Abstract
The PHylogenetic Analysis with Space/Time models (PHAST) software package consists of a collection of command-line programs and supporting libraries for comparative genomics. PHAST is best known as the engine behind the Conservation tracks in the University of California, Santa Cruz (UCSC) Genome Browser. However, it also includes several other tools for phylogenetic modeling and functional element identification, as well as utilities for manipulating alignments, trees and genomic annotations. PHAST has been in development since 2002 and has now been downloaded more than 1000 times, but so far it has been released only as provisional (‘beta’) software. Here, we describe the first official release (v1.0) of PHAST, with improved stability, portability and documentation and several new features. We outline the components of the package and detail recent improvements. In addition, we introduce a new interface to the PHAST libraries from the R statistical computing environment, called RPHAST, and illustrate its use in a series of vignettes. We demonstrate that RPHAST can be particularly useful in applications involving both large-scale phylogenomics and complex statistical analyses. The R interface also makes the PHAST libraries acccessible to non-C programmers, and is useful for rapid prototyping. PHAST v1.0 and RPHAST v1.0 are available for download at http://compgen.bscb.cornell.edu/phast, under the terms of an unrestrictive BSD-style license. RPHAST can also be obtained from the Comprehensive R Archive Network (CRAN; http://cran.r-project.org).
Keywords: statistical phylogenetics, functional element identification
INTRODUCTION
As complete genome sequences have become available for large numbers of closely related organisms, interest has steadily grown in improved computational methods for comparative genomics. Of particular interest are statistical, phylogenetic methods for characterizing rates and patterns of molecular evolution and for identifying sequences under natural selection against a background of neutral evolution. Methods of this kind have been used to identify evolutionary conserved elements [1–3], novel protein-coding genes [4–6], fast-evolving noncoding sequences [7–9], transcription factor binding sites [10], noncoding RNAs [11] and other types of functional elements.
Since 2002, we have been developing a software package, called PHylogenetic Analysis with Space/Time models (PHAST), that consists of a collection of programs and supporting libraries for statistical phylogenetic modeling and functional element identification. (The phrase ‘space/time models’, borrowed from Yang [12], reflects the prominent role of phylogenetic hidden Markov models in the package.) Our initial goal in developing PHAST was to support our own research in comparative genomics. Over time, however, as the package has expanded in functionality, it has gradually been adopted by a fairly large group of researchers from the broader comparative genomics community. PHAST is best known as the engine behind several popular tracks in the UCSC Genome Browser [13] (including, most notably, the Conservation track), but it can also be downloaded and installed for use in custom analyses not available through the browser. As of September 2010, the package has been downloaded more than 1000 times (counting unique IP addresses). More than two-thirds of those downloads have occurred since November 2008, when the PHAST web site became available (http://compgen.bscb.cornell.edu/phast).
PHAST has some overlap with the popular phylogenetic modeling package PAML [14] as well as with other packages for phylogenetics such as HYPHY [15] and MEGA [16], tools for conservation analysis such as GERP [2] and SCONE [17], and comparative gene finders such as N-SCAN [18]. However, PHAST is unique in that it combines phylogenetic modeling and functional element identification. In addition, it supports some phylogenetic modeling features not available in other packages, such as context-dependent subsitution models and model fitting by expectation–maximization. PHAST also has a particularly rich collection of methods for detecting departures from neutrality in rates and patterns of molecular evolution, with the ability to detect both consevation and acceleration, either across the branches of a phylogeny or on individual branches or clades. Finally, PHAST is well-suited for large-scale phylogenomics, with the ability to process entire mammalian genomes efficiently and native support for a variety of file formats used by the UCSC Genome Browser.
