Abstract
The direction of migration of a cell invading a host population is assumed to be controlled by the magnitude of the strains in the host medium (cells plus extracellular matrix) that arise as the host medium deforms to accommodate the invader. The single assumption that invaders are cued by strains external to themselves is sufficient to generate network structures. The strain induced by a line of invaders is greatest at the extremity of the line and thus the strain field breaks symmetry, stabilizing branch formation. The strain cue also triggers sprouting from existing branches, with no further model assumption. Network characteristics depend primarily on the ratio of the rate of advance of the invaders to the rate of relaxation of the host cells after their initial deformation. Intra-cell mechanisms that govern these two rates control network morphology. The strain field that cues an individual invader is a collective response of the combined cell populations, involving the nearest 100 cells, to order of magnitude, to any invader. The mechanism does not rely on the pre-existence of the entire host medium prior to invasion; the host cells need only maintain a layer several cells thick around each invader. Consistent with recent experiments, networks result only from a strain cue that is based on strain magnitudes. Spatial strain gradients do not break symmetry and therefore cannot stabilize branch formation. The theory recreates most of the geometrical features of the nervous network in the mouse gut when the most influential adjustable parameter takes a value consistent with one inferred from human and mouse amelogenesis. Because of similarity in the guiding local strain fields, strain cues could also be a participating factor in the formation of vascular networks.
Keywords: nervous network, angiogenesis, strain stimulus, morphogenesis, symmetry breaking
1. Introduction
The question of what directs invading cells to migrate and proliferate only along discrete, separated paths, rather than distributing themselves among the host cells in a statistically uniform spatial pattern, is the key to the formation of network structures. We conjecture that the mechanism that guides invading cells to follow discrete paths or branches is their response to spatial variations in the strain or stress fields that arise around them. A strain-cue rule for migration is consistent with considerable experimental evidence that strain is a major factor in cell migration, proliferation and differentiation (e.g. [1–16]) and that many of the responses seen to strain are shared universally by different cells [17].
Consider a cell that has just invaded a space that was previously densely occupied by host cells and their extracellular matrix (ECM). To accommodate the invader, the host cells and ECM must displace (figure 1a), generating stress and strain fields. Stresses and strains are computed in this paper assuming that the host cells and ECM are homogenized. If the host cells and ECM displace together, the computed strains will be the same as those in either the host cells or the ECM. This is not true for the stresses, because the host cells and ECM differ in stiffness.
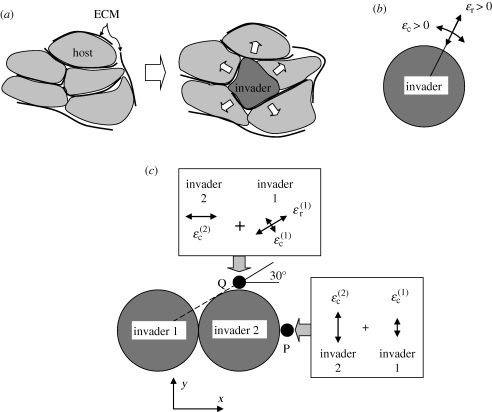
Figure 1.
(a) When invaded, the host cells and their associated ECM must deform to accommodate the invader. (b) A spherical invader creates a compressive strain in any radial direction and tensile strain in any direction perpendicular to a radius. (c) The strain generated at any point in the host by two invaders is the sum of the strains owing to each invader.
To be guided by the strain-cue rule, an invading cell must sense the strain in the host at locations to which it might migrate either directly, e.g. using lamellipodia or filipodia, or indirectly via chemical signals released by the host cells when they are strained. Lamellipodia and filipodia are very thin cell features that are ideal for probing among host cells and could sense strains or released chemical signals; and detailed calculations below show that significant strains arise around an invader over distances that are similar to the reach of lamellipodia and filipodia. But chemical signals released by the host cells under strain might also stimulate an invader by diffusing to the nearest part of its membrane. These alternative routes for the strain cue need not be resolved to understand how strain field patterns can control network formation; the key information is in the spatial variations of the strain fields themselves. In the present theory, the effect of strain patterns on morphogenesis can be investigated separately from other factors.
1.1. How a strain cue can break symmetry
When a single spherical cell invades a homogeneous, isotropic host population, the host cells and their ECM displace away from the invader and therefore develop compressive strain in all radial directions (figure 1b). They must also stretch, i.e. develop tensile strain, around the periphery of the invader. If the host cells have no time to relax or rearrange, stresses arise that are proportional to the strains, being compressive in the radial direction and tensile in the circumferential directions.
Invasion deeper into the host population might occur from this single invader by migration into new territory accompanied by proliferation to sustain the invader density. We conjecture that the direction of further invasion depends on the strain state already induced in the surrounding host population by the invasion that has occurred so far.
One possibility is that invasion in a given direction is favoured by the presence of tensile strains in the directions perpendicular to that direction; intuitively, one might expect that a body of cells is more easily invaded if it is already stretched. Indeed, in agglomerates of mesendoderm cells migrating across a surface present either naturally or in culture, cells that are initially located away from the surface accelerate the rate of migration of the agglomerate by intercalating themselves between cells abutting the surface when the latter cells are strained [5]. Analogous biomechanical mechanisms of migration and intercalation are implicit in the morphogenesis of many organs [5]. In the present problem, there are two mutually orthogonal directions, 1 and 2, that are tangential to the domain occupied by invaders at any point on the surface. We assume that the strength of the cue for migration increases when the sum of the strain components in the tangential directions, ɛ1 + ɛ2, increases, where the sum is evaluated on the candidate migration path.
For further invasion from a single spherical invader, the strain cue favours no particular direction; the strain sum ɛ1 + ɛ2 is invariant over the surface of a sphere (but see below for the case of a moving invader). However, once the next invasion step has occurred, in some randomly chosen direction, the picture changes. Now there are two invaders present and the strain field in the surrounding host cells can be computed as the sum of the strain fields generated by each invader (we assume that the host cells and their ECM constitute a linear elastic medium, at least briefly, which is reasonable until the host cells have time to relax). The summation of strain contributions breaks symmetry, favouring the formation of long thin branches, as follows.
Under the strain-cue hypothesis, migration from invader 2 in the direction of point P in figure 1c is controlled by the components of strain in the y-direction, which is the tangential direction at P. (To develop intuition, the concept is described referring to only one of the tangential strain components. The other tangential strain component is considered in full calculations of an invasion, but its inclusion does not change the qualitative outcome of the present argument.) Because P is in line with the centres of both invaders, the tangential strain, ɛ2, at P is simply the sum of the two circumferential strains, 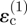 and
and 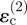 , induced by invaders 1 and 2. Because it is further away from point P, the contribution from invader 1 is smaller than that from invader 2; but it has the same sign, both being tensile, and therefore the tangential strain component
, induced by invaders 1 and 2. Because it is further away from point P, the contribution from invader 1 is smaller than that from invader 2; but it has the same sign, both being tensile, and therefore the tangential strain component 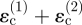 is increased over the strain
is increased over the strain 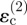 that would arise from invader 2 alone.
that would arise from invader 2 alone.
Consider next the possibility of migration from invader 2 in the direction of point Q in figure 1c. Under the strain-cue hypothesis, this candidate migration is controlled by the strain component in the x-direction, which is now the tangential direction. The contribution from invader 2 remains the circumferential strain, 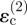 . However, the radial and circumferential strain components owing to invader 1 at point Q are not aligned with the (x, y)-axes; their contribution to the tangential strain in the x-direction must therefore be obtained from the rules for strain transformation from one axis system to another, yielding the sum
. However, the radial and circumferential strain components owing to invader 1 at point Q are not aligned with the (x, y)-axes; their contribution to the tangential strain in the x-direction must therefore be obtained from the rules for strain transformation from one axis system to another, yielding the sum 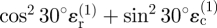 . The radial strain
. The radial strain 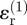 , which is compressive (
, which is compressive (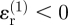 ), contributes most strongly to this sum; the presence of invader 1 therefore decreases the strength of the strain cue at point Q.
), contributes most strongly to this sum; the presence of invader 1 therefore decreases the strength of the strain cue at point Q.
Thus, the strain-cue hypothesis favours continued migration along the line that has already been formed by invaders 1 and 2 and disfavours migration from invader 2 in an orthogonal direction. This tendency, which exists even if the line is only two invaders long, breaks symmetry and underpins network formation. For example, if a population of invaders is migrating along a uniform, broad front and one invader moves ahead of the front, the strain cue will favour the extension of the spur into a branch that tends to increase in length, perhaps indefinitely.
The strain-cue rule is all that is needed to generate life-like network structures, including the formation and maintenance of an invasion front, the sprouting of new branches and the union of branches to form a system whose topology is similar to that in some nervous and capillary networks. No other physical assumption is necessary.
Alternative concepts have been proposed in the past for branching morphogenesis based on viscous fingering or diffusion processes. Comparison with these is deferred to §5, when the characteristics of the present theory have been established.
For the particular case of innervation of the gut, prior continuum and cellular automata theories have dealt successfully with prediction of the rate of advance of the invasion front and aspects of cell trajectories in both the non-growing and growing gut tissues, but have not addressed the formation of the network morphology [18–23].
The ability of strain variations in developing organs to generate dendritic fractal structures has been previously suggested, but without demonstration of branch stabilization by the strain field variations specific to different cell configurations, or consideration of the role of time dependence in the fields [24].
2. Mechanics model
2.1. Overview
We present simulations of the penetration of a three-dimensional domain of host cells and their ECM by an army of invading cells. The simulation is formulated as though the entire host cell domain is already established before the invasion begins. However, the results of the simulations show that this can be regarded merely as a computational convenience; it is sufficient that the host cell domain expands sufficiently rapidly to maintain a layer at least several cells thick around the invaders (see below).
The invasion causes spatial variations in the density of cells as invaders intercalate themselves among the hosts. The density variations create spatially varying and time-dependent strain and stress fields. The fields are re-computed continually as the invasion progresses. The local values of strain components are used as cues to control the migration of invaders from domains that have already been invaded. The strain-cue rule is applied equally to all invaders on every iteration; all variations in migration patterns result solely from variations in the strain fields. No other control over branch stability, sprouting or any other phenomenon is introduced.
