Abstract
Genetic variability at the 3′ region of SNCA locus has been repeatedly associated with susceptibility to sporadic Parkinson’s disease (PD). Accumulated evidence emphasizes the importance of SNCA dosage and expression levels in PD pathogenesis. However, the mechanism through which the 3′ region of SNCA gene modulates the risk to develop sporadic PD remained elusive. We studied the effect of PD risk-associated variants at SNCA 3′ regions on SNCA112-mRNA (exon 5 in-frame skipping) levels in vivo in 117 neuropathologically normal, human brain frontal cortex samples. SNPs tagging the SNCA 3′ showed significant effects on the relative levels of SNCA112-mRNA from total SNCA transcripts levels. The “risk” alleles were correlated with increased expression ratio of SNCA112-mRNA from total. We provide evidence for functional consequences of PD-associated SNCA gene variants at the 3′ region, suggesting that genetic regulation of SNCA splicing plays an important role in the development of the disease. Further studies to determine the definite functional variant/s within SNCA 3′ and to establish their association with PD pathology are necessary.
Keywords: SNCA-mRNA, SNCA112, Splicing, Parkinson’s disease, Functional variants, eSNP
Introduction
Alpha-synuclein (SNCA) (Ensembl—ENSG00000145335; OMIM, Online Mendelian Inheritance in Man—MIM 163890) was the first gene implicated in Parkinson’s disease (PD [MIM 168600]) [1]. The identification of SNCA gene multiplication as PD-causative [2] suggests that elevated levels of wild-type SNCA lead to neuronal dysfunction and are sufficient to cause early onset familial PD in a dose-dependent manner [3–8]. Furthermore, elevated levels of SNCA-mRNA have been reported in midbrain tissues [9] and in individual substantia nigra dopaminergic neurons from sporadic PD post mortem brains compared to controls [10]. Collectively, these observations emphasize the importance of SNCA dosage and expression levels in PD pathogenesis and suggest that the regulation of SNCA expression could have an important role in the development of the disease.
Genetic studies demonstrated that variants within the SNCA 5′ and 3′ linkage disequilibrium (LD) blocks are associated independently with increased susceptibility to sporadic PD in different populations [11–16]. The association of the SNCA locus with PD risk was further established in two recent genome-wide association studies [17, 18]. Recently, we and others studied the functional effect of the PD-associated genetic variability on the expression of full-length SNCA [19, 20]. In the 5′ region, we found that the Rep1 alleles’ effect on total levels of SNCA-mRNA is consistent with the common hypothesis that an increase in the expression of SNCA contributes to the common, “idiopathic” PD phenotype, while decreased SNCA levels reduce the risk of developing this devastating disease [20]. In contrast, the effect observed at the 3′ region runs contrary to the conventional hypothesis that higher SNCA expression confers PD risk, i.e., the “risk” alleles at the 3′ region were associated with lower total SNCA-mRNA levels. Therefore, we hypothesized that variant/s in the 3′ LD block mediates a regulatory effect not simply by changing total SNCA-mRNA levels but by a different molecular mechanism, such as splicing, to change the relative levels of the different splice forms.
SNCA post-transcriptional regulation, in particular alternative splicing, has shown disease-related differential expression [21]. Alternative SNCA splicing gives rise to three major isoforms: the full-length SNCA 140, and the truncated transcripts SNCA126 and SNCA112, resulting from in-frame deletions lacking exon 3 (SNCA 126) [22] or exon 5 (SNCA 112), respectively [23]. In the current study, we aimed to understand the cis- regulatory effect of variants within the 3′ region of SNCA gene; hence, we focused on exon 5 deletion splicing form. Interestingly, deletion of exon 5 predicts functional consequences relevant to Lewy body pathology, a hallmark of PD and related diseases [24]. The deletion results in significant shortening of the unstructured C-terminus and has been proposed to enhanced SNCA aggregation, which may lead to Lewy body formation [25, 26].
We aim to reveal the functional consequence of genetic variations in the SNCA 3′ region associated with PD. Specifically, we studied the effect of the PD risk-associated variants tagging the SNCA 3′ LD block on SNCA112-mRNA steady-state levels in vivo in human frontal cortex samples from unaffected individuals to directly assess the genetic contribution to the regulation of SNCA splicing, avoiding other confounding factors arising from the neuro-degeneration associated with PD.