Here we describe the first ‘official’ release of PHAST, denoted v1.0. While most of the key algorithmic and modeling ideas behind PHAST have been published, this is the first article summarizing all components of the package and showing how they fit together and complement one another. We provide an overview of the programs and libraries in PHAST, and describe several recent improvements. In addition, we introduce a new interface to the PHAST libraries from the R statistical computing environment [19], called RPHAST. The combination of PHAST and R is particularly powerful, especially in applications requiring a mixture of comparative genomic and downstream statistical analyses, and for rapid prototyping of new phylogenomic methods. We expect the improved usability of PHAST v1.0 (with RPHAST) to increase interest in the package among comparative genomics researchers.
MAIN FEATURES
PHAST
The command-line programs in PHAST currently include six major applications and roughly two dozen supporting utilities. The most heavily used programs are the phastCons, phyloFit and phyloP applications. The other three major programs—dless, exoniphy and prequel—are also substantial in scope but somewhat less widely used. The utilities include general-purpose file manipulation programs (e.g. msa_view, maf_parse, refeature, tree_doctor), programs for scoring predictions (phastOdds), generating simulated data and assessing statistical significance (phyloBoot, base_evolve) and various tools for more specialized purposes (indelHistory, clean_genes). The major applications and several representative utilities are summarized in Table 1. When a program in PHAST is invoked with the --help(-h) option, a message is printed to the terminal including a high-level description, the expected form of a command-line call, a list of optional arguments and (in some cases) a list of examples of specific command-line calls.
Table 1:
Selected programs in PHAST
| Program | Description | RPHASTa | References |
|---|---|---|---|
| dless | Prediction of elements under lineage-specific selection | [28] | |
| exoniphy | Phylogenetic exon prediction | [29] | |
| phastCons | Conservation scoring and identification of conserved elements | ✓ | [3] |
| phyloFit | Fitting of phylogenetic models to aligned DNA sequences | ✓ | [20] |
| phyloP | Computation of P-values for conservation or acceleration, either lineage specific or across all branches | ✓ | [23] |
| prequel | Probabilistic reconstruction of ancestral sequences | – | |
| base_evolve | Simulation of synthetic alignments from phylogenetic models | ✓ | – |
| clean_genes | Filtering of gene annotations based on conservation properties | – | |
| indelHistory | Inference of phylogenetic indel histories for alignments, based on a simple algorithm | – | |
| maf_parse | Efficient manipulation of large alignments in MAF format | – | |
| msa_view | Manipulation of multiple alignments, including format conversion and extraction of sub alignments | ✓ | – |
| phastOdds | Log-odds scoring for phylogenetic models or phylo-HMMs | ✓ | – |
| phyloBoot | Parametric and nonparametric bootstrapping for phylogenetic models | ✓ | – |
| refeature | Manipulation of genomic features, including format conversion, filtering, grouping | ✓ | – |
| tree_doctor | Manipulation of phylogenetic trees, including scaling, pruning, rerooting, relabeling | ✓ | – |
aCheckmark indicates that majority of functionality is currently supported in RPHAST.
Most of the general modeling and algorithmic ideas that are implemented in PHAST have been published (see references in Table 1), but v1.0 incorporates several improvements to these methods as well as new programs that have not yet been published. For example, the phyloP program now allows for tests of lineage-specific selection on any arbitrary subset of branches, not just the branches in a clade. In addition, the phyloFit program has evolved from an application focused on context-dependent nucleotide substitution [20] into a full-featured program for fitting phylogenetic models to sequence alignments by maximum likelihood. Finally, the new prequel program supports probabilistic reconstruction of ancestral sequences given an alignment and phylogenetic model, and the new maf_parse utility allows efficient manipulation of large-scale multiple alignment format (MAF) files, without storage of entire alignments in memory (as with msa_view).
RPHAST
RPHAST is meant to address two major goals. First, it provides a flexible and convenient programming environment for both large-scale phylogenomics and computational statistics, allowing users to perform analyses in which these two components are closely intertwined—such as bootstrapping analyses, computation of empirical P-values or permutation tests. Second, RPHAST makes the functionality of the PHAST libraries accessible from a scripting environment. This enables non-C programmers to make use of the libraries and provides an environment for rapid prototyping of new models and algorithms.