The strain-cue rule for migration includes a probabilistic factor, representing the influence of random factors such as variations in the positions or shapes of cells. The statistical characteristics of the invasion are analysed by the Monte Carlo method: the probabilistic factor is evaluated each time the strain-cue rule is invoked using a pseudo-random number generator and the simulations are repeated with different initial seed values for the random number sequence until the network statistics are established.
Continuum elasticity theory is used to compute the strain and stress fields at any time. For accuracy and computational efficiency, the fields are calculated using analytical results for the mechanics of inclusions, rather than the finite-element method. This allows the treatment of much larger populations.
2.2. Spatial and temporal discretization
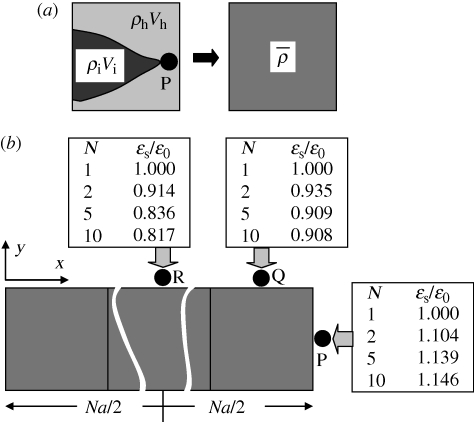
The problem is solved numerically on a simple cubic lattice with element size a, aligned with a Cartesian coordinate system (x, y, z). Each cubic element may be occupied by a combination of hosts and invaders, with numbers nh and ni, respectively, which need not be integers (figure 2a). An element is defined to be somewhat larger than a single cell. The host and invader cell number densities in any element are ρh = nh/a3 and ρi = ni/a3, respectively. The cell densities are smeared out to be uniform within each element, but they differ from element to element.
Figure 2.
(a) A system of irregularly shaped invader and host cells is discretized into cubic elements within each of which the cell densities are uniform. (b) Values of the strain measure, ɛs = ɛ1 + ɛ2, the sum of the two tangential strain components, at three points in the host cell domain adjacent to a line of N invaded elements.
In the absence of mechanical stress, a single host and a single invader cell are assumed to have volumes Vh and Vi, respectively. The volume that the cells in a single element would occupy in the absence of stress is V = nhVh + niVi. The normalized quantities
| 2.1a |
| 2.1b |
| 2.1c |
summarize information about cell number density and the stress-free volume of the cells occupying an element. If 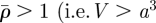 ), a mechanical stress will arise if the element is confined among other elements.
), a mechanical stress will arise if the element is confined among other elements.
2.3. Strain and stress calculation
To simplify computation of the stresses and strains, consider the composite material of the host cells and their ECM to be a homogeneous and isotropic elastic medium. The assumption of homogeneity is valid if stresses and strains are averaged over a gauge length that is somewhat larger than a single host cell. Stresses and strains discussed in this work refer to smoothly varying fields defined over such a gauge length.
The stress field generated by a single element for which 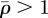 is computed using analytical formulae derived for an ‘inclusion’ in an elastic medium that contains a ‘stress-free eigenstrain’ [25,26]. The volume eigenstrain,
is computed using analytical formulae derived for an ‘inclusion’ in an elastic medium that contains a ‘stress-free eigenstrain’ [25,26]. The volume eigenstrain, 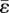 , measures the difference between the stress-free volume of the cells that occupy an element and the volume of the element:
, measures the difference between the stress-free volume of the cells that occupy an element and the volume of the element:
| 2.2a |
The eigenstrain is assumed to be volumetric within an element, with components
| 2.2b |
The method of computing these strains during simulations is detailed in the electronic supplementary material, appendix A.
When a line of N elements have been invaded, key variations of the strain fields are as shown in figure 2b for three locations P, Q and R just outside the line. The strain measure, ɛs = ɛ1 + ɛ2, is tabulated, which is the sum of the components of strain in the two directions, 1 and 2, that are parallel to the nearest surface of the line of elements. Trends in the ratio, ɛs/ɛ0, are shown for different line lengths, N, where ɛ0, the ‘reference strain’, is the value ɛs takes when N = 1. As the line lengthens, ɛs/ɛ0 increases beyond its extremity (point P) and decreases along its flanks (points Q and R). The maximum variation of ɛs is approximately 30 per cent of ɛ0. Variations in ɛs/ɛ0 are independent of the magnitude, 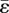 , of the eigenstrain, provided it is uniform, or the element size, a. Therefore, predictions of network formation that depend only on ɛs/ɛ0 are not affected by the choices made for
, of the eigenstrain, provided it is uniform, or the element size, a. Therefore, predictions of network formation that depend only on ɛs/ɛ0 are not affected by the choices made for 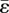 and a. They are also independent of Young's modulus, E, and only weakly dependent on Poisson's ratio, which has been assigned the value 0.3. The fact that ɛ0 ≈ 0.267 for eigenstrain
and a. They are also independent of Young's modulus, E, and only weakly dependent on Poisson's ratio, which has been assigned the value 0.3. The fact that ɛ0 ≈ 0.267 for eigenstrain 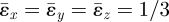 will be used to make rough estimates of the stress and strain magnitudes expected in nature (see below).
will be used to make rough estimates of the stress and strain magnitudes expected in nature (see below).
The variations in ɛs shown in figure 2b, which concur qualitatively with those for a spherical inclusion (figure 1b,c), break symmetry and stabilize branches. When a branch bends by a random variation in its direction to form an elbow, a strain enhancement also arises at the point of the elbow. This trend generates sprouting. These two effects together generate networks.
2.4. Migration
We hypothesize that the probability of migration is governed by a strain cue. Whether migration occurs in the mth time step from element i to neighbour j is determined by first evaluating the variables
| 2.3a |
for each of the neighbours j of cell i, where Sij is a strain-cue factor defined in the electronic supplementary material, appendix B, X is a pseudo-random number (chosen separately for each j) that is uniformly distributed in the interval [0,1], and p > 0 is a velocity factor. The strain-cue factor is a function of the normalized strain measure ɛs/ɛ0 and a strain sensitivity parameter, c1, and is bounded by 0 < Sij < 1 (electronic supplementary material, appendix B).
Denote by k the value of j for which fj is maximum. Migration is only allowed on the current iteration from element i to neighbour k and occurs if and only if
| 2.3b |
Restricting migration from element i to at most one neighbour allows the change in the local strain field owing to the migration step to be computed before a second possible migration from element i can occur. If the local strain field remains conducive, migration from element i can possibly occur to any number up to all of its vertex-sharing neighbours in subsequent iterations.
Migration is favoured by higher positive values of 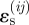 relative to the reference strain ɛ0, becoming certain for very large positive
relative to the reference strain ɛ0, becoming certain for very large positive 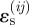 and impossible for very large negative
and impossible for very large negative 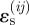 . When
. When 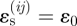 , the strain-cue factor takes the mid-value Si j = 0.5. The strain-sensitivity factor, c1, determines how far
, the strain-cue factor takes the mid-value Si j = 0.5. The strain-sensitivity factor, c1, determines how far 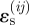 needs to differ from ɛ0 before migration attains certainty or impossibility.
needs to differ from ɛ0 before migration attains certainty or impossibility.
The probabilistic nature of equation (2.3) represents the effects of random variations in the local character of the host/invader system that are not represented by variations in the local strain fields. These might consist, for example, of variations in the shapes of an invader, which will modify the strain fields created by the invader, or in the positions of host cells relative to an invader, which must find a gap between them. The random fluctuations introduced by equation (2.3) have no spatial order; they introduce no spatial bias that might favour branch formation or stabilization, branch bifurcation or other topological features of a network.
The velocity factor, p, a constant, is introduced to provide control over the average velocity of the invasion front. It is assigned a value p < 1. As p decreases, the front moves more slowly. For a single, long line of elements that have been invaded, analytical results can be derived for the average rate of extension of the branch, v0 (electronic supplementary material, appendix C).
The velocity parameter, p, is included to allow tests of mesh independence, which reveal an essential scaling property of the mechanics model.
2.5. Host relaxation
Where invading cells have migrated into regions already occupied by host cells, the local cell density rises and the host cells will tend to relax away from the invaders to restore uniformity. This motion is introduced into the simulations as a diffusion-like process in which only the host cells relax. Whether the relaxation is mediated by host cell migration away from the invaders, or by changes in the volume of individual host cells, e.g. the flow of water or other matter through the host cell membranes, need not be stated; the relaxation rule is valid if the host cell density is modified by any kind of mass transport and the rate of relaxation is proportional to the density gradient.
The relaxation calculation used in simulations is presented in the electronic supplementary material, appendix D. The degree of relaxation permitted after each time step in a simulation is determined by a constant relaxation factor β and the number nr of relaxation iterations applied at that time step.
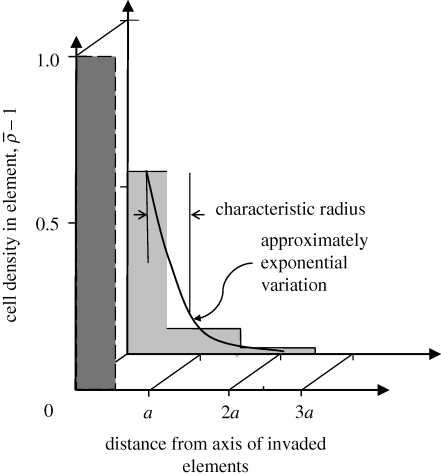
The relaxation process can be illustrated by considering its effect on the environs of a single line of invaded elements. The invaded elements are assigned initial densities 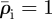 and
and 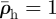 for invaders and hosts, respectively, so that
for invaders and hosts, respectively, so that 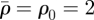 . All other elements are assigned initial densities
. All other elements are assigned initial densities 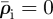 and
and 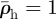 . Successive applications of the relaxation step cause the density spike to spread radially away from the line of invaded elements: the density in the invaded line falls, while the density in the neighbouring elements rises (figure 3). After some steps, the density decreases approximately exponentially with distance from the invasion branch, with some characteristic decay length, λr. Because λr is proportional to the element size, a, and increases approximately linearly with the number of relaxation iterations, nr, per time step, δt, the relaxation process can be characterized by a velocity
. Successive applications of the relaxation step cause the density spike to spread radially away from the line of invaded elements: the density in the invaded line falls, while the density in the neighbouring elements rises (figure 3). After some steps, the density decreases approximately exponentially with distance from the invasion branch, with some characteristic decay length, λr. Because λr is proportional to the element size, a, and increases approximately linearly with the number of relaxation iterations, nr, per time step, δt, the relaxation process can be characterized by a velocity
| 2.4 |
This quantity is proportional to, but not equal to, dλr/dt.
