Abstract
The Lamin B receptor (LBR) is a pivotal architectural protein in the nuclear envelope. Mutations in the Lamin B receptor lead to nuclear hyposegmentation (Pelger-Huët anomaly). We have exactly quantified the nuclear lobulation in neutrophils from individuals with 0, 1, 2 and 3 functional copies of the lamin B receptor gene and analyzed the effect of different mutation types. Our data demonstrate that there is a highly significant gene-dosage effect between the gene copy number and the nuclear segmentation index of neutrophils. This finding is paralleled by a dose-dependent increase in LBR protein and staining intensity of the nuclear membrane in corresponding lymphoblastoid cell lines, which demonstrates a significant correlation on the protein level as well. We further show that LBR expression continually increases during granulopoiesis in vitro from human precursor cells with ovoid nuclei to multi-segmented neutrophil nuclei 11 days later, indicating relevance for regular human granulopoiesis. Altogether, LBR is a unique model that will allow the systematic study of gene-dosage effects and of modifying endogeneous and exogeneous factors on granulopoiesis.
Key words: lamin B receptor, LBR, pelger anomaly, Pelger-Huët anomaly, granulopoiesis, neutrophil, nucleus, gene-dosage effect, dosage compensation
Introduction
Pelger-Huët anomaly (PHA, MIM 169400) is an autosomal dominant trait. Heterozygous persons have incompletely segmented neutrophil nuclei, while homozygous individuals show ovoid nuclei. The interfamilial variability between carriers, however, is considerable.1 Apart from hereditary Pelger-Huët anomaly, there are also acquired forms (“Pseudo-Pelger”). Thus, hyposegmentation is observed as a transient phenomenon in inflammatory processes (“left shift”), myelodysplastic syndromes, some leukemias, and toxic drug reactions.2–5 Hypersegmentation (“right shift”) is associated with defects in vitamin B12 and folate metabolism, iron deficiency, and cases of myelodysplasia.2,6–8
The hereditary forms are caused by mutations in the lamin B receptor (LBR), a multifunctional protein of the inner nuclear membrane.9 The nucleoplasmic part interacts with lamin B, heterochromatin protein 1 (HP1) and other nuclear components, explaining its structural impact on nuclear shape, while the transmembrane part belongs to the sterol reductase family.10–14
It has also been shown in the mouse model that adequate levels of LBR are required for neutrophil nuclear lobulation. Mice that are homozygous for null mutations at the ichthyosis (ic) locus, which codes for LBR, present abnormalities in neutrophil nuclear morphology quite similar to those observed in human Pelger anomaly.15,16 If progenitor EML cells are generated from these mice and induced to mature neutrophils, the cells also exhibit nuclear hyposegmentation.17,18
The typical nuclear structure of human neutrophils is an evolutionarily evolved and conserved characteristic. Interspecies comparison of nuclear morphology in neutrophils revealed ovoid nuclei in most non-vertebrates, followed by rodlike or bilobed nuclei in fish and birds, and multi-segmented nuclei in neutrophils of most mammals.19 This is reminiscent of the stages in human granulopoiesis since CD34+ progenitor cells in the bone marrow also have ovoid nuclei progressing to rod-like (or bilobed) forms and ending in a multi-segmented nuclear shape in mature neutrophils.19–21
In this study our goal was to quantify the gene-dosage effect for the lamin B receptor in human neutrophils. We analyzed LBR expression during in vitro granulopoiesis of human neutrophils. Furthermore, we analyzed blood smears and, where available, lymphoblastoid cell lines from individuals with zero to three functional copies of the LBR-gene (Table 1): two patients with homozygous Pelger anomaly (0 functional LBR copies), nine persons with heterozygous Pelger anomaly (1 functional LBR copy), 12 controls (2 functional LBR copies), and 3 individuals with a triplication of the LBR-gene (3 functional LBR copies). We show that (1) the degree of nuclear segmentation depends in a quantitative way on the type of mutation, (2) a highly significant correlation exists between the amount of LBR protein, as determined by the number of 0 to three functional LBR copies, and the degree of nuclear segmentation, and (3) that also during granulopoiesis in vitro, changes in neutrophil nuclear morphology are correlated with an increase in the expression of the LBR-gene. This is a unique situation in humans, which allows a systematic study of the gene-dosage effect of a series from null to three functional copies of an autosomal structural protein on a distinct cellular phenotype in vivo.
Table 1.
