Abstract
Erythromycin and related macrolide antibiotics are widely used polyketide natural products. We have evolved an engineered biosynthetic pathway in Escherichia coli that yields erythromycin analogs from simple synthetic precursors. Multiple rounds of mutagenesis and screening led to the identification of new mutant strains with improved efficiency for precursor directed biosynthesis. Genetic and biochemical analysis suggested that the phenotypically relevant alterations in these mutant strains were localized exclusively to the host-vector system, and not to the polyketide synthase. We also demonstrate the utility of this improved system through engineered biosynthesis of a novel alkynyl erythromycin derivative with comparable antibacterial activity to its natural counterpart. In addition to reinforcing the power of directed evolution for engineering macrolide biosynthesis, our studies have identified a new lead substance for investigating structure-function relationships in the bacterial ribosome.
Keywords: erythromycin, 6-deoxyerythronolide B synthase, precursor directed biosynthesis, polyketide, directed evolution
Introduction
Polyketides form an important class of natural products having a wide range of uses including as anti-infective agents, antitumor drugs, immunosuppressants and veterinary products 1,2. The emergence of antibiotic resistance is a serious problem 3, and is fueling the demand for new antibiotics. The multi-modular structure of certain bacterial polyketide synthases (PKS) has made them an attractive target for protein engineering to produce improved drug candidates 4.
Precursor directed biosynthesis 5–7 is a powerful approach to expand the range of engineered macrolide antibiotics, especially in conjunction with semisynthesis 8 and hybrid PKS construction 9,10. Precursor directed biosynthesis combines the complementary power of synthetic chemistry, which allows a virtually unlimited range of simple compounds, and biosynthesis, which routinely creates complex molecules 11 from simple precursors. While the range of natural precursors for polyketide biosynthesis is relatively limited 12, precursor directed biosynthesis enables the incorporation of unnatural precursors with diverse functional groups, expanding the pool of “unnatural” natural products. Additionally, regio- and chemo-selectively modified and modifiable functional groups can be introduced easily using this approach.
Notwithstanding its promise, there are significant limitations to precursor directed biosynthesis. The feeding of labile precursors to Streptomyces or Saccharopolyspora bacteria that naturally produce bioactive polyketides is fraught with challenges 11,13–15. By contrast, incorporation of precursors into heterologous polyketide biosynthetic pathways in E. coli is relatively efficient 16, but such systems typically yield inactive polyketide intermediates which have not undergone post-PKS modifications. Furthermore, due to the low affinity of the acceptor PKS module for unnatural thioester substrates 16, precursors are often required to be fed in high concentration to the engineered strains to effectively transform precursors into polyketide products. The substrate specificity of one or more downstream PKS modules may also cause kinetic bottlenecks in the production of polyketides by precursor directed biosynthesis 11,14.
Recently, the entire erythromycin biosynthetic pathway has been reconstituted in E. coli 17,18, enabling the production of active antibacterial agents in this heterologous host. A rapid and sensitive screen for detecting erythromycin from precursor directed biosynthesis was also established 17. Here, we have modified the erythromycin biosynthetic pathway in a directed fashion increasing titers in E. coli.
