Abstract
Nuclear pore complexes (NPCs) function as selective gates for nucleocytoplasmic transport. Although the NPC was discovered more than half a century ago, our knowledge of NPC components in atomic detail has exploded only over the past few years. Recent structural, biochemical, and in vivo studies of NPC components, in particular the membrane-coating heptameric Nup84 complex, have shed light onto the NPC architecture as well as onto its dynamic nature. Striking similarities were revealed between the components of the NPC and of coat protein complexes in the endocytic and secretory pathways, supporting their common evolutionary origin in a progenitor protocoatomer. Here, we summarize these findings and discuss emerging concepts that underlie the molecular architecture and the dynamics of the NPC. We conclude that the uncovered principles are not limited to the NPC, but are likely to extend to other macromolecular assemblies.
Key words: nucleoporin, nucleocytoplasmic transport, coat protein complex, macromolecular assembly, dynamic interaction, binding promiscuity, crystallography, electron microscopy, vesicle coats, coatomer
The evolution of the eukaryotic cell has entailed the formation of intricate intracellular membrane systems that enable it to concurrently carry out a diverse set of processes in various chemical milieus. Among the dozen or so compartments, the nucleus harbors the majority of the genetic information that is encoded in the DNA and protects it from the potentially harmful environment of the cytosol. Moreover, the physical separation of transcription in the nucleus from translation in the cytosol enables the cell to regulate the flow of genetic information from DNA to RNA to protein at various stages. As a result of this compartmentalization, sophisticated transport systems have evolved which mediate and regulate macromolecular transport between different subcellular locations.1 In transmembrane transport, membrane-embedded protein translocators directly transport proteins from the cytosol across an intracellular membrane. In vesicular transport, cargo is ferried between membrane-bound organelles via membrane-enclosed intermediates, such as spherical vesicles, which are stabilized by protein coats that are reminiscent of cages. A third major transport system refers to the selective macromolecular transport in and out of the nucleus, which is exclusively mediated by nuclear pore complexes (NPCs). NPCs are embedded into circular openings of the nuclear envelope (NE), thus resembling “biological grommets”.2,3 The NE is formed by two closely juxtaposed membranes, termed the inner nuclear membrane (INM) and outer nuclear membrane (ONM). The INM contains a unique set of membrane proteins that bind to chromatin and the nuclear lamina inside the nucleus.4,5 The INM proteins establish connections with the cytoskeleton via interactions with ONM proteins inside the lumen of the NE.6,7 While the proteins in the INM generally attach to densely-packed heterochromatin, NPCs are typically associated with actively transcribed, loosely-packed euchromatic regions.8 Thus, the NE, together with the embedded NPCs, is an important interface between the nucleus and the cytosol that is involved in nucleocytoplasmic transport, chromatin organization, replication-coupled DNA repair, regulation of gene expression, nuclear positioning, and nuclear migration among other cellular processes.9–12
With a molecular weight of ∼60 MDa in yeast and ∼120 MDa in humans, the NPC represents one of the largest proteinaceous assemblies in the eukaryotic cell.13 The NPC is composed of ∼30 different proteins collectively termed nucleoporins or nups that are organized in several subcomplexes.3,14,15 Cryo-electron microscopy revealed that the core of the NPC contains eightfold rotational symmetry along the nucleocytoplasmic axis and twofold rotational symmetry in the plane of the NE.16 Due to the high degree of internal symmetry of the NPC, each nucleoporin occurs in multiple copies. Thus, the fully-assembled NPC contains hundreds of protein molecules that are held together by an even larger number of interactions.17 Despite these numerous interactions, NPCs readily dismantle during prometaphase into distinct subcomplexes from which they reassemble at the completion of telophase in eukaryotes that undergo open mitosis.1
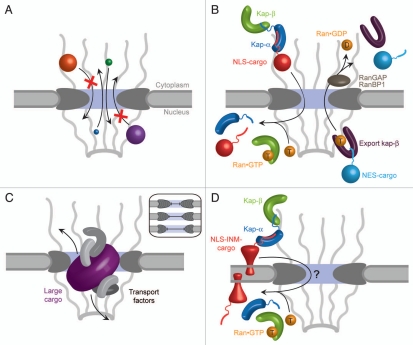
The major function of the NPC is nucleocytoplasmic transport. Ions and molecules smaller than ∼40 kDa can freely diffuse through the aqueous channel of the NPC (Fig. 1A), while the transport of larger macromolecules relies on signal sequences within the cargo molecules, which are recognized by receptors termed karyopherins. The directionality of transport is governed by a gradient of RanGTP, which is maintained at a high level inside the nucleus, but at a low level in the cytoplasm (Fig. 1B).18–21 To allow the passage of large cargo such as pre-ribosomal particles, the central channel has to dilate substantially (Fig. 1C).22,23 Structural rearrangements are probably also necessary for the import of integral membrane proteins destined for the INM. These proteins are thought to remain anchored in the pore membrane during import, while the attached karyopherin transport factors pass through the central channel (Fig. 1D).16,24–26
Figure 1.