Figure 3.
Relaxation of host density around a hypothetical line or branch of invaded cells. The invasion creates a density spike which is at first confined to the invasion branch (element at 0). After five relaxation iterations, the density exceeds unity (the initial host density) significantly in elements at distance a and 2a from the branch, while the excess density  in the invasion branch has diminished by almost half. For relaxation parameters (nr = 1, β = 0.02) and initial densities
in the invasion branch has diminished by almost half. For relaxation parameters (nr = 1, β = 0.02) and initial densities 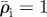 in the invaded elements and
in the invaded elements and 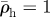 everywhere. Dark grey, zero iterations; light grey, five iterations.
everywhere. Dark grey, zero iterations; light grey, five iterations.
Relaxation reduces the stress and strain fields around an advancing branch of invaders (electronic supplementary material, appendix C). If the branch does not advance on a time increment, further strain reduction will occur; therefore, the longer a branch lingers at a given length, the lower the probability of its further extension. One possible outcome is that the branch will arrest permanently as the strains become evanescent. The probability of arrest, parrest, per unit length of advance of the branch, with length normalized by the element size, a, can be estimated analytically for a straight branch (electronic supplementary material, appendix C).
A network consisting of many branches can only continue to advance indefinitely with undiminished density if new branches are formed from existing branches by bifurcation or sprouting at a rate per unit length of advance of each branch that at least equals parrest.
2.6. Replenishing the supply of invaders
When a line of invader cells extends into a host domain that was not previously invaded, the density of invaders in the wake of the invasion point will tend to be diminished. To maintain their density and support continued invasion, the invaders must re-supply their ranks by a combination of migration from the rear and proliferation. Both the migration from the rear and proliferation may be influenced by strain signalling (as well as chemical signalling). In this first study of strain stimulus effects on network formation, a simplifying assumption is made, that re-supply of invaders occurs rapidly relative to other processes. After migration from invaded to previously non-invaded elements has been computed during a time increment, the density of invaders is immediately set to a constant value, 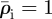 , in all elements that have been invaded, remaining zero in all other elements. At least in the case of innervation of the mouse gut, the limit of perfect re-supply is a reasonable assumption for a first model: branches maintain approximately constant invader cell densities during the invasion [27].
, in all elements that have been invaded, remaining zero in all other elements. At least in the case of innervation of the mouse gut, the limit of perfect re-supply is a reasonable assumption for a first model: branches maintain approximately constant invader cell densities during the invasion [27].
3. Simulations of network formation
The simulations reported in this paper are of the formation of planar networks: the invading cells are restricted to migrate within a single layer of elements whose centres lie on z = 0 in the Cartesian system (x, y, z). This restriction is appropriate for the formation of some nerve networks, such as in the gut and retina, and some instances of angiogenesis. While the invaders are confined to a plane, the simulation space has three dimensions, to permit relaxation of the host cells in three dimensions, which is necessary to achieve realistic density variations.
Invasion begins from some initial set of invaders with an assigned spatial distribution. Two cases will be considered: first, a relatively dense population of invaders that are distributed randomly in space, with statistically uniform density, over a narrow band −5a < x < 0, and second, an initial invader population that consists of a single invaded cell, centrally located in the host domain. In the case of the initial band, the invasion tends to progress towards positive large x, while invasion from the single element radiates out in a roughly circular pattern. These growth characteristics are outcomes, not stipulations: the rule for invader migration has no built-in bias for any global direction.
Invaded elements are assigned the invader cell density, 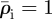 , both for the initial conditions and following re-supply. The host cell density is initially
, both for the initial conditions and following re-supply. The host cell density is initially 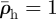 everywhere and changes as a continuous variable during relaxation.
everywhere and changes as a continuous variable during relaxation.
The remaining parameters that must be specified for a simulation are the set {β, n, p, c1, ɛ0} appearing in equations (2.3) and (B 1). Of these, nr was set to 1, the reference strain ɛ0 was set to the value of the strain measure ɛs outside a single invaded element and the other parameters were varied.
Electronic supplementary material, appendix E, presents further details of the computations; a discussion of invariances in the underlying equations, which have the important effect of making the predicted network morphology mesh independent and also independent of the size of cells, the choice of initial densities and the values of elastic constants; a possible multi-scale strategy for the future inclusion of details such as cell shape; an approximate isomorphism between the model of cells invading the interior of a domain of host cells and cells invading superficially across the surface of a body of host cells; and a discussion of the expected effects of delays in the re-supply of invaders.
3.1. The range of strain effects
The elastic strain at any point is the sum of the contributions arising from all elements in which 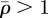 . However, since the magnitude of an element's contribution falls asymptotically as the cube of its distance, calculations show that the sum can be cut off at some distance rcut. A few tests reveal that the computed strain fields do not change significantly when rcut is increased beyond rcut = 2.1a. One result that partly explains the short range of strain interactions is visible in figure 2b. The quantity ɛs varies only weakly as the number of invaded elements in the line increases beyond N = 2 (see also [25]); and the influence of the more distant elements on ɛs is lessened even further by host relaxation effects. The limited range of strain influences is an important result (not assumption) of the theory, demonstrating that the strain field surrounding any invader is determined by the cell densities in the nearest 32 neighbouring elements. If a computational element is somewhat bigger than a living cell (figure 2a), these 32 elements represent 100 cells, to order of magnitude. Network formation is governed by the collective behaviour of groups of 100 cells, comprising 10 invaders, to order of magnitude, and the host cells that relax around them.
. However, since the magnitude of an element's contribution falls asymptotically as the cube of its distance, calculations show that the sum can be cut off at some distance rcut. A few tests reveal that the computed strain fields do not change significantly when rcut is increased beyond rcut = 2.1a. One result that partly explains the short range of strain interactions is visible in figure 2b. The quantity ɛs varies only weakly as the number of invaded elements in the line increases beyond N = 2 (see also [25]); and the influence of the more distant elements on ɛs is lessened even further by host relaxation effects. The limited range of strain influences is an important result (not assumption) of the theory, demonstrating that the strain field surrounding any invader is determined by the cell densities in the nearest 32 neighbouring elements. If a computational element is somewhat bigger than a living cell (figure 2a), these 32 elements represent 100 cells, to order of magnitude. Network formation is governed by the collective behaviour of groups of 100 cells, comprising 10 invaders, to order of magnitude, and the host cells that relax around them.
The fact that the range of strain effects is limited has two important biological implications.
— Networks can form in a growing organ. Since only local cell clusters influence the strain-cue rule, the same network structures would be predicted in a simulation in which host cells were added to fill the space somewhat ahead of the forming network as the network advanced. Thus, the strain-cue rule can operate in an organ in which host cells are being recruited and their ECM is being expressed simultaneously with the invasion. (Such a simulation is not presented here. To pursue it in detail, the strains generated by the accumulating host cells would also have to be considered, an additional modelling effort. However, since the key invader-generated strains are local and large in magnitude, longer range strains arising from organ (or tumour) growth would constitute a relatively small and uniform contribution that would not significantly perturb the symmetry-breaking character of the local invader-generated strains.)
— The strain mechanism is robust. The local origin of strains allows the network formation process to be robust. Long-range stress and strain fields in a developing organ could vary owing to extrinsic factors and therefore be unreliable cues for determining spatially varying organ structure. For example, in the case of planar invasions, such as innervation of the gut, long-range compression fields owing to the invasion could be relaxed by global buckling of the gut wall. However, the short-range strain fields generated by an invader would remain minimally affected. The local strain fields depend mainly on local cell positions and local rate processes and are relatively immune to variability in the global geometry of the organ.
3.2. Characteristics of networks grown from an initial band of invaders
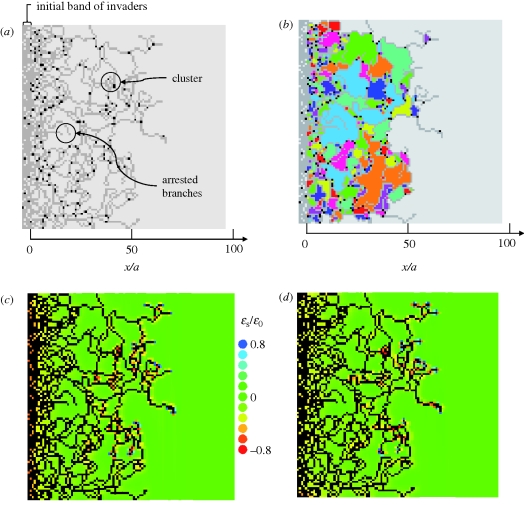
As a consequence of the symmetry-breaking property of strain cues, the simulations generate networks. One network is shown in figure 4a, grown from a band of initial invaders at x < 0. Invaded elements are marked in grey. Elements from which migration has occurred into two or more neighbours are marked in black. These are bifurcation points or the origins of sprouts. The invasion advances across a diffuse front, towards positive x.
Figure 4.
A network grown from an initial band of invaders for parameter values β = 0.04, nr = 1, p = 1, c1 = 0.3. (a) Branches of invader cells (grey) are punctuated by occasional bifurcation or sprout points (black). Permanently arrested branches and clusters of invaders occur occasionally. (b) Colour coding highlights distinct void areas between branches (the simple algorithm for colour choice does not forbid neighbouring voids from having the same colour). (c,d) Strain measures ɛs = ɛy + ɛz and ɛs = ɛz + ɛx, respectively, normalized by ɛ0. The colour assigned to each computational element refers to the value of the plotted quantity next to the face of the element through which invasion is most favoured (the face for which the value of the strain-cue factor, Sij, of equation (2.3a) is maximum). In (c,d), all invaded elements are black.