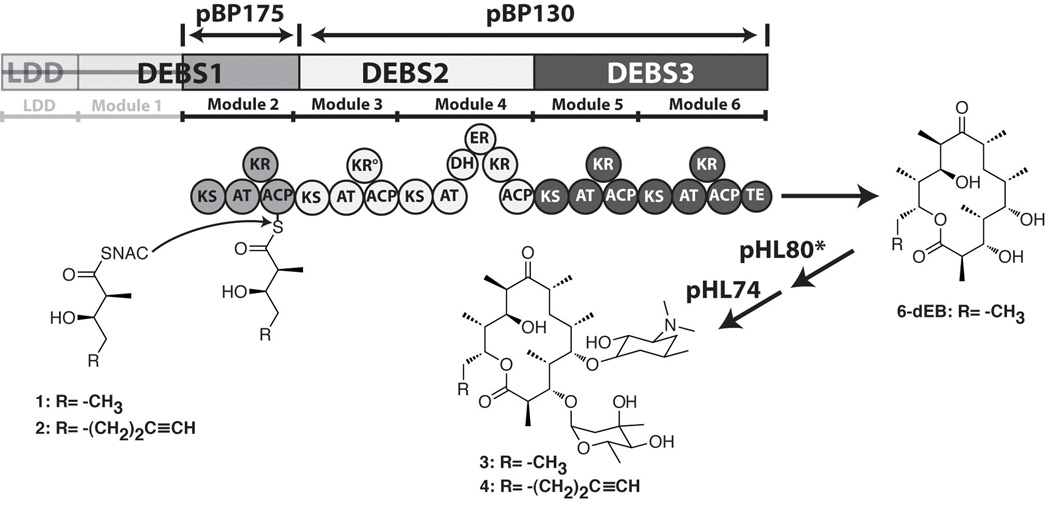
The feasibility of producing inactive macrolides aglycones in E. coli by precursor directed biosynthesis was originally demonstrated using an engineered host BAP1 containing plasmids pBP175, which encodes for DEBS module 2 and propionyl-CoA carboxylase, and pBP130, which encodes for DEBS2 and DEBS3 (Figure 1; Table 1) 16. Subsequently, we engineered two additional compatible plasmids pHL80, which encodes mycarose biosynthetic and glycosyl transfer functions, and pHL74, which encodes desosamine biosynthetic and glycosyl transfer functions (Table 1) to produce bioactive glycosylated macrolides 17. E. coli BAP1/pBP175/pBP130/pHL80/pHL74 produces the antibiotic 6-deoxyerythromycin D (6d-EryD) when fed with compound 1 (Figure 1), a synthetic cell permeable mimic of the natural diketide intermediate in erythromycin biosynthesis 17. Through directed evolution, we also isolated a mutant plasmid pHL80*, which substantially improves mycarosylation of the aglycone compared to wild-type pHL80, facilitating improved precursor directed biosynthesis 17. A simple, visual single colony screening assay was used to isolate a mutant of E. coli BAP1/pBP175/pBP130/pHL80*/pHL74 (designated Mutant D) which showed a >3-fold improvement in the amount of 6-deoxyerythromycin D production with high (>0.3 mM) concentrations of diketide 1 17. Here we have analyzed Mutant D with the goal of understanding the molecular basis of this phenotype, and have extended this finding through additional rounds of directed evolution toward improved mutants. We identified and analyzed several new mutants to explain the improved erythromycin biosynthesis from simple precursors, and demonstrated their utilities for the engineered biosynthesis of a novel derivative of erythromycin with promising antibacterial properties.
Figure 1. Precursor directed biosynthesis of 6-deoxyerythromycin D and analogs.
A truncated derivative of the 6-deoxyerythronolide B synthase (DEBS) was constructed lacking the loading module and module 1 of DEBS114. E. coli cells harboring this truncated DEBS, along with plasmids responsible for mycarosyl and desosaminyl sugar biosynthesis and transfer, are able to synthesize 6-deoxyerythromycin D (3) or its analog (4) when fed with an appropriate diketide analog precursor (1 or 2).
Table 1.
Plasmids Used
| Plasmid | Encoded Proteins | Function | Vector |
|---|---|---|---|
| pBP130 | DEBS2-DEBS3 | Biosynthesis of 6dEB | pET21 |
| pBP144 | PccAB-DEBS1 | pET28 | |
| pBP175 | PccAB-DEBS Mod2 | pET28 | |
| pBP190 | thioesterase II (TEII) | pGZ119 | |
| pHL74 | EryCIII-EryCII-TylAI-DesIV-DesI-DesII-DesV-DesVI | TDP-desosamine biosynthesis and transfer | pGZ119 |
| pHL80* | GroES-GroEL-TylCVII-EryBIV-EryBVI-EryBV-TylCIII-EryBII | TDP-mycarose biosynthesis and transfer + Chaperone | pCDF-Duet |
Methods
Analysis of mutant D
Mutants showing improved conversion of diketide 1 into erythromycin were grown in liquid LB medium containing kanamycin, carbenicillin, streptomycin and chloramphenicol, and the plasmids were isolated from each culture. Each plasmid was separated by re-transforming a dilute sample of the plasmid mixture into E. coli XL1-Blue and isolating transformants that expressed only the desired antibiotic resistance gene. Each purified plasmid was then co-transformed along with other wild-type plasmids to obtain E. coli HYL3/pBP175(*)/pBP130(*)/pHL80*(*)/pHL74(*) (* refers to plasmid derived from the mutant strain). These transformants were analyzed by plate-based assays to identify the plasmid responsible for the desired phenotype.
