Abstract
The function of the human voltage-gated sodium channel NaV1.5 is regulated in part by intracellular calcium signals. The ubiquitous calcium sensor protein calmodulin (CaM) is an important part of the complex calcium-sensing apparatus in NaV1.5. CaM interacts with an IQ motif in the large intracellular C-terminal domain of the channel. Using co-expression and co-purification, we have been able to isolate a CaM-IQ motif complex and to determine its high-resolution structure in absence of calcium using multi-dimensional solution NMR. Under these conditions, the NaV1.5 IQ motif interacts with the C-terminal domain (C-lobe) of CaM, with the N-terminal domain remaining free in solution. The structure reveals that the C-lobe adopts a semi-open conformation with the IQ motif bound in a narrow hydrophobic groove. Sequence similarities between voltage-gated sodium channels and voltage-gated calcium channels suggest that the structure of the CaM-NaV1.5 IQ motif complex can serve as a general model for the interaction between CaM and ion channels IQ motifs under conditions of low calcium. The structure also provides insight into the biochemical basis for disease-associated mutations that map to the IQ motif in NaV1.5.
Keywords: Calmodulin, protein-protein interaction, calcium signaling, calcium sensor, hydrophobic anchors, Long QT syndrome
Introduction
The cardiac voltage-gated sodium channel NaV1.5 is an essential element of regulation of the beating of the heart, as it is involved in the initial phase of the action potential. Upon activation, NaV1.5 opens and allows sodium ions to pass through the cell membrane according to their electrochemical gradient. Nav1.5 will then close and stop sodium ion flow. This channel is mainly activated by change in membrane potential, but recent studies showed that intracellular calcium also plays an important role in its gating mechanism. The structure of NaV1.5 is composed of one alpha chain divided in four homologous domains (DI, DII, DIII, DIV) with distinct N and C-termini. The domains, each containing six trans-membrane helices, are linked by intracellular loops. The loop between domains DIII and DIV has a very important role in the channel function as it is involved in the phenomenon of fast-inactivation of the channel.1 The N- and C-terminal domains are also intracellular. Several mutations located in these domains are associated with diseases such as long QT syndrome, Brugada syndrome and atrial or ventricular fibrillations.2-7
Although membrane potential variations are the primary stimulus for NaV1.5 activation, calcium ions are also able to modulate channel gating. Recent studies from our lab and others8-13 have shown that calcium sensitivity involves three domains of NaV1.5: the DIII-DIV linker and two regions in the C-terminal domain- an IQ motif and an EF-hand domain (CTD-EF). The IQ motif is a key element in the calcium sensing apparatus of NaV1.5 as it serves as a molecular switch that couples the actions of the Ca2+ sensing modules calmodulin (CaM) and CTD-EF.8,12
IQ motifs are well-characterized CaM binding motifs that are associated with localization of CaM to sites of action in the cell. CaM, a member of the EF-hand family of calcium-binding proteins, is a ubiquitous protein that serves as a primary calcium signal transducer, and it is involved in a multitude of biological processes. CaM is composed of two homologous structural domains (commonly referred to as lobes). The N-lobe (residues 1-76) and C-lobe (residues 80-148) are tethered together by a short linker, and each contains a pair of helix-loop-helix EF-hands (Ca2+ binding motifs). CaM has been shown to interact with the NaV1.5 IQ motif in the absence and presence of calcium,8,10 and it serves as an extrinsic calcium sensor for NaV1.5 that complements the intrinsic CTD-EF calcium sensor.
In this manuscript we present the NMR solution structure of human calmodulin in complex with IQ motif of human NaV1.5 in the absence of calcium. While several structures of calmodulin in complex with ion channel IQ motifs exist in the presence of calcium, to our knowledge this is the first structure of such a complex in the apo-state.
Results
Structure of the calmodulin-NaV1.5 IQ motif complex
The three-dimensional solution structure of human apo-calmodulin in complex with the human voltage-gated sodium channel NaV1.5 IQ motif (residues Glu1901-Ser1927) was determined using multi-dimensional heteronuclear NMR spectroscopy. Nearly complete 1H, 13C, and 15N resonance assignments were obtained using a series of standard double and triple resonance experiments as described in Methods. This resulted in >98% of all protons assigned. The majority of the missing assignments were in the third calcium binding loop of CaM as a result of significant line broadening.
The combination of specific patterns of medium range NOEs, a Chemical Shift Index (CSI) analysis,14 and {1H}-15N heteronuclear NOEs (Figures 1, S1) indicate that CaM residues Gln3-Lys147 and IQ motif residues Glu1901-Lys1922 form stable structures with regular secondary structure. The N- and C-termini (Asp2 and Lys148) of CaM and of the IQ motif (Gly -2, Ser -1, His1923-Leu1927) are disordered. Residues located in the CaM calcium binding loops (Lys21, Asp22, Gly23, Asp24, Asp56, Ala57, Asp58, Asn60, Lys94, Asp95 and Ile130) and those located in the linker (Asp78, Thr79 and Asp80) between N- and C- lobes give rise to lower NOE values that are indicative of above average flexibility (Figure 1). Qualitative analysis of the pattern of medium range NOEs combined with the chemical shift index (CSI) revealed the presence of the expected eight α helices and 2 cross-strand β-type interactions between paired calcium binding loops in CaM and one α helix in the IQ motif (Figure S1). Helical structure in the NaV1.5 IQ motif is consistent with observations made for many CaM binding peptides isolated from target proteins, for example as seen in the structures of Ca2+-CaM bound to IQ motif peptides from CaV1.1 (PDB ID: 2VAY),15 CaV1.2 (PDB IDs: 2F3Y 2BE6),16,17 Cav2.1 (PDB ID: 3BXK),18 Cav2.2 (PDB ID: 3DVE),19 and CaV2.3 (PDB IDs : 3DVK,3BXL),18,19 as well as peptides from calmodulin kinase II (PDB ID : 3GP2) and myosin light chain kinase (PDB ID : 2BBM)20. Helical conformation is observed even with larger fragments and intact target proteins such as the edema factor from Bacillus anthracis (PDB ID: 1XFV)21 and Death Associated Protein kinase 1(PDB ID: 2X0G),22 and in the apo-CaM complex with myosin (PDB ID: 2IX7).23
Figure 1.
Distribution of proton NOEs and correlation with heteronuclear NOEs for the complex of human CaM with the NaV1.5 IQ motif. Histogram of 15N NOE (top) and all protein NOEs restraints (bottom). Intraresidue NOEs are in white, sequential in light gray, medium-range in dark gray, and long-range in black. Calmodulin and IQ motif regions are labeled.
The three-dimensional structure of the complex of human CaM with the NaV1.5 IQ motif was calculated using a two-step protocol consisting of the generation of initial structures using CYANA24 followed by rMD refinement using AMBER,25 as described previously.12,26,27 The 3054 NOE based distance restraints generated (Table 1), including 175 intermolecular NOE restraints between CaM and IQ motif, are well distributed along the sequence of CaM and IQ motif, except for the third calcium binding loop of calmodulin (for which several residues are exchange broadened) and the C-terminus of the IQ motif, which is highly dynamic (Figure 1). 248 restraints for psi and phi angles from TALOS were included, and CYANA provided stereo-specific assignments for 5 pairs of Leu and Val methyl groups and 18 pairs of Cβ,γ protons. The final ensemble of 20 conformers was selected based on lowest restraint violation. The input and refinement statistics are shown in Table 1. The final ensemble has no violations of NOE restraints > 0.2 A and no angle violations > 5 degrees. The large number of restraints (>18 per residue) produces a high precision ensemble with an RMSD of 0.50 Å for the backbone and 0.97 Å for all heavy atoms for CaM N-lobe (Leu4-Lys75), values of 0.53 Å for the backbone and 1.06 Å for heavy atoms for the C-lobe (Glu82-Met145 excluding the unstructured loop Asp131-Gly134), and values of 0.58 Å for the backbone and 1.09 Å for all heavy atoms in the IQ motif (Glu1902-Arg1919). Secondary structure analysis using PROCHECK-NMR28 revealed a high quality structure with, for example, 98.9% of backbone torsion angles occupying favored regions of the Ramachandran plot and a Molprobity29 score of 0.89 (100 percentile) for the lowest restraint energy structure (Table 1).