Materials and methods
Brain samples
Frontal cortex (n=117) from neurologically healthy controls (Table 1) were obtained through the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University, the Brain and Tissue Bank for developmental Disorders at the University of Maryland, the Layton Aging & Alzheimer’s Disease Center at Oregon Health and Science University, and the National NeuroAIDS Tissue Consortium (NNTC). All post mortem intervals (PMI) were <24 h. Demographics for these samples are included in Table 1. All brain samples were collected from clinically and neuropathologically healthy cadavers who had no evidence of PD, AD, or other neurodegenerative disorder at post mortem examination.
Table 1.
Demographic description of the brain samples
FC frontal cortex
PMI post mortem interval
Other ethnicities: African American, Asian
DNA extraction and genotyping
Genomic DNA was extracted from brain tissues by the standard Qiagen protocol. Genotype determination of each single-nucleotide polymorphism (SNP) was performed by allelic discrimination using TaqMan SNP genotyping assays (Applied Biosystems, Foster City, CA, USA). Each genomic DNA sample (20 ng) was amplified using TaqMan Universal PCR master mix reagent (Applied Biosystems) combined with the specific TaqMan SNP genotyping assay mix corresponding to the genotyped SNP. The assays were carried out using the ABI 7900HT and the following conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles: 15 s at 95°C, and 1 min at 60°C. Genotype determination was performed automatically using the SDS version 2.2 Enterprise Edition Software Suite (Applied Biosystems). All genotypes were tested for Hardy–Weinberg equilibrium (Table 2).
Table 2.
The analyzed SNPs at SNCA 3′ region
| rs Number | Map position (bp) | Distance from previous (bp) | SNP type | Allele (minor/major) | MAFa | HWb (p) | 112/total (p) |
|---|---|---|---|---|---|---|---|
| rs2736990 | 90897564 | Intron4 | G/A | 0.442 | 0.6594 | 0.03 | |
| rs3857059 | 90894261 | 3303 | Intron4 | G/A | 0.111 | 1 | 0.20 |
| rs17016074 | 90866301 | 27,960 | 3′UTR | A/G | 0.016 | 1 | 0.56 |
| rs356165 | 90865909 | 392 | 3′UTR | G/A | 0.421 | 0.7409 | 0.01 |
| rs356219 | 90856624 | 9,285 | Downstream | G/A | 0.368 | 0.5747 | 0.009 |
MAF minor allele frequency
HW p values for Hardy–Weinberg Equilibrium
RNA extraction and cDNA synthesis
Total RNA was extracted from brain samples (100 mg) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) followed by purification with an RNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. RNA concentration was determined spectrophotometrically at 260 nm, while the quality of the purification was determined by 260 nm/280 nm ratio that showed values between 1.9 and 2.1, indicating high RNA quality. Additionally, quality of sample and lack of significant degradation products was confirmed on an Agilent Bio-analyzer. The RNA integrity number (RIN) measurements were greater than 7, validating the RNA quality control. Next, cDNA was synthesized using MultiScribe RT enzyme (Applied Biosystems) under the following conditions: 10 min at 25°C and 120 min at 37°C.
Real-time PCR
Real-time PCR was used to quantify the ratio levels of human SNCA112-mRNA [9, 27]. Briefly, duplicates of each sample were assayed by relative quantitative real-time PCR using the ABI 7900HT for analysis of the level of SNCA112 message as compared in brain tissues to mRNAs encoding the human neuronal proteins enolase 2 (ENO2) and synaptophysin (SYP). ABI MGB probe and primer set assays were used to amplify SNCA112 cDNA (custom assay, 81 bp) and the two RNA reference controls, ENO2 (ID Hs00157360_m1, 77 bp) and SYP (ID Hs00300531_m1, 63 bp) (Applied Biosystems). The custom assay to detect specifically the alternative splicing variant, SNCA112, was designed to target the novel exons 4 and 6 junction. Each cDNA (10 ng) was amplified in duplicate in at least two independent runs (overall ≥4 repeats), using TaqMan Universal PCR master mix reagent (Applied Biosystems) and the following conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles: 15 s at 95°C, and 1 min at 60°C. As a negative control for the specificity of the amplification, we used RNA control samples that were not converted to cDNA (no-RT) and no-cDNA/RNA samples (no-template) in each plate. No observable amplification was detected. Data were analyzed with a threshold set in the linear range of amplification. The cycle number at which any particular sample crossed that threshold (Ct) was then used to determine fold difference, whereas the geometric mean of the two control genes served as a reference for normalization. Fold difference was calculated as 2−ΔΔCt; ΔCt=[Ct (SNCA112)−Ct (reference)]. ΔΔCt=[ΔCt (sample)]−[ΔCt (calibrator)]. The calibrator was a particular brain RNA sample used repeatedly in each plate for normalization within and across runs. The variation of the ΔCt values among the calibrator replicates was smaller than 10%. Total SNCA-mRNA levels were measured as previously described (ID Hs00240906_m1) [27]. The relative ratios of the 112 transcripts from total SNCA-mRNA levels were then determined by calculating for each sample the ratio 2−ΔΔCt (SNCA112)/2−ΔΔCt (total SNCA).