Quantifying relative protein expression in BAP1 vs HYL3
To compare the relative expression levels of DEBS module 2 in the wild-type (BAP1) and evolved (HYL3) hosts, pBP175 was introduced into each via transformation. As described earlier 19, cells were grown at 37°C in LB medium with 50 µg/ml kanamycin to an OD600 = 0.6, then chilled on ice for 10 min, and induced at 20°C with 0.2 mM IPTG for 15 h. Thereafter, the cells were harvested by centrifugation (4,500g, 15 min) and disrupted by sonication (10 × 30 sec, Buffer: 50 mM sodium phosphate pH 8.0, 10 mM imidazole, 0.3 M NaCl, 1 mM DTT, 20% glycerol). Cellular debris was removed by centrifugation (17,000g, 45 min), and DEBS module 2 as well as PccA and PccB (both have similar molecular masses and therefore migrate as a single band) were purified using nickel affinity chromatography, with protein elution at 200 mM imidazole (Buffer: 50 mM sodium phosphate pH 8.0, 200 mM imidazole, 0.3 M NaCl, 1 mM DTT, 20% glycerol).
Generation of pBP175 Library
Plasmid pBP175 was mutagenized using a mutator strain XL1-Red. The wild-type plasmid was transformed to XL1-Red, and selected with kanamycin (50 mg/L) on LB plate. Approximately 200–2000 colonies were collected and grown to stationary phase in LB media with kanamycin. The culture was used to re-inoculate LB medium with kanamycin (1% inoculum volume), and also to prepare pBP175 plasmid DNA. These steps were repeated 7 times. At the end of the entire procedure, individual plasmid preparations from each step were combined in equal quantities to yield a mutagenized pBP175 sample, which was transformed into HYL3/pHL80*/pHL74 along with pBP130. Approximately 103 transformants were screened as described below.
Screening of pBP175 Library
The library of E. coli HYL3/pBP175/pBP130/pHL80*/pHL74 was plated on a cellophane disk placed on LB agar containing 1 mg/ml sodium propionate and 10 µM diketide 1. The plates were incubated at 30°C for 60–68 h. The cellophane was then transferred onto a second plate while soft agar containing Bacillus subtilis was overlaid onto the original plate. After overnight incubation at 30°C, halos of growth inhibition were observed. Colonies producing the biggest halos were isolated from the secondary plate, and re-analyzed to verify the phenotype.
Biosynthesis of macrolides in shake-flask cultures
E. coli BAP1/pBP175/pBP130/pHL80*/pHL74 (or any derivative mutant) was grown in LB medium containing 50 µg/ml kanamycin, 100 µg/ml carbenicillin, 50 µg/ml streptomycin and 34 µg/ml chloramphenicol to OD600 = 0.6. The cells were chilled on ice for 10 min and centrifuged briefly. After washing with LB, the cell pellet was resuspended in 5% of the original volume of LB (i.e., 20-fold concentration) without antibiotics. Following addition of 0.1–0.3 mM IPTG and 0.1–1 mM diketide precursor, cell growth was allowed to proceed at 30°C for additional 72 h. To quantify 6-deoxyerythromycin D (or analogue) in the culture medium, serial (two-fold) dilutions of the 0.2 µm filter sterilized culture medium were added to freshly inoculated cultures of B. subtilis. No exogenous antibiotics were used during the growth of the E. coli culture to be tested. The growth inhibition of B. subtilis, monitored by measuring OD490 after ~20 h at 30°C in a 96-well plate reader, was used to determine the titer of antibiotic. Any apparent discrepancy between the titers shown here in Figure 3 and those in previous reports (17) is a result of the lack, until recently, of an authentic standard of 6d-Ery D. All previously reported titers were calculated based on a similar, more potent, antibiotic.
Figure 3. Characterization of Mutant E.coli strains.
