattle, WA 98195-9460, USA. Department of Biostatistics, University of Washington, Seattle, WA, USA
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos CI. Robust variance-components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- Bates TC, Luciano M, Castles A, Coltheart M, Wright MJ, Martin NG. Replication of reported linkages for dyslexia and spelling and suggestive evidence for novel regions on chromosomes 4 and 17. Eur J Hum Genet. 2007;15:194–203. doi: 10.1038/sj.ejhg.5201739. [DOI] [PubMed] [Google Scholar]
- Berninger VW, Abbott RD, Thomson JB, Raskind WH. Language phenotype for reading and writing disability: a life span approach. Sci Stud Read. 2001;5:59–105. [Google Scholar]
- Berninger VW, Abbott RD, Thomson J, Wagner R, Swanson HL, Wijsman E, Raskind W. Modeling phonological core deficits within a working memory architecture in children and adults with developmental dyslexia. Sci Stud Read. 2006;10:165–198. [Google Scholar]
- Berninger VW, et al. Writing problems in developmental dyslexia: under-recognized and under-treated. J Sch Psychol. 2008a;46:1–21. doi: 10.1016/j.jsp.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger VW, Nielsen KH, Abbott RD, Wijsman E, Raskind W. Gender differences in severity of writing and reading disabilities. J Sch Psychol. 2008b;46:151–172. doi: 10.1016/j.jsp.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Boehnke M, Moll P. Identifiying pedigrees segregating at a major locus for a quantitative trait: an efficient strategy for linkage analysis. Am J Hum Genet. 1989;44:216–224. [PMC free article] [PubMed] [Google Scholar]
- Brkanac Z, Chapman NH, Igo RP, Matsushita MM, Nielsen K, Berninger VW, Wijsman E. Genome scan of a Nonword Repetition phenotype in families with dyslexia: evidence for multiple loci. Behav Genet. 2008;38:462–475. doi: 10.1007/s10519-008-9215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck M. Word-recognition skills of adults with childhood diagnoses of dyslexia. Dev Psychol. 1990;26:439–454. [Google Scholar]
- Bruck M. Persistence of Dyslexics phonological awareness deficits. Dev Psychol. 1992;28:874–886. [Google Scholar]
- Bruck M. Component spelling skills of college-students with childhood diagnoses of dyslexia. Learn Disabil Q. 1993a;16:171–184. [Google Scholar]
- Bruck M. Word recognition and component phonological processing skills of adults with childhood diagnosis of dyslexia. Dev Rev. 1993b;13:258–268. [Google Scholar]
- Cannings SC, Thompson EA, Skolnick MH. Probability functions on complex pedigrees. Adv Appl Probab. 1978;10:26–61. [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading-disability on chromosome-6. Science. 1994;266:276–279. doi: 10.1126/science.7939663. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Smith SD, Fulker DW, Kimberling WJ, Pennington BF, DeFries JC. Quantitative trait locus for reading disability: correction. Science. 1995;268:1553. doi: 10.1126/science.7777847. [DOI] [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton D. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003a;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Chapman NH, Raskind WH, Thomson JB, Berninger VW, Wijsman EM. Segregation analysis of phenotypic components of learning disabilities. II. Phonological decoding. Am J Med Genet B. 2003b;121B:60–70. doi: 10.1002/ajmg.b.20068. [DOI] [PubMed] [Google Scholar]
- Chapman NH, Igo RPJ, Thomson JB, Matsushita MM, Brkanac Z, Holzman T, Berninger VW, Wijsman E, Raskind WH. Linkage analyses of four regions previously implicated in dyslexia: confirmation of a locus on chromosome 15q. Am J Med Genet B. 2004;131B:67–75. doi: 10.1002/ajmg.b.30018. [DOI] [PubMed] [Google Scholar]
- Daw EW, Chen SN, Czernuszewicz G, Lombardi R, Lu Y, Ma J, Roberts R, Shete S, Marian AJ. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Hum Mol Genet. 2007;16:2463–2471. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kovel CGF, Hol FA, Heister J, Willemen J, Sandkuijl LA, Franke B, Padberg GW. Genomewide scan identifies susceptibility locus for dyslexia on Xq27 in an extended Dutch family. J Med Genet. 2004;41:652–657. doi: 10.1136/jmg.2003.012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFries JC, Olson RK, Pennington BF, Smith SD. Colorado reading project: past, present, and future. Learning Disabilities. 1991;2:37–46. [Google Scholar]