For assay validation, we generated standard curves for SNCA112 and each reference assay, ENO2 and SYP, using different amounts of human brain total RNA (0.1–100 ng). In addition, the slope of the relative efficiency plot for SNCA112 with each internal control (ENO2 or SYP) was determined to validate the assays. The slope in the relative efficiency plot for SNCA112 and the reference genes were <0.1, showing a standard value required for the validation of the relative quantitative method.
Statistical analysis
All analyses were carried out using SAS statistical software, version 9.1 (SAS Institutes, Cary, NC, USA). Ratio values of SNCA112 mRNA expression from total SNCA of each sample were analyzed repetitively and the results of all replicates were averaged. All average values were expressed as mean±SE. Correlations of SNCA genotypes with expression trait (SNCA112/total) were assessed using the Generalized Linear Models procedure (PROC GLM). A log transformation (log2) was performed on all mRNA levels to assure normal distribution [28]. Genotypes were coded in the additive model, and for SNPs with low minor allele frequency (rs3857059 rs17016074) genotypes were also coded as a dominant model pooling homozygous for minor alleles and heterozygous genotypes. All models adjusted for gender, age, ethnicity, PMI, and tissue source. Correction for multiple testing employed the Bonferroni method.
Linkage disequilibrium (r2) within the studied 3′ region of SNCA was determined using Haploview software (version 4.2) [29].
Results
SNCA112-mRNA ratio levels (112/total) were measured in 117 frontal cortex tissue samples obtained from neurologically healthy subjects (81% white, 53.5% males; Table 1).
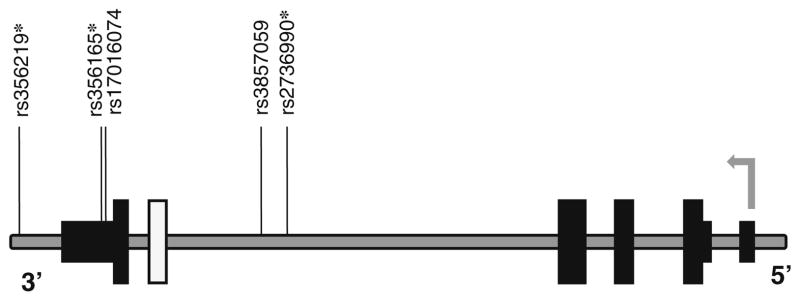
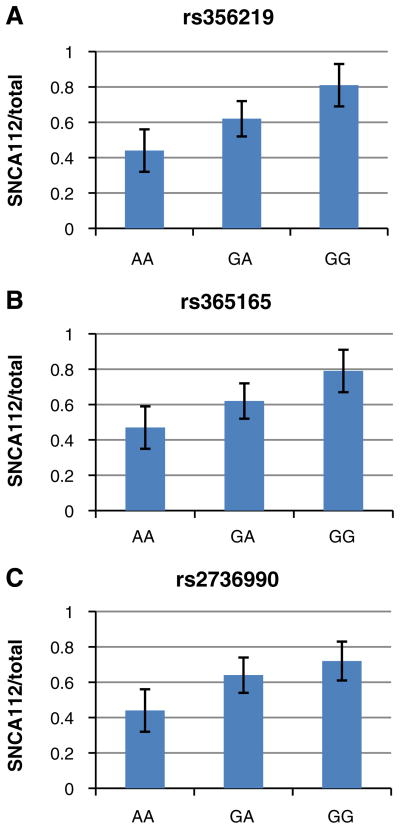
The effect of the SNCA-3′ region on the relative levels of SNCA 112-mRNA was tested with five tagging SNPs: two SNPs, rs2736990 and rs3857059, positioned within the 3′ end of intron 4; rs17016074 and rs356165 at the 3′UTR; and rs356219 located 3′ of SNCA gene (Fig. 1 and Table 2). The three SNPs rs2736990, rs356165, and rs356219 showed a significant effect on SNCA112-mRNA ratio levels in the frontal cortex (p=0.03, 0.01, and 0.009; Table 2 and Fig. 3). The two SNPs rs356165 and rs356219 are in high LD (r2=0.76), hence likely tagging a common genetic effector (Fig. 2). The LD of SNP rs2736990 with both rs356165 and rs356219 was not as high (r2=0.61 and 0.45, respectively; Fig. 2); however, it may also indicate the same splicing effector variant. In adjusted analysis, the PD “risk” allele “G” at SNP rs356219 correlated in an additive manner with higher ratio levels of 112SNCA-mRNA to total SNCA-mRNA than the PD-“protective” allele “A”, amounting to a nearly two-fold overall difference between the “risk” and the “protective” homozygous genotypes (GG vs. AA). The correlation of the different genotypes with the relative