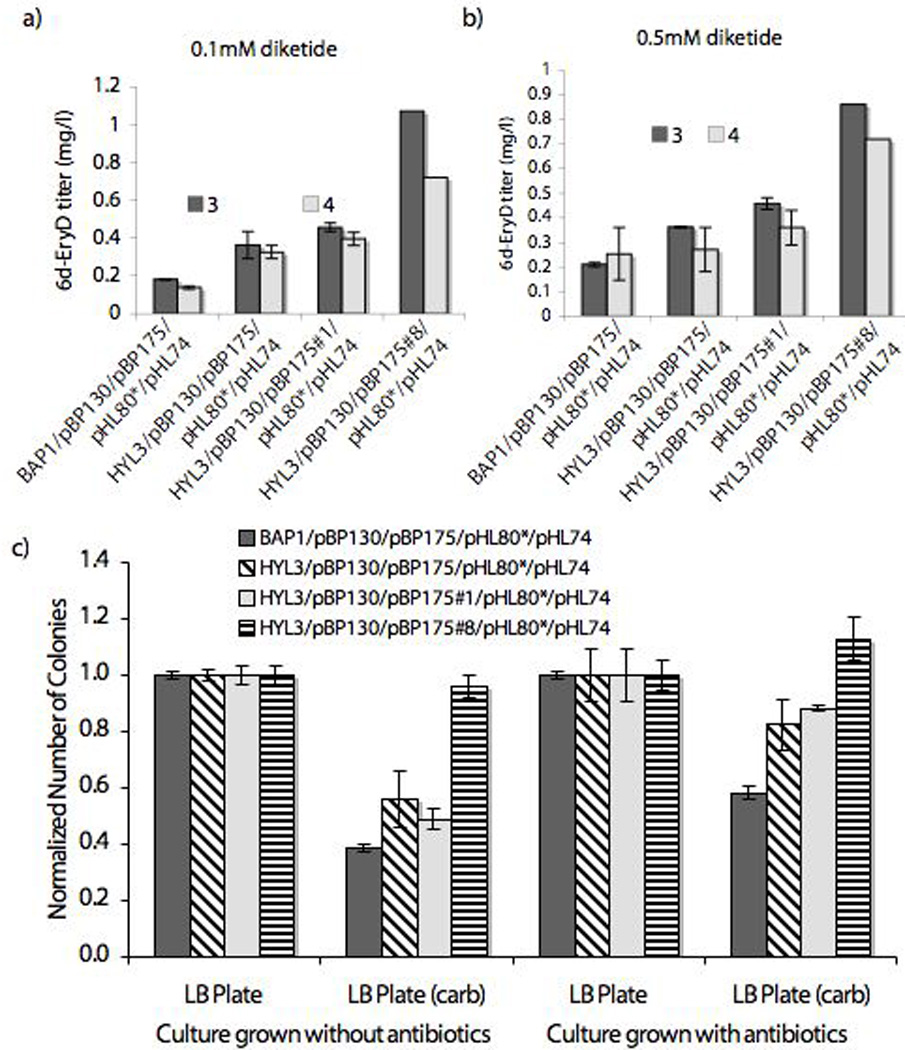
a),b) Comparison of macrolide production by wild-type and mutant E. coli cell lines: Biosynthesis of 6-deoxyerythromycin D (3) and alkynyl-6-deoxyerythromycin D (4) by E. coli BAP1/pBP175/pBP130/pHL80*/pHL74, HYL3/pBP175/pBP130/pHL80*/pHL74, HYL3/pBP175 #1/pBP130/pHL80*/pHL74 and HYL3/pBP175 #8/pBP130/pHL80*/pHL74 when fed 1 and 2, respectively at 100 µM in a) and 500 µM in b). For experimental details, see text.
c) Plasmid stability of pBP130 in E. coli BAP1/pBP175/pBP130/pHL80*/pHL74, HYL3/pBP175/pBP130/pHL80*/pHL74, HYL3/pBP175 #1/pBP130/pHL80*/pHL74 and HYL3/pBP175 #8/pBP130/pHL80*/pHL74 was compared by counting the number of cells having pBP130 (CarbR) after grown for 12 h at 37 °C.
Plasmid stability analysis
To quantify plasmid stability in a recombinant E. coli strain, the cell line was grown overnight in LB medium containing 50 µg/ml kanamycin, 100 µg/ml carbenicillin, 50 µg/ml streptomycin and 34 µg/ml chloramphenicol. The culture was diluted 106-fold, and 100 µl was spread on an LB agar petri-dish with containing each antibiotic individually as well as control plates containing all antibiotics or lacking any antibiotic. Plasmid stability was estimated by comparing the number of colonies on plates with individual antibiotics to the control plates.
Construction of hybrid plasmids
To localize mutations on mutant plasmids pBP175#1 and pBP175#8, three different hybrid plasmids were constructed in each case using the unique PacI, PstI and/or XbaI sites on this plasmid 17. The first hybrid contained the vector from wild-type pBP175 and the DEBS module 2 and pccAB genes from the mutant plasmid, whereas the second hybrid had the vector from the mutant plasmid, and the wild-type genes. In the third hybrid, the pccAB genes alone from the mutant plasmid were swapped with the corresponding genes from the wild-type plasmid. The phenotype of each hybrid plasmid was analyzed in E. coli HYL3 along with wild-type plasmids pBP130, pHL80* and pHL74.