TABLE 1.
Statistics for the apo-CaM/Nav1.5 IQ structural ensemble
| Restraints used for structure calculation | |||
| Distance restraints | |||
| Total | 3054 | ||
| CaM | Intraresidue | 665 | |
| Sequential | 680 | ||
| Medium range | 642 | ||
| Long range | 552 | ||
| IQ motif | Intraresidue | 113 | |
| Sequential | 122 | ||
| Medium range | 105 | ||
| Long range | 0 | ||
| Inter molecular | 175 | ||
| Dihedral angle restraints | |||
| CaM | 210 | ||
| IQ motif | 38 | ||
| Constraint violations, mean ± S.D. | |||
| Distance violations | |||
| 0.1 Å < d < 0.2 Å | 2.3 ± 1.3 | ||
| d > 0.2 Å | 0 | ||
| Average maximum distance violations (Å) | 0.13 ± 0.02 | ||
| Torsion angle violations | |||
| θ < 5.0° | 0.35 ± 0.67 | ||
| θ > 5.0° | 0 | ||
| Average maximum torsion angle violations (degree) | 0.87 ± 1.63 | ||
| AMBER energiesa, mean ± S.D. (kcal.mol-1) | |||
| Restraint | 3.6 ± 0.6 | ||
| Van der Waals | -1419 ± 18 | ||
| Total molecular | -7599 ± 18 | ||
| Precision, root mean square deviation from the mean (Å) | |||
| N-lobe CaM (residues Leu4-Lys75) | |||
| Backbone | 0.50 ± 0.12 | ||
| All heavy atoms | 0.97 ± 0.11 | ||
| C-lobe CaM (residues Glu82-Ile130, Gln135-Met145) | |||
| Backbone | 0.53 ± 0.09 | ||
| All heavy atoms | 1.06 ± 0.11 | ||
| IQ motif (residues Glu1902-Arg1919) | |||
| Backbone | 0.39 ± 0.13 | ||
| All heavy atoms | 1.03 ± 0.16 | ||
| Complex (residues Glu82-Ile130, Gln135-Met145, Glu1902-Arg1919) | |||
| Backbone | 0.58 ± 0.11 | ||
| All heavy atoms | 1.09 ± 0.12 | ||
| Ramachandran statisticsb (%) | |||
| Most favoured | 92.6 | ||
| Additionally allowed | 6.3 | ||
| Generously allowed | 0.6 | ||
| Disallowed | 0.5 | ||
| Molprobityc scores (percentiles) for the lowest restraint energy structure | |||
| Molprobity score | 0.89 (100th), | ||
| all-atom clash score | 0 (100th) | ||
The overall structure of CaM is in accord with previously published structures of CaM in complex with various binding partners (Figure 2a, b). There are two EF-hand domains (Ala1-Arg74, Glu82-Lys148) each formed by four α-helices (N-Lobe: α1 Glu6-Phe19, α2 Thr29-Ser38, α3 Glu45-Val55, α4 Phe65-Lys75; C-lobe: α5 Glu82-Phe92, α6 Ala102-Asn111, α7 Asp118-Asp129, α8 Tyr138-Thr146) and one cross-stranded β-type interaction between the calcium binding loops in each lobe (N-lobe: β1 Thr26-Thr28, β2 Thr62-Asp64; C-lobe: β3 Ile100-Ser101, β4 Gln135-Val136). The IQ motif is a relatively long and straight α helix (Glu1901-Ser1920) (Figure 2). The {1H}-15N heteronuclear NOEs show that the four calcium binding loops between the pairs of helices within each EF-hand motif have above average flexibility, which is reflected by lower precision in the structure (Figure 2a, b). Importantly, the linker between N- and C-lobes (residues Lys75-Ser81) has low {1H}-15N NOEs, indicating a higher than average conformational flexibility, which implies that the two lobes retain their independent motion even in the complex (Figure 2a, b).
Figure 2.
Three-dimensional solution structure of the complex of human CaM with the NaV1.5 IQ motif. (a,b) Representation of the final ensemble of 20 conformers representing the solution structure depicted with all backbone atoms and superimposed on the N-lobe of CaM (a) and the C-lobe of CaM (b). α helices and the IQ motif are labeled. (c) Ribbon representation of the single representative conformer with α helices and β strands labeled. (d) Schematic drawing of the interactions between apo-CaM and the IQ motif. IQ motif residues exhibiting inter-molecular NOE with CaM residues (boxed) are indicated. Hydrophobic residues are colored yellow, acidic in red, basic in blue and polar in green. (e) Representation of the residues at the binding interface in the complex. CaM residues are labeled with transparent white fonts and IQ motif residues are labeled in black font. Hydrophobic residues are colored yellow, acidic in red, basic in blue and polar in green.
Interaction between CaM and IQ motif
As seen in Figure 2c, the IQ motif interacts only with the C-lobe of calmodulin: all 175 intermolecular NOEs detected involve residues of the C-lobe. The key residues at the interface include CaM residues Phe92 (25 intermolecular NOEs), Leu112 (21), Met124 (17), Ile85 (14), Ala88 (13), and Leu116 (12) in CaM and IQ motif residues Ile1908 (33), Phe1912 (31), Ala1905 (26), Glu1901 (19), and Glu1902 (12). Figure 2d summarizes the contacts observed between residues from CaM and the IQ motif. These data highlight the involvement of hydrophobic residues at the binding interface. The average total buried surface area is 2063 Å2, and approximately half (1013 Å2) arises from hydrophobic residues. A substantial contribution (758 Å2) is also provided by contact between charged residues (Asp, Glu, Arg, Lys, His).
IQ motifs are defined by the consensus sequence (I/L/V)Q(X)3R(X)4(R/K), with X being any amino-acid.30,31 The NaV1.5 IQ motif does not perfectly match the consensus because it contains one extra residue (IQRAFRRHLLQR, i.e. IQ(X)3R(X)5R). As found in other CaM-IQ motif complexes, the Ile1908 side chain of the IQ motif is deeply buried (solvent exposed surface area (SSA) of 0.0 Å2) in a hydrophobic pocket of CaM created by Ala88, Phe89, Phe92, Met109 and Met124 (Figure 2e). Gln1909 is also buried (SSA of 1.8 Å2); its side chain interacts with the loop between α7 and α8 of CaM by making hydrogen bonds with the side chain of Glu114. Arg1913 in the middle of the IQ and Arg1919 at the C-terminus of the IQ motif helix are not buried (SSA of 36.3 Å2 and 45.2 Å2 respectively). Their side-chains interact via hydrogen bonds with acidic residues Glu120, Glu123 and Glu127 from CaM α7.
In other CaM-IQ motif complexes, hydrophobic residues besides the IQ motif consensus residues serve as additional anchors in the interaction with CaM.16-18 In our complex, only two hydrophobic residues (Ile1908 and Phe1912) are deeply buried in hydrophobic pockets, one of which is the consensus Ile1908. The other is Phe1912, which has a SSA of only 1.05 Å2 and is nestled in a CaM pocket formed by Met124, Phe141 and Met145 (Figure 2e).
In-silico alanine scanning mutagenesis
The structure of the complex was analyzed using Rosetta to calculate the contributions to the free energy of binding of residues from CaM and the IQ motif. An alanine scanning approach was utilized in which each contact residue is substituted by an alanine and the binding energy is re-evaluated. To optimize the results, the calculation was performed on each conformer of the final representative ensemble. The results for the IQ motif are summarized in Table 2. Hot spots were assigned for residues predicted to have a ΔΔG greater than 2 kcal.mol-1. The greatest contributors to the stability of the complex are predicted to be mutations of residues Ile1908, Gln1909, Arg1910, Phe1912, Arg1913, Arg1914 and Arg1919 (Figure 2e). These correspond remarkably well to the IQ motif consensus residues and the structural anchoring residues. This conclusion is fully consistent with the observation of hydrophobic residues Ile1908 and Phe1912 having the most intermolecular NOEs and being deeply buried in hydrophobic pockets and with the arginine residues being involved in electrostatic interactions with CaM. It is interesting that none of the greatest contributors were from CaM. While the exact origin of this effect is uncertain, it presumably reflects the larger number of CaM residues involved in creating the convoluted binding interface for the IQ motif.