Overview on sample IDs according to type of mutation or chromosomal rearrangement respectively
| Patient ID | Mutation/Karyotype | Status | Reference/Name |
| 02P316 | (R) unknown regulatory mutation | homozygous | znar et al.32 |
| 8387 | (S) IVS12-5-10del | homozygous | Hoffmann et al.9 |
| 8388 | (S) IVS12-5-10del | heterozygous | mother of 8387 |
| 8386 | (S) IVS12-5-10del | heterozygous | father of 8387 |
| 8114 | (S) IVS12-5-10del | heterozygous | Hoffmann et al.9 |
| 6295 | (S) IVS12-5-10del | heterozygous | Hoffmann et al.9 |
| 8198 | (S) IVS12-5-10del | heterozygous | Hoffmann et al.9 |
| 11747 | (N) c.32-35del4; p.V11E fs X14 | heterozygous | this report |
| 11904 | (N) c.265C>T; p.R89X | heterozygous | this report |
| 11300 | (N) c.1129C>T; p.R377X | heterozygous | mutation described in9 |
| 11299 | (N) c.1129C>T; p.R377X | heterozygous | mutation described in9 |
| A | control | wild type | |
| B | control | wild type | |
| C | control | wild type | |
| D | control | wild type | |
| E | control | wild type | |
| F | control | wild type | |
| G | control | wild type | |
| H | control | wild type | |
| I | control | wild type | |
| J | control | wild type | |
| K | control | wild type | |
| L | control | wild type | |
| 03P394 | 46,XY, dup(1)(q32q44) | duplication | Nowaczyk et al.33 |
| 01P233 | 46,XY, der(1)(1qter→1q42.3::1p36.3→1qter) | duplication | this report |
| 01P146 | 46,XY, der(3)(3qter→3p26.1::1q25→1qter) | duplication | this report |
We analyzed data from 26 individuals. Two were homozygous for LBR mutations (one with a splice-site mutation and another one with a putative regulatory mutation, (see materials and methods section), and 9 were heterozygous (5 with splice mutation IVS12-5-10del and 4 with null mutations). We further analyzed 12 controls and three individuals with three copies of the LBR-gene due to a cytogenetic rearrangement with duplication of the LBR-gene region at chromosome 1q42 on one allele. (N, null mutation; S, splice site mutation; R, regulatory mutation).
Results
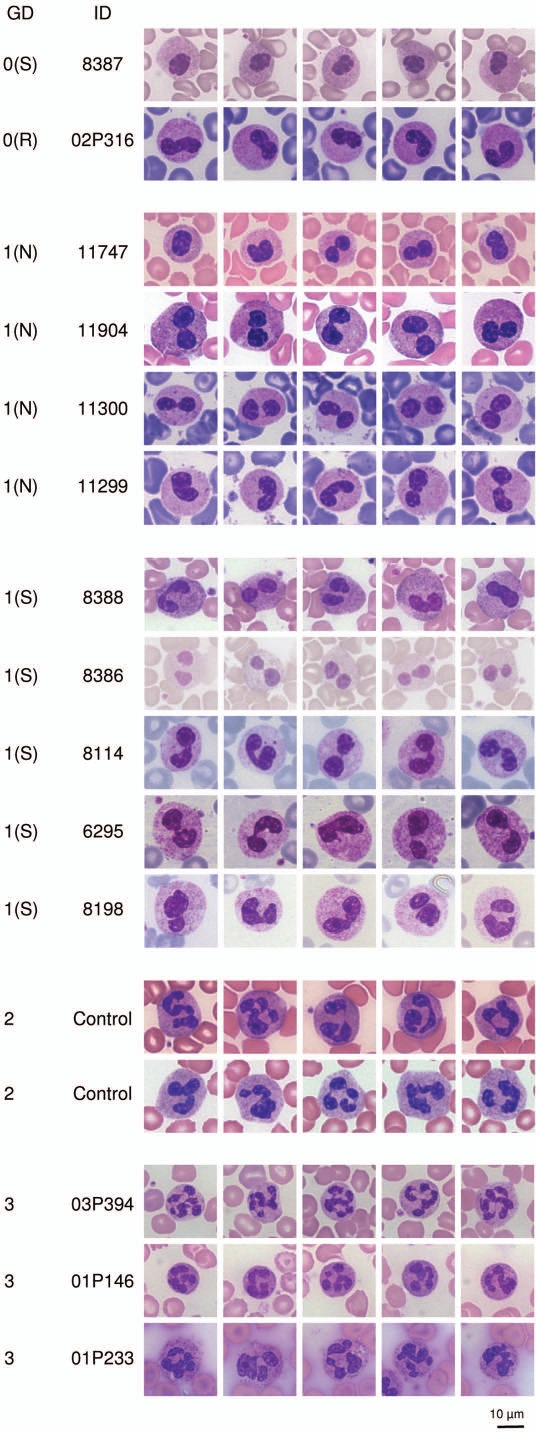
We analyzed blood smears from 2 homozygous and 9 heterozygous PHA persons, 12 controls, and 3 individuals with triplication of the LBR-gene. The number of functional copies of the LBR-gene (0–3) was strongly correlated with the segmentation of neutrophil nuclei in native blood smears (Fig. 1).
Figure 1.
Effects of 0 to 3 functional (wild type) LBR copies on nuclear segmentation in neutrophils. The number of functional LBR copies correlates with the nuclear segmentation of neutrophils in blood smears. A representative panel of 5 neutrophils per individual is shown. In total, 50 neutrophils per individual were analyzed.