Preparative production and isolation of 6-deoxyerythromycin D
E. coli HYL3/pBP130/pBP144,/pHL74/pHL80* cells were grown in LB media containing 50 µg/ml kanamycin, 100 µg/ml carbenicillin, 50 µg/ml streptomycin and 34 µg/ml chloramphenicol at 37°C to OD600 = 0.4–0.5. The cell pellets were collected by centrifugation and resuspended in 5% of the original volume of 4°C LB media supplemented with antibiotics at the concentrations mentioned above. The cells were induced with 500 µM IPTG along with the addition of 2.5 g/l sodium propionate and left to shake at 30°C for 72h.
After the 72h production phase, the culture broth was clarified by centrifugation for 1h at 4420 g. The supernatant was basified to pH=12 with 2 M NaOH, and extracted with EtOAc. The organic phase was dried over Na2SO4, and the solvent was removed in vacuo. Preliminary purification through an acid-base back extraction was performed based on the presence of a tertiary amine on the target molecule. The crude extract resuspended in 10 ml water was acidified to pH=2 with 2 M HCl, and washed with 10 ml Et2O 5 times. The aqueous phase was then basified to pH=11 with 2 M NaOH, and extracted with 10 ml EtOAc 5 times. The EtOAc extracts were combined, dried over Na2SO4 and the solvent was removed by evaporation. For the final purification, HPLC was used with an Applikon Zorbax RX-C8 column (9.4 × 250 mm) with a solvent system of CH3CN/H2O each with 0.1% formic acid. The desired product eluted at 45–50 min (CH3CN gradient: 0–25% in 10 min, 25–40% in 80 min, 40–95% in 10 min, 95% for 10 min; detection at 210 nm). The identity of 6-deoxyerythromcin D was confirmed by high resolution mass spectrometric and NMR analysis (Table 2 and supporting information). The MIC of this compound was determined to be 0.04 µg/ml against B. subtilis.
Results and Discussion
Analysis of the Mutant D
Individual plasmids were isolated from Mutant D and re-transformed into E. coli BAP1 along with the remaining complement of wild-type plasmids. In addition, Mutant D was cured of all plasmids by propagation in the absence of selection markers, and was subsequently transformed with a full complement of wild-type plasmids. At least four clones of each lineage were independently isolated and compared to both Mutant D and wild-type E. coli BAP1/pBP175/pBP130/pHL80*/pHL74 using single colony assays on plates that included 10 µM diketide 1. Unexpectedly, the overproduction phenotype could be exclusively assigned to the re-transformed derivative of the cured host cell derived from Mutant D (data not shown). This new cured host, designated E. coli HYL3, was therefore assumed to carry the phenotypically relevant genetic change identified in Mutant D, and was investigated further, as described below.
To determine if the mutant strain only improved diketide directed biosynthesis of 6-deoxyerythromyin D, or whether it could also enhance macrolide production upon expression of the complete biosynthetic pathway, E. coli HYL3/pBP144/pBP130/pHL80*/pHL74 was compared to its BAP1 counterpart using single colony assays. No statistical difference was observed between the two host cells. We alsocompared two related aglycone-producing systems, HYL3/pBP175/pBP130/pBP190 versus BAP1/pBP175/pBP130/pBP190. Plasmid pBP190 encoding the thioesterase II from S. erythraea was included in these cell lines 19 (Table 1). The former strain produced more 6-deoxyerythronolide B than the latter strain in the presence of diketide 1 (Figure 2a). Taken together, these results highlight HYL3’s ability to increase polyketide production while suggesting that the phenotypically relevant mutation functions either through increasing the efficiency of acylation of module 2 by compound 1 or by increasing the intracellular concentrations of stand alone module 2.
Figure 2. Analysis of E. coli HYL3 mutant.

a) Biosynthesis of 6-deoxyerythronolide B by E. coli strains BAP1/pBP175/pBP130/pBP190 and HYL3/pBP175/pBP130/pBP190. (b) Densitometric quantitation of DEBS module 2 and PccAB expressed in BAP1/pBP175 and HYL3/pBP175.