Transport functions of the NPC. (A) Overall, the NPC consists of a cylindrical, symmetric core, which is asymmetrically decorated with filaments and a nuclear basket structure on the cytoplasmic and nucleoplasmic sides, respectively.53 Molecules smaller than ∼40 kDa freely diffuse through the NPC. (B) Active import and export of cargoes are facilitated by nuclear localization and nuclear export sequences (NLS and NES, respectively) that are recognized by transport factors, collectively termed karyopherins (kaps). The NLS of import cargoes is recognized either directly by an import karyopherin-β (kap-β) or via an adapter karyopherin (kap-α). Ran•GTP binding inside the nucleus leads to dissociation of the import complex (left). By contrast, the assembly of an NES-cargo kap-β export complex requires Ran•GTP binding. In the cytosol, this export complex is dissociated by GTP hydrolysis, which is catalyzed by Ran GTPase-activating protein (RanGAP) or Ran binding protein 1 (RanBP1). (C) The transport of large cargoes is thought to require the dilation of the central channel of the NPC (inset). (D) Inner nuclear membrane (INM) proteins are co-translationally integrated into the endoplasmic reticulum membrane, which is continuous with the outer nuclear membrane (ONM), and then imported to the INM. Similar to (B), the transport of INM proteins is also dependent on the Ran-cycle and karyopherins that likely travel through the central channel, while the cargo protein is anchored in the membrane and somehow pulled through. Substantial structural changes within the NPC would be necessary to facilitate this transport event.
These diverse transport tasks suggest that the NPC is a dynamic assembly. Indeed, electron microscopy (EM) has revealed structural plasticity during the transport of cargo, consistent with a flexible and dynamic structure.26,27 The marked flexibility of the NPC combined with its sheer size makes the determination of the entire structure at high resolution by x-ray crystallography a formidable goal.28 In this context, “high resolution” refers to ∼3 Å or better, where the amino acid sequence can unambiguously be assigned. Therefore, to tackle the atomic structure of the NPC, an interdisciplinary “divide-and-conquer” strategy must be employed. According to this approach, high-resolution crystal structures of individual nucleoporins and of subcomplexes would be fitted into lower-resolution envelopes obtained from three-dimensional EM reconstructions. However, the current resolution limit (∼60 Å) does not allow for the docking of crystal structures into EM maps of the entire NPC,26 since atomic models can generally only be fitted into EM maps at resolutions of ∼15 Å or better with high confidence.