levels of alternative splicing was linear, i.e., rs356219 homozygous of the “risk” GG genotype (n=18) showed the highest expression levels of SNCA112-mRNA, followed by the heterozygotes GA (n=58), and the “protective” homozygote genotype AA resulted in the lowest levels (n=41) (Fig. 3a). A similar effect in the frontal cortex was observed with SNPs rs356165 and rs2736990. Mean adjusted SNCA 112 mRNA ratio levels were correlated in an additive mode with SNP rs356165 (G/A) and SNP rs2736990 (G/A) genotypes (Fig. 3b, c), following the same linear correlations of genotypes to SNCA 112 mRNA expression ratio observed for rs356219. Accordingly, the rs365165 and rs2736990 homozygotes of the “risk” allele (GG, n=25, 29) were correlated with higher SNCA 112 mRNA ratio levels than the heterozygotes (GA, n=54, 53), and the homozygotes of the “protective” allele (AA, n=38, 35) correlated with the lowest expression levels (Fig. 3b, c). No correlation with the ratio expression levels of the alternative splicing form—SNCA112 mRNA was observed in the frontal cortex for either SNP rs3857059 or the rare (<5%) SNP rs17016074 (p=0.20 and p=0.56, respectively). A summary of the results is listed in Table 3.
Fig. 1.
A schematic representation of the human SNCA gene with the relative positions of the SNPs. Organization of the human SNCA locus: the alternative exon 5, wide white solid box; other translated exons, wide black solid boxes; 5′ and 3′UTR, narrow black solid boxes; introns and intergenic regions, gray line. The SNCA gene is in the minus strand, the 5′ and 3′ indicate the gene’s orientation. The gray arrow marks the transcription start site and direction. The relative positions of the genetic variants are indicated above; asterisks designate variants that were associated with ratio of SNCA112-mRNA
Fig. 3.
Effect of SNP genotypes at SNCA 3′ region on human SNCA112-mRNA expression levels relative to from total SNCA-mRNA levels in human frontal cortex. Individuals were genotyped for SNPs rs356219 (a), rs356165 (b), and rs2736990 (c). Fold levels of human SNCA112-mRNA in the frontal cortex were assayed by real-time RT–PCR using TaqMan technology and calculated relative the geometric mean of SYP and ENO mRNAs reference control using the 2−ΔΔCt method (i.e., results presented are relative to a specific brain RNA sample). The values presented here are mean ratios from total SNCA-mRNA levels, adjusted for age, gender, ethnicity, PMI, and source. a Analysis of rs356219 showed that the risk allele G significantly correlates in an additive manner with higher ratio of SNCA112-mRNA levels (p= 0.009). b Analysis of rs356165 showed that the risk allele G significantly correlates in an additive manner with higher ratio of SNCA112-mRNA levels (p=0.01). c Analysis of rs2736990 showed that the risk allele G significantly correlates in an additive manner with higher ratio of SNCA112-mRNA levels (p=0.03)
Fig. 2.