- Fagerheim T, Raeymaekers P, Tønnessen FE, Pedersen M, Tranebjærg L, Lubs HA. A new gene (DYX3) for dyslexia is located on chromosome 2. J Med Genet. 1999;36:664–669. [PMC free article] [PubMed] [Google Scholar]
- Felton RH, Naylor CE, Wood FB. Neuropsychological profile of adult Dyslexics. Brain Lang. 1990;39:485–497. doi: 10.1016/0093-934x(90)90157-c. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Marlow AJ, Lamb J, Maestrini E, Williams DF, Richardson AJ, Weeks DE, Stein JF, Monaco AP. A quantitative-trait locus on chromosome 6p influences different aspects of developmental dyslexia. Am J Hum Genet. 1999;64:146–156. doi: 10.1086/302190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE, Francks C, Marlow AJ, MacPhie IL, Newbury DF, Cardon LR, Ishikawa-Brush Y, Richardson AJ, Talcott JB, Gayan J, Olson RK, Pennington BF, Smith SD, DeFries JC, Stein JF, Monaco AP. Independent genome-wide scans identify a chromosome 18 quantitative-trait locus influencing dyslexia. Nat Genet. 2002;30:86–91. doi: 10.1038/ng792. [DOI] [PubMed] [Google Scholar]
- Flannery KA, Liederman J, Daly L, Schultz J. Male prevalence for reading disability is found in a large sample of Black and White children free from ascertainment bias. J Int Neuropsychol Soc. 2000;6:433–442. doi: 10.1017/s1355617700644016. [DOI] [PubMed] [Google Scholar]
- Forrest WF, Feingold E. Composite statistics for QTL mapping with moderately discordant sibling pairs. Am J Hum Genet. 2000;66:1642–1660. doi: 10.1086/302897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon F, Jarvik GP, Badzioch MD, Motulsky AG, Brunzell JD, Wijsman EM. Genome scan for quantitative trait loci influencing HDL levels: evidence for multilocus inheritance in familial combined hyperlipidemia. Hum Genet. 2005;117:494–505. doi: 10.1007/s00439-005-1338-4. [DOI] [PubMed] [Google Scholar]
- Gayan J, Smith SD, Cherny SS, Cardon LR, Fulker DW, Brower AM, Olson RK, Pennington BF, DeFries JC. Quantitative-trait locus for specific language and reading deficits on chromosome 6p. Am J Hum Genet. 1999;64:157–164. doi: 10.1086/302191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilger JW, Borecki IB, DeFries JC, Pennington BF. Commingling and segregation analysis of reading performance in families of normal reading probands. Behav Genet. 1994;24:345–355. doi: 10.1007/BF01067536. [DOI] [PubMed] [Google Scholar]
- Grigorenko EL, Wood FB, Meyer MS, Hart LA, Speed WC, Shuster A, Pauls DL. Susceptibility loci for distinct components of developmental dyslexia on chromosomes 6 and 15. Am J Hum Genet. 1997;60:27–29. [PMC free article] [PubMed] [Google Scholar]
- Grossglenn K, Lewis DC, Smith SD, Lubs HA. Phenotype of adult familial dyslexia—reading of visually transformed texts and nonsense passages. Int J Neurosci. 1985;28:49–59. doi: 10.3109/00207458509070819. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. The combination of linkage values, and the calculation of distance between the loci of linked factors. J Genet. 1919;8:299–309. [Google Scholar]
- Hatcher J, Snowling MJ, Griffiths YM. Cognitive assessment of dyslexic students in higher education. Br J Educ Psychol. 2002;72:119–133. doi: 10.1348/000709902158801. [DOI] [PubMed] [Google Scholar]
- Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L, Wijsman EM, Berninger VW, Thomson JB, Raskind WH. Familial aggregation of dyslexia phenotypes. II: Paired correlated measures. Am J Med Genet. 2002;114:471–478. doi: 10.1002/ajmg.10523. [DOI] [PubMed] [Google Scholar]
- Huang S, Ballard D, Zhao H. The role of heritability in mapping expression quantitative trait loci. BMC Proc. 2007;1(Suppl 1):S86. doi: 10.1186/1753-6561-1-s1-s86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo RP, Wijsman EM. Empirical significance values for linkage analysis: trait simulation using posterior model distributions from MCMC oligogenic segregation analysis. Genet Epidemiol. 2008;32:119–131. doi: 10.1002/gepi.20267. [DOI] [PubMed] [Google Scholar]