To evaluate these options, the expression levels of the biosynthetically relevant proteins were compared between the two hosts. E. coli HYL3/pBP175 showed an approximately 3-fold higher expression of soluble module 2 than BAP1/pBP175 (Figure 2b). Owing to their large size and codon usage differences between the native and heterologous hosts, type I polyketide synthase proteins are generally not expressed well in E. coli 20. Previous work has shown the loading of diketide thioester substrates onto DEBS module 2 to be slow compared to subsequent steps, suggesting the increased concentration of module 2 likely relieves a kinetic bottleneck in precursor directed biosynthesis of erythromycin antibiotics.
As shown below, the ability of E. coli HYL3 to effect increased erythromycin production by precursor directed biosynthesis is not exclusive to the wild-type system as it also led to increased titers of 4, an unnatural alkynyl derivative. (Figure 3a,b).
Directed evolution of plasmid pBP175
The above result suggested that DEBS module 2 could be an attractive target for evolution aimed at further enhancing precursor directed biosynthesis of macrolides. In particular, due to the high cost of synthetic precursors and the strong dependence of polyketide production on exogenous precursor concentrations 21 we sought to identify mutants capable of more efficient transformation of low concentrations of synthetic precursors into macrolides. We therefore subjected plasmid pBP175 to random mutagenesis by passing it through the mutator strain of E. coli, XL1-Red 22, for multiple generations. Plasmid DNA was isolated and co-transformed into HYL3 along with unmutated plasmids pBP130, pHL80* and pHL74. Approximately 500–1000 colonies from the resulting library of transformants were screened in a single-colony assay for mutants that showed larger zones of growth inhibition of B. subtilis than HYL3/pBP175/pBP130/pHL80*/pHL74 in the presence of limiting (10µM) concentrations of diketide 1. Two mutants, designated mutant #1 and mutant #8, were isolated and purified by re-streaking and re-testing. As discussed below, their enhanced ability to convert synthetic precursors into corresponding macrolides is not limited to diketide 1 alone (Figure 3a). By isolating and characterizing the host cell and each plasmid from mutant #1 and mutant #8, it was deduced that both mutants contained phenotypically relevant changes in plasmid pBP175, which were designated pBP175 #1 and pBP175 #8, respectively.
Analysis of mutant plasmids pBP175 #1 and pBP175 #8
Because of the large size of plasmid pBP175, sequencing was not considered a practical strategy for decoding the mechanistic basis for enhanced precursor directed biosynthesis of pBP175 #1 or pBP175 #8. Alternatively, we investigated several properties of each mutant plasmid, including copy number, stability and level of module 2 expression. While these represented the pertinent altered phenotypes within previous studies of pHL80* and the HYL3 host 17,, no statistical differences in these properties were observed here between wild-type pBP175 and either mutant plasmid. We therefore constructed three hybrid plasmids each from pBP175 #1 and pBP175 #8, as described in the Methods section. Each hybrid plasmid contained a portion of the mutant plasmid with the remainder being derived from wild-type pBP175. In the case of both mutant #1 and mutant #8, the phenotypically relevant mutation was exclusively localized to the vector portion, as opposed to either the DEBS module 2 or pccAB genes.
Given the apparently contradictory observations that the vector portion of plasmid pBP175 had undergone mutation albeit without affecting its own copy number, stability, or protein expression level, we speculated that these genetic changes may induce a phenotypic benefit for one or more of the other plasmids in the recombinant cell lines. Among plasmids pBP130, pBP175, pHL80* and pHL74, plasmid pBP130 has the lowest stability with only ca. 40% retention when a culture of E. coli BAP1/pBP175/pBP130/pHYL80*/pHYL74 is grown for 12 h in the absence of antibiotic (Figure 3c). Plasmids pBP130 and pBP175 have the same origin of replication (pBR322), a feature that is known to induce plasmid incompatibility 23–25. The larger size of pBP130 presumably makes it less stable than pBP175 due to a slower rate of replication 26. As shown in Figure 3c, the stability of pBP130 is enhanced significantly in the presence of pBP175 #1 and to an even greater extent when co-expressed with pBP175 #8. Taken together, these results suggested the phenotypically relevant mutations were related to the origin of replication of mutant plasmids such that the competition between pBP175 and pBP130 is relieved 27.
Sequencing of the origin of replication of pBP175 #8 revealed a single T to C mutation as compared to the wild-type pBR322 origin, supporting that the phenotypically relevant mutation is in this region of the plasmid (Figure S4).