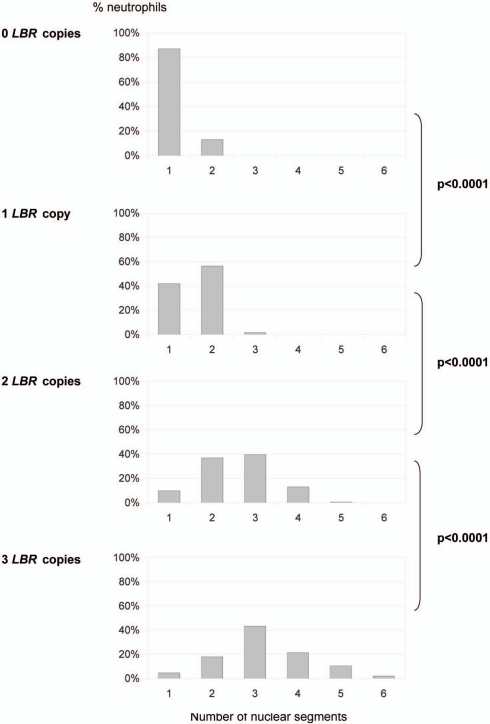
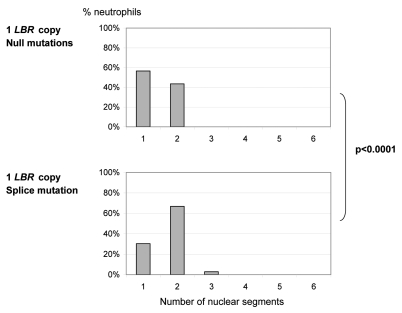
This gene dosage effect was confirmed objectively by the lobulation index, as assessed either through the “Fadenregel” (Rule of Threads) (data not shown) or the “Drittelregel” (Rule of Thirds) (Fig. 2A, Suppl. Table 1). According to the “Fadenregel”, a segment is defined by lobes which are connected only via a thin chromatin thread. According to the “Drittelregel”, a segment is defined by an indentation with width <1/3 of length.22,23 The differences between 0, 1, 2 and 3 functional genes were highly significant in terms of mean values and standard deviations (p < 0.0001). Additionally, the neutrophil nuclei of the five heterozygous persons with the splice mutation IVS12-5-10del had more segments (mean 1.72) than those of the four heterozygous persons with null mutations (mean 1.44; p < 0.0001) (Suppl. Table 1, Fig. 2B), supporting again the gene-dosage effect. We noticed that with increasing gene dose the phenotypic variation increased as well (“norm of variation”, Suppl. Fig. 1).
Figure 2.
Significant dose dependent effect of LBR on the number of nuclear segments in neutrophils, calculated by the “Drittelregel”. (A) The differences between 0, 1, 2 and 3 functional LBR copies are highly significant (p < 0.0001) in terms of mean values and standard deviation (0 LBR-genes: 1.130 ± 0.338; 1 LBR-gene: 1.596 ± 0.522; 2 LBR-genes: 2.573 ± 0.856; 3 LBR-genes: 3.213 ± 1.072). The graph is based on the analysis of 1,300 neutrophil nuclei derived from 26 individuals (2 patients with no wild type allele, 9 individuals with one wild type allele, 12 controls with two wild type alleles and 3 patients with three wild type alleles). 50 neutrophils were analyzed per individual.
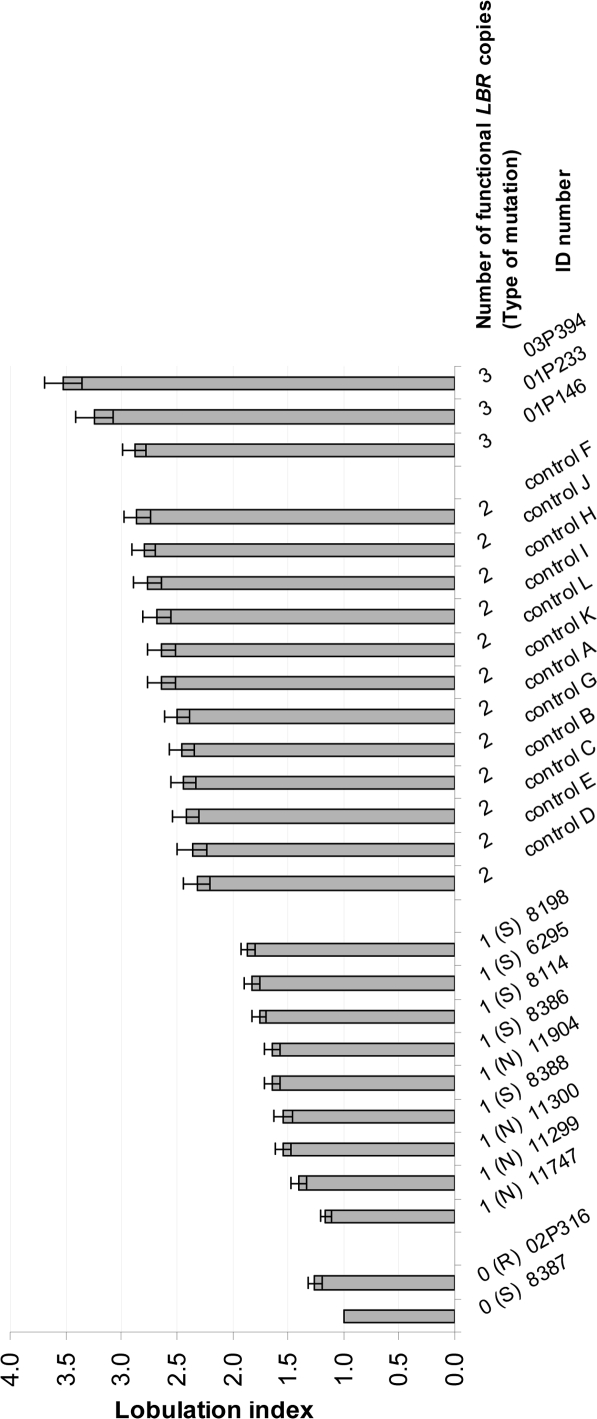
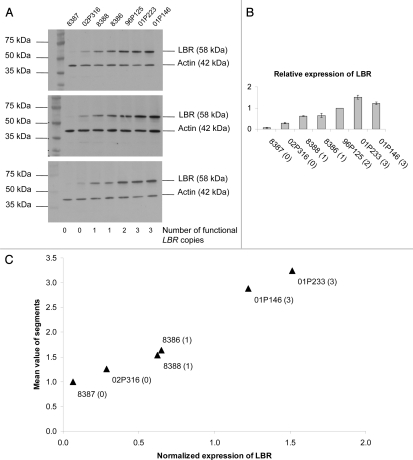
When we listed the lobulation index of each individual in ascending order (Fig. 3), we found a correlation between the segmentation grade and the amount of wild type LBR within the zero and one category. Thus, the homozygous patient with the suspected regulatory mutation, who had more wild type LBR protein than the patient with the homozygous splice mutation also showed a higher lobulation index (for details see the materials and methods section, Suppl. Table 1, Figs. 3 and 4). However, we also observed variability, as the lobulation index of one of the carriers was not different than for the homozygous individuals and one of the patients with three copies had a lobulation index almost within the range of normal diploid individuals. The clearest difference was observed between the class with one and two wild type alleles (Fig. 3, Suppl. Fig. 1).