Table 2.
Complete data of the in-silico alanine scanning of Navi.5 IQ motif. ΔΔG value are in kcal.mol-1.
| Structures number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IQ residues number | Average | Stdev | ||||||||||||||||||||
| -4 | 0.7 | 0.8 | 0.4 | 1.1 | 1.8 | 0.3 | 0.8 | 1 | 2 | 1 | 0.8 | 0.2 | 0.1 | 1.5 | 0.6 | 0.1 | 1.3 | 1 | 0.2 | 0.2 | 0.8 | 0.6 |
| -3 | 0.8 | 0.8 | 0.8 | 0.9 | 1.4 | 0.8 | 0.8 | 0.8 | 0.9 | 0.8 | 0.8 | 0.6 | 0.2 | 1.5 | 0.9 | 0.2 | 0.8 | 0.8 | 0.6 | 0.7 | 0.8 | 0.3 |
| -2 | -0.6 | -0.4 | -0.3 | -0.5 | 0.1 | -0.4 | -0.2 | -0.3 | 0 | -0.4 | -0.5 | 0.1 | 1.1 | 0.6 | 2 | 1.3 | -0.5 | 2.3 | -0.2 | 0 | 0.2 | 0.9 |
| -1 | 0.3 | 0.2 | 0.3 | 0.2 | 1.2 | 0.3 | 0.6 | 0.2 | 0.2 | 0.3 | 0.3 | 0.2 | 0.6 | 0.3 | 0.7 | 0.7 | 0.2 | 0.3 | 0.3 | 0.3 | 0.4 | 0.3 |
| 1901 | 1.1 | 1.3 | 1.2 | 1.2 | 1.2 | 1.5 | 1.2 | 1.4 | 1.5 | 1.9 | 1.4 | 1.3 | 1.2 | 1.6 | 2.1 | 1.9 | 1.3 | 1.2 | 1.2 | 1.8 | 1.4 | 0.3 |
| 1902 | 1.6 | 2.5 | 2.8 | 1.3 | 1.8 | 1.3 | 1.8 | 2.5 | 1.2 | 1.1 | 1.3 | 1.4 | 1.4 | 1.6 | 1.7 | 1.4 | 1.5 | 1.3 | 1.5 | 1.2 | 1.6 | 0.5 |
| 1903 | 0.6 | 0.7 | 0.7 | 0.7 | 0.5 | 0.4 | 0.6 | 0.9 | 0.5 | 0.5 | 0.4 | 0.5 | 0.4 | 0.5 | 0.4 | 0.4 | 0.7 | 0.5 | 0.6 | 0.4 | 0.5 | 0.1 |
| 1904 | -0.1 | -0.1 | -0.1 | -0.2 | 0 | -0.1 | -0.2 | -0.1 | -0.1 | 0 | -0.1 | -0.2 | -0.1 | -0.1 | 0 | -0.1 | -0.1 | -0.1 | -0.1 | -0.1 | -0.1 | 0.1 |
| 1905 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| 1906 | 1.9 | 2 | 2.3 | 1.7 | 2 | 2.1 | 2.1 | 2 | 1.7 | 2.1 | 1.9 | 2 | 2.2 | 1.8 | 2.1 | 2.1 | 1.9 | 2.1 | 1.8 | 1.7 | 2.0 | 0.2 |
| 1907 | 0.7 | 0.8 | 1.1 | 0.8 | 0.9 | 1.3 | 0.8 | 0.7 | 1.1 | 1.1 | 1 | 1.3 | 1 | 1.1 | 1 | 0.8 | 1.1 | 1.4 | 1.3 | 0.8 | 1.0 | 0.2 |
| 1908 | 3 | 2.9 | 2.9 | 3.1 | 3.1 | 2.9 | 2.8 | 2.9 | 3 | 3 | 2.9 | 2.7 | 3 | 2.9 | 2.8 | 2.9 | 3 | 3 | 2.8 | 3 | 2.9 | 0.1 |
| 1909 | 3.5 | 3 | 3.1 | 2.8 | 3.9 | 3.7 | 4 | 3.1 | 4.9 | 4.4 | 3.3 | 2.8 | 3 | 2.7 | 3 | 3.9 | 4.9 | 5.1 | 3.9 | 3.9 | 3.6 | 0.7 |
| 1910 | 1.3 | 4 | 2.4 | 3.3 | 2.3 | 1.6 | 2.9 | 3.1 | 3 | 3.8 | 3.3 | 2.3 | 2.7 | 2.5 | 2 | 2.6 | 1.1 | 2.4 | 2.8 | 2.9 | 2.6 | 0.7 |
| 1911 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| 1912 | 3.3 | 3.6 | 3.3 | 3.3 | 3.1 | 3.9 | 3.8 | 3.4 | 3.2 | 3.5 | 2.9 | 3.9 | 3.4 | 3.4 | 3.1 | 3.5 | 3.6 | 3.5 | 3.5 | 3.3 | 3.4 | 0.3 |
| 1913 | 2.9 | 4 | 1.8 | 4 | 3.5 | 2.5 | 4.7 | 3.5 | 3 | 3.7 | 4.3 | 3.2 | 2.7 | 3.2 | 4.3 | 3.5 | 3 | 3.1 | 4 | 4.4 | 3.5 | |
| 1914 | 2.3 | 2 | 1.5 | 3.6 | 4.2 | 2.8 | 2.6 | 4.4 | 3.1 | 3.1 | 1.2 | 2.4 | 1.5 | 1.5 | 2.1 | 3 | 2.4 | 1.9 | 3.9 | 1.5 | 2.6 | 1.0 |
| 1915 | 1.3 | 1.2 | 0.5 | 0.8 | 0.8 | 1.6 | 0.5 | 1 | 1 | 1.5 | 0.7 | 1 | 0.9 | 0.5 | 1.2 | 1.1 | 0.8 | 0.4 | 0.5 | 1.1 | 0.9 | 0.3 |
| 1916 | 1.6 | 2.2 | 1.7 | 1.8 | 1.6 | 1.6 | 2.2 | 1.9 | 1.7 | 1.6 | 1.5 | 2 | 2 | 1.9 | 1.6 | 1.6 | 2 | 1.9 | 2 | 2 | 1.8 | 0.2 |
| 1917 | 0.4 | 1.3 | 0.4 | 0.8 | 0.5 | 1.1 | 0.7 | 0.7 | 0.9 | 1 | 1.4 | 0.5 | 0.8 | 0.5 | 0.6 | 0.7 | 0.3 | 0.7 | 0.8 | 0.8 | 0.7 | 03 |
| 1918 | 1.4 | 1.9 | 1.6 | 1.4 | 2.6 | 1.7 | 2.4 | 2.3 | 1.7 | 1.4 | 1.5 | 1.5 | 1.7 | 2.3 | 1.9 | 1.5 | 1.6 | 1.3 | 1.6 | 1.4 | 1.7 | 0.4 |
| 1919 | 2.8 | 2.4 | 1.3 | 2.3 | 3.1 | 3.1 | 3.1 | 2.4 | 2.2 | 3.4 | 2.2 | 3.3 | 3.1 | 2.1 | 2.1 | 3.7 | 2.7 | 2.4 | 2 | 3 | 2.6 | 0.6 |
| 1920 | 0.2 | 0.5 | 0.7 | 0.5 | 0.5 | 1.5 | 0.6 | 0.5 | 0.8 | 0.5 | 0.6 | 1 | 0.7 | 0.4 | 0.8 | 0.6 | 0.5 | 0.3 | 0.6 | 0.6 | 0.6 | 0.3 |
| 1921 | 1.9 | 1.5 | 2.5 | 1.1 | 1.1 | 0.5 | 1.4 | 1.7 | 0.9 | 1.3 | 2.3 | 1.5 | 0.8 | 2.4 | 0.7 | 2.8 | 1.3 | 2.5 | 1.5 | 1.5 | 1.6 | 0.7 |
| 1922 | 0.8 | 1.5 | 1.1 | 1.3 | 1.1 | 1.2 | 1.1 | 1 | 2.2 | 0.8 | 1 | 1.1 | 0.9 | 1.1 | 1.4 | 1.5 | 1.4 | 1.2 | 2.1 | 1.5 | 1.3 | 0.4 |
| 1923 | -0.2 | 0.6 | -0.2 | -0.3 | 0.4 | 0.7 | -0.4 | -0.1 | 0.4 | 0 | 1.1 | 0.3 | 1 | 0 | 0.3 | 0.7 | 0.8 | 0.7 | -0.1 | 0.8 | 0.3 | 0.5 |
| 1924 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| 1925 | 0.8 | 0.6 | 0.9 | 0.5 | 0.5 | 0.6 | 0.4 | 0.7 | 0.6 | 0.4 | 0.7 | 0.7 | 0.5 | 0.6 | 1 | 0.7 | 0.6 | 0.7 | 0.6 | 0.5 | 0.6 | 0.2 |
| 1926 | 1.2 | -0.5 | 0 | 1.2 | 1.3 | 0 | 1.7 | 1.8 | 0.1 | -0.2 | 0.6 | 1.4 | 2.6 | -0.1 | -0.3 | -0.4 | 1.6 | 0.4 | -0.3 | 1.7 | 0.7 | 0.9 |
| 1927 | 0.8 | 0.3 | 1 | 0.4 | 0.4 | 0.3 | 0.2 | 0.3 | 0.7 | 0.3 | 0.6 | 1.3 | 1.2 | 0.7 | 0.3 | 0.3 | 1.3 | 0.8 | 0.4 | 0.2 | 0.6 | 0.4 |
Comparison with other CaM/IQ complexes
Numerous structures of peptide complexes of Ca2+-loaded CaM have been determined either by NMR or x-ray crystallography,15-19,32,33 and several among them are for CaM in complex with IQ motifs of voltage gated calcium (Cav) channels.15-19 However, few are of complexes in the absence of calcium (e.g. Ref. 23) and none for an ion channel IQ motif. In the complexes with apo-CaM, the IQ motifs are found to occupy positions similar to our structure (Figure 3). Moreover, their relative orientations are identical, with the N-terminus of the IQ motif in close proximity to the N-terminal EF-hand of the CaM C-lobe and the C-terminus of the IQ motif facing the C-terminal EF-hand of the C-lobe.