Depending on the parameter choices, the network formed has some overall density that is usually much less than the density of the elements initially set as invaded (x < 0). Branches emerge from just a few locations on the initial array. Most branches propagate towards positive x, but branch extension also occurs in the y-direction and diagonally, and towards negative x if non-invaded space is left behind the front. The variable direction of branch extension causes branch intersection (coalescence) and the formation of wholly enclosed areas of host cells (figure 4b). The statistics of the enclosed areas or ‘voids’ are useful metrics of the network. The branches can increase in number by sprouting or bifurcation and diminish in number by coalescence or arrest. The relative rates of these processes determine the network density.
As the branches of the network advance, the strain fields around them evolve. Figure 4c,d shows spatial variations of the strain measure 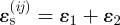 . In the colour code for figure 4c,d, tensile strains are blue, and compressive strains are yellow or red. Since figure 4c plots ɛs = ɛy + ɛz, migration is likely from any black element into a blue element that is to the left or right of it. Similarly, migration is likely from any black element in figure 4d into any blue element that is above or below it.
. In the colour code for figure 4c,d, tensile strains are blue, and compressive strains are yellow or red. Since figure 4c plots ɛs = ɛy + ɛz, migration is likely from any black element into a blue element that is to the left or right of it. Similarly, migration is likely from any black element in figure 4d into any blue element that is above or below it.
Thus, the blue elements in figure 4c,d indicate invader cells that will tend to be active (migrate) on the next time increment; but whether they will be active also depends on the random factor introduced in equation (2.3), which is not represented in these plots. The blue elements indicate the diffuse front of the invasion. In the example illustrated, some future invasion is likely into areas that are already enclosed by branches of the network.
Tensile strain concentrations are present not only at the tip of a branch, but also at locations behind the tip. Application of the strain-cue rule at these locations can therefore generate sprouting as a natural consequence of strain effects, requiring no additional special rule. Sprouting is especially likely wherever the branch has deviated in its path to form an elbow feature, which tends to generate strain concentrations.
Figure 4c,d also shows compressive strains (yellow to red colour range) surrounding branches at locations well behind their tips. The compressive strains reflect the increase in host cell density in branch-neighbouring elements, owing to relaxation of host cells out of the invaded elements (as in figure 3). The compressive strains tend to switch off sprouting from the older segments of branches. As branches continue to age, the compressive strains also fade away owing to continuing host relaxation.
The cumulative probability distribution of the area of voids can be fitted approximately by a lognormal cumulative distribution function
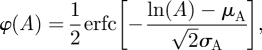 |
3.1 |
with erfc the complementary Gauss error function (figure 5a). This distribution has median = exp[μA]. The lognormal distribution fits well in some cases, but tends to under-represent the probability of large area voids in others, including that of figure 5a. Log–log plots of histograms of the void area indicate an approximately linear region, with slope denoted −1/λA, for void areas between approximately 3a2 and 20a2, suggesting a power-law distribution. The fitted parameter λA indicates the width of the distribution. It is used below to compare the void area distributions from different simulations.
Figure 5.
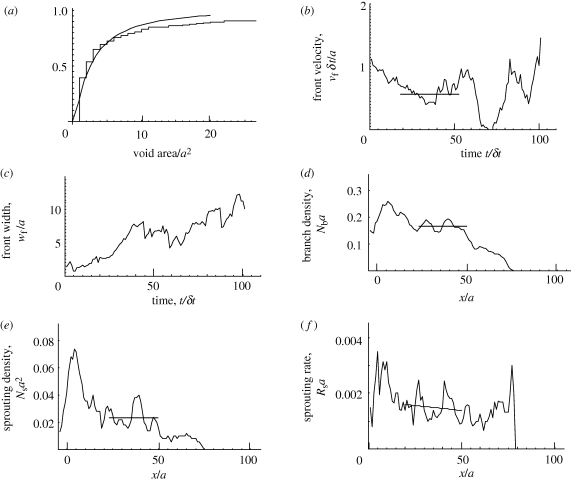
Some statistics of the simulation shown in figure 4. (a) Distribution of void areas. (b,c) The velocity and the width of the front, respectively, versus time. The velocity is a moving average over 11 time increments. (d) The average density of branches per unit length normalized by the element width along a line of constant x (moving average over five values of x). (e) The density of sprouts per unit area normalized by the element area along a line of constant x (moving average over five values of x). Mean values over intervals of uniform network characteristics are indicated by horizontal lines in (b), (d) and (e). (f) Rate of sprouting per length of advance of a branch in the x-direction normalized by a. Smoothed trend over interval following transient (x > 25) shown by fitted line.
The set of active elements, defined as those from which migration occurs at any time step, constitutes the invasion front. The average velocity of the front and its width can be determined by statistical analysis of the set of x-coordinates of the active elements, {xa}. The front position and front width are defined as the mean and the root mean square deviation of {xa}, respectively. The front velocity is the time derivative of the mean. The front velocity and width are plotted for the simulation of figure 4 in figure 5b,c. Other statistics of interest include the number of branches per unit length parallel to the front, Nb (figure 5d) and the number of bifurcations or sprouts per unit area, Ns (figure 5e). The sprouting rate, defined as the probability that a branch forms a sprout per unit length of its advance in the x-direction, is given by the ratio of the sprout density to the branch density (figure 5f).
Three regimes can be distinguished in the network of figure 4 and the related statistical characteristics of figure 5. In the early invasion (x/a < 20, t/δt < 20), the branch and sprout densities are high (figure 5d,e), void areas small (figure 4b) and the front width small (figure 5c). In this regime, the spatial distribution of invaders is transitioning from the arbitrarily chosen initial distribution (x < 0) to the distribution of the network structure that corresponds to the particular values of the parameters {β, p, c1}.
The initial transient is followed by a regime of approximately uniform network formation (20 < x/a < 50, 20 < t/δt < 70), where the front velocity and width, the branch density and the sprout density are all approximately constant (figure 5b–e).
The third regime (x/a > 50, t/δt > 70) extends from the uniform regime to the time where the simulation was stopped. In this regime, the network is only partially formed (figure 4a), the front velocity shows very large fluctuations (figure 5b), the front width increases and the branch and sprout densities decay to zero at the extremity of the advance (figure 5d,e). On the other hand, the sprouting rate remains approximately constant after the transient regime, even as the sprout density falls to zero at the extremity of the invasion. The sprouting rate depends only on local attributes of a branch and is uninfluenced by the presence of other branches.
The statistics of a single simulation exhibit substantial fluctuations, especially in the front velocity, which is the derivative of a stochastic quantity. The fluctuations arise from randomness in the rate and direction of branch extension. Moving averages have been used in some parts of figure 5 to smooth the fluctuations and make trends more easily visible. Relationships between model parameters and the characteristics of predicted networks can be analysed using multiple Monte Carlo runs to reduce uncertainty to acceptable levels.
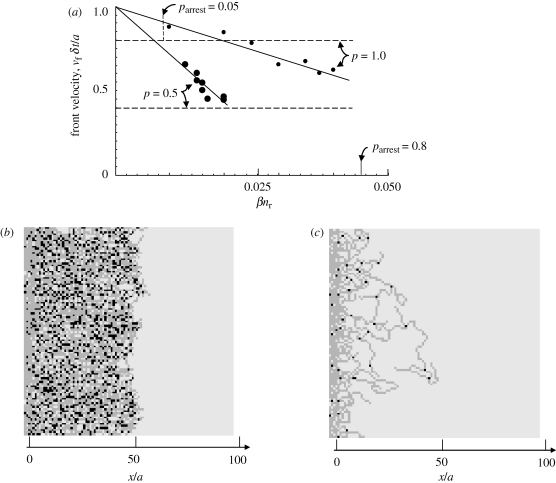
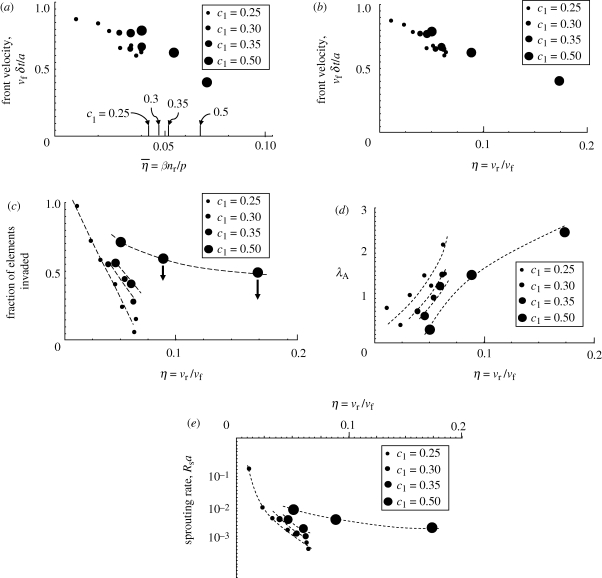
The main parameter dependences of the front velocity are depicted in figure 6a. For fixed p and c1, the velocity declines with increasing βnr, the relaxation rate of the host cells. The velocity is roughly indicated by the expected rate of advance of a single long branch (equation (C 1b) of the electronic supplementary material), which is marked by horizontal lines in figure 6a.
Figure 6.
(a) The average front velocity in the regime of approximately uniform network characteristics versus the host relaxation rate, βnr. For c1 = 0.25 and two marked values of the velocity parameter, p. The horizontal lines show the upper-bound estimates for the front velocity given by equation (3.2b). The short vertical lines show the values of βnr for which parrest takes the marked values when p = 1, from figure 2c. (b) A high-density network generated under slow host relaxation (βnr = 0.02, p = 1, c1 = 0.25). (c) A low-density arresting network generated under rapid host relaxation (βnr = 0.04, p = 1, c1 = 0.25).
As βnr increases, branch arrest is more frequent. For certain values of βnr, an approximate balance is achieved between the sprouting frequency, which increases branch density, and the sum of branch arrest and branch coalescence, which decreases branch density, and networks exemplified by figures 4 and 5 are generated.
Above a critical value of βnr, branch arrest is so probable that the front ceases to advance altogether; the front velocity shows extreme fluctuations during the transient regime before settling to zero, and no regime of uniform network formation is achieved. A typical arrested network is shown in figure 6c.