Figure 2.
Significant dose dependent effect of LBR on the number of nuclear segments in neutrophils, calculated by the “Drittelregel”. (B) The mutation type (null or splicing defect) significantly influences nuclear segmentation in neutrophils of heterozygous Pelger individuals. We analyzed four individuals heterozygous for null mutations (p.V11E fs X14, p.R89X and two individuals with mutation p.R377X, respectively) and five individuals heterozygous for splice mutation IVS12-5-10del.
Figure 3.
Average number of nuclear segments (lobulation index), calculated by the “Drittelregel” for individuals with zero to three functional (wild type) LBR-genes. The bar chart is based on the analysis of 26 individuals. 50 neutrophil nuclei were analyzed each. The error bar refers to the standard error of the mean. Two individuals had no functional (wild type) LBR-gene. One was homozygous for the splice mutation IVS12-5-10del (S), for the other homozygous the mutation was not identified thus far, but is probably regulatory ((R); detailed description in the materials and methods section). Nine individuals had one functional (wild type) LBR copy. Four of them were heterozygous for null mutations (N), five were heterozygous for a splice mutation (S). Twelve individuals with two functional (wild type) LBR copies were analyzed as controls. Three individuals had a triplication of the LBR-gene.
In lymphoblastoid cells, we observed a significant LBR dosage effect with respect to the amount of LBR in western blots and also a clear correlation with respect to the LBR staining intensity of the nuclear envelope (Figs. 4 and 5). To quantify the amount of LBR protein, we measured the staining intensities for LBR in western blots adjusted for actin expression (Fig. 4, Suppl. Table 2). The amount of LBR protein increased with the number of functional LBR copies and was significantly correlated with the segmentation grade in neutrophils (Fig. 4; p = 0.0001). In contrast to neutrophils, the nuclei in lymphoblastoid cells were always round or ovoid (one segment per definition). We never observed two or more nuclear segments in any of the lymphoblastoid cells despite the demonstrated correlation between the number of functional LBR copies, the amount of LBR protein and the LBR staining intensity. Thus, neutrophils appear to be especially sensitive to the dosage of LBR with respect to nuclear shape.
Figure 4.
Effects of 0 to 3 functional (wild type) copies of the LBR-gene on the LBR protein level. (A) The number of functional LBR copies correlates with the amount of LBR protein in western blots from lymphoblastoid cells. Three representative blots are shown. (B) Quantification for the determination of the relative expression of LBR revealed highly significant (p < 0.0001) inter-group differences (0–1, 1–2, 2–3). Error bars indicate the standard deviation of three independent experiments. (C) The correlation between LBR protein level and the segmentation index is highly significant (p = 0.0001).
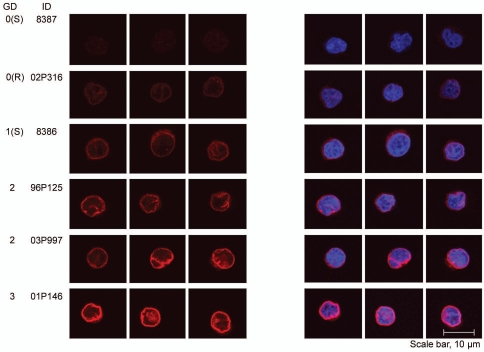
The immunofluorescence data showed that independent of the number of functional LBR copies, the protein was always correctly targeted to the nuclear envelope, even in the presence of an extra third copy of LBR (Fig. 5). As expected, the amount of LBR mRNA, based on Chip-analysis (Human Genome U133A 2.0 Array) was rather constant in the cell lines with up to two functional copies of the gene and increased in the two lymphoblastoid cell lines with three gene copies (data not shown).