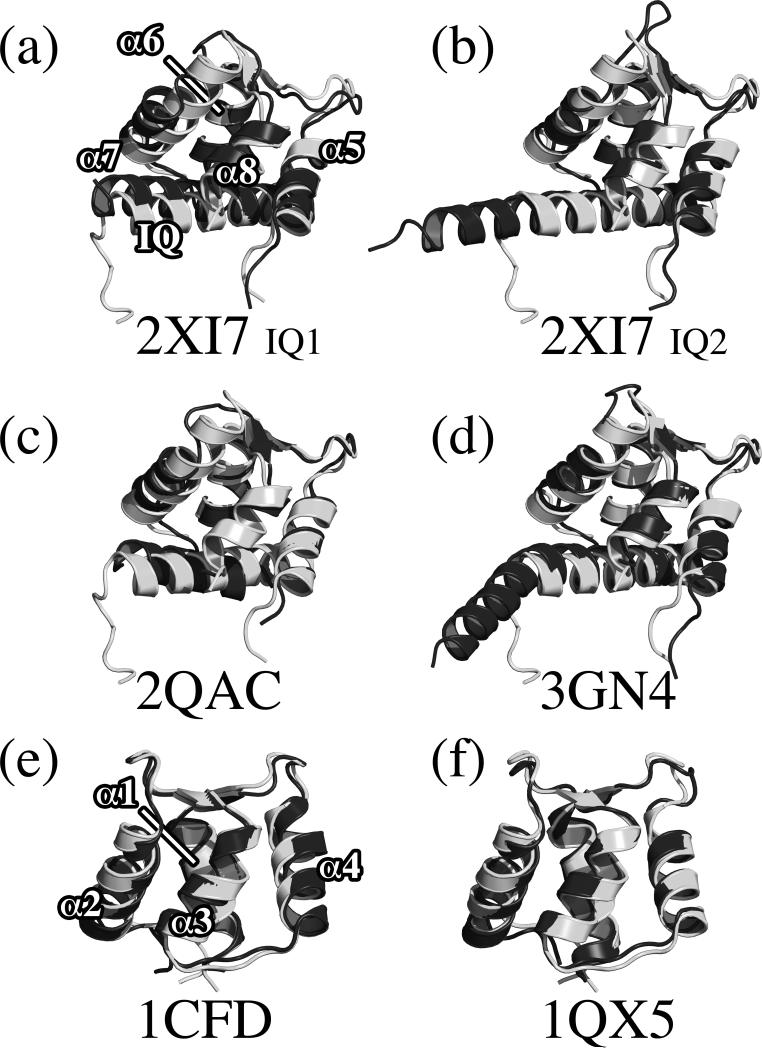
Figure 3.
Comparison of the CaM-human NaV1.5 IQ motif complex with structures of apo-CaM and other IQ motif complexes. (a) CaM/ IQ motif 1 of myosin V (PDB code 2IX7), (b) CaM/ IQ motif 2 of myosin V (2IX7), (c) myosin A/myosin A tail interacting protein (2QAC), (d) CaM/myosin VI (3GN4), (e) apo-CAM (1CFD, NMR), and (f) apo-CaM (1QX5, x-ray). In each comparison the CaM/NaV1.5 IQ complex is white. For clarity, only the α helices and IQ motif of the CaM/NaV1.5 IQ motif complex in the first overlay are labeled. In panels a, b, c and d, only the C-lobe of CaM is shown. In panels e and f, only the N-lobe of CaM is shown.
A Dali34 search for structural neighbors of C-lobe of apo-CaM bound to NaV1.5 IQ motif reveals myosin-A tail domain interacting protein (MTIP) bound to myosin-A and calmodulin in complex with myosin as closest neighbors. Overlays with MTIP (PDB ID: 2QAC),35 CaM/myosin-VI (PDB ID: 3GN4)36 and with CaM/myosin-V IQ motifs 1 and 2 (PDB ID: 2IX7)23 gave root mean square deviation (RMSD) values of 1.6 Å over 65 residues, 1.4 Å over 67 residues, 1.5 Å over 63 residues, and 1.4 Å over 64 residues, respectively (Figure 3). In our complex, CaM is found to adopt the same orientation as found in the myosin-A structure and in the structure of apo-CaM bound to myosin IQ motif.
Some differences are also evident, for example, between our structure and the complex with myosin-V IQ1 (Figure 3a), in which helix α7 is slightly shifted due to an aromatic residue present in myosin-V IQ1. Also, calcium-binding loops L3 and L4 occupy different conformations. Loop 4 in particular is seen to adopt several different conformations in different structures. In our study, the NMR resonances of residues in this loop were not observable, presumably due to an intermediate rate of exchange between conformations on the NMR time scale. Perturbations of L3 and L4 upon binding of the homologous NaV1.2 IQ motif to apo-CaM have been reported.37 These observations are all consistent with the well known flexibility of calcium binding loops in EF-hand proteins in the absence of calcium.
As anticipated, the structure of the N-lobe of CaM, which is not engaged in binding to the NaV1.5 IQ motif, is similar to the structure of the N-lobe in free apo-calmodulin (determined by NMR (PDB ID: 1CFD)38 or x-ray crystallography (PDB ID: 1QX5)).39 The RMSD over 72 N-lobe residues is of 0.93 Å for the NMR structure and 1.13 Å over 72 residues for the crystal structure (Figure 3e,f).
DISCUSSION
Evidence has accumulated showing that intracellular calcium modulates inactivation gating of ion channels including Nav1.5.40 The molecular basis for this regulation is complex and involves the NaV1.5 DIII-DIV linker, C-terminal domain EF-hand (CTD-EF), and the downstream IQ motif as well as calmodulin.8,9,41,42 Knowledge of the mechanism for Ca2+-dependent modulation of NaV1.5 is of high importance because it will provide insight into the origin of certain Brugada syndrome and Long QT syndrome diseases. The structure reported here represents an extension of our effort to define the action of the Ca2+ sensing apparatus of NaV1.5, following upon the determination of the structure of the unique CTD-EF. 12
Semi-open conformation
Calmodulin is a highly flexible molecule that has substantial variation in the relative orientation of its two lobes and very significant conformational flexibility within the lobes, which fluctuate between “closed” and “open” states. Calcium serves to shift this equilibrium towards the “open” state.43,44 The “closed” state is a relatively common compact four helix-bundle conformation; the hydrophobic residues of calmodulin are buried in the hydrophobic core of each bundle. In the “open” state, there is an opening of the interhelical angle between the helices within each EF-hand of each domain, which results in the creation of a substantial hydrophobic accessible surface (Figure 4a,b). This surface provides the interaction site for the many CaM binding partners. When both lobes bind to a single target molecule, there is a significant reduction in interdomain flexibility,45 in some cases leading to the so-called “wrap around” binding mode with very substantial compaction and very tight binding to the target.