In general, RPHAST parallels the PHAST libraries fairly closely, with R versions of major PHAST classes and R wrappers for selected functions in PHAST, which make use of the .Call interface from R to C. However, in some cases RPHAST avoids particularly complex details in PHAST and operates at a slightly higher level of abstraction. One particular design challenge in RPHAST relates to very large PHAST objects, such as multiple alignments for entire mammalian chromosomes, which are better manipulated in C but still need to be inspected in R. This problem is addressed by providing an option to represent certain newly created objects only by ‘external pointers’ in R, effectively allowing them to be passed by reference rather than by value. The essential properties of referenced objects—such as the sequence names or number of columns in a multiple alignment—can still be accessed within R. The initial release of RPHAST does not provide access to the entire PHAST libraries, but many key functions are supported, and others will be added as time goes on.
In the standard manner for R packages, we have developed a series of detailed ‘vignettes’ that illustrate the use of RPHAST in realistic phylogenomic analyses. We describe the package by summarizing these vignettes, including code snippets that highlight key features of RPHAST. The complete vignettes are available from the RPHAST website (http://compgen.bscb.cornell.edu/rphast) and from the Comprehensive R Archive Network (CRAN; http://cran.r-project.org).
Vignette #1: conservation analysis
This vignette illustrates how RPHAST can be used to produce conservation scores and conserved elements for aligned genomic sequences, similar to those shown in the UCSC Genome Browser. It also demonstrates the use of RPHAST in analyzing the predicted conserved elements. The vignette makes use of alignments and gene annotations from a recent study of a 105 kb conserved syntentic segment in five Solanaceae species [21] (tomato, potato, eggplant, pepper and petunia), a fairly typical comparative genomic analysis of organisms for which few ‘off-the-shelf’ bioinformatic resources are available.
Several key steps of the vignette are detailed in Figure 1. Briefly, the alignment, gene annotations and the assumed tree topology are read (lines 2–4), a neutral phylogenetic model is estimated from fourfold degenerate (4D) sites in coding regions (lines 5–6), and then conservation scores and predicted conserved elements are predicted using phastCons (line 7) and phyloP (line 8). These scores and elements are then displayed, along with gene annotations, using plotting functions in RPHAST (lines 10–16; Figure 2A). In addition, the length distributions of phastCons elements are examined, and the distributions for elements that primarily overlap coding and noncoding regions are contrasted (lines 17–22; Figure 2B). Finally, the enrichment of predicted conserved elements in genomic regions of different types (coding, intronic and noncoding regions) is examined, as is the composition of the conserved elements based on these same annotation types (lines 23–29; Figure 2C). The full version of the vignette also shows an alternative phastCons run, with parameters estimated by expectation–maximization. Many other downstream analyses can be easily performed in R, including tests for enrichment based on gene ontology categories (e.g. using the ‘topGO’ package) or the generation of Venn diagrams showing the degree of overlap between, say, conserved elements and coding exons (e.g. using the ‘venn’ package).
Figure 1:
Code snippets showing the use of RPHAST in vignette #1, which describes a conservation analysis of a conserved syntenic segment in five Solanaceae genomes. The plots generated by these commands are shown in Figure 2. The complete vignette is available at http://compgen.bscb.cornell.edu/rphast.
Figure 2:
Plots generated by RPHAST in vignette #1. (A) Browser-like plot showing genes, conserved elements and conservation scores in a region of interest. (B) Length distributions for coding, noncoding and all elements. (C) Enrichment and composition of conserved elements by annotation type.