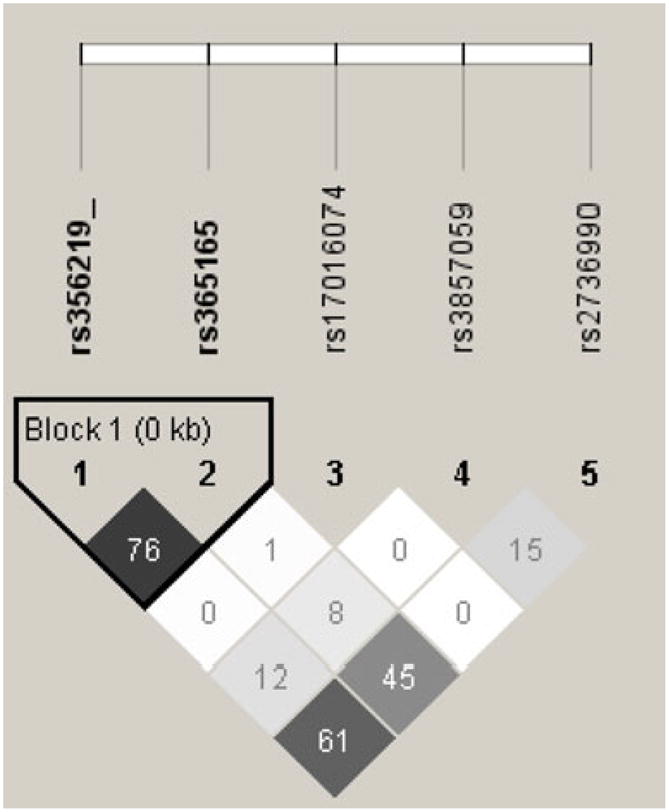
Linkage disequilibrium (LD) structure of the 3′ region of SNCA gene. Genotypes of five SNPs from the sample set of this study (N=117) were used to determine LD (r2) within the SNCA 3′ region using Haploview software. r2 values are shown within cells. Black cells, strong LD; shaded of gray cells, intermediate; white cells, evidence of recombination
Table 3.
Summary of the genetic correlations between genetic variants at SNCA locus and SNCA112-mRNA levels
Significant correlation
No correlation
Discussion
In this report, we attempted to elucidate the molecular mechanism underlying the reported genetic association between SNCA 3′ region and PD by examining variation in RNA splicing of SNCA as a function of genotype in the brains of unaffected people (age matched to late-onset PD). In particular, our study looked for variations in alternative splicing ratio levels of exon 5 deletion in correlations to PD-associated variants that are likely to tag putative splicing regulatory regions upstream and downstream of the alternative exon 5. All variants chosen for the present study had been repeatedly reported to confer increased risk for developing PD [12–16]. Moreover, the studied SNPs were also associated with another known synucleinopathy, multiple system atrophy (MSA) [30], suggesting a possible common molecular mechanism for this group of diseases.
We found evidence for a regulatory role for three of the five tested SNPs (Table 2). The “risk” allele at the 3′ region defined by each of these three tagging SNPs was associated with higher SNCA112-mRNA ratio levels in the human brain tissues, while the “protective” allele correlated with lower relative 112 transcripts levels. The effect was dose dependent with the copy number of the “risk” alleles, i.e., homozygotes genotype for the “risk” allele showed higher expression levels than the heterozygous genotype. Interestingly, it has been suggested that exon 5 deletion (SNCA112) results in enhanced SNCA aggregation due to a significant shortening of the unstructured C-terminus [25, 26] and therefore higher proportion of the 112 isoform may increase PD risk. Our current study demonstrated for the first time the disease-relevant biological consequence of the 3′ region of SNCA gene and showed that the PD genetic association at this region correlates with regulation of SNCA exon 5 skipping levels which may influence SNCA protein aggregation.
Recently, we and others reported that these SNPs at SNCA 3′ region correlate with overall SNCA-mRNA levels; however, these findings ran contrary to the conventional hypothesis that higher SNCA expression confers PD risk [19, 20]. We therefore hypothesized that genetic variability in the SNCA 3′ region exerts a regulatory effect not simply by changing total SNCA-mRNA levels but by a different molecular mechanism, such as splicing, to change the relative levels of the different splice forms [20]. In this study, we tested this hypothesis and demonstrated that alteration in specific SNCA-mRNA isoform levels might contribute in some extent to “idiopathic” PD pathogenesis.