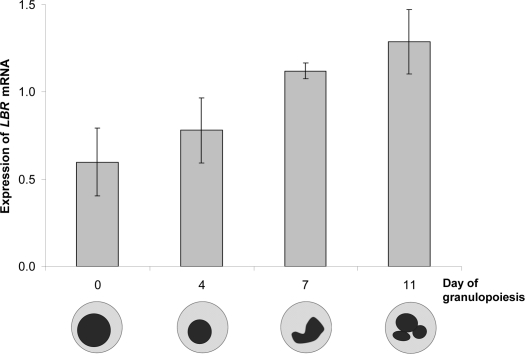
In addition, we found that during in vitro granulopoiesis, LBR mRNA was rather low in the CD34+ progenitor cells with ovoid nuclei and then increased steadily until day 11, when polymorph nuclear cells became visible (Fig. 6). The progenitor cells were obtained from unused portions of bone marrow transplants; there was no additional material left for protein analysis.
Figure 5.
Dose dependent effect on the staining intensity of the nuclear membrane in lymphoblastoid cells in immunofluorescence microscopy using an anti-LBR antibody. A representative set of LCLs is shown. To evaluate and compare LBR staining intensities, confocal images were collected with identical laser intensities and scanning parameters.
Discussion
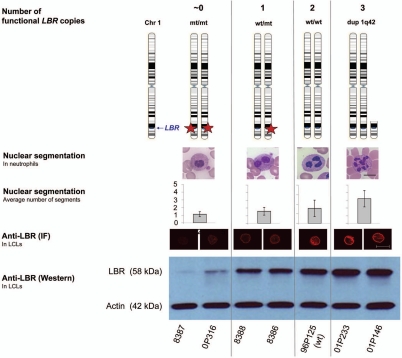
The peculiar shape of the neutrophil nucleus is an evolutionarily ancient trait and reminiscent of the stages in human granulopoiesis whereby the nucleus progresses from ovoid over bisegmented to a multilobed shape with 3 to 4 segments.19 The same situation was found in individuals with zero to two functional copies of the LBR-gene.9 We have shown here that a third LBR copy leads to a further increase in the amount of LBR protein and to a hypersegmentation of neutrophil nuclei. This model allows the systematic study of gene-dosage effects on several levels (Summarized in Fig. 7).
We quantified the gene-dosage-effect for LBR on human neutrophils and showed (1) that the amount of LBR protein and degree of nuclear segmentation is significantly correlated with the number of functional LBR copies, (2) that the degree of nuclear segmentation depends quantitatively on the class of the LBR mutation, and (3) that also during granulopoiesis in vitro nuclear morphology of neutrophils is correlated with LBR-gene expression.
The expression of LBR mRNA increased continually from precursor cells with ovoid nuclei, to multi-segmented neutrophil nuclei 11 days later during human granulopoiesis. This is consistent with previous data from a mouse model, where progenitor EML cells were induced to mature neutrophils,17,18 which suggests that increasing amounts of lamin B receptor on RNA and protein level play a major role in granulopoiesis.
In blood smears from individuals with zero to three LBR copies, we found a significant dose-dependent correlation between the number of LBR copies and the segmentation grade of neutrophil nuclei. Further, the analyses revealed significantly different segmentation pattern between splice-site and null mutations. A significant gene-dosage effect could also be demonstrated on the LBR protein level in lymphoblastoid cells. Moreover, the immunofluorescence data showed that LBR reached its place in the nuclear membrane, even if three doses were synthesized, suggesting that also a third copy can exercise an effect on nuclear shape. Thus, the lobulation index seems to be a rather sensitive indicator of allelic differences in LBR expression revealing even differences between mutations types. The significant correlation between lobulation of neutrophil nuclei and both the LBR copy number and the LBR protein level is consistent with the concept of a gene-dosage effect.
The concept of gene-dosage effects is based on the assumption that the amount of a primary gene product is directly proportional to the number of genes coding for the respective protein. In the 1980s, Epstein reviewed all relevant human and mouse studies, affecting over forty different loci that almost all coded for enzymes or enzyme products. The data provided convincing support for a quantitatively precise gene-dosage effect.24 However, whereas Epstein's data showed a gene-dosage effect for enzymes or enzyme products, our analyses provide evidence for gene-dosage effects for a structural protein.
As a constituent of the inner nuclear membrane LBR belongs to the structural class of proteins. Thus, to the best of our knowledge, this series from zero to three LBR copies represents a unique situation in human genetics, in which a gene-dosage effect over such a range of gene doses of a structural protein revealed distinct quantitative cellular phenotypes.
Despite this significant correlation, the relationship was not absolutely linear: The transition from zero to one and from two to three functional copies was much less pronounced than for between one and two wild type alleles (even if keeping in mind that the patients with no wild type allele still synthesized a small amount of LBR). This situation speaks in favor of a control mechanism (partial dosage compensation) that could be acting at the level of transcription and translation, the rate of protein turnover and degradation, or the processing from the endoplasmatic reticulum to the inner nuclear membrane.25
Furthermore, despite the significant correlation of LBR and the nuclear segmentation in neutrophils, the data presented here as well as in previous observations1,26 revealed slight inter- and intra-familial variations, which indicates modifying effects of endogeneous and exogeneous factors. Clearly, LBR expression is necessary for normal neutrophil nuclear differentiation, but that alone is not sufficient. Heterochromatinization during myeloid differentiation is another prerequisite.17 Moreover, other proteins such as lamin A/C and lamin B also control nuclear shape.27,28 The paucity of lamins A/C makes the neutrophil nuclear envelope more deformable than other nuclei. This has been demonstrated in the human myeloid leukemia cell line HL-60, which can be differentiated to both granulocytic and macrophage/monocyte forms in vitro. Both cell types show an increased expression of LBR compared to undifferentiated cells. Only the neutrophilic forms, however, lack lamin A/C.29 As examples for exogeneous factors, substances disrupting microtubules, such as nocodazole or colchicine, impaired nuclear lobulation in differentiating HL-60 cells and mouse neutrophils in vivo, respectively.30,31 Our observation that the number of LBR copies affected the number of nuclear segments in neutrophils but not in lymphoblastoid cells, fibroblasts and lymphocytes (data not shown) confirms that polymorphonuclear blood cells are especially sensitive to the loss or gain of LBR.