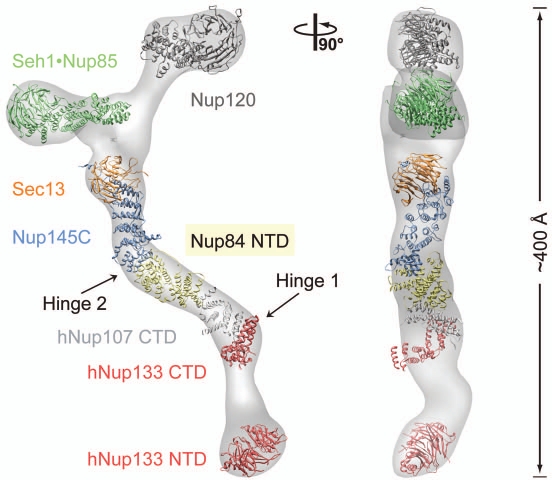
Although the divide-and-conquer strategy has not yet been applied on the whole NPC, this approach proved successful for the structural characterization of the evolutionarily-conserved Nup84 complex, a subcomplex of the NPC core that is located close to the pore membrane. In yeast, the Nup84 complex consists of seven nucleoporins: Seh1, Nup85, Nup120, Sec13, Nup145C, Nup84 and Nup133 (Fig. 2).29–31 Two-dimensional negative-stain EM on the heptamer assembled from recombinant proteins revealed a ∼400-Å-long Y-shaped complex and established the relative position of its members.31 To date, the following structural snapshots of Nup84 complex components have been obtained: (1) the human Nup133 N-terminal domain (NTD),32 (2) the Nup107 (human homolog of Nup84) C-terminal domain (CTD)•Nup133 CTD heterodimer,33 (3) the Sec13•Nup145C heterodimer,34 (4) the Seh1•Nup85 heterodimer,35,36 (5) the Sec13•Nup145C•Nup84 heterotrimer,37 Nup120 NTD (Fig. 2).38,39 The structure of the whole heptameric building block was recently determined by three-dimensional negative-stain EM to ∼35 Å resolution, which allowed the approximate fitting of the available crystal structures into the EM maps (Fig. 2).37,40 Thus, the structural work on the Nup84 complex constitutes a paradigm for the determination of the whole NPC architecture.
Figure 2.
Structural analysis of the heptameric Nup84 complex. Approximately 85% of the protein mass of the heptamer has been determined by x-ray crystallography. High-resolution crystal structures of the yeast nucleoporins Seh1•Nup85, Nup120, Sec13•Nup145C•Nup84, and the human nucleoporins Nup107 (homolog of Nup84) and Nup133 were fitted into a three-dimensional electron microscopy (EM) reconstruction.37 The crystallized N-terminal and C-terminal domains (NTD and CTD, respectively) are indicated. Two hinge regions are indicated at which the heptamer displayed marked flexibility.40 A 90°-rotated view is shown on the right, with the dimension of the heptamer indicated.
Despite this tremendous progress, the molecular architecture of the NPC core remains enigmatic. One of the major obstacles to reach this goal lies in the lack of an in vitro assay to assemble the NPC core, which would provide a specimen for its experimental structure determination by EM and, importantly, may define the components required to create such an assembly. Although, at present, the detailed architectural organization of the NPC core cannot be directly obtained by established structural methods such as EM, various approaches have been applied to arrive at three different models of the structural organization of the NPC: (1) In a “computational model”, a diverse set of biophysical and proteomic data as well as a fixed stoichiometry was used as input to obtain a crude blueprint of the entire NPC.17 (2) The “lattice model” is based on the homology between nucleoporins of the heptamer and proteins of the clathrin and COPII coats.36 (3) We have proposed an architecture of a “coat for the nuclear pore membrane” that is composed of 32 heptamers.34,35,37,38 This “NPC coat” is deduced from several crystal structures, biochemical studies, in vivo data, homologies between COPII components and nucleoporins, as well as symmetry and size considerations derived from EM. A prominent feature of this proposal refers to the homo-oligomerization of several heptamer components, which was observed in the crystal and in solution. Although the three models markedly differ in the arrangement and the stoichiometry of the Nup84 complex in the NPC,41 they more or less assume that the heptamer constitutes a rigid bona fide building block of the NPC. Clearly, more crystal structures combined with biochemical and in vivo data need to be obtained to resolve these ambiguities.