For the simulations represented in figure 6a, approximate balance between branch arrest, branch coalescence and sprouting occurs when parrest = 0.05 per (unit length/a); and network arrest occurs when parrest = 0.6–0.8 per (unit length/a). The value of βnr corresponding to parrest = 0.8 (obtained from figure 3c) is marked on figure 6a for the case p = 1.
When the velocity parameter p is decreased, the advance of individual branches is slowed and host relaxation becomes more effective for a given value of βnr. Both the front velocity and the critical value of βnr for front arrest are therefore reduced (figure 6a). The change in front velocity follows quite closely the linear dependence on p that is predicted by equation (C 1b) of the electronic supplementary material, even though the absolute velocity is only roughly predicted.
3.3. Network development from a single invader
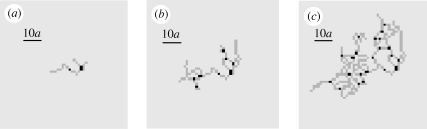
The characteristics of the developed network do not depend on the spatial configuration assigned to the initial invaders. Figure 7 shows network development from a single centrally located invader cell. The problem definition in this simulation is identical to that in figure 4, with the sole difference being the initial distribution of invaders. Snapshots after increasing time intervals show a network expanding in a roughly circular shape from the single initial invader, eventually filling the host domain (at times later than those shown). In the fully developed network, the branch density, sprout density and distribution of enclosed void areas are statistically similar to those calculated from figure 4.
Figure 7.
A network grown from a single centrally located invader (for parameter values β = 0.04, nr = 1, p = 1, c1 = 0.3). Branches of invader cells (grey) are punctuated by occasional bifurcation or sprout points (black). Networks shown for t/δt = 10, 20 and 40 in (a), (b) and (c), respectively. Initial invader is the black element near the centre of the network in (a).
3.4. Parameters that control the network
While five parameters {β, nr, p, c1, ɛ0} have been defined, statistical analysis confirms that most of these parameters do not affect network morphology independently. Parameter ɛ0 is determined from the strains around a single invaded element; strain magnitudes then appear only via the ratio ɛ/ɛ0. The remaining four parameters influence morphology only via two parameters, the first being the strain sensitivity parameter, c1, and the second a dimensionless combination of β, nr and p. Parameters β and nr act solely through their product βnr, which governs the rate of host relaxation. The parameter p influences the rate of advance of the invasion front, vf; and since time does not appear explicitly in the governing equations, only the ratio of this rate to that of host relaxation can matter. One possible choice for the second parameter is the ratio
| 3.2a |
with vr = βnra/δt the relaxation velocity of equation (2.4). However, statistical analysis shows better correlations if the second parameter is chosen to be
| 3.2b |
with vf(βnr, p) the front velocity in the regime of uniform network formation.
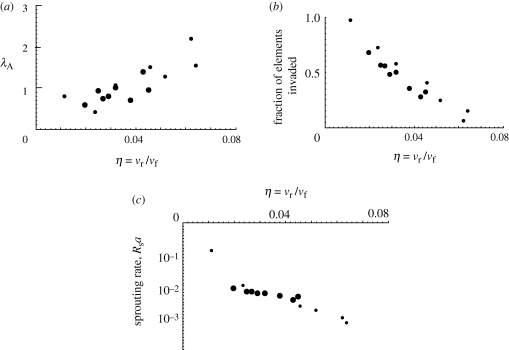
Figure 8 exemplifies the role of η through three network characteristics: the parameter λA, which indicates the width of the distribution of void areas (figure 5a); the fraction of all elements in the regime of uniform propagation that have been invaded; and the sprouting rate in the uniform regime. Results from simulations for p = 0.5 and p = 1 are plotted against η. The two datasets approximately coincide for each quantity. Thus β, nr and p are not independent factors: any combinations of them that yield the same rate ratio, η, will generate the same network characteristics.
Figure 8.
Characteristics of simulations are statistically independent of p for fixed βnr/vf. (a) The parameter λA, indicating the width of the void area distribution. (b) The fraction of elements invaded for the uniform propagation regime of figure 5d. (c) The sprouting rate in the uniform propagation regime. Small filled circles, p = 1; large filled circles, p = 0.5.
Slightly inferior correlations are obtained when network characteristics are plotted against 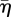 instead of η, because the front velocity is almost but not quite proportional to η (the ratio of the slopes of the fitted lines in figure 6a is not quite two).
instead of η, because the front velocity is almost but not quite proportional to η (the ratio of the slopes of the fitted lines in figure 6a is not quite two).
The main effect of the rate ratio, η, is illustrated by the simulations of figure 6b,c. The only difference between these simulations is a change in η (βnr varied, p fixed); yet in one case, the invaders advance indefinitely to form a dense network that occupies a large fraction of the space initially assigned to the host cells, while in the other they form a sparse network that extends only a finite distance before the invasion front arrests permanently.
The effect of the strain sensitivity parameter, c1, is illustrated in figure 9. As c1 increases, the strain cue triggers migration for strain measures, ɛs, that depart increasingly from ɛ0, the reference strain. Therefore, branch extension can persist after increasing levels of host relaxation; and higher values of the host relaxation rate, βnr, are required to cause the invasion front to arrest. Figure 9a shows the front velocity versus 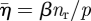 for c1 ranging from 0.25 to 0.5. The datum with the highest value of
for c1 ranging from 0.25 to 0.5. The datum with the highest value of 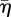 in each case is close to the value at which arrest occurs. The marks on the abscissa show the value of
in each case is close to the value at which arrest occurs. The marks on the abscissa show the value of 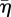 for which parrest = 0.8, and predict the invasion arrest reasonably well. Inspection of the electronic supplementary material, figure C.1, suggests that arrest can be deferred to high values of
for which parrest = 0.8, and predict the invasion arrest reasonably well. Inspection of the electronic supplementary material, figure C.1, suggests that arrest can be deferred to high values of 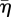 by further increases in c1.
by further increases in c1.
Figure 9.
Influence of the strain sensitivity parameter c1 on network characteristics, with the velocity parameter set to p = 1. Front velocity plotted against (a) η̄ and (b) η. (c) Fraction of all elements that are invaded. (d) A metric of the void size distribution. (e) Sprouting rate per unit of normalized advance of a branch. The arrows in (c) indicate points that are high because they refer to the early part of an invasion (see text). The dashed curves have been added by hand to highlight trends for each of the four values of c1.
The front velocity shows mild increase with c1 when plotted against 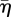 (figure 9a), but is almost independent of c1 when plotted against η = vr/vf (figure 9b). Although the front velocity is influenced only mildly, other network characteristics change more substantially (figure 9c–e). As expected, the sprouting rate increases with c1, because milder strain concentrations trigger new migration (figure 9e). Consequently, the networks are denser (figure 9c) with narrower distributions of void area (figure 9d).
(figure 9a), but is almost independent of c1 when plotted against η = vr/vf (figure 9b). Although the front velocity is influenced only mildly, other network characteristics change more substantially (figure 9c–e). As expected, the sprouting rate increases with c1, because milder strain concentrations trigger new migration (figure 9e). Consequently, the networks are denser (figure 9c) with narrower distributions of void area (figure 9d).
4. Innervation of the gut
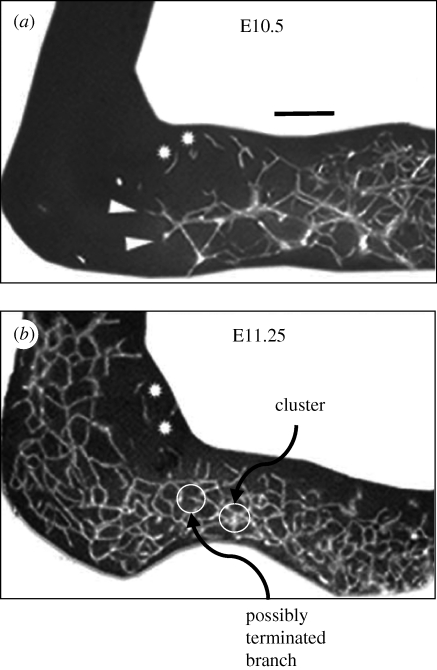
Simulations based on the strain-cue rule for invasion predict a number of features observed in the nerve networks that are formed by the invasion of the ileum and caecum of a mouse by neural crest cells. These successes can be seen by comparing figure 4 (simulation) and figure 10 (nature). The simulation parameters chosen in figure 4, i.e. the values of βnr and c1, can be regarded as best-fit parameters that optimize agreement with figure 10. However, the particular values that achieve success can also be justified a priori at least approximately by independent considerations, which has interesting implications for the possible universality of cell behaviour (§5).
Figure 10.
Nerve networks (white) formed in the mouse ileum and caecum, imaged at two different times during the invasion by neural crest cells (from [38]). The invasion is progressing from right to left. (a) Scale bar, 0.1 mm.
Suppose that the element size in the simulation of figure 4 has been chosen to be slightly larger than the size of an invading cell. Then the proportion of the width of a branch of the network to the width of typical voids can be compared directly between figures 4 and 10; for the chosen model parameters, the proportions are approximately equal. Some other characteristics are also similar. First, the front of the invasion in figure 10 is comparable to the width of the larger voids; figure 5a,c shows a similar characteristic for the simulation. Second, comparison of the earlier and later images in figure 10 shows some backfilling as the invasion proceeds, i.e. densification of the branch structure behind the leading extremity of the invasion. Such backfilling is common in simulations, sometimes affecting relatively large areas, as exemplified by figure 11. Third, branches occasionally appear to terminate permanently in the image of figure 10. Terminated branches in figure 4 occur at an approximately similar frequency, in a small percentage of enclosed void areas. Fourth, invading cells occasionally appear in clusters (figures 4 and 10). In the simulations, clusters arise as fluctuations in the outcome of inserting the local strain field values into the rule for advance. Fifth, where sprouting forms three-branch junctions in either figure 4 or 10, no two of the three branches involved are collinear. This is consistent with the prediction of the simulations that sprouting is triggered by the strain concentration that arises at an elbow when a branch's extension randomly changes direction. Both figures 4 and 10 occasionally show four-branch junctions, but, in the images of figure 10, some of these appear to involve two branches lying slightly above and below one another, which cannot occur in the simulations, because of the restriction of the invasion to a single plane.