Vignette #2: identification of rodent accelerated regions
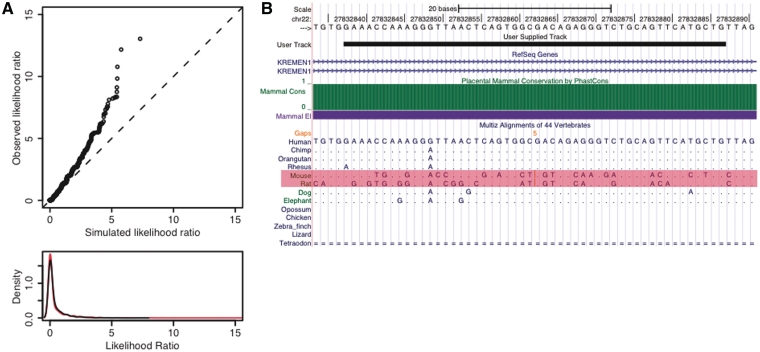
The second vignette illustrates the use of RPHAST in an analysis like the ones used to identify ‘human accelerated regions’ (HARs) [7] and similarly defined regions in other species [22, 23]. In this case, elements displaying indications of accelerated evolution in rat and mouse (denoted ‘rodent accelerated regions’ or RARs) are identified by a likelihood ratio test (LRT), using multiple alignments corresponding to human chromosome 22. This is a good example of an analysis with both phylogenetic and computational statistical components, for which RPHAST is particularly well suited. This example also demonstrates the convenience of using RPHAST together with the UCSC Genome Browser.
Several key steps of vignette #2 are detailed in Figure 3. First, a set of precomputed conserved elements is read from a file downloaded from UCSC, and split into fragments of a fixed size (50 bp), to simplify the subsequent analysis (lines 2–4). Next, the alignment columns corresponding to these elements and a precomputed neutral model are also read into memory (lines 5–7). The alignment columns are then randomly sampled with replacement (nonparametric bootstrapping), to generate a large number of ‘null’ alignments that can be used in characterizing the null distribution of the LRT. For reasons of efficiency, this is accomplished by simulating one large alignment (lines 8–9) and producing a feature set that allows it to be interpreted as a series of short alignments (lines 10–11). The LRT for acceleration in rodents is applied to each of these ‘null’ alignments using the RPHAST interface to phyloP (line 12). The same LRT is then applied to the real elements of interest (line 13), and P-values are computed based on the empirical null distribution (lines 14–15).
Figure 3:
Code snippets showing the use of RPHAST in vignette #2, in which ‘accelerated regions’ in the rodent (rat and mouse) genomes are identified and analyzed. The plots generated here are shown in Figure 4. The complete vignette is available at http://compgen.bscb.cornell.edu/rphast.
As has been described elsewhere [7, 23], this LRT compares a null hypothesis of an overall change in evolutionary rate, represented by a phylogenetic model with a single branch-length scaling parameter, against an alternative hypothesis of accelerated evolution in the subtree of interest (here, the rodents), represented by a phylogenetic model with separate scaling factors for the subtree and for the rest of the phylogeny. In this case, we make use of phyloP, which has an efficient implementation of the test of interest. However, a similar LRT based on any of the models implemented in PHAST could be performed by making two calls to the phyloFit function (see example in full vignette).
Next, quantile–quantile (Q–Q) and density plots are generated to compare the distributions of log likelihood ratios for the real and simulated elements (lines 16–19; Figure 4A). Finally, false discovery rates (FDRs) are estimated from these P-values using the Benjamini and Hochberg procedure [24] (line 20), and elements with FDR <0.05 are written to a file (lines 21–23) for subsequent display as a custom track in the UCSC Genome Browser (Figure 4B). These elements could also be analyzed using the Galaxy system [25]. The full vignette also shows an alternative method for characterizing the null distribution, based on parametric simulations of alignments.