Improved precursor directed biosynthesis of novel macrolide antibiotics
To test the relevance of the above results for precursor directed biosynthesis of unnatural erythromycin analogs, we targeted preparation of an unnatural macrolide bearing an alkynyl functional group. Alkynes can be used as particularly convenient handles for further semi-synthetic modification using cycloaddition chemistry 28–30. An alkynyl diketide substrate (2, Figure 1) was prepared and fed to the various cell lines as detailed in the methods section. The presence of macrolide 4 and absence of compound 3 in liquid cultures that were fed diketide 2 was confirmed by high-resolution mass spectrometry. This result suggests that the bioactivity observed in these experiments does arise from the production of a novel alkynyl erythromycin analog (see supporting information). The bioconversion efficiency of diketide 2 was comparable to that of diketide 1, and the relative productivity trends observed between BAP1/pBP175/pBP130/pHL80*/pHL74 and various mutant strains characterized in this study were also similar for the two precursors (Figure 3a, b). These results reinforce the utility of E. coli HYL3/pBP175#8/pBP130/pHL80*/pHL74 as a superior cellular biocatalyst for precursor directed biosynthesis of erythromycin analogs.
In summary, we have evolved a system capable of improved production of erythromycin-type macrolide antibiotics by precursor directed biosynthesis in E. coli. The parent producer of 6-deoxyerythromycin D was subjected to two rounds of random mutagenesis and with the improved mutants identified via activity-based single colony screening procedures. Phenotypic characterization revealed no changes of the biosynthetic proteins themselves, but rather a genetically more stable system in which altered protein expression relieved a kinetic bottleneck present in the wild-type system. The best of these mutants is not only able to produce four-fold more antibiotic, but also does so at a relatively low precursor concentration. Finally, we have demonstrated the utility of this system by, for the first time, reporting the production of a novel erythromycin analog derived from a simple synthetic precursor in a single E. coli cell line. This new antibiotic not only displays promising antibacterial activity, but also possesses an attractive handle for semi-synthetic modification.
Supplementary Material
Acknowledgements
This research was supported by grants from the National Institutes of Health to C.K. (GM 087934) and D.E.C. (GM 22172). C.J.B.H. is supported by a fellowship from the Stanford Center for Molecular Analysis and Design (CMAD).
Footnotes
Dedicated to the late Dr. C. Richard Hutchinson for his exceptional contributions to natural product biosynthesis, engineering, and drug discovery.
* Supporting Information available: This material is available free of charge via the Internet.
References
- 1.Cane DE, Walsh CT, Khosla C. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 2.Khosla C, Gokhale RS, Jacobsen JR, Cane DE. Tolerance and specificity of polyketide synthases. Annual review of biochemistry. 1999;68:219–253. doi: 10.1146/annurev.biochem.68.1.219. [DOI] [PubMed] [Google Scholar]
- 3.Tenover FC, et al. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrobial agents and chemotherapy. 2004;48:275–280. doi: 10.1128/AAC.48.1.275-280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh CT. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science (New York, N.Y. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsen JR, Hutchinson CR, Cane DE, Khosla C. Precursor-directed biosynthesis of erythromycin analogs by an engineered polyketide synthase. Science (New York, N.Y. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 6.Cane DE, Kudo F, Kinoshita K, Khosla C. Precursor-directed biosynthesis: biochemical basis of the remarkable selectivity of the erythromycin polyketide synthase toward unsaturated triketides. Chemistry & biology. 2002;9:131–142. doi: 10.1016/s1074-5521(02)00089-3. [DOI] [PubMed] [Google Scholar]
- 7.Boddy CN, Hotta K, Tse ML, Watts RE, Khosla C. Precursor-directed biosynthesis of epothilone in Escherichia coli. Journal of the American Chemical Society. 2004;126:7436–7437. doi: 10.1021/ja048108s. [DOI] [PubMed] [Google Scholar]
- 8.Elliott RL, et al. Anhydrolide macrolides. 1. Synthesis and antibacterial activity of 2,3-anhydro-6-O-methyl 11,12-carbamate erythromycin A analogues. Journal of medicinal chemistry. 1998;41:1651–1659. doi: 10.1021/jm970547x. [DOI] [PubMed] [Google Scholar]