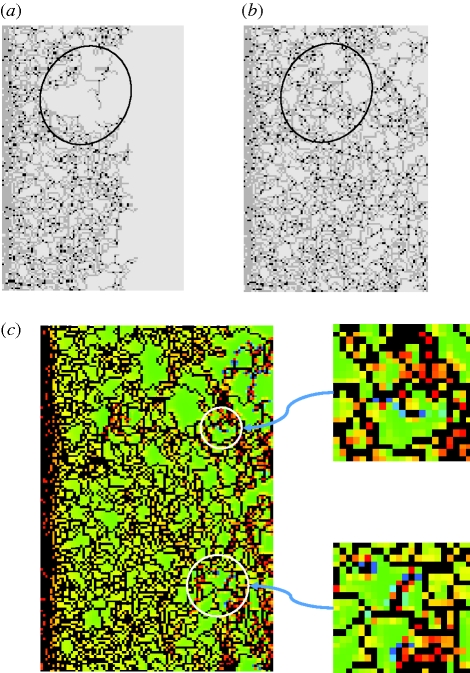
Figure 11.
A simulation initiated from a band of invaders at left (similar to figure 4) with βnr = 0.035 and c1 = 0.3 shows backfilling between (a) time step 100 and (b) time step 150 in the encircled area. (c) The distribution of the strain measure ɛ1 = ɛy + ɛz after 150 time increments shows that backfilling is likely to continue into already-closed voids in other areas (encircled in white). Scale in (c) is the same as in figure 4c.
A characteristic of the networks in the mouse gut (figure 10) that is not replicated in any of figures 4, 11, etc. is the appearance of small islands of incipient network structures, disconnected from and a little ahead of the main invasion front. A possible explanation of such islands is that single nerve cells break away from the invasion front and migrate in isolation, before beginning to proliferate. Consistent with the network formation from an isolated cell that is simulated in figure 7, such cells nucleate new network contributions that have the same statistical characteristics as the network formed by the main invasion front, with which union eventually occurs.
5. Discussion
5.1. Consistency in density relaxation rates across species and organs
The morphology of predicted networks depends on the values of just two parameters, the strain sensitivity parameter c1 and the velocity ratio η. If one assumes that cells have evolved to maximize their ability to respond to the strain variations that are available for guiding morphogenesis, then a reasonable value for c1 is the maximum fractional variation that arises in the strain fields created during an invasion. Thus, the value c1 = 0.3 has been selected in figure 4 (echoing the strain variations of 30% in figure 2b).
The value of η should be the ratio of the observed migration velocity, vf, to the observed host relaxation velocity, vr. For the innervation of the mouse gut shown in figure 9, vf ≈ 1 mm d−1 ≈ 1.2 × 10−2 µm s−1. The relaxation velocity, vr, has not been measured directly. However, a related cell motion has been quantified in studies of amelogenesis [28]. A new analysis of results from Cox [28] yields the following estimates for relaxation velocities (see electronic supplementary material, appendix F, for details). For ameloblasts in the human molar, vr=1.1 ± 0.6 × 10−3 µm s−1 and remains approximately constant during amelogenesis, while the migration velocity increases by over 50 per cent. For ameloblasts in the mouse incisor, vr = 2.4 ± 1.0 × 10−3 µm s−1.
Figure 4 and other results show that, with c1 = 0.3, qualitative agreement of nerve network predictions with observations for innervation of the mouse gut requires η ≈ 0.03–0.06. With vf = 1.2 × 10−2 µm s−1, this implies required values for the host relaxation velocity of 3.6–7.2 × 10−4 µm s−1. The upper end of this range is within a factor of 2 of the relaxation velocity for mouse ameloblasts, and within 50 per cent of that for human ameloblasts. In contrast, the migration velocity of human ameloblasts (3–6 × 10−5 µm s−1) and mouse ameloblasts (15 × 10−5 µm s−1) is two to three orders of magnitude slower than the migration velocity of neural cells during innervation of the mouse gut (1.2 × 10−2 µm s−1). The morphogenic outcomes of amelogenesis and innervation are of course very different. Nature is inferred to control morphogenesis partly by controlling migration velocities, while the velocity of cell relaxation associated with migratory strains appears to be approximately invariant.
A host relaxation velocity in the range 3.6–7.2 × 10−4 µm s−1 is approximately one order of magnitude less than the rate of propagation of the stimulus wave associated with the release of the signalling molecule SRC that is triggered in single cells by force stimulus [29]. One would expect the rate of propagation of chemical signal release to exceed the rate of propagation of cell density relaxations, which require cytoskeletal re-organization. On the other hand, the migration speed of nerve cells in innervation of the gut, vf = 1.2 × 10−2 µm s−1, is an order of magnitude higher than the velocity of the aforementioned stimulus wave.
5.2. The largest strain gradients do not break symmetry
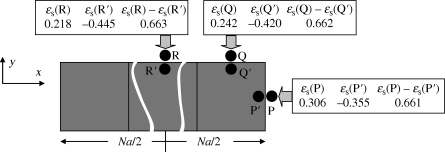
The strain-cue function used in this study has referred to the magnitude of strain components just beyond the boundary of the element through which the next migration of an invader might occur. One might alternatively consider a strain cue that refers to spatial gradients in the strain fields. The largest gradients occur across the boundary of an invaded element, where, if the element bears an eigenstrain while its neighbour does not, the elastic strain field is discontinuous. Two strain measures summarize trends in the strain gradient across the boundary. First, consider the elastic volumetric strain, ɛ = ɛx + ɛy + ɛz. For a line (branch) of N invaded elements each bearing eigenstrain 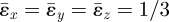 , the elastic volumetric strain inside the elements takes the value −0.381 at all locations; ɛ is spatially uniform. Furthermore, the volumetric strain is also uniform outside the line of elements, taking the value zero everywhere. (These results also hold for ellipsoidal inclusions [25].) Therefore, the jump in the volumetric strain in passing across the boundary of the line of elements is the same at all locations; symmetry is unbroken. Second, consider the strain measure ɛs = ɛ1 + ɛ2 at points just inside and just outside the line of invaded elements (figure 12). For the points P, Q and R ahead of the tip and to the sides of the line, ɛs takes different values. It also takes different values at the points P′, Q′ and R′ just inside the line of elements. However, the jump in ɛs is the same, whether crossing the boundary from P′ to P, or Q′ to Q or R′ to R; the jump does not break symmetry. (This result follows analytically from the compatibility of the tangential displacements across the boundary of an inclusion. The small deviances in figure 12 arise because the strains were evaluated at a distance δ = 0.005a from the boundary.)
, the elastic volumetric strain inside the elements takes the value −0.381 at all locations; ɛ is spatially uniform. Furthermore, the volumetric strain is also uniform outside the line of elements, taking the value zero everywhere. (These results also hold for ellipsoidal inclusions [25].) Therefore, the jump in the volumetric strain in passing across the boundary of the line of elements is the same at all locations; symmetry is unbroken. Second, consider the strain measure ɛs = ɛ1 + ɛ2 at points just inside and just outside the line of invaded elements (figure 12). For the points P, Q and R ahead of the tip and to the sides of the line, ɛs takes different values. It also takes different values at the points P′, Q′ and R′ just inside the line of elements. However, the jump in ɛs is the same, whether crossing the boundary from P′ to P, or Q′ to Q or R′ to R; the jump does not break symmetry. (This result follows analytically from the compatibility of the tangential displacements across the boundary of an inclusion. The small deviances in figure 12 arise because the strains were evaluated at a distance δ = 0.005a from the boundary.)
Figure 12.
The jump in the strain measure ɛs (sum of strain components tangential to the element surface) on crossing the invasion boundary is the same for an invader migrating from a line of elements to any of points P, Q and R. Numbers computed from equation (2.3) for a line of length N = 10.
Thus, gradients in neither the volume strain nor the strain measure ɛs break symmetry around the surface of the branch. A strain-cue rule based on strain gradients would generate uniform growth of the branch in all directions and it therefore could not provide a mechanism for branch stabilization or network formation. Consistently, the rates of proliferation and differentiation of individual cells in populations of fibroblasts have been found to correlate with the local strain magnitude, but not spatial gradients of strain [7].
5.3. Strain magnitudes and strain rates
In executing the current calculations, a doubling of density (eigenstrain 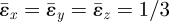 ) was assumed in an invaded computational element. If invader and host cells have equal stress-free volumes and the invader is spherical and centred within the computational element, this choice implies that the invader's boundaries will lie very close to the element boundaries at the points P and Q of figure 2 (at x/(a/2) = (3/π)1/3 ≈ 0.985 for an element centred at x = 0). Therefore, the magnitudes of the strain measure ɛs calculated just outside computational elements (e.g. at points P, Q and R of figure 2) will be approximately representative of the strains expected just outside an invader cell in a biological system. For a single invaded element, ɛs = ɛ0 = 0.267, corresponding to individual strain components ɛx = ɛy = ɛ0/2 ≈ 13 per cent. Because the strains outside an invaded element fall quite quickly with distance from the element (asymptotically as the cube of the distance), the average strain that could be sensed by a typical lamellipodium or filipodium is perhaps half of this. The variations in the strain cue, ɛs/ɛ0, of figure 2 therefore correspond to variations in individual strain components from roughly 4–5% at point Q (mid-branch) to 7–8% at point P (tip extremity). These strain amplitude variations coincide with the range over which substantial changes have been measured in the rates of proliferation and differentiation of fibroblasts [7].
) was assumed in an invaded computational element. If invader and host cells have equal stress-free volumes and the invader is spherical and centred within the computational element, this choice implies that the invader's boundaries will lie very close to the element boundaries at the points P and Q of figure 2 (at x/(a/2) = (3/π)1/3 ≈ 0.985 for an element centred at x = 0). Therefore, the magnitudes of the strain measure ɛs calculated just outside computational elements (e.g. at points P, Q and R of figure 2) will be approximately representative of the strains expected just outside an invader cell in a biological system. For a single invaded element, ɛs = ɛ0 = 0.267, corresponding to individual strain components ɛx = ɛy = ɛ0/2 ≈ 13 per cent. Because the strains outside an invaded element fall quite quickly with distance from the element (asymptotically as the cube of the distance), the average strain that could be sensed by a typical lamellipodium or filipodium is perhaps half of this. The variations in the strain cue, ɛs/ɛ0, of figure 2 therefore correspond to variations in individual strain components from roughly 4–5% at point Q (mid-branch) to 7–8% at point P (tip extremity). These strain amplitude variations coincide with the range over which substantial changes have been measured in the rates of proliferation and differentiation of fibroblasts [7].