Our study examined several SNPs for association with expression and, as such, is subject to false-positive associations. Therefore, p values should be interpreted with caution. After Bonferroni adjustment for five hypotheses (five variants), the associations with rs356219, rs365165 variants remained significant while the correlation with rs2736990 became non-significant. However, given that these were not independent hypotheses (SNPs in linkage disequilibrium), this may be an overly conservative interpretation. Nevertheless, the results of this study suggest an association between specific variants in the 3′ region of SNCA and expression of SNCA splicing isoform that warrants further investigation in follow-up functional studies. The five analyzed SNPs are located within evolutionary highly conserved regions; however, none of them is positioned at splice sites and hence does not disrupt splicing consensus sequences. Nevertheless, a search using ESEfinder 3.0 found potential splicing enhancer/silencer cis- elements within the sequences surrounding the SNPs. In addition, this search identified SR protein binding motifs (in particular for SRp40) that might be created or disrupted by the SNPs (data not shown). Therefore, we suggest using deep re-sequencing analysis of the expression associated region defined in this study to identify additional candidate causal variant/s that will be pursued in follow-up studies, including cell-based reporter system and animal models, in order to elucidate their effect on splicing regulation. Better understanding of the molecular mechanisms modulating SNCA gene splicing will further our understanding of the underlying biology of the genetic associations with PD and other synucleinopathies and may lead to novel therapeutic approaches targeting specific SNCA isoforms.
Acknowledgments
This work was supported in part by the Ellison Medical Foundation New Scholar award AG-NS-0441-08 (O.C.) and the Institute for Genome Sciences and Policy at Duke University (O.C.). We thank the Kathleen Price Bryan Brain Bank (KPBBB) at Duke University funded by NIA AG028377, the National NeuroAIDS Tissue Consortium (NNTC), Layton Aging & Alzheimer’s Disease Center at Oregon Health and Science University funded by NIA AG008017, and the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore (NICHD contract no. HHSN275200900011C, Ref. No. NO1-HD-9-0011), for providing us with the brain tissues. We would also like to thank Dr. Randy Woltjer, Dr. Kathleen Hayden, Dr. Lauren Warren, Mari Szymanski, and John Ervin for their assistance in obtaining the required brain samples for the study.
Footnotes
Ethical standards The experiments comply with the current laws of the USA.
Conflict of interest The authors declare that they have no conflict of interest.
Contributor Information
Jeanette J. McCarthy, Institute for Genome Sciences & Policy, Duke University, DUMC, Box 3445, Durham, NC 27710, USA
Colton Linnertz, Institute for Genome Sciences & Policy, Duke University, DUMC, Box 3445, Durham, NC 27710, USA.
Laura Saucier, Institute for Genome Sciences & Policy, Duke University, DUMC, Box 3445, Durham, NC 27710, USA.
James R. Burke, Division of Neurology, Department of Medicine, Duke University Medical Center, Durham, NC 27710, USA. Joseph and Kathleen Bryan Alzheimer’s Disease Research Center, Duke University, Durham, NC 27705, USA
Christine M. Hulette, Joseph and Kathleen Bryan Alzheimer’s Disease Research Center, Duke University, Durham, NC 27705, USA
Kathleen A. Welsh-Bohmer, Joseph and Kathleen Bryan Alzheimer’s Disease Research Center, Duke University, Durham, NC 27705, USA
Ornit Chiba-Falek, Email: o.chibafalek@duke.edu, Institute for Genome Sciences & Policy, Duke University, DUMC, Box 3445, Durham, NC 27710, USA, Division of Neurology, Department of Medicine, Duke University Medical Center, Durham, NC 27710, USA. Joseph and Kathleen Bryan Alzheimer’s Disease Research Center, Duke University, Durham, NC 27705, USA.