- Igo RP, Chapman NH, Berninger VW, Matsushita M, Brkanac Z, Rothstein JH, Holzman T, Nielsen K, Raskind WH, Wijsman EM. Genomewide scan for real-word reading subphenotypes of dyslexia: novel chromosome 13 locus and genetic complexity. Am J Med Genet B. 2006a;141B:15–27. doi: 10.1002/ajmg.b.30245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo RP, Chapman NH, Wijsman EM. Segregation analysis of a complex quantitative trait: approaches for identifying influential data points. Hum Hered. 2006b;61:80–86. doi: 10.1159/000093085. [DOI] [PubMed] [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestila M, Lahermo P, Muller K, Kaaranen M, Myllyluoma B, Voutilainen A, Lyytinen H, Nopola-Hemmi J, Kere J. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40:340–345. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan D, Cooper R, Zhu X. A genome wide linkage study of GAW15 gene expression data. BMC Proc. 2007;1(Suppl 1):S87. doi: 10.1186/1753-6561-1-s1-s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R, Raftery A. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- Katusic SK, Colligan RC, Barbaresi WJ, Schaid DJ, Jacobsen SJ. Incidence of reading disability in a population-based birth cohort, 1976–1982, Rochester, Minn. Mayo Clin Proc. 2001;76:1081–1092. doi: 10.4065/76.11.1081. [DOI] [PubMed] [Google Scholar]
- Knopik VS, Smith SD, Cardon L, Pennington B, Gayan J, Olson RK, DeFries JC. Differential genetic etiology of reading component processes as a function of IQ. Behav Genet. 2002;32:181–198. doi: 10.1023/a:1016069012111. [DOI] [PubMed] [Google Scholar]
- Marchani EE, Di Y, Choi Y, Cheung C, Su M, Boehm F, Thompson EA, Wijsman EM. Contrasting IBD estimators, association studies, and linkage analysis using the Framingham Heart Study. BMC Proc. 2009;3:S102. doi: 10.1186/1753-6561-3-s7-s102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchani EE, Bird TD, Steinbart EJ, Rosenthal E, Yu CE, Schellenberg GD, Wijsman EM. Evidence for three loci modifying age-at-onset of Alzheimer’s disease in early-onset PSEN2 families. Am J Med Genet B. 2010;153B:1031–1041. doi: 10.1002/ajmg.b.31072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise TC, Chen F, Chen WW, De la Vega FM, Hansen M, He CS, Hyland FCL, Kennedy GC, Kong XY, Murray SS, Ziegle JS, Stewart WCL, Buyske S. A second-generation combined linkage-physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naples AJ, Chang JT, Katz L, Grigorenko EL. Same or different? Insights into the etiology of phonological awareness and rapid naming. Biol Psychol. 2009;80:226–239. doi: 10.1016/j.biopsycho.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopola-Hemmi J, Myllyluoma B, Haltia T, Taipale M, Ollikainen V, Ahonen T, Voutilainen A, Kere J, Widen E. A dominant gene for developmental dyslexia on chromosome 3. J Med Genet. 2001;38:658–664. doi: 10.1136/jmg.38.10.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöthen MM, Schulte-Körne G, Grimm T, Cichon S, Vogt IR, Muller-Myhsok B, Propping P, Remschmidt H. Genetic linkage analysis with dyslexia: Evidence for linkage of spelling disability to chromosome 15. Eur Child Adolesc Psychiatry. 1999;8:56–59. doi: 10.1007/pl00010696. [DOI] [PubMed] [Google Scholar]
- Pennington BF, McCabe LL, Smith SD, Lefly DL, Bookman MO, Kimberling WJ, Lubs HA. Spelling-errors in adults with a form of familial Dyslexia. Child Dev. 1986;57:1001–1013. doi: 10.1111/j.1467-8624.1986.tb00261.x. [DOI] [PubMed] [Google Scholar]
- Pennington B, Van Orden G, Smith S, Green P, Haith M. Phonological processing skills and deficits in adult dyslexics. Child Dev. 1990;61:1753–1778. [PubMed] [Google Scholar]
- Pennington BF, Gilger JW, Pauls D, Smith SA, Smith SD, DeFries JC. Evidence for major gene transmission of developmental dyslexia. JAMA. 1991;18:1527–1534. [PubMed] [Google Scholar]
- Ramus F, Rosen S, Dakin SC, Day BL, Castellote JM, White S, Frith U. Theories of developmental dyslexia: insights from a multiple case study of dyslexic adults. Brain. 2003;126:841–865. doi: 10.1093/brain/awg076. [DOI] [PubMed] [Google Scholar]