Figure 9 shows that invasion of the gut by neural cells advances at a rate of approximately 1 mm d−1 or 10−2 µm s−1. Assuming a cell diameter of 10 µm and an approximately equal element width (corresponding to the choice of equal invader and host cell densities in an invaded element—see above), the invasion of an element will be accomplished in approximately 1000 s. The strain in the host medium (cells plus ECM) just beyond the element being invaded will therefore rise from zero to approximately 13 per cent in this time, implying a strain rate during loading of the host of 10−4 s−1 to order of magnitude. After the host medium has been strained by the approaching invasion, its relaxation occurs one to two orders of magnitude more slowly (because vr/vf = 0.03–0.06 for simulations that yield life-like networks), implying strain rates for unloading of 10−6–10−5 s−1.
Even the highest strain rates (during host loading) are four orders of magnitude lower than the strain rates chosen in most in vitro tests of cells. For example, the cyclic strain experiments of Kaunas et al. [4] were run at 1 Hz with strain amplitudes up to 10 per cent, so that, assuming sinusoidal strain cycling, 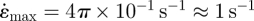 . The low strain rates occurring during natural organogenesis raise questions about whether the constitutive laws deduced for cells from relatively rapid in vitro experiments are accurate predictors of natural cell behaviour.
. The low strain rates occurring during natural organogenesis raise questions about whether the constitutive laws deduced for cells from relatively rapid in vitro experiments are accurate predictors of natural cell behaviour.
5.4. Distinction from other concepts of network formation
Existing models of branching structures and other shape instabilities that mark organogenesis have primarily exploited concepts that originated in the physical sciences. Viscous fingering (explained by Saffman & Taylor [30], with earlier recognition in the oil industry) arises when a less viscous fluid is injected into a more viscous fluid in the confines of a porous medium. Repeated finger formation can result in fractal tree-like structures. While the formative shape instability depends on the viscosity ratio and the local pressure at branch tips, the driving force is ultimately the far-field pressure, whose transmission and diminution along the existing structure are important to the morphological outcome. Diffusion-limited aggregation (first conceived to account for the aggregation of colloids [31]) generates shape instabilities in an expanding domain whose growth depends on the availability of some species that diffuse towards it from surrounding material. Enhanced particle collection by nascent protrusions favours their extension while shadowing the adjacent growth front, which therefore tends not to advance, resulting in fractal tree structures very similar to those caused by viscous fingering, in spite of the very different physics involved [32]. Many adaptations of these physical principles have been applied to organogenesis, addressing the great observed variety of shape instabilities in nature (e.g. [33]). Mechanics has been noted to influence morphology in different ways, e.g. via fluid dynamics in angiogenesis, which partly controls local branching conditions, or via surface tension arising from cell adhesion, which tends to maintain a smooth growth front in either a growing organ or a tumour (e.g. [34,35]).
Some essential distinctions separate the present strain-cue mechanism from this body of prior work. First, the strains that guide the migrating cell are generated locally and may be only weakly influenced by far-field conditions. No far-field pressure gradient is required to drive cells; they employ their own motility, using the strain cue only to determine their direction. An extreme illustration of this locality can be obtained by simulating the migration of a single cell guided by strain cues, with no proliferation permitted to add any other cells. Once moving, such an isolated cell will tend to follow its current migration direction, because the strain measure ɛs is largest ahead of it (newly strained host material) and lower behind it (host partially relaxed); its own motion breaks symmetry in the surrounding strain field. The rule of equation (2.3) leads to the cell having a probability for continuing straight ahead, rather than deviating, that increases with increasing η and decreasing c1. This result for an isolated cell, or group of cells moving together, may have implications, for example, in metastasis, guiding a cell that breaks from a tumour to continue outwards to invade the surrounding tissue.
Second, whereas both the viscous fingering mechanism and other organogenesis models refer to pressure differences [30] or gradients [35] as motive forces, the strain-cue rule cannot generate networks if the strain (or stress) referred to is the volumetric strain (or hydrostatic stress). These quantities do not break symmetry (see above).
Third, diffusion-based mechanisms for symmetry breaking tend to generate tree-like structures, in which branches do not intersect to form closed networks. When two branches approach, the diffusing quantity becomes depleted between them, discouraging contact. In contrast, the strain-cue mechanism readily permits or even encourages branch coalescence.
Fourth, while diffusion-based mechanisms readily generate shape instabilities with long spatial wavelength [35], the strain-cue mechanism readily generates long narrow branches. If the strain-cue mechanism were to act in conjunction with diffusion-based mechanisms and long-range chemical or pressure gradients, a wide variety of morphological outcomes could be possible.
5.5. Cellular automata models of network formation
The theoretical strategy of this paper also resembles cellular automata models of cell behaviour. However, the present work is distinct from most cellular automata models in that the occupancy of elements is treated by continuous rather than discrete variables; and stress and strain fields are calculated exactly for a continuous elastica.
Furthermore, unlike the present theory, cellular automata models have invoked special rules to induce branches to extend at fixed width and trigger and control sprouting. For example, in a mechano-biological model of capillary network formation, a ‘persistence ratio’ was arbitrarily introduced into the model definition, to bias for capillary extension in the direction already established [36]. In the same work, an arbitrarily chosen probability for branching was specified to control sprouting frequency. In a theory of angiogenesis and vascular remodelling, a rule was imposed that defined an exclusion radius around the base of each sprout or incipient branch, inhibiting further spreading of the cells into that space [37]. This rule has the effect of ensuring that branches and sprouts extend without thickening, an essential characteristic for achieving a network. The physical origin of such a rule is suggested to be in the actions of Delta-Notch signalling, but it is unclear why such signalling should operate more effectively at the base of a sprout than elsewhere around a branch. In the same work, only the tip of a branch is allowed to extend, a modelling choice that automatically ensures branch stability. Furthermore, a time is chosen for a sprout to connect to some other branch in the network, before it is made to die and disappear. The choice of the time is arbitrary.
5.6. Qualitative effects of finite-rate replenishment
As discussed more fully in the electronic supplementary material, appendix E, if invader re-supply is limited by finite-rate invader migration and proliferation in the wake of branches, the rate of sprouting will tend to increase. Furthermore, detachment of invaders from existing branches becomes possible, potentially leading to detached segments of network similar to the islands of network seen ahead of the main invasion front during innervation of the mouse gut (figure 9).
5.7. Networks in three dimensions
If the restriction that migration can occur only on a plane is relaxed, the rules of equations (2.3) and (B 1), without modification, will generate three-dimensional networks. The strain concentrations around either branch tips or branch elbows are independent of permutations of the global directions (x, y, z). Therefore, both the sprouting rate and the probability of branch arrest per unit length of advance of a branch will be the same in two- and three-dimensional networks for given model parameters. The rate of branch coalescence will drop, implying a tendency towards open tree rather than closed network structures.
5.8. Capillary formation
The theory as formulated in this paper simulates the stress and strain fields that are caused by the intercalation of an invader cell into a domain already occupied by host cells. The stress fields arise because of variations in cell density. In a blood vessel, stress fields are generated largely by the hydrostatic pressure of the blood. This is a different loading configuration and therefore the stress fields are not the same. However, further calculations for the eigenstrain problem of figure 2 show that the differences are relatively small. The stress state inside a cuboid bearing a uniform eigenstrain (the problem solved above) can be separated into hydrostatic and deviatoric contributions. For the uniform eigenstrain assumed in this paper, the hydrostatic stress developed is uniform throughout the cuboid. The deviatoric stresses are small over most of the interior region of the cuboid and only depart significantly from zero in a relatively thin layer near the surface of the cuboid. Therefore, over most of the cuboid, the stress state is close to that induced by a pressurized non-viscous fluid. Therefore, the stress and strain fields predicted outside a branch of invaded elements in the present simulations approximate those that would be found around a blood vessel bearing a pressurized fluid.
In particular, the strain measure ɛs will break symmetry around a pressurized blood vessel in much the same way that it does around a line of invaded elements. Therefore, the same strain-cue rule, applied to the proliferation and migration of cells that are required to extend the blood vessel, will stabilize branch and network formation.
6. Concluding remarks
The mechanics theory is built on the single physical assumption that an invading cell is cued by the strain fields, external to itself, that its own presence creates in the host medium that it is invading. The strain fields break symmetry, favouring the extension of a nascent branch over recession of the branch into the invasion front. As well as stabilizing branch formation, the strain-cue rule causes spontaneous sprouting, especially where the strain is enhanced at any bend in a branch.
A strain-cue rule based on the spatial gradient in the strain field or the jump in hydrostatic pressure (or volumetric strain) across the growth front does not break symmetry and therefore could not generate networks.
Other network characteristics accounted for include the presence of clusters of invader cells, terminated branches and a tendency for backfilling where empty domains have been left behind the main invasion front. A simple elaboration would account for island networks that form ahead of the main invasion front.
The probability of branch arrest and therefore the density of the branch structure in the network are governed by the ratio of two rates, namely the migration velocity of the invaders and the relaxation velocity of the host cells. A similar rate ratio has been identified as a governing factor for the microstructure of dental enamel during amelogenesis [28]. These two pieces of theoretical work imply that factors that influence the rate of motion of cells are critical to successful organogenesis.
The relaxation velocities inferred for ameloblasts during mouse and human amelogenesis are within a factor of 2 and 50 per cent, respectively, of that needed to account for nerve network characteristics in the mouse gut, while the migration velocities differ by two to three orders of magnitude (amelogenesis being very slow). Migration velocities are inferred to be much more widely variable in nature than relaxation rates.
The predicted strains referenced by the strain-cue rule have magnitudes recently associated with enhanced differentiation and proliferation in in vitro tests of fibroblasts.
The strain rate for host cells induced by the invasion of a neural cell is approximately two orders of magnitude greater than the strain rate for the same cells as they subsequently relax: 10−4 versus 10−6 s−1. Both these rates are much lower than the strain rates used in common in vitro tests.