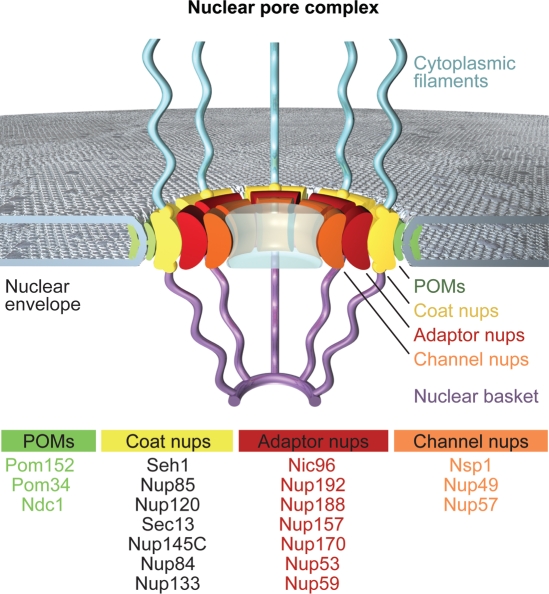
Conceptually, our proposed NPC coat can be envisioned to represent one out of four concentric cylinders (Fig. 3). This cylinder is sandwiched between the pore membrane protein (POM) and the adaptor cylinders. The POMs, which are embedded in the nuclear envelope, are thought to anchor the NPC to the pore membrane, while the adaptor cylinder would cushion dynamic changes in the pore diameter during the nucleocytoplasmic transport of cargo. The innermost channel cylinder contains the phenylalanine-glycine (FG)-repeat nucleoporins that contribute to the transport barrier and provide the docking sites for karyopherins. These cylinders should not be mistaken for solid walls. Instead, interdigitation of adjacent cylinders is likely to occur to a large extent, with only the principal mass of the nucleoporins being localized in the respective cylinders. Moreover, large-scale structural rearrangements of the cylinders may be necessary to fulfill the diverse transport tasks of the NPC.
Figure 3.
A schematic model of the NPC. The model includes four concentric cylinders composed of integral pore membrane proteins (POMs), coat nups, adaptor nups and channel nups. Natively unfolded FG-repeats of a number of nups make up the transport barrier in the central channel and are indicated by a transparent plug. Note that the surface topology of the pore membrane involves both convex and concave curvatures.
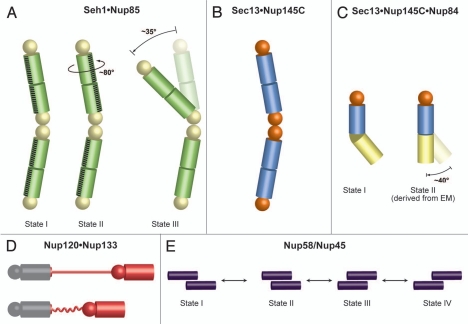
Interestingly, the numerous structural snapshots of nucleoporins and subcomplexes also offer insight into the flexibility of NPC components in atomic detail. This is particularly noteworthy, since, until recently, the dynamics of the NPC was only described on a phenomenological level, but mechanistic details remained elusive.26,27 Crystal structures of the hetero-octameric Seh1•Nup85 pole revealed three different conformational states which are related by a simple hinge motion and a rotation, respectively (Fig. 4A).35 On the molecular level, these motions are accomplished by slight rearrangements of two neighboring Seh1 β-propellers at the center of the Seh1•Nup85 pole. The observed movements provide examples of how individual parts of the NPC may rearrange. Comparison of the Sec13•Nup145C and Sec13•Nup145C•Nup84 structures revealed that Nup145C can promiscuously interact either with another copy of Nup145C or with Nup84 (Fig. 4B and C)34,37 and suggests that the binding partners may switch in the NPC. Furthermore, the interaction of Nup120 with Nup133 is mediated by a flexible, anchor-like element. The interaction between Nup120 and Nup133, located at opposite ends of the heptamer (Fig. 2), supports a head-totail arrangement of heptamers, consistent with the “computational”17 and “NPC coat”34,35 models, but inconsistent with the “lattice model”.17,34,36 As the attachment site for Nup120 resides at the N-terminal end of a ∼55-residue unstructured region in Nup133, this flexible segment may allow for a varying diameter of an eightmembered ring of heptamers (Fig. 4D).38 A dynamic behavior was also observed for the whole Nup84 complex at low resolution (∼35 Å) by EM.40 This EM analysis described two specific hinge regions at which the heptamer shows great flexibility (Fig. 2).40 While the dynamics of the coat cylinder, including the heptameric complex, is thought to be important for the import of membrane-embedded INM proteins, the dynamics of the adaptor and channel cylinders is critical for the passage of cargo through the central channel. Circumferential sliding of helical elements in the nucleoporins Nup58 and Nup45 offers a plausible mechanism for the dilation of the central transport channel in response to cargo translocation (Fig. 4E).42
Figure 4.