- Raskind W, Hsu L, Berninger V, Thomson J, Wijsman E. Familial aggregation of dyslexia phenotypes. Behav Genet. 2000;30:385–395. doi: 10.1023/a:1002700605187. [DOI] [PubMed] [Google Scholar]
- Raskind WH, Igo RP, Jr, Chapman N, Berninger VW, Matsushita M, Brkanac Z, Holzman T, Brown M, Thomson J, Wijsman EM. A genome scan in multigenerational families with dyslexia: identification of a novel locus on chromosome 2q that contributes to phonological decoding efficiency. Mol Psychiatry. 2005;10:699–711. doi: 10.1038/sj.mp.4001657. [DOI] [PubMed] [Google Scholar]
- Remschmidt H, Hennighausen K, Schulte-Korne G, Deimel W, Warnke A. The influence of different diagnostic approaches on familial aggregation of spelling disability. Eur Child Adolesc Psychiatry. 1999;8:13–20. doi: 10.1007/s007870050122. [DOI] [PubMed] [Google Scholar]
- Roeske D, Ludwig KU, Neuhoff N, Becker J, Bartling J, Bruder J, Brockschmidt FF, Warnke A, Remschmidt H, Hoffman P, Müller-Myhsok B, Nöthen MM, Schulte-Körne G. First genome-wide association scan on neurophysiological endophenotypes points to trans-regulation effects on SLC2A3 in dyslexic children. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.102. [DOI] [PubMed] [Google Scholar]
- Rosenthal EA, Wijsman EM. Joint linkage and segregation analysis under multiallelic trait inheritance: simplifying interpretations for complex traits. Genet Epidemiol. 2010;34:344–353. doi: 10.1002/gepi.20490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Caspi A, Fergusson D, Horwood LJ, Goodman R, Maughan B, Moffitt TE, Meltzer H, Carroll J. Sex differences in developmental reading disability—new findings from 4 epidemiological studies. JAMA. 2004;291:2007–2012. doi: 10.1001/jama.291.16.2007. [DOI] [PubMed] [Google Scholar]
- Schulte-Korne G, Grimm T, Nothen MM, Muller-Myhsok B, Cichon S, Vogt IR, Propping P, Remschmidt H. Evidence for linkage of spelling disability to chromosome 15. Am J Hum Genet. 1998;63:279–282. doi: 10.1086/301919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Korne G, Ziegler A, Deimel W, Schumacher J, Plume E, Bachmann C, Kleensang A, Propping P, Nothen MM, Warnke A, Remschmidt H, Konig IR. Interrelationship and familiality of dyslexia related quantitative measures. Ann Hum Genet. 2007;71:160–175. doi: 10.1111/j.1469-1809.2006.00312.x. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Konig IR, Plume E, Propping P, Warnke A, Manthey M, Duell M, Kleensang A, Repsilber D, Preis M, Remschmidt H, Ziegler A, Nothen MM, Schulte-Korne G. Linkage analyses of chromosomal region 18p11–q12 in dyslexia. J Neural Transm. 2006;113:417–423. doi: 10.1007/s00702-005-0336-y. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Hoffmann P, Schmal C, Schulte-Korne G, Nothen MM. Genetics of dyslexia: the evolving landscape. J Med Genet. 2007;44:289–297. doi: 10.1136/jmg.2006.046516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz S, Shaywitz B, Fletcher J, Esobar M. Prevalence of reading disability in boys and girls. JAMA. 1990;264:998–1002. [PubMed] [Google Scholar]
- Shaywitz S, Fletcher J, Holahan J, Schneider A, Marchione K, Stuebing K, Francis D, Pugh K, Shaywitz B. Persistence of dyslexia: the Connecticut longitudinal study at adolescence. Pediatrics. 1999;104:1351–1359. doi: 10.1542/peds.104.6.1351. [DOI] [PubMed] [Google Scholar]
- Smith S, Kimberling W, Pennington B, Lubs H. Specific reading disability: identification of an inherited form through linkage analysis. Science. 1983;219:1345–1347. doi: 10.1126/science.6828864. [DOI] [PubMed] [Google Scholar]
- Stevenson J, Graham P, Fredman G, McLoughlin V. A twin study of genetic influences on reading and spelling ability and disability. J Child Psychol Psychiatry. 1987;28:229–247. doi: 10.1111/j.1469-7610.1987.tb00207.x. [DOI] [PubMed] [Google Scholar]