- 9.Sheehan LS, et al. Engineering of the spinosyn PKS: directing starter unit incorporation. Journal of natural products. 2006;69:1702–1710. doi: 10.1021/np0602517. [DOI] [PubMed] [Google Scholar]
- 10.Donadio S, McAlpine JB, Sheldon PJ, Jackson M, Katz L. An erythromycin analog produced by reprogramming of polyketide synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:7119–7123. doi: 10.1073/pnas.90.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobsen JR, Keatinge-Clay AT, Cane DE, Khosla C. Precursor-directed biosynthesis of 12-ethyl erythromycin. Bioorganic & medicinal chemistry. 1998;6:1171–1177. doi: 10.1016/s0968-0896(98)00081-9. [DOI] [PubMed] [Google Scholar]
- 12.Dayem LC, et al. Metabolic engineering of a methylmalonyl-CoA mutase-epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry. 2002;41:5193–5201. doi: 10.1021/bi015593k. [DOI] [PubMed] [Google Scholar]
- 13.Lowden PA, Bohm GA, Metcalfe S, Staunton J, Leadlay PF. New rapamycin derivatives by precursor-directed biosynthesis. Chembiochem. 2004;5:535–538. doi: 10.1002/cbic.200300758. [DOI] [PubMed] [Google Scholar]
- 14.Kinoshita K, Williard PG, Khosla C, Cane DE. Precursor-directed biosynthesis of 16-membered macrolides by the erythromycin polyketide synthase. Journal of the American Chemical Society. 2001;123:2495–2502. doi: 10.1021/ja004139l. [DOI] [PubMed] [Google Scholar]
- 15.Frykman S, Leaf T, Carreras C, Licari P. Precursor-directed production of erythromycin analogs by Saccharopolyspora erythraea. Biotechnology and bioengineering. 2001;76:303–310. doi: 10.1002/bit.10086. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita K, Pfeifer BA, Khosla C, Cane DE. Precursor-Directed polyketide biosynthesis in Escherichia coli. Bioorganic & medicinal chemistry letters. 2003;13:3701–3704. doi: 10.1016/j.bmcl.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Lee HY, Khosla C. Bioassay-guided evolution of glycosylated macrolide antibiotics in Escherichia coli. PLoS biology. 2007;5:e45. doi: 10.1371/journal.pbio.0050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peiru S, Menzella HG, Rodriguez E, Carney J, Gramajo H. Production of the potent antibacterial polyketide erythromycin C in Escherichia coli. Applied and environmental microbiology. 2005;71:2539–2547. doi: 10.1128/AEM.71.5.2539-2547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer B, Hu Z, Licari P, Khosla C. Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Applied and environmental microbiology. 2002;68:3287–3292. doi: 10.1128/AEM.68.7.3287-3292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer BA, Khosla C. Biosynthesis of polyketides in heterologous hosts. Microbiol Mol Biol Rev. 2001;65:106–118. doi: 10.1128/MMBR.65.1.106-118.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaf T, et al. Precursor-directed biosynthesis of 6-deoxyerythronolide B analogs in Streptomyces coelicolor: understanding precursor effects. Biotechnology progress. 2000;16:553–556. doi: 10.1021/bp000063l. [DOI] [PubMed] [Google Scholar]
- 22.Greener A, Callahan M, Jerpseth B. An efficient random mutagenesis technique using an E. coli mutator strain. Molecular biotechnology. 1997;7:189–195. doi: 10.1007/BF02761755. [DOI] [PubMed] [Google Scholar]
- 23.Novick RP. Plasmid incompatibility. Microbiological reviews. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordstrom K, Austin SJ. Mechanisms that contribute to the stable segregation of plasmids. Annual review of genetics. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 25.Austin S, Nordstrom K. Partition-mediated incompatibility of bacterial plasmids. Cell. 1990;60:351–354. doi: 10.1016/0092-8674(90)90584-2. [DOI] [PubMed] [Google Scholar]
- 26.Smith MA, Bidochka MJ. Bacterial fitness and plasmid loss: the importance of culture conditions and plasmid size. Canadian journal of microbiology. 1998;44:351–355. [PubMed] [Google Scholar]
- 27.Arini A, Tuscan M, Churchward G. Replication origin mutations affecting binding of pSC101 plasmid-encoded Rep initiator protein. Journal of bacteriology. 1992;174:456–463. doi: 10.1128/jb.174.2.456-463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 29.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective "ligation" of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Himo F, et al. Copper(I)-catalyzed synthesis of azoles. DFT study predicts unprecedented reactivity and intermediates. Journal of the American Chemical Society. 2005;127:210–216. doi: 10.1021/ja0471525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.