Flexibility and dynamics of NPC components. Spheres represent β-propeller domains, while cylinders represent α-helical solenoid domains (A) Crystal structures of the Seh1•Nup85 hetero-octamer (yellow and green, respectively) in three different conformational states revealed a rotation and a hinge motion.35 (B) Sec13•Nup145C (orange and blue, respectively) forms a hetero-octamer.34 (C) Sec13•Nup145C•Nup84 (colored as in panel B, Nup84 in yellow) forms a subcomplex that matches a hinge observed in one conformation of the heptameric Nup84 complex as determined by EM.37 A straight arrangement of this trimer would correspond to the second conformation of this subcomplex as revealed by EM.40 Notably, the homodimerization and heterodimerization surfaces of Nup145C are overlapping and therefore promiscuous. (D) Nup133 binds to Nup120 via a short N-terminal segment at the end of an unstructured region.38 Since the two proteins are localized at opposite ends of the Nup84 complex, this finding suggests a head-to-tail arrangement of the heptamers. Contraction and expansion of the unstructured region may allow for the flexible tethering of Nup120 and Nup133. (E) Four different states of Nup58/Nup45 tetramers have been crystallographically observed.42 The sliding of two Nup58/Nup45 dimers (purple) alters the overall dimensions of the tetramer. Nup58/Nup45 refers to two alternatively spliced variants (Nup58 and Nup45) and the determined structure is common to both proteins.
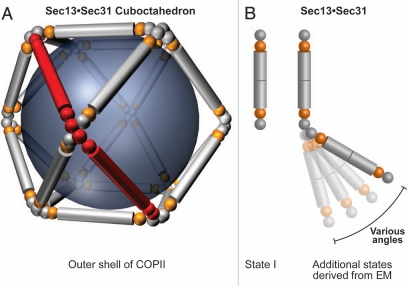
Besides these insights into the molecular architecture and dynamics of the NPC, the recent structural work has uncovered numerous remarkable similarities between the NPC and coated vesicles34,43 and supports the hypothesis for their common evolutionary origin in a progenitor protocoatomer.44 According to the protocoatomer proposal, the heptamers serve as “membrane curving modules,” similar to the coat protein complexes I and II (COPI and COPII) and clathrin complexes, which play critical roles in cargo trafficking between different compartments of the secretory and endocytic pathways.45–47 In detail, the proteins of both systems typically contain a β-propeller fold, an α-solenoid fold, or a distinctive arrangement of both. Sec13 is a common component of the NPC and COPII (Figs. 3 and 5) and promiscuously interacts with both Nup145C and Sec31 in the NPC and the COPII, respectively, forming related hetero-octameric complexes. In the outer shell of the COPII complex, the heterooctameric Sec13•Sec31 complex forms the basic unit, twelve copies of which homo-oligomerize to generate a complete cuboctahedral cage (Fig. 6A). In the NPC, the hetero-octameric Sec13•Nup145C complexes are thought to form eight vertical rods in a fence-like coat.34 Consistent with the more complicated surface topology of the pore membrane with respect to the curvature of membrane-bound vesicles, additional nucleoporins are required to form a complete coat for the pore in the nuclear envelope.
Figure 5.

The architecture of the COPII coat. The vesicle-coating COPII complex consists of only five proteins. Sec13 and Sec31 assemble the outer shell, while Sec23, Sec24 and the membrane bound Sar1•GTP assemble the inner shell of COPII. In contrast to the pore membrane domain, the curvature of membrane-bound vesicles is convex in all directions.
Figure 6.
The COPII coat is dynamic. (A) The Sec13•Sec31 hetero-octamer (red) is the repeating unit of the outer shell of the COPII complex. A cuboctahedral arrangement made of 12 modules is depicted, but other larger geometries of COPII cages were also observed by EM.48 Sec13 and Sec31 are shown in orange and grey, respectively. The inner shell of the COPII complex has been omitted for clarity. (B) With increasing cage size, the decreasing curvature of the vesicle is accounted for by a hinge motion that leads to more extended conformations of the Sec13•Sec31 hetero-octamer.49 Spheres represent β-propeller domains, while cylinders represent α-helical solenoid domains. Although Sec13 interacts with Sec31 and Nup145C in an analogous fashion, the higher-order organization differs and is facilitated by an additional β-propeller domain in Sec31 that is absent in Nup145C. The Sec13 binding site in Sec31 is located between the N-terminal β-propeller domain and the C-terminal α-helical solenoid domain.