In conclusion, we have demonstrated a direct link between the gene product, the lamin B receptor, and its phenotype, the lobulation of the neutrophil nucleus. LBR is therefore an ideal model for systematically studying the fundamental questions of autosomal gene-dosage effects and even for analyzing the consequences of perturbations by endo- and exogenous factors. Moreover, the availability of an in vitro model for granulopoiesis in man and of an in vivo mouse model offers promising experimental approaches.
Materials and Methods
The local ethical committee approved the study and written informed consent was obtained from all participants.
Subjects.
We obtained blood smears from 2 homozygous and 9 heterozygous PHA persons, 12 controls and 3 individuals with triplication of the LBR-gene. In addition, 10 ml of blood were obtained from one homozygous individual (8387), his heterozygous parents, another homozygous person (02P316), and two individuals with three doses of the LBR-gene to establish lymphoblastoid cell lines. The clinical and molecular data of one of the homozygous individuals (8387) is published.9 Clinical data from the other homozygous person (02P316) is published;32 however, no mutations in the exons of LBR, its flanking introns, and the promoter region were identified. Both parents originated from the same village and the daughter 02P316 was not only homozygous with respects to the neutrophil phenotype but also for microsatellite markers and SNPs in this region due to inheritance of the identical haplotype from both parents (data not shown). The finding indicates that the Pelger trait in the family is consistent with linkage to the LBR region on chromosome 1q42. The affection of the LBR-gene is underscored by the presence of some wild type LBR protein, pointing to a regulatory mutation. In western blots, the amount of LBR in this homozygote individual 02P316 is higher than in the other homozygote patient 8387, who has a splice-site mutation (IVS12-5-10del) and synthesizes only a trace amount of wild type protein (Fig. 4). His parents (8388 and 8386) are included as heterozygous carriers for mutation IVS12-5-10del in this study, as well as three other persons with the same splice mutation (Table 1). The other four heterozygous persons had null mutations, a deletion of four base pairs in exon 2 (c.32-35del4), a stop mutation in exon 3 (c.265C>T), and two carried a nonsense mutation in exon 9 (c.1129C>T) (Table 1). From the three cases with triplication of the LBR-gene at 1q42.1, one patient was already published.33 The two new patients are from Germany and from the Sultanate of Oman, respectively (Table 1). Both came to clinical attention because of severe mental and motoric retardation. The duplications were identified by standard karyotyping and confirmed by comparative genomic hybridization (CGH) or FISH, respectively.
Characterization of nuclear segmentation in neutrophils.
Two investigators independently analyzed the nuclear segmentation on coded slides. The segmentation was scored according to the “Fadenregel” (Rule of Threads) and the “Drittelregel” (Rule of Thirds) described elsewhere.22,23 In the first case, a segment is defined by lobes, which are connected only via a thin chromatin thread, in the latter case, by an indentation with width <1/3 of length. For each individual, 50 neutrophils were analyzed and statistically compared by means of the nonparametric Wilcoxon-Man-Whitney test.
Protein analysis.
Lymphoblastoid cells in the logarithmic growth phase were counted, washed, and proteins extracted as previously described.9 For western blotting the samples were adjusted to identical protein concentrations and loaded on a 4–12% Bis-Tris-Gel (Invitrogen). Gel electrophoresis was performed under denaturing conditions using an XCell Blot Module (Invitrogen). Guinea pig anti-LBR serum was used at 1:500 dilution; an antibody against β-actin confirmed equal protein loading. Dr. Harald Herrmann (DKFZ, Heidelberg) provided the antibodies. Each experiment was repeated at least three times.
Densitometry.
Blots with a linear response range of the photographic film were scanned using the ScanMaker scanner (Microtek, Hsinchu, Taiwan) and the images quantified using ImageQuaNT software (Molecular Dynamics, version 5.2, GE Healthcare, UK). LBR signal intensities for each sample were normalized to the corresponding β-actin signal intensity. Error bars indicate the standard deviation of three independent experiments.
Regression analysis.
(Expression of LBR protein versus grade of nuclear segmentation). The expression of LBR protein in lymphoblastoid cell lines (LCLs) from 7 subjects was quantified three times (2 subjects each with 0, 1 or 3 functional LBR copies; 1 control subject with 2 functional LBR copies). The relationship between normalized LBR expression (independent variable) and neutrophil segmentation as estimated by means of the “Rule of Thirds” from 50 cells per subject (dependent variable) was modeled through standard regression analysis (SAS/STAT Statistical Analysis Package; SAS Institute).
Immunofluorescence microscopy.