References
- 1.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 2.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 3.Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 4.Miller DW, Hague SM, Clarimon J, Baptista M, Gwinn-Hardy K, et al. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62:1835–1838. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 5.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 6.Ibanez P, Bonnet AM, Debarges B, Lohmann E, Tison F, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364:1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs J, Nilsson C, Kachergus J, Munz M, Larsson EM, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68:916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 8.Ross OA, Braithwaite AT, Skipper LM, Kachergus J, Hulihan MM, et al. Genomic investigation of alpha-synuclein multiplication and parkinsonism. Ann Neurol. 2008;63:743–750. doi: 10.1002/ana.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiba-Falek O, Lopez GJ, Nussbaum RL. Levels of alpha-synuclein mRNA in sporadic Parkinson disease patients. Mov Disord. 2006;21:1703–1708. doi: 10.1002/mds.21007. [DOI] [PubMed] [Google Scholar]
- 10.Grundemann J, Schlaudraff F, Haeckel O, Liss B. Elevated alpha-synuclein mRNA levels in individual UV-laser-microdissected dopaminergic substantia nigra neurons in idiopathic Parkinson’s disease. Nucleic Acids Res. 2008;36:e38. doi: 10.1093/nar/gkn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. Jama. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 12.Pals P, Lincoln S, Manning J, Heckman M, Skipper L, et al. alpha-Synuclein promoter confers susceptibility to Parkinson’s disease. Ann Neurol. 2004;56:591–595. doi: 10.1002/ana.20268. [DOI] [PubMed] [Google Scholar]
- 13.Mueller JC, Fuchs J, Hofer A, Zimprich A, Lichtner P, et al. Multiple regions of alpha-synuclein are associated with Parkinson’s disease. Ann Neurol. 2005;57:535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 14.Mizuta I, Satake W, Nakabayashi Y, Ito C, Suzuki S, et al. Multiple candidate gene analysis identifies alpha-synuclein as a susceptibility gene for sporadic Parkinson’s disease. Hum Mol Genet. 2006;15:1151–1158. doi: 10.1093/hmg/ddl030. [DOI] [PubMed] [Google Scholar]
- 15.Winkler S, Hagenah J, Lincoln S, Heckman M, Haugarvoll K, et al. alpha-Synuclein and Parkinson disease susceptibility. Neurology. 2007;69:1745–1750. doi: 10.1212/01.wnl.0000275524.15125.f4. [DOI] [PubMed] [Google Scholar]
- 16.Myhre R, Toft M, Kachergus J, Hulihan MM, Aasly JO, et al. Multiple alpha-synuclein gene polymorphisms are associated with Parkinson’s disease in a Norwegian population. Acta Neurol Scand. 2008;118:320–327. doi: 10.1111/j.1600-0404.2008.01019.x. [DOI] [PubMed] [Google Scholar]
- 17.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 18.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs J, Tichopad A, Golub Y, Munz M, Schweitzer KJ, et al. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 20.Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS ONE. 2009;4:e7480. doi: 10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beyer K, Domingo-Sabat M, Humbert J, Carrato C, Ferrer I, et al. Differential expression of alpha-synuclein, parkin, and synphilin-1 isoforms in Lewy body disease. Neurogenetics. 2008;9:163–172. doi: 10.1007/s10048-008-0124-6. [DOI] [PubMed] [Google Scholar]
- 22.Campion D, Martin C, Heilig R, Charbonnier F, Moreau V, et al. The NACP/synuclein gene: chromosomal assignment and screening for alterations in Alzheimer disease. Genomics. 1995;26:254–257. doi: 10.1016/0888-7543(95)80208-4. [DOI] [PubMed] [Google Scholar]
- 23.Ueda K, Saitoh T, Mori H. Tissue-dependent alternative splicing of mRNA for NACP, the precursor of non-A beta component of Alzheimer’s disease amyloid. Biochem Biophys Res Commun. 1994;205:1366–1372. doi: 10.1006/bbrc.1994.2816. [DOI] [PubMed] [Google Scholar]
- 24.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, et al. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 25.Beyer K. Alpha-synuclein structure, posttranslational modification and alternative splicing as aggregation enhancers. Acta Neuropathol. 2006;112:237–251. doi: 10.1007/s00401-006-0104-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277:671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 27.Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18:3274–3285. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Scholz SW, Houlden H, Schulte C, Sharma M, Li A, et al. SNCA variants are associated with increased risk for multiple system atrophy. Ann Neurol. 2009;65:610–614. doi: 10.1002/ana.21685. [DOI] [PMC free article] [PubMed] [Google Scholar]