In addition to the structural parallels, Sec13•Sec31 of COPII and the homologous Sec13•Nup145C and Seh1•Nup85 nucleoporin pairs in the NPC display a similar oligomerization behavior in solution. In the COPII coat, the mammalian Sec13•Sec31 protein pair exists as a heterotetramer in solution, which synergistically assembles into closed cages under certain buffer conditions, enabling the structure determination of the COPII cage by EM.48 Notably, the hetero-octameric species is not detectable in solution, although it forms the repeating unit of the cuboctahedral COPII cage (Fig. 6A).48 In the case of the NPC components, Sec13•Nup145C and Seh1•Nup85 predominantly exist as heterotetramers and heterodimers in solution, respectively, which are in equilibrium with smaller populations of higher-order species, including Sec13•Nup145C hetero-octamers.34,35 Although the interactions between the isolated units are relatively weak in solution, their association is most likely dramatically enhanced in the cellular context. In particular, the recruitment of the units to their subcellular destinations, i.e., the nuclear pores and the prebudding membranes, respectively, leads to a high local concentration of the units and, thus, to their energetically-favored assembly.
Similarities between NPC and COPII components extend to the dynamic nature of these macromolecular assemblies. To accommodate vesicles of various sizes and shapes, the COPII cage adopts various geometries.49 Specifically, a flexible hinge region stemming from interactions between the Sec31 β-propeller domains was observed at the vertices (Fig. 6B).49 The observed hinge motion between Seh1 β-propellers in two Seh1•Nup85 conformations35 is analogous to the movement postulated for the Sec31 β-propellers.49 The cuboctahedron represents the smallest of various cage geometries. By incorporation of additional Sec13•Sec31 units in more extended conformations (Fig. 6B), the COPII cage can expand to coat larger vesicles.
While considerable parallels have been revealed between the COPII and the NPC coats, the interaction with the membrane remains an unresolved question in the case of the NPC. Notably, the molecular surfaces of the heptamer nucleoporins are predominantly negatively charged.32,34,35,37 Since the membrane surface is also negatively charged, we note that the membraneanchored, positively-charged POMs would be prime candidates for providing the link between the pore membrane and the heptamers (NPC coat cylinder), reminiscent of the membrane-anchored form of Sar1 in the COPII assembly.47 A further fundamental issue refers to the conformation(s) of the heptamer in the NPC. Due to the observed flexibility of the heptamer by EM, it is conceivable that the conformation of the heptamer is affected considerably by the additional nucleoporins in the assembled NPC. These neighboring nucleoporins may even alter the interactions between the heptamer components and lead to markedly different associations than those observed in the isolated heptamer. This idea is supported by the finding that Nup145C has promiscuous binding partners.34,37 The question then arises whether both of these interactions occur in the NPC and, if so, at which stage they would occur, for example during NPC formation, during various NPC functions, or during NPC disassembly. Alternatively, some nucleoporins may act as assembly factors or chaperones that may prevent premature assembly of NPC structures in the cytosol. Lastly, promiscuity may provide redundant interactions in the NPC, which would confer robustness to this complex transport organelle.
Despite these burning questions, several architectural principles are emerging that appear to underlie not only the NPC, but also other macromolecular assemblies. First, weak and promiscuous interactions between stable units seem to be a critical design feature of protein coats that allow them to be malleable and readily disassembled. At the same time, weak interactions ensure that assembly is triggered only at the proper subcellular locations, where high local concentrations of the building blocks are achieved, while their premature assembly at other sites is disfavored. Second, homo-oligomerization represents a key principle with regard to the molecular architectures of COPII and clathrin coats.47 In the NPC, a similar oligomerization behavior of Sec13•Nup145C and Seh1•Nup85 is observed and provided the basis of the “NPC coat” model.34,35 Such homotypic interactions may not be limited to vesicle coats and potentially the NPC. Viral coats provide numerous additional examples of this assembly principle and illustrate how proteins interact with each other in different ways to form the typical icosahedral symmetry of viral capsids.50 Third, the occurrence of alternate binding partners of a specific nucleoporin offers the intriguing possibility that the NPC can adopt markedly different conformations and states, depending on the specific cellular function that the NPC performs in a particular instance. This “protean behavior”, i.e., ability to assume different forms, appears to be a general theme for macromolecular machines.51,52
In conclusion, recent structural and functional data strongly suggest that the NPC is a dynamic entity, which is necessary to fulfill its many diverse functions. Clearly, more crystal structures are needed to obtain a high-resolution architecture of the NPC. The divide-and-conquer approach that was successfully applied to the membrane-coating Nup84 complex has to be expanded to the entire NPC. The key challenge will be the combination of the structural snapshots into a movie that portrays the interconversion of various NPC states. This movie will substantially advance our understanding of the dynamic structure and function of this fundamental transport organelle in four dimensions.