Figure 4.
Comparison of helix organization (a) and exposure of hydrophobic surface (b) in the CaM C-lobe structure in different conformational states. From left to right: apo-CaM (PDB code 1DMO), Ca2+-loaded CaM (1CLL) and the apo-CaM complex with the NaV1.5 IQ motif. Coloring is based on a hydrophobicity scale ranging from white to red for hydrophilic to hydrophobic residues, respectively.
Substantially less is known about the interactions of apo-CaM with target proteins. The first atomic resolution structure revealing interactions between apo-CaM and a physiological binding partner came in the form of the structure of two CaM molecules bound to myosin V.23 In the present study, we determined the first structure of apo-CaM in complex with an IQ motif from one of the many ion channels that are known to interact with CaM. The concept of apo-CaM binding to targets in a “semi-open” conformation was first described by Houdusse et al. in a model of CaM bound to myosin.23,46 This conformational state is intermediate between the “open” and “closed” conformations, although not at the geometric average between the two.47 Rather, the domain is only slightly open, allowing the target to bind in a narrow exposed hydrophobic pocket (groove) (Figure 4b, right). In this study, we show that apo-CaM binds to human NaV1.5 channel IQ-motif exclusively through the C-lobe in a “semi-open” conformation. Complete analysis of the N-lobe reveals a structure that is fully consistent with the corresponding N-lobe of the free protein. The main difference between our structure and the apo-CaM/myosin-V and apo-CaM/myosin-VI complexes is the absence of contact between the N-lobe of CaM and NaV1.5 IQ motif. We note that our structure has been determined in solution. It is conceivable that a very weak interaction between CaM N-lobe and myosin could have been stabilized by packing interactions in the crystal.
Structural conservation in the IQ motif
Much of the IQ motif sequence is conserved in NaV channels, and the conservation even extends to CaV channels. Figure 5 shows the sequence alignment of the IQ motifs of NaV and CaV channels and the corresponding the NaV1.5 IQ motif secondary structure. As seen, Glu1901, Glu1902, Ala1905, Ile1908, Gln1909, Phe1912 and Arg1913 residues are the most conserved residues in the NaV family. Notably, these are also the residues providing the most intermolecular NOEs (123 of 175 NOEs) and correspondingly, the majority of the interaction interface in the complex. These findings suggest that the CaM-NaV1.5 IQ motif structure can serve as an accurate template for modeling the interaction of apo-CaM with other NaV channels.
Figure 5.
Conservation of residues in the IQ motif of NaV and CaV channels. NaV and CaV sequence alignments using the sequences (from top to bottom): NaV1.5 (SWISSPROT Q14524), NaV1.1 (P35498), NaV1.2 (Q99250), NaV1.3 (Q9NY46), NaV1.4 (P35499), NaV1.6 (Q9UQD0), NaV1.7 (Q15858), NaV1.8 (Q9Y5Y9), NaV1.9 (Q9UI33), Cav1.1 (Q13698), Cav1.2 (Q13936), CaV2.1 (O00555), and Cav2.2 (Q00975). Residues are colored based on extent of sequence identity. The NaV1.5 IQ motif structure is represented on the top of the alignment. The zigzag line represents α helix, straight lines non-helical regions, and dashed lines residues not present in the IQ motif construct. Stars highlight residues predicted to be important for CaM/NaV1.5 IQ motif interaction. An open circle highlights Ser1904.
Residue conservation with CaV channels is not quite as extensive. In particular, Glu1902 is not conserved, and Glu1901 and Arg1910 are of similar length but opposite charge. Glu1901 and Glu1902 exhibit a substantial number of intermolecular NOEs with hydrophobic residues Val91, Phe92 and Leu112 through the non-charged part of their side chains. In fact, the Glu1901, Glu1902 and Arg1910 side chains point away from CaM in the structure, suggesting that their charge does not directly contribute to the interaction interface and that loss or reversal of charge will not drastically reduce affinity. Importantly, the critical Ile1908, Phe1912 and Arg1913 residues in the NaV1.5 IQ motif are conserved in CaV channels (Ile, Phe/Tyr and Arg, respectively). Moreover, apo-CaM has been shown to bind CaV1.2 through its C-lobe.48 Based on these many similarities, we propose that the apo-CaM-NaV1.5 IQ motif structure provides a viable template for modeling the interaction of apo-CaM with CaV channels.
Insights into the biochemical basis of disease-associated mutations
Mutations within the NaV1.5 IQ motif map to single nucleotide polymorphisms associated with cardiac arrhythmia syndromes. In particular, the Ala1924Thr mutation gives rise to a Brugada syndrome,49 and a recently discovered Ser1904Leu mutation has been associated with a Long QT syndrome.50 It was previously predicted that the residue Ala1924 was a key element in the interaction of CaM with the IQ motif, and shown that the Ala-Thr mutant channel failed to exhibit the significant calcium-dependent shift in steady-state availability observed in wild-type Nav1.5.8 Surprisingly, the structure, heteronuclear NOEs and in-silico alanine-scanning calculations reported here reveal that Ala1924 is in a dynamic region (Figure 1) and is not directly involved in the interaction with apo-CaM (Figure 2d). These results suggest that the mechanistic basis for the dysfunction of this mutant does not involve direct interaction with apo-CaM, despite our previous report of an ~10-fold reduction in the affinity of CaM for an Ala1924Thr mutant IQ peptide.8 While the origin of this discrepancy is unknown, the effect of this mutation on the interactions of Ca2+-CaM with the IQ motif must also be factored into the analysis, in particular because Ca2+-CaM appears to engage the IQ motif in a drastically different manner than apo-CaM, wrapping both lobes around the IQ motif. Moreover, the critical role for the IQ motif in stimulating the binding of calcium ions to the C-terminal domain EF-hand (CTD-EF) needs to also be considered.8,12 The uncertainty arising from inconsistencies in the limited available data underscores the importance of comprehensive investigation of both biochemical and electrophysiological properties to be able to make an accurate analysis of the mechanistic basis for mutated channels. Such a detailed study is beyond the scope of this manuscript and will be published separately as part of a larger study of mutations in the NaV1.5 C-terminus.
The Long QT syndrome Ser1904Leu mutation within the IQ motif has been shown to perturb channel inactivation and promote reopening of the channel.50 As Ser1904 is located at the interface between CaM and the NaV1.5 IQ motif, the leucine mutation likely affects their affinity. Sequence alignment (Figure 5) reveals that the position corresponding to NaV1.5 Ser1904 is always occupied by a relatively small side-chain residue (glycine, alanine, serine, cysteine). Our structure predicts that the Ser-Leu mutation would create a steric clash that would perturb the interaction between CaM and the IQ motif (Figure 6). To obtain further insight into the potential defect associated with this mutation, we used Rosetta software to predict the ΔΔG associated with a switch from serine to leucine in our structure. The analysis showed that the addition of a large hydrophobic residue at residue 1904 was highly destabilizing, with an average ΔΔG value of +6.3 ±3.5 kcal.mol-1. Together, these findings lead us to speculate that the mutation causes a reduced ability to recruit CaM to the CTD or more facile release of CaM.
Figure 6.
Potential steric clash in the interface between CaM and the NaV1.5 IQ motif in the Ser1904Leu mutation associated with a Long QT syndrome. Packing of Ser1904 in the complex of NaV1.5 IQ motif with CaM (left) and Ser1904Leu mutant (right). Ser1904 is packed in a CaM hydrophobic pocket formed by Ala88, Val91 and Phe92. Substitution of serine for leucine in the structure leads to a substantial steric clash with the leucine side-chain protruding into Val91 sidechain and the CaM backbone. Secondary structures and the key residues in the packing interaction are labeled.