Figure 4:
Plots for vignette #2. (A) Quantile-quantile (Q-Q) [top] and density [bottom] plots of the distribution of log likelihood ratios (LLRs) for conserved elements from chromosome 22 (‘observed’) versus the null distribution estimated by nonparametric bootstrapping (‘simulated’). In the density plot, the observed distribution is shown in red and the null distribution in black. They appear to be very similar, but the Q-Q plot reveals a clear excess of large values for the observed elements. (B) One of the identified rodent accelerated regions (RARs), as displayed in the UCSC Genome Browser (the element is shown in the ‘User Supplied Track’). This RAR is part of a 597-bp highly conserved but eutherian-specific element in the third intron of the gene KREMEN1, which encodes a type I transmembrane protein important in embryonic development [27]. In the alignment, the bases that match the human reference genome are shown as periods (‘.’), and unaligned regions are shown as blank spaces or ‘=’ symbols. Notice the large number of rodent-specific substitutions (rows highlighted in pink). This is the fourth highest-scoring RAR on chromosome 22.
We note that this example makes use of several short-cuts in the interest of brevity. For example, one might wish to re-estimate conserved elements by excluding the foreground species, or to allow for distributions of element lengths rather than forcing all elements to have the same length (see [7]). These steps can easily be performed using RPHAST as well.
Vignette #3: custom phylo-HMM for binding sites
The third vignette illustrates the use of RPHAST in creating a custom phylogenetic hidden Markov model (phylo-HMM) for use in functional element identification. Here, we design a phylo-HMM to detect binding sites for a particular transcription factor of interest, neuron-restrictive silencer factor (NRSF). The 21 bp long motif model is trained on a set of putative binding sites identified in previous work [23], and then its performance is evaluated on a simulated data set. This example shows how RPHAST can be used to prototype new models, as well as to apply existing methods.
Key steps from vignette #3 are shown in Figure 5. First, a set of alignment fragments representing the predicted binding sites are read into memory and aggregated into one large alignment (lines 2–5). Next, a separate phylogenetic model is fitted to the subset of columns associated with each of the 21 motif positions (lines 6–14). These models are fitted by estimating both equilibrium base frequencies and a branch-length scaling factor, so they capture both the base preferences and conservation patterns at each motif position. The estimated models are summarized by plotting a sequence logo (using the ‘seqLogo’ package in Bioconductor [26]) along with a per-position likelihood ratio (lines 15–16; Figure 6). To complete the definition of the phylo-HMM, a matrix of state-transition probabilities is then created, using a simple parameterization (lines 17–21). The model has 22 states—one for each of the 21 positions in the motif (associated with the estimated phylogenetic models), plus a ‘neutral’ or ‘background’ state. Next, a large synthetic alignment is generated, and binding sites are predicted in this alignment using the phylo-HMM (lines 22–23). The model is shown to have excellent sensitivity and specificity on simulated data (lines 24–29). These predictions can also be displayed as a track, along with the true binding sites (see full vignette). While more work would be required to make such a model useful with real data, this example illustrates the general principles involved in building a custom phylo-HMM using RPHAST.
Figure 5:
Code snippets showing the use of RPHAST in vignette #3, in which a custom phylo-HMM for prediction of NRSF binding sites is defined. The plots generated here are shown in Figure 6. The complete vignette is available at http://compgen.bscb.cornell.edu/rphast.
Figure 6:
Plots for vignette #3. (A) Logo plot of NRSF motif reflecting the nucleotide frequencies in the provided training data (from Pollard et al. [23]). This plot was generated using the ‘seqLogo’ package from Bioconductor [26]. (B) Likelihood ratios of position-specific phylogenetic models versus the background model, also based on the training data.
AVAILABILITY
The PHAST source code (consisting of ∼60 000 lines of C code) is freely downloadable from http://compgen.bscb.cornell.edu/phast under the terms of a Berkeley Software Distribution (BSD) license. The source code can be downloaded as a compressed tar (*.tgz) file or accessed directly from a subversion server. It compiles cleanly under linux, MacOS X and most UNIX implementations and under Windows in the presence of the Cygwin linux-like environment (http://www.cygwin.com). Binaries are also provided for linux (as RPM and DEB packages), MacOS X and 32- and 64-bit Windows platforms. PHAST is self-contained except that it requires the (free) LAPACK linear algebra package (http://www.netlib.org/clapack) for certain matrix operations. It also makes use of the the PCRE (Perl Compatible Regular Expressions) library, which is included in the PHAST distribution (as permitted under a BSD license) and does not need to be separately installed. Documentation for the command-line programs in PHAST can be viewed at http://compgen.bscb.cornell.edu/phast or accessed by running each program with the --help (-h) option. Questions and bug-reports may be sent to the phast-help-l@cornell.edu mailing list. Interested users may also join the phast-users-l@cornell.edu mailing list to receive updates about new releases and new features.