For immunofluorescence microscopy, suspensions of lymphoblastoid cells were counted and diluted with PBS (Phosphate Buffered Saline) to a concentration of 2 × 105 cells/ml. 0.5 ml of cell suspension was pipetted onto poly-lysine coated cover slips and allowed to settle for 30 min. The cells were then washed with PBS, fixed with 4% formaldehyde in PBS for 10 min, washed three times with PBS, and permeabilized by 0.2% Triton-X 100/PBS for 10 min at −20°C. The cover slips were washed three times in PBS, and then blocked with bovine serum albumin. Primary LBR antibody was diluted 1:500 in PBS and incubated for 20 min. After washing with PBS (3 × 2 min), incubation followed with the species-specific secondary antibody for 20 min (1:500 dilution in PBS, Cy3-goat anti-guinea pig, Dianova, Hamburg, Germany). Finally, the cells were stained with DAPI (10 min, 1:10,000, Serva, Heidelberg), washed in PBS (3 × 2 min), briefly rinsed in distilled water, and dehydrated in 100% ethanol for 1 min. The cells were air dried and mounted in Fluoromount G (Southern Biotechnologies Associates INC, Birmingham, AL). To evaluate and compare LBR staining intensities, confocal images were collected with identical laser intensities and scanning parameters. Each set was replicated at least twice.
Number of nuclear segments in lymphoblastoid cells, fibroblasts and lymphocytes.
Lymphoblastoid cells and fibroblasts were prepared as described for immunofluorescence microscopy; lymphocytes were analyzed in blood smears. We analyzed at least 25 lymphoblastoid cells, at least 30 fibroblasts and at least 25 lymphocytes per individual. All nuclei in lymphoblastoid cells, fibroblasts and lymphocytes were round or ovoid; none of them matched the criteria of two segments according to the Faden- or the Drittelregel (described above under “characterization of nuclear segmentation in neutrophils”).
The experiments for in vitro granulocyte differentiation and transcriptional profiling.
The experiments for in vitro granulocyte differentiation and transcriptional profiling were described elsewhere.34 Briefly, CD34+ cells were separated from heparinized bone marrow samples from healthy individuals (donors of bone marrow transplants) after Ficoll-Hypaque separation of mononuclear cells by high-gradient magnetic cell separation. At least 5 × 105 CD34+ cells were cultured in 3 ml of granulopoietic differentiation medium and harvested at days 4, 7 and 11. Total RNA was extracted using TRIzol. Oligonucleotide microarray hybridization was performed on Affymetrix chips (HG-U133A). Data analysis was performed with GeneSpring software version 4.2 and microarray analysis suites 5.0. The expression of IRCI and FCGR2B, known to be specific for the granulocytic lineage and not expressed in CD34+ cells, was analyzed by real-time PCR to verify the in vitro differentiation.
Figure 6.
In vitro granulocyte differentiation is associated with increasing amounts of LBR mRNA after 0, 4, 7 and 11 days of culture. Oligonucleotide microarray hybridization (Affymetrix, probe 201795_at for LBR).
Figure 7.
Effects of 0 to 3 functional (wild type) copies of the LBR-gene on nuclear segmentation and LBR protein level. The number of functional LBR copies correlates with the nuclear segmentation of neutrophils in blood smears, the staining intensity of the nuclear membrane in lymphoblastoid cells in immunofluorescence microscopy with an anti-LBR antibody, and the amount of LBR protein in western blots from lymphoblastoid cells.
Acknowledgements
We thank the patients and their families. Tom H. Lindner, Friedrich C. Luft and Pamela Winchester critically read the manuscript. The Deutsche Forschungsgemeinschaft (DFG, SFB 577, project A4) supported K.H., who also received a Rahel Hirsch fellowship from the Charité Medical Faculty.