Inactivation gating of the channel is mediated by the DIII-DIV linker. It has been shown that an interaction between DIII-DIV linker and CTD is needed to stabilize NaV1.5 inactivation gate.51 Notably, functional interaction requires the presence of CaM,10 so the interaction between DIII-DIV linker and CTD may be direct or mediated by CaM. These results support the idea that the Ser1094Leu leads to reduced interaction between DIII-DIV linker and CTD, facilitating reopening of the channel. Recent studies have shown that CaM binds directly to the DIII-DIV liner.11,13 In particular, Tyr1494 from the DIII-DIV linker is purported to interact with other parts of the channel pore region and lock the gate, but when the C-lobe of CaM binds to the linker, it is unable to lock the gate and cannot promote channel inactivation.13 Binding of CaM to the inactivation gate suggests that destabilization of the CaM-IQ motif interaction by the Ser1094Leu mutation could increase the availability of CaM to interact with the DIII-DIV linker and prevent inactivation gate formation, thereby increasing reopening propensity. Clearly, elucidation of the mechanistic basis for the functional defects of Ser1904Leu NaV1.5 channels will require considerable further analysis. The IQ motif is a keystone in the regulation of NaV1.5 by intracellular calcium signals. This short sequence, located in the C-terminal intracellular region of the channel, functions as a molecular switch.8 It couples the intrinsic CTD-EF domain and extrinsic CaM calcium sensors actions.8,12 In this report, we show how CaM is physically linked to IQ motif in the absence of calcium. We believe this is the molecular basis for how CaM is localized to the channel to be ready to participate in the sensing of intracellular Ca2+ signals by the channel. The absence of any significant interaction with the CaM N-lobe is highly intriguing as this means there is a fully functional calcium sensor available in the immediate vicinity of the remainder of the NaV1.5 calcium sensing apparatus. A general model with independent function of the two lobes of CaM has been hypothesized,52 similar to a previous proposal for the homologous and structurally similar EF-hand protein centrin.53 The mechanism of action of CaM in the complex NaV1.5 Ca2+ sensing apparatus seems to provide yet another example of the remarkable adaptability and functional diversity of CaM. While remaining a considerable challenge, the elucidation of the structural basis of this mechanism holds significant promise for understanding the functional basis for specific cardiac arrhythmia syndromes mapped to the NaV1.5 channel.
Materials and Methods
Expression and purification
Human calmodulin (CaM) was subcloned between NcoI and BamHI restriction sites of a pET15b vector (ampicillin resistant, Novagen). The human NaV1.5 IQ motif (E1901-L1927) was subcloned between BamHI and XhoI restrictions sites of a pET27 derivative in-house pBG102 plasmid (kanamycin resistant, Dr. L. Mizoue, Center for Structural Biology, Vanderbilt University). To facilitate purification and solubility, this vector codes for a SUMO-His6 tag and a flexible linker containing a 3C protease cleavage site. The IQ motif construct is expressed as a 31-residue polypeptide that contains Gly-Pro-Gly-Ser fused to its N terminus after cleavage of the His6 tag.
Escherichia coli BL21 (DE3) cells (Novagen) were cotransformed with both plasmids (pET15b coding for CaM and pBG102 coding for IQ motif expressions). Colonies containing both plasmids were selected after overnight growth on the Petri dish containing kanamycin and ampicillin antibiotics. Cells were grown at 37 °C up to 0.6 OD600, then isopropyl 1-thio-beta-D-galactopyranoside (1mM) was added and the culture induced at 20 °C overnight. Production of unlabelled and 13C,15N-enriched proteins were carried out by growth on lysogeny broth or minimal medium, respectively. The minimal medium was supplemented with 15NH Cl (0.5 g.L-1) and glucose or 13C6-glucose (2.0 g.L-1 as the sole nitrogen and carbon sources. Cell pellets were resuspended in 50 mM Tris at pH 7.5 and 300 mM NaCl and lysed using sonication. The solution was centrifuged at 20,000 g for 20 min. Supernatant was filtered and loaded onto a Ni-affinity chromatography column (Amersham Pharmacia Biosciences) pre-equilibrated with 50 mM Tris at pH 7.5 and 300 mM NaCl. Non-specifically bound proteins and excess CaM were removed by washing the column with 50 mM Tris at pH 7.5 and 300 mM NaCl, and the bound protein complex (CaM/His6-SUMO-IQ) was eluted using a 0 – 500 mM imidazole gradient in 50 mM Tris, pH 7.5, 300 mM NaCl. Fractions containing CaM/ His6-SUMO-IQ complex were pooled, and rhinovirus His6-tagged 3C protease was added. This solution was dialyzed at 4 °C overnight against 50 mM Tris at pH 7.5, 300 mM NaCl. The solution was then loaded onto a Ni-affinity chromatography column to separate CaM/IQ complex from the His6-SUMO-tag and His6-tagged protease. The fractions containing CaM/IQ motif complex were concentrated in 3K-Centricon devices to ~0.5 ml and run over an S75 gel-filtration column (Amersham Pharmacia Biosciences).
NMR sample preparation
CaM/IQ complex was dialyzed into 100 mM KCl, 1 mM EDTA, and 0.01% NaN3 and the pH adjusted to 6.3. CaM/IQ motif concentration was determined by A276, using the published extinction coefficient for CaM of 3,006 M-1 cm-1. The concentration of the complex was 1.0 mM for structure determination and 0.1 mM for dynamic studies. To minimize ionic strength effects on sensitivity, 300 μL of protein solution was loaded into a 4 mm NMR tube, and this tube was inserted into a 5 mm NMR tube containing 150 μL D2O.54
NMR spectroscopy
NMR data were recorded at 25 °C on Bruker DRX600 and DRX800 spectrometers equipped with cryoprobes. Backbone resonance assignments were obtained by the combined use of 2D 15N-1H HSQC and 3D HNCACB, CBCA(CO)NH and HNCO experiments (reviewed in ref 55). Aliphatic side chain resonance assignments were obtained from 3D (H)CC(CO)NH, H(CCCO)NH, HCCH-TOCSY, HCCH-COSY and HBHANH experiments. 1H chemical shift assignments of aromatic side chains were based on 2D heteronuclear (HB)CB(CGCD)HD and (HB)CB(CGCDCE)HE and 2D homonuclear COSY, TOCSY and NOESY experiments. The chemical shifts have been deposited in BMRB databank under accession code 17264. To assign NOE-based distance restraints, a 2D 1H homonuclear NOESY experiment was recorded on an unlabeled sample, and 3D 15N NOESY-HSQC and 13C NOESY-HSQC were recorded on the uniformly 13C,15N-enriched sample. The mixing time used in all NOESY experiments was set to 120 ms, and the experiments were recorded on the DRX800.
A steady-state {1H}-15N heteronuclear NOE relaxation experiment was recorded at 800 MHz as described previously56 using a 3 s delay for 1H saturation. Saturated and non-saturated spectra were collected in an interleaved manner and the NOE values were extracted as the ratio of peak intensities with and without proton saturation for 170 well-resolved resonances.
Data were processed and analyzed with Topspin 2.0b (Bruker) and Sparky57 software, respectively.
Structure Calculations
The first stage of calculation used CYANA 2.1.24 Starting from a set of manually assigned NOEs, the standard CYANA protocol of 7 iterative cycles of calculations was performed with NOE assignment by the embedded CANDID58 routine combined with torsion angle dynamics structure calculation. In each cycle, 100 structures starting from random torsion angle values were calculated with 15,000 steps of torsion angle dynamics-driven simulated annealing. 3054 NOE-based distance and 248 backbone angle restraints were used for the final calculations. The angle restraints were obtained from 13Cα, 13Cβ, 13C’ and 15N chemical shifts using TALOS59 with an assigned minimum range of ± 30°. No hydrogen bond restraints were used for structure calculation. The second stage consisted of the refinement of the 50 lowest CYANA target function conformers by restrained molecular dynamics (rMD) simulations in AMBER25 10 using the generalized Born solvent model. The starting CYANA structures were quickly regularized by a 1 ps energy minimization in the AMBER force field and then 20 ps of rMD were performed with the following protocol: the system was heated for 5 ps at 600 K with tight coupling for heating and equilibration (TAUTP = 0.4); 15 ps of cooling to 0 K with 13 ps of slow cooling (loose coupling, TAUTP = 4.0–1.0) followed by 1 ps of faster cooling (TAUTP = 1.0) and a final 1 ps of very fast cooling (TAUTP = 0.1–0.05). The restraints were slowly ramped from 10% to 100% of their final values over the first 3 ps. Force constants for distances and angles restraints were set to 32 kcal.mol-1.Å-2 and 50 kcal.mol-1.radians-2 respectively. The representative ensemble corresponds to the 20 conformers with the lowest restraint and AMBER energy terms.