RPHAST is freely available from http://compgen.bscb.cornell.edu/rphast. The full vignettes can be downloaded in portable document format (PDF) from the same URL. RPHAST is also available from the Comprehensive R Archive Network (CRAN) (http://cran.r-project.org).
DISCUSSION
Many good software tools are now available for phylogenetics and comparative genomics. In addition, resources such as the UCSC Genome Browser and Galaxy allow researchers to visualize and analyze comparative genomic data using only a web browser. We see PHAST as a valuable addition to these resources. Its particular niche is at the intersection of large-scale comparative genomics, statistical phylogenetic modeling and functional element identification. PHAST is especially well-suited for analyzing patterns of conservation and acceleration in aligned sequences, and for extracting data from or exporting data to the UCSC Genome Browser and related resources, such as Galaxy. The RPHAST package significantly broadens the usefulness of PHAST by allowing access to its libraries from the R programming environment. As we have shown, RPHAST is a powerful environment for combining comparative genomic analysis and computational statistics and for prototyping of new models.
While PHAST and RPHAST are now fairly stable and feature-rich packages, several opportunities for improvement remain. For example, many features in the PHAST libraries are not yet available through RPHAST. We plan to gradually expand RPHAST's functionality, giving priority to features most likely to be useful within R. The PHAST documentation has recently improved considerably, but the API-level documentation is still incomplete. Work is under way to improve this documentation and to provide examples and code templates that will make it easier for C programmers to use the PHAST libraries. In addition, we are in the process of adding several new models and algorithms to PHAST and RPHAST, including phylogenetic models of biased gene conversion (BGC) and statistical tests that distinguish positive selection from BGC (D. Kostka et al., submitted for publication). Continued development of PHAST and RPHAST will support our own research programs at the same time as it makes the package more useful to the broader research community.
Key Points.
The PHAST package includes a rich collection of command-line tools and supporting libraries for comparative genomics.
PHAST is unique in that it combines phylogenetic modeling and functional element identification. It is particularly well-suited for large-scale applications, with support for the genome annotation and alignment formats used by the UCSC Genome Browser.
RPHAST enables access to the PHAST libraries from R statistical computing environment, providing a powerful, flexible environment for statistical phylogenomics.
PHAST is freely available from http://compgen.bscb.cornell.edu/ phast. RPHAST is available from http://compgen.bscb.cornell.edu/rphast and the CRAN.
FUNDING
National Science Foundation (grant DBI-0644111 to A.S.); the National Institutes of Health (grant R01-GM082901 to K.S.P.); and a David and Lucile Packard Fellowship for Science and Engineering (to A.S.). Past support has come from an Achievement Rewards for College Scientists (ARCS) scholarship (to A.S.) and a Graduate Research and Education in Adaptive bio-Technology (GREAT) fellowship from the University of California Biotechnology Research and Education Program (to A.S.).
Acknowledgements
We thank David Haussler, Jim Kent, Kate Rosenbloom, Hiram Clawson, Mark Diekhans, Elliott Margulies, James Taylor, Andre Luis Martins, Nick Peterson, Duncan Temple Lang and many users of PHAST for support and advice.
Biographies
Melissa J. Hubisz is a programmer/analyst in the Department of Biological Statistics and Computational Biology at Cornell University. She has been the lead software developer for PHAST and RPHAST since 2008.
Katherine S. Pollard is an associate investigator at the Gladstone Institutes and an associate professor in the Division of Biostatistics at the University of California, San Francisco. She has been a contributor to the PHAST project since 2005 and has a particular interest in RPHAST.