- Turic D, Robinson L, Duke M, Morris DW, Webb V, Hamshere M, Milham C, Hopkin E, Pound K, Fernando S, Grierson A, Easton M, Williams N, Van Den Bree M, Chowdhury R, Gruen J, Stevenson J, Krawczak M, Owen MJ, O’Donovan MC, Williams J. Linkage disequilibrium mapping provides further evidence of a gene for reading disability on chromosome 6p21.3–22. Mol Psychiatry. 2003;8:176–185. doi: 10.1038/sj.mp.4001216. [DOI] [PubMed] [Google Scholar]
- Tzenova J, Kaplan BJ, Petryshen TL, Field LL. Confirmation of a dyslexia susceptibility locus on chromosome 1p34–p36 in a set of 100 Canadian families. Am J Med Genet B. 2004;127B:117–124. doi: 10.1002/ajmg.b.20139. [DOI] [PubMed] [Google Scholar]
- Wadsworth SJ, Defries JC, Stevenson J, Gilger JW, Pennington BF. Gender ratios among reading-disabled children and their siblings as a function of parental impairment. J Child Psychol Psychiatry. 1992;33:1229–1239. doi: 10.1111/j.1469-7610.1992.tb00941.x. [DOI] [PubMed] [Google Scholar]
- Wadsworth SJ, Olson RK, Pennington BF, DeFries JC. Differential genetic etiology of reading disability as a function of IQ. J Learn Disabil. 2000;33:192–199. doi: 10.1177/002221940003300207. [DOI] [PubMed] [Google Scholar]
- Wagner R, Torgesen J. The nature of phonological processing and its causal role in the acquisition of reading skills. Psychol Bull. 1987;101:192–212. [Google Scholar]
- Wechsler D. Wechsler adult intelligence scale—revised (WAIS-R) The Psychological Corporation; San Antonio: 1981. [Google Scholar]
- Wechsler D. Wechsler intelligence scale for children—third edition (WISC-III) The Psychological Corporation; San Antonio: 1992. [Google Scholar]
- Wechsler D. Wechsler individual achievement test. 2. The Psychological Corporation; San Antonio: 2002. [Google Scholar]
- Wijsman EM, Amos CI. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet Epidemiol. 1997;14:719–735. doi: 10.1002/(SICI)1098-2272(1997)14:6<719::AID-GEPI28>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Yu D. Joint oligogenic segregation and linkage analysis using Bayesian Markov chain Monte Carlo methods. Mol Biotechnol. 2004;28:205–226. doi: 10.1385/MB:28:3:205. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Peterson D, Leutenegger A-L, Thomson JB, Goddard KAB, Hsu L, Berninger VB, Raskind WH. Segregation analysis of phenotypic components of learning disabilities, I. Nonword memory and digit span. Am J Hum Genet. 2000;67:631–646. doi: 10.1086/303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM, Rothstein JH, Thompson EA. Multipoint linkage analysis with many multiallelic or dense diallelic markers: MCMC provides practical approaches for genome scans on general pedigrees. Am J Hum Genet. 2006;79:846–858. doi: 10.1086/508472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson G. Wide range achievement tests—revised (WRAT-R) Wide Range, Inc; Wilmington, DE: 1993. [Google Scholar]
- Willcutt EG, Pennington BF, DeFries JC. Twin study of the etiology of comorbidity between reading disability and attention-deficit/hyperactivity disorder. Am J Med Genet. 2000;96:293–301. doi: 10.1002/1096-8628(20000612)96:3<293::aid-ajmg12>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Williams J, Duggirala R, Blangero J. Statistical properties of a variance components method for quantitative trait linkage analysis in nuclear families and extended pedigrees. Genet Epidemiol. 1997;14:1065–1070. doi: 10.1002/(SICI)1098-2272(1997)14:6<1065::AID-GEPI84>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Wilson AM, Lesaux NK. Persistence of phonological processing deficits in college students with dyslexia who have age-appropriate reading skills. J Learn Disabil. 2001;34:394–400. doi: 10.1177/002221940103400501. [DOI] [PubMed] [Google Scholar]
- Wolff PH, Melngailis I. Family patterns of developmental dyslexia: clinical findings. Am J Med Genet B. 1994;54:122–131. doi: 10.1002/ajmg.1320540207. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Konig IR, Deimel W, Plume E, Nothen MM, Propping P, Kleensang A, Muller-Myhsok B, Warnke A, Remschmidt H, Schulte-Korne G. Developmental dyslexia—recurrence risk estimates from a German bi-center study using the single proband sib pair design. Hum Hered. 2005;59:136–143. doi: 10.1159/000085572. [DOI] [PubMed] [Google Scholar]