Computational Analysis
Multiple sequence alignments were performed with T-Coffee60 through the phylogeny web server.61 Graphical analyses of the structures and figure preparation were carried out with the programs MOLMOL62 and PyMOL.63 Chemical Shift Index (CSI) was calculated with PREDITOR software64 based on Hα, Cα, Cβ and C’ chemical shifts. The geometric quality, stereochemistry and structure quality of CaM/IQ motif complex structural ensemble was assessed using PROCHECK-NMR software28 and Molprobity server.29 Electrostatic surface potentials were calculated with the Adaptive Poisson-Boltzmann Solver (APBS) software.65 Amino-acid hydropathy was based on Hopp/Woods and Kyte/Doolittle scales.66,67 Surface accessibility was calculated using a 1.4 Å probe radius with the NACCESS software.68 In-silico alanine scanning, single point mutation (S1904L) and ΔΔG calculations were performed using Rosetta2.3.69,70
Accession numbers
1H, 13C, and 15N chemical shift assignments are deposited in the BioMagResBank71 under BMRB ID: 17264. The coordinates of the final ensemble of 20 structures and the NMR restraints used for structure determination are deposited in the Protein Data Bank72 under PDB ID: 2l53.
Supplementary Material
Figure S1. Distribution of sequential and medium-range NOEs and chemical shift index. Strong, medium and weak NOE intensities are indicated by thick, medium, and thin lines, respectively. The chemical shift index is shown as vertical bars.
Acknowledgements
We thank Marie-Eve Chagot for expert technical assistance and Dr. Steven M. Damo for critical reading of the manuscript. This work was supported by an operating grant (R01 GM56307) from the National Institutes of Health.
Abbreviations
The abbreviations used are:
- CaM
calmodulin
- IQ motif
Isoleucine-Glutamine (I-Q) motif
- Tris
tris(hydroxymethyl)aminomethane
- HSQC
heteronuclear single quantum correlation
- NOESY
nuclear Overhauser effect (NOE) spectroscopy
- EDTA
ethylenediaminetetraacetic acid
- RMSD
root mean square deviation
- CTD-EF
NaV1.5 C-terminal domain EF-hand
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An RH, Wang XL, Kerem B, Benhorin J, Medina A, Goldmit M, Kass RS. Novel LQT-3 mutation affects Na+ channel activity through interactions between alpha- and beta1-subunits. Circ Res. 1998;83:141–6. doi: 10.1161/01.res.83.2.141. [DOI] [PubMed] [Google Scholar]
- 3.Smits JP, Eckardt L, Probst V, Bezzina CR, Schott JJ, Remme CA, Haverkamp W, Breithardt G, Escande D, Schulze-Bahr E, LeMarec H, Wilde AA. Genotype-phenotype relationship in Brugada syndrome: electrocardiographic features differentiate SCN5A-related patients from non-SCN5A-related patients. J Am Coll Cardiol. 2002;40:350–6. doi: 10.1016/s0735-1097(02)01962-9. [DOI] [PubMed] [Google Scholar]
- 4.Petitprez S, Jespersen T, Pruvot E, Keller DI, Corbaz C, Schlapfer J, Abriel H, Kucera JP. Analyses of a novel SCN5A mutation (C1850S): conduction vs. repolarization disorder hypotheses in the Brugada syndrome. Cardiovasc Res. 2008;78:494–504. doi: 10.1093/cvr/cvn023. [DOI] [PubMed] [Google Scholar]
- 5.Millat G, Chevalier P, Restier-Miron L, Da Costa A, Bouvagnet P, Kugener B, Fayol L, Gonzalez Armengod C, Oddou B, Chanavat V, Froidefond E, Perraudin R, Rousson R, Rodriguez-Lafrasse C. Spectrum of pathogenic mutations and associated polymorphisms in a cohort of 44 unrelated patients with long QT syndrome. Clin Genet. 2006;70:214–27. doi: 10.1111/j.1399-0004.2006.00671.x. [DOI] [PubMed] [Google Scholar]
- 6.Priori SG, Napolitano C, Gasparini M, Pappone C, Della Bella P, Giordano U, Bloise R, Giustetto C, De Nardis R, Grillo M, Ronchetti E, Faggiano G, Nastoli J. Natural history of Brugada syndrome: insights for risk stratification and management. Circulation. 2002;105:1342–7. doi: 10.1161/hc1102.105288. [DOI] [PubMed] [Google Scholar]
- 7.Darbar D, Kannankeril PJ, Donahue BS, Kucera G, Stubblefield T, Haines JL, George AL, Jr., Roden DM. Cardiac sodium channel (SCN5A) variants associated with atrial fibrillation. Circulation. 2008;117:1927–35. doi: 10.1161/CIRCULATIONAHA.107.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah VN, Wingo TL, Weiss KL, Williams CK, Balser JR, Chazin WJ. Calcium-dependent regulation of the voltage-gated sodium channel hH1: intrinsic and extrinsic sensors use a common molecular switch. Proc Natl Acad Sci U S A. 2006;103:3592–7. doi: 10.1073/pnas.0507397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingo TL, Shah VN, Anderson ME, Lybrand TP, Chazin WJ, Balser JR. An EF-hand in the sodium channel couples intracellular calcium to cardiac excitability. Nat Struct Mol Biol. 2004;11:219–25. doi: 10.1038/nsmb737. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Ghosh S, Liu H, Tateyama M, Kass RS, Pitt GS. Calmodulin mediates Ca2+ sensitivity of sodium channels. J Biol Chem. 2004;279:45004–12. doi: 10.1074/jbc.M407286200. [DOI] [PubMed] [Google Scholar]
- 11.Sarhan MF, Van Petegem F, Ahern CA. A double tyrosine motif in the cardiac sodium channel domain III-IV linker couples calcium-dependent calmodulin binding to inactivation gating. J Biol Chem. 2009;284:33265–74. doi: 10.1074/jbc.M109.052910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chagot B, Potet F, Balser JR, Chazin WJ. Solution NMR structure of the C-terminal EF-hand domain of human cardiac sodium channel NaV1.5. J Biol Chem. 2009;284:6436–45. doi: 10.1074/jbc.M807747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Potet F, Chagot B, Anghelescu M, Viswanathan PC, Stepanovic SZ, Kupershmidt S, Chazin WJ, Balser JR. Functional Interactions between Distinct Sodium Channel Cytoplasmic Domains through the Action of Calmodulin. J Biol Chem. 2009;284:8846–54. doi: 10.1074/jbc.M806871200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–80. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 15.Halling DB, Georgiou DK, Black DJ, Yang G, Fallon JL, Quiocho FA, Pedersen SE, Hamilton SL. Determinants in CaV1 channels that regulate the Ca2+ sensitivity of bound calmodulin. J Biol Chem. 2009;284:20041–51. doi: 10.1074/jbc.M109.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fallon JL, Halling DB, Hamilton SL, Quiocho FA. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Ca(v)1.2 calcium channel. Structure. 2005;13:1881–6. doi: 10.1016/j.str.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Van Petegem F, Chatelain FC, Minor DL., Jr. Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat Struct Mol Biol. 2005;12:1108–15. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori MX, Vander Kooi CW, Leahy DJ, Yue DT. Crystal structure of the CaV2 IQ domain in complex with Ca2+/calmodulin: high-resolution mechanistic implications for channel regulation by Ca2+. Structure. 2008;16:607–20. doi: 10.1016/j.str.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim EY, Rumpf CH, Fujiwara Y, Cooley ES, Van Petegem F, Minor DL., Jr. Structures of CaV2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure. 2008;16:1455–67. doi: 10.1016/j.str.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikura M, Clore GM, Gronenborn AM, Zhu G, Klee CB, Bax A. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–8. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 21.Shen Y, Zhukovskaya NL, Guo Q, Florian J, Tang WJ. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J. 2005;24:929–41. doi: 10.1038/sj.emboj.7600574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Diego I, Kuper J, Bakalova N, Kursula P, Wilmanns M. Molecular basis of the death-associated protein kinase-calcium/calmodulin regulator complex. Sci Signal. 2010;3:ra6. doi: 10.1126/scisignal.2000552. [DOI] [PubMed] [Google Scholar]