Adam Siepel is an associate professor in the Department of Biological Statistics and Computational Biology at Cornell University. He initiated the PHAST project in 2002, as a graduate student at the University of California, Santa Cruz, and now oversees the project.
References
- 1.Margulies EH, Blanchette M, NISC Comparative Sequencing Program et al. Identification and Characterization of Multi-Species Conserved Sequences. Genome Res. 2003;13:2507–18. doi: 10.1101/gr.1602203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper GM, Stone EA, Asimenos G, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guigó R, Dermitzakis ET, Agarwal P, et al. Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes. Proc Natl Acad Sci USA. 2003;100:1140–45. doi: 10.1073/pnas.0337561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kellis M, Patterson N, Endrizzi M, et al. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–54. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 6.Siepel A, Diekhans M, Brejova B, et al. Targeted discovery of novel human exons by comparative genomics. Genome Res. 2007;17:1763–73. doi: 10.1101/gr.7128207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollard KS, Salama SR, Lambert N, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–72. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 8.Prabhakar S, Noonan JP, Paabo S, et al. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- 9.Kim SY, Pritcchard JK. Adaptive evolution of conserved noncoding elements in mammals. PLoS Genet. 2007;3:1572–86. doi: 10.1371/journal.pgen.0030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moses AM, Chiang DY, Pollard DA, et al. MONKEY: identifying conserved transcriptionfactor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol. 2004;5:R98. doi: 10.1186/gb-2004-5-12-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen JS, Bejerano G, Siepel A, et al. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z. A space-time process model for the evolution of DNA sequences. Genetics. 1995;139:993–1005. doi: 10.1093/genetics/139.2.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhead B, Karolchik D, Kuhn RM, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–9. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:l586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 15.Pond S, Muse S. HyPhy: hypothesis testing using phylogenies. In: Nielsen R, editor. Statistical Methods in Molecular Evolution. Springer; 2005. pp. 125–81. [Google Scholar]
- 16.Kumar S, Nei M, Dudley J, et al. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asthana S, Roytberg M, Stamatoyannopoulos JA, et al. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol. 2007;3:e254. doi: 10.1371/journal.pcbi.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross SS, Brent MR. Using multiple alignments to improve gene prediction. J Comput Biol. 2006;13:379–93. doi: 10.1089/cmb.2006.13.379. [DOI] [PubMed] [Google Scholar]
- 19. R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2010. http://www.R-project.org. ISBN 3-900051-07-0. [Google Scholar]
- 20.Siepel A, Haussler D. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 2004;21:468–88. doi: 10.1093/molbev/msh039. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Diehl A, Wu F, et al. Sequencing and comparative analysis of a conserved syntenic segment in the Solanacea. Genetics. 2008;180:391–408. doi: 10.1534/genetics.108.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holloway AK, Begun DJ, Siepel A, et al. Accelerated sequence divergence of conserved genomic elements in Drosophila melanogaster. Genome Res. 2008;18:1592–601. doi: 10.1101/gr.077131.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pollard KS, Hubisz MJ, Rosenbloom KR, et al. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 25.Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura T, Aoki S, Kitajirma K, et al. Molecular cloning and characterization of Kremen, a novel kringle-containing transmembrane protein. Biochim Biophys Acta. 2001;1518:163–72. doi: 10.1016/s0167-4781(01)00168-3. [DOI] [PubMed] [Google Scholar]
- 28.Siepel A, Pollard K, Haussler D. New methods for detecting lineage-specific selection. In: Proceedings of the 10th International Conference on Research in Computational Molecular Biology; Berlin: Springer; 2006. pp. 190–205. [Google Scholar]
- 29.Siepel A, Haussler D. Computational identification of evolutionarily conserved exons. In: Proceedings of the 8th International Conference on Research in Computational Molecular Biology; New York, NY: ACM Press; 2004. pp. 177–86. [Google Scholar]