Acknowledgements
We thank Oliver Dreesen, Marc Elsliger, Thomas Huber, Stephan Kutik, Johanna Napetschnig, Alina Patke, Pete Stavropoulos, Till Strowig and Deniz Top for comments on the manuscript, Stephanie Etherton for help with editing of the manuscript, and Gunter Blobel for continuing support and advice. E.W.D. is the Dale F. and Betty Ann Frey Fellow of the Damon Runyon Cancer Research Foundation, DRG-1977-08, and A.H. is supported by a grant from the Leukemia and Lymphoma Society.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11120
References
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular biology of the cell. New York: Garland Science. 2008 [Google Scholar]
- 2.Green NM. The nuclear pore—a biological grommet? Nature. 1982;297:287–288. doi: 10.1038/297287a0. [DOI] [PubMed] [Google Scholar]
- 3.Suntharalingam M, Wente SR. Peering through the pore: nuclear pore complex structure, assembly and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 4.Vlcek S, Foisner R. Lamins and lamin-associated proteins in aging and disease. Curr Opin Cell Biol. 2007;19:298–304. doi: 10.1016/j.ceb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet. 2006;7:369–405. doi: 10.1146/annurev.genom.7.080505.115732. [DOI] [PubMed] [Google Scholar]
- 6.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 9.Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- 10.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, et al. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khadaroo B, Teixeira MT, Luciano P, Eckert-Boulet N, Germann SM, Simon MN, et al. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat Cell Biol. 2009;11:980–987. doi: 10.1038/ncb1910. [DOI] [PubMed] [Google Scholar]
- 12.Towbin BD, Meister P, Gasser SM. The nuclear envelope—a scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Reichelt R, Holzenburg A, Buhle EL, Jr, Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 17.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 18.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 19.Hoelz A, Blobel G. Cell biology: popping out of the nucleus. Nature. 2004;432:815–816. doi: 10.1038/432815a. [DOI] [PubMed] [Google Scholar]
- 20.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 21.Debler EW, Blobel G, Hoelz A. Nuclear transport comes full circle. Nat Struct Mol Biol. 2009;16:457–459. doi: 10.1038/nsmb0509-457. [DOI] [PubMed] [Google Scholar]
- 22.Kiseleva E, Goldberg MW, Allen TD, Akey CW. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci. 1998;111:223–236. doi: 10.1242/jcs.111.2.223. [DOI] [PubMed] [Google Scholar]
- 23.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- 25.Saksena S, Summers MD, Burks JK, Johnson AE, Braunagel SC. Importin-alpha-16 is a transloconassociated protein involved in sorting membrane proteins to the nuclear envelope. Nat Struct Mol Biol. 2006;13:500–508. doi: 10.1038/nsmb1098. [DOI] [PubMed] [Google Scholar]
- 26.Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 27.Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya A. Protein structures: Structures of desire. Nature. 2009;459:24–27. doi: 10.1038/459024a. [DOI] [PubMed] [Google Scholar]
- 29.Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, et al. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 31.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, et al. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci USA. 2009;106:17693–17698. doi: 10.1073/pnas.0909373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci USA. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leksa NC, Brohawn SG, Schwartz TU. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brohawn SG, Partridge JR, Whittle JR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melcák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 43.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 44.Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–198. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- 46.Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Stagg SM, Gurkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, et al. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- 49.Stagg SM, LaPointe P, Razvi A, Gurkan C, Potter CS, Carragher B, et al. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 51.Bamford DH, Gilbert RJ, Grimes JM, Stuart DI. Macromolecular assemblies: greater than their parts. Curr Opin Struct Biol. 2001;11:107–113. doi: 10.1016/s0959-440x(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 52.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fahrenkrog B, Koser J, Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]