- 23.Houdusse A, Gaucher JF, Krementsova E, Mui S, Trybus KM, Cohen C. Crystal structure of apo-calmodulin bound to the first two IQ motifs of myosin V reveals essential recognition features. Proc Natl Acad Sci U S A. 2006;103:19326–31. doi: 10.1073/pnas.0609436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–78. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 25.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YT, Dimitrova YN, Schneider G, Ridenour WB, Bhattacharya S, Soss SE, Caprioli RM, Filipek A, Chazin WJ. Structure of the S100A6 complex with a fragment from the C-terminal domain of Siah-1 interacting protein: a novel mode for S100 protein target recognition. Biochemistry. 2008;47:10921–32. doi: 10.1021/bi801233z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nordquist KA, Dimitrova YN, Brzovic PS, Ridenour WB, Munro KA, Soss SE, Caprioli RM, Klevit RE, Chazin WJ. Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry. 2010;49:347–55. doi: 10.1021/bi901620v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–40. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 31.Bahler M, Rhoads A. Calmodulin signaling via the IQ motif. FEBS Lett. 2002;513:107–13. doi: 10.1016/s0014-5793(01)03239-2. [DOI] [PubMed] [Google Scholar]
- 32.Osawa M, Tokumitsu H, Swindells MB, Kurihara H, Orita M, Shibanuma T, Furuya T, Ikura M. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–24. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 33.Ishida H, Rainaldi M, Vogel HJ. Structural studies of soybean calmodulin isoform 4 bound to the calmodulin-binding domain of tobacco mitogen-activated protein kinase phosphatase-1 provide insights into a sequential target binding mode. J Biol Chem. 2009;284:28292–305. doi: 10.1074/jbc.M109.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bosch J, Turley S, Roach CM, Daly TM, Bergman LW, Hol WG. The closed MTIP-myosin A-tail complex from the malaria parasite invasion machinery. J Mol Biol. 2007;372:77–88. doi: 10.1016/j.jmb.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukherjea M, Llinas P, Kim H, Travaglia M, Safer D, Menetrey J, Franzini-Armstrong C, Selvin PR, Houdusse A, Sweeney HL. Myosin VI dimerization triggers an unfolding of a three-helix bundle in order to extend its reach. Mol Cell. 2009;35:305–15. doi: 10.1016/j.molcel.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theoharis NT, Sorensen BR, Theisen-Toupal J, Shea MA. The neuronal voltage-dependent sodium channel type II IQ motif lowers the calcium affinity of the C-domain of calmodulin. Biochemistry. 2008;47:112–23. doi: 10.1021/bi7013129. [DOI] [PubMed] [Google Scholar]
- 38.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Solution structure of calcium-free calmodulin. Nat Struct Biol. 1995;2:768–76. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 39.Schumacher MA, Crum M, Miller MC. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12:849–60. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Saimi Y, Kung C. Calmodulin as an ion channel subunit. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 41.Young KA, Caldwell JH. Modulation of skeletal and cardiac voltage-gated sodium channels by calmodulin. J Physiol. 2005;565:349–70. doi: 10.1113/jphysiol.2004.081422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan HL, Kupershmidt S, Zhang R, Stepanovic S, Roden DM, Wilde AA, Anderson ME, Balser JR. A calcium sensor in the sodium channel modulates cardiac excitability. Nature. 2002;415:442–7. doi: 10.1038/415442a. [DOI] [PubMed] [Google Scholar]
- 43.Malmendal A, Evenas J, Forsen S, Akke M. Structural dynamics in the C-terminal domain of calmodulin at low calcium levels. J Mol Biol. 1999;293:883–99. doi: 10.1006/jmbi.1999.3188. [DOI] [PubMed] [Google Scholar]
- 44.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–72. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 45.Trewhella J, Blumenthal DK, Rokop SE, Seeger PA. Small-angle scattering studies show distinct conformations of calmodulin in its complexes with two peptides based on the regulatory domain of the catalytic subunit of phosphorylase kinase. Biochemistry. 1990;29:9316–24. doi: 10.1021/bi00492a003. [DOI] [PubMed] [Google Scholar]
- 46.Houdusse A, Silver M, Cohen C. A model of Ca(2+)-free calmodulin binding to unconventional myosins reveals how calmodulin acts as a regulatory switch. Structure. 1996;4:1475–90. doi: 10.1016/s0969-2126(96)00154-2. [DOI] [PubMed] [Google Scholar]
- 47.Nelson MR, Chazin WJ. An interaction-based analysis of calcium-induced conformational changes in Ca2+ sensor proteins. Protein Sci. 1998;7:270–82. doi: 10.1002/pro.5560070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lian LY, Myatt D, Kitmitto A. Apo calmodulin binding to the L-type voltage-gated calcium channel Cav1.2 IQ peptide. Biochem Biophys Res Commun. 2007;353:565–70. doi: 10.1016/j.bbrc.2006.12.070. [DOI] [PubMed] [Google Scholar]
- 49.Rook MB, Bezzina Alshinawi C, Groenewegen WA, van Gelder IC, van Ginneken AC, Jongsma HJ, Mannens MM, Wilde AA. Human SCN5A gene mutations alter cardiac sodium channel kinetics and are associated with the Brugada syndrome. Cardiovasc Res. 1999;44:507–17. doi: 10.1016/s0008-6363(99)00350-8. [DOI] [PubMed] [Google Scholar]
- 50.Bankston JR, Sampson KJ, Kateriya S, Glaaser IW, Malito DL, Chung WK, Kass RS. A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels (Austin) 2007;1:273–80. doi: 10.4161/chan.4956. [DOI] [PubMed] [Google Scholar]
- 51.Motoike HK, Liu H, Glaaser IW, Yang AS, Tateyama M, Kass RS. The Na+ channel inactivation gate is a molecular complex: a novel role of the COOH-terminal domain. J Gen Physiol. 2004;123:155–65. doi: 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamniuk AP, Rainaldi M, Vogel HJ. Calmodulin has the Potential to Function as a Ca-Dependent Adaptor Protein. Plant Signal Behav. 2007;2:354–7. doi: 10.4161/psb.2.5.4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheehan JH, Bunick CG, Hu H, Fagan PA, Meyn SM, Chazin WJ. Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function. J Biol Chem. 2006;281:2876–81. doi: 10.1074/jbc.M509886200. [DOI] [PubMed] [Google Scholar]
- 54.Voehler MW, Collier G, Young JK, Stone MP, Germann MW. Performance of cryogenic probes as a function of ionic strength and sample tube geometry. J Magn Reson. 2006;183:102–9. doi: 10.1016/j.jmr.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavanagh J, Fairbrother WJ, Palmer AGI, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic Press Inc.; New York: 1996. [Google Scholar]
- 56.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4:727–34. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 57.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 58.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 59.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 60.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 61.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–5. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 63.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 64.Berjanskii MV, Neal S, Wishart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–9. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci U S A. 1981;78:3824–8. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 68.Hubbard S, Thornton J. NACCESS; London: 1993. [Google Scholar]
- 69.Kortemme T, Kim DE, Baker D. Computational Alanine Scanning of Protein-Protein Interfaces. Sci. STKE. 20042004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 70.Keeble AH, Joachimiak LA, Mate MJ, Meenan N, Kirkpatrick N, Baker D, Kleanthous C. Experimental and computational analyses of the energetic basis for dual recognition of immunity proteins by colicin endonucleases. J Mol Biol. 2008;379:745–59. doi: 10.1016/j.jmb.2008.03.055. [DOI] [PubMed] [Google Scholar]
- 71.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–8. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Distribution of sequential and medium-range NOEs and chemical shift index. Strong, medium and weak NOE intensities are indicated by thick, medium, and thin lines, respectively. The chemical shift index is shown as vertical bars.