Abbreviations
- GD
gene dosage
- HP1
heterochromatin protein 1
- LBR
lamin B receptor (protein)
- LBR
lamin B receptor (gene)
- PHA
Pelger-Huët anomaly
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/11120
Supplementary Material
References
- 1.Harm H. Zur Klassifizierung und Vererbung der Neutrophilen-Kernform bei Mensch und Kaninchen. Blut. 1955;1:3–25. (Ger). [Google Scholar]
- 2.Sainty D, Liso V, Cantu-Rajnoldi A, Head D, Mozziconacci MJ, Arnoulet C, et al. A new morphologic classification system for acute promyelocytic leukemia distinguishes cases with underlying PLZF/RARA gene rearrangements. Group Francais de Cytogenetique Hematologique, UK Cancer Cytogenetics Group and BIOMED 1 European Coomunity-Concerted Acion “Molecular Cytogenetic Diagnosis in Haematological Malignancies. Blood. 2000;96:1287–1296. [PubMed] [Google Scholar]
- 3.Shenkenberg TD, Rice L, Waddell CC. Acquired Pelger-Huet nuclear anomaly with tuberculosis. Arch Intern Med. 1982;142:153–154. [PubMed] [Google Scholar]
- 4.Juneja SK, Matthews JP, Luzinat R, Fan Y, Michael M, Rischin D, et al. Association of acquired Pelger-Huet anomaly with taxoid therapy. Br J Haematol. 1996;93:139–141. doi: 10.1046/j.1365-2141.1996.4701020.x. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 6.Bills T, Spatz L. Neutrophilic hypersegmentation as an indicator of incipient folic acid deficiency. Am J Clin Pathol. 1977;68:263–267. doi: 10.1093/ajcp/68.2.263. [DOI] [PubMed] [Google Scholar]
- 7.Davenport J. Macrocytic anemia. Am Fam Physician. 1996;53:155–162. [PubMed] [Google Scholar]
- 8.Westerman DA, Evans D, Metz J. Neutrophil hypersegmentation in iron deficiency anaemia: a case-control study. Br J Haematol. 1999;107:512–515. doi: 10.1046/j.1365-2141.1999.01756.x. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly) Nat Genet. 2002;31:410–414. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 10.Worman HJ, Yuan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Worman HJ, Evans CD, Blobel G. The lamin B receptor of the nuclear envelope inner membrane: a polytopic protein with eight potential transmembrane domains. J Cell Biol. 1990;111:1535–1542. doi: 10.1083/jcb.111.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Q, Callebaut I, Pezhman A, Courvalin JC, Worman HJ. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 13.Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- 14.Holmer L, Pezhman A, Worman HJ. The human lamin B receptor/sterol reductase multigene family. Genomics. 1998;54:469–476. doi: 10.1006/geno.1998.5615. [DOI] [PubMed] [Google Scholar]
- 15.Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, et al. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet. 2003;12:61–69. doi: 10.1093/hmg/ddg003. [DOI] [PubMed] [Google Scholar]
- 16.Cohen TV, Klarmann KD, Sakchaisri K, Cooper JP, Kuhns D, Anver M, et al. The lamin B receptor under transcriptional control of C/EBPepsilon is required for morphological but not functional maturation of neutrophils. Hum Mol Gen. 2008;17:2921–2933. doi: 10.1093/hmg/ddn191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, et al. Mouse neutrophils lacking lamin B-receptor expression exhibit aberrant development and lack critical functional responses. Exp Hematol. 2008;36:965–976. doi: 10.1016/j.exphem.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zwerger M, Herrmann H, Gaines P, Olins AL, Olins DE. Granulocytic nuclear differentiation of lamin B receptor-deficient mouse EPRO cells. Exp Hematol. 2008;36:977–987. doi: 10.1016/j.exphem.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Undritz E. Das Pelger-Huet'sche Blutbild beim Tier und seine Bedeutung für die Entwicklungsgeschichte des Blutes. Schweiz Med Wochenschr. 1939;69:1177–1186. (Ger). [Google Scholar]
- 20.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169:338–346. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, et al. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bucher U. Labormethoden in der Hämatologie. Bern, Stuttgart, Toronto: Hans Huber Vlg; 1988. (Ger). [Google Scholar]
- 23.Begemann H, Rastetter J. Atlas of Clinical Hematology. Berlin, Heidelberg, New York, London, Paris, Tokyo, Hong Kong: Springer-Verlag; 1989. [Google Scholar]
- 24.Epstein CJ. The Consequences of Chromosome. Imbalance: Principles, Mechanisms and Models: Cambridge University Press; 1986. [Google Scholar]
- 25.Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 26.Harm H. Beiträge zur Morphologie und Genetik der Pelger Anomalie bei Mensch und Kaninchen. Z menschl Vererb-u Konstitutionslehre. 1952;30:501–539. (Ger). [Google Scholar]
- 27.Schirmer EC, Guan T, Gerace L. Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J Cell Biol. 2001;153:479–489. doi: 10.1083/jcb.153.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenz-Bohme B, Wismar J, Fuchs S, Reifegerste R, Buchner E, Betz H, et al. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J Cell Biol. 1997;137:1001–1016. doi: 10.1083/jcb.137.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olins AL, Herrmann H, Lichter P, Kratzmeier M, Doenecke D, Olins DE. Nuclear envelope and chromatin compositional differences comparing undifferentiated and retinoic acid- and phorbol ester-treated HL-60 cells. Exp Cell Res. 2001;268:115–127. doi: 10.1006/excr.2001.5269. [DOI] [PubMed] [Google Scholar]
- 30.Olins AL, Olins DE. Cytoskeletal influences on nuclear shape in granulocytic HL-60 cells. BMC Cell Biol. 2004;5:30. doi: 10.1186/1471-2121-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harm H. Beeinflussung des weissen Blutbildes von Pelger- und Nicht-Pelger-Kaninchen durch Colchicin. Acta Haemat. 1953;10:95–105. (Ger). [PubMed] [Google Scholar]
- 32.Aznar J, Vaya A. Homozygous form of the Pelger-Huet leukocyte anomaly in man. Acta Haematol. 1981;66:59–62. doi: 10.1159/000207095. [DOI] [PubMed] [Google Scholar]
- 33.Nowaczyk MJ, Bayani J, Freeman V, Watts J, Squire J, Xu J. De novo 1q32q44 duplication and distal 1q trisomy syndrome. Am J Med Genet A. 2003;120:229–233. doi: 10.1002/ajmg.a.20028. [DOI] [PubMed] [Google Scholar]
- 34.Komor M, Guller S, Baldus CD, de Vos S, Hoelzer D, Ottmann OG, et al. Transcriptional profiling of human hematopoiesis during in vitro lineage-specific differentiation. Stem Cells. 2005;23:1154–1169. doi: 10.1634/stemcells.2004-0171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.