The key strain variations and the host relaxation mechanisms involve the interactions of 100 cells to order of magnitude; the influence on strain fields of cells beyond the nearest 100 cells in a branch falls rapidly with their distance. Thus, the network-forming mechanism has the robust characteristic of not depending on the maintenance of long-range order in the population. The host cell population provides sufficient mechanical context if it is recruited fast enough to maintain a layer that is a few cells thick ahead of each advancing branch tip in the network.
Network formation requires that the strain stimulus mechanism be switched off at some point. If strain stimulus continues to operate indefinitely around a newly formed network branch, the branch disappears quite quickly owing to the formation of an ever-increasing density of newly formed branches emerging from it. The switching off is effected by relaxation owing to host cell motions, which removes the signalling strains.
With appropriate minor modifications to the details of stress and strain fields, the strain-cue rule might apply to both nervous and vascular networks. The symmetry-breaking property of the strain fields around an incipient branch might also be involved in the stabilization of invaginations developing from a previously smooth mesenchymal–epithelial bilayer; in shape instabilities in developing tumours; and in metastasis.
Acknowledgements
The author is indebted to Drs T. Bromage, L. Iruela-Arispe, R. Lacruz, K. A. Landman, D. Newgreen, C. E. Smith and M. L. Snead, and an anonymous reviewer, for many instructive discussions.
References
- 1.Discher D. E., Janmey P., Wang Y. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 10.1126/science.1116995 (doi:10.1126/science.1116995) [DOI] [PubMed] [Google Scholar]
- 2.Folkman J., Moscona A. 1978. Role of cell shape in growth control. Nature 273, 345–349 10.1038/273345a0 (doi:10.1038/273345a0) [DOI] [PubMed] [Google Scholar]
- 3.Hamant O., et al. 2008. Developmental patterning by mechanical signals in Arabidopsis. Science 322, 1650–1655 10.1126/science.1165594 (doi:10.1126/science.1165594) [DOI] [PubMed] [Google Scholar]
- 4.Kaunas R., Nguyen P., Usami S., Chien S. 2005. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc. Natl Acad. Sci. USA 102, 15 895–15 900 10.1073/pnas.0506041102 (doi:10.1073/pnas.0506041102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller R., Davidson L. A., Shook D. R. 2003. How we are shaped: the biomechanics of gastrulation. Differentiation 71, 171–205 10.1046/j.1432-0436.2003.710301.x (doi:10.1046/j.1432-0436.2003.710301.x) [DOI] [PubMed] [Google Scholar]
- 6.Kolega J. 1986. Effects of mechanical tension on protrusive activity and microfilament and intermediate filament organization in an epidermal epithelium moving in culture. J. Cell Biol. 102, 1400–1411 10.1083/jcb.102.4.1400 (doi:10.1083/jcb.102.4.1400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B., Li F., Puskar K. M., Wang J. H.-C. 2009. Spatial patterning of cell proliferation and differentiation depends on mechanical stress magnitude. J. Biomech. 42, 1622–1627 10.1016/j.jbiomech.2009.04.033 (doi:10.1016/j.jbiomech.2009.04.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo C.-M., Wang H.-B., Dembo M., Wang Y.-L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144–152 10.1016/S0006-3495(00)76279-5 (doi:10.1016/S0006-3495(00)76279-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson C. M., Jean R. P., Tan J. L., Liu W. F., Sniadecki N. J., Spector A. A., Chen C. S. 2005. Emergent patterns of growth controlled by multicellular form and mechanics. Proc. Natl Acad. Sci. USA 102, 11 594–11 599 10.1073/pnas.0502575102 (doi:10.1073/pnas.0502575102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oster G. F., Murray J. D., Harris A. K. 1983. Mechanical aspects of mesenchymal morphogenesis. J. Embryol. Exp. Morphol. 78, 83–125 [PubMed] [Google Scholar]
- 11.Parker K. K., et al. 2002. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB J. 16, 1195–1204 10.1096/fj.02-0038com (doi:10.1096/fj.02-0038com) [DOI] [PubMed] [Google Scholar]
- 12.Pelham R. J., Jr, Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94, 13 661–13 665 10.1073/pnas.94.25.13661 (doi:10.1073/pnas.94.25.13661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rot-Nikcevic I., Reddy T., Downing K. J., Belliveau A. C., Hallgrimson B., Hall B. K., Kahlar B. 2006. Myf5-/-:MyoD-/- amyogenic fetuses reveal the importance of early contraction and static loading by striated muscle in mouse skeletogenesis. Dev. Genes Evol. 216, 1–9 10.1007/s00427-005-0024-9 (doi:10.1007/s00427-005-0024-9) [DOI] [PubMed] [Google Scholar]
- 14.Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., Chen C. S. 2003. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl Acad. Sci. USA 100, 1484–1489 10.1073/pnas.0235407100 (doi:10.1073/pnas.0235407100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeung T., et al. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton 60, 24–34 10.1002/cm.20041 (doi:10.1002/cm.20041) [DOI] [PubMed] [Google Scholar]
- 16.Zeichen J., van Griensven M., Bosch U. 2000. The proliferative response of isolated human tendon fibroblasts to cyclic biaxial mechanical strain. Am. J. Sports Med. 28, 888–892 [DOI] [PubMed] [Google Scholar]
- 17.Trepat X., Deng L., An S. S., Navajas D., Tschumperlin D. J., Gerthoffer W. T., Butler J. P., Fredberg J. J. 2007. Universal physical responses to stretch in the living cell. Nature 447, 592–595 10.1038/nature05824 (doi:10.1038/nature05824) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binder B. J., Landman K. A., Simpson M. J., Mariani M., Newgreen D. F. 2008. Modeling proliferative tissue growth: a general approach and an avian case study. Phys. Rev. E 78, 031912. 10.1103/PhysRevE.78.031912 (doi:10.1103/PhysRevE.78.031912) [DOI] [PubMed] [Google Scholar]
- 19.Landman K. A., Simpson M. J., Newgreen D. F. 2007. Mathematical and experimental insights into the development of the enteric nervous system and Hirschsprung's disease. Dev. Growth Differ. 49, 277–286 [DOI] [PubMed] [Google Scholar]
- 20.Simpson M. J., Landman K. A., Hughes B. D., Newgreen D. F. 2006. Looking inside an invasion wave of cells using continuum models: proliferation is the key. J. Theor. Biol. 243, 343–360 10.1016/j.jtbi.2006.06.021 (doi:10.1016/j.jtbi.2006.06.021) [DOI] [PubMed] [Google Scholar]
- 21.Simpson M. J., Mariani M., Zhang D., Landman K. A., Newgreen D. F. 2007. Cell proliferation drives neural crest cell invasion of the intestine. Dev. Biol. 302, 553–568 10.1016/j.ydbio.2006.10.017 (doi:10.1016/j.ydbio.2006.10.017) [DOI] [PubMed] [Google Scholar]
- 22.Simpson M. J., Merrifield A., Landman K. A., Hughes B. D. 2007. Simulating invasion with cellular automata: connecting individual and macroscopic properties. Phys. Rev. E 76, 021 918. 10.1103/PhysRevE.76.021918 (doi:10.1103/PhysRevE.76.021918) [DOI] [PubMed] [Google Scholar]
- 23.Trewenack A. J., Landman K. A. 2009. A travelling wave model for invasion by precursor and differentiated cells. Bull. Math. Biol. 71, 291–317 doi:10.1007/s11538-008-9362-x) [DOI] [PubMed] [Google Scholar]
- 24.Ingber D. E. 2006. Mechanical control of tissue morphogenesis during embryological development. Int. J. Dev. Biol. 50, 255–266 10.1387/ijdb.052044di (doi:10.1387/ijdb.052044di) [DOI] [PubMed] [Google Scholar]
- 25.Eshelby J. D. 1957. The determination of the elastic field of an ellipsoidal inclusion, and related problems. Proc. R. Soc. Lond. A 241, 376–396 10.1098/rspa.1957.0133 (doi:10.1098/rspa.1957.0133) [DOI] [Google Scholar]
- 26.Mura T. 1982. Micromechanics of defects in solids. The Hague, The Netherlands: Martinus Nijhoff [Google Scholar]
- 27.Young H. M., Bergmer A. J., Anderson R. B., Enomoto H., Milbrandt J., Newgreen D. F., Whitington P. M. 2004. Dynamics of neural crest-derived cell migration in the embryonic mouse gut. Dev. Biol. 270, 455–473 10.1016/j.ydbio.2004.03.015 (doi:10.1016/j.ydbio.2004.03.015) [DOI] [PubMed] [Google Scholar]
- 28.Cox B. N. 2010. A multi-scale, discrete-cell simulation of organogenesis: application to the effects of strain stimulus on collective cell behavior during ameloblast migration. J. Theor. Biol. 262, 58–72 10.1016/j.jtbi.2009.09.010 (doi:10.1016/j.jtbi.2009.09.010) [DOI] [PubMed] [Google Scholar]
- 29.Pathak A., Deshpande V. S., McMeeking R. M., Evans A. G. 2008. The simulation of stress fibre and focal adhesion development in cells on patterned substrates. J. R. Soc. Interface 5, 507–524 10.1098/rsif.2007.1182 (doi:10.1098/rsif.2007.1182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saffman P. G., Taylor G. I. 1958. The penetration of a fluid into a porous medium or Hele-Shaw cell containing a more viscous liquid. Proc. R. Soc. Lond. A 245, 312–329 10.1098/rspa.1958.0085 (doi:10.1098/rspa.1958.0085) [DOI] [Google Scholar]
- 31.Witten T. A., Jr, Sander L. M. 1981. Diffusion-limited aggregation, a kinetic critical phenomenon. Phys. Rev. Lett. 47, 1400–1403 10.1103/PhysRevLett.47.1400 (doi:10.1103/PhysRevLett.47.1400) [DOI] [Google Scholar]
- 32.Halsey T. C. 2000. Diffusion-limited aggregation: a model for pattern formation. Phys. Today 53, 36–41 10.1063/1.1333284 (doi:10.1063/1.1333284) [DOI] [Google Scholar]
- 33.Fleury V., Gouyet J.-F., Leonetti M. (eds) 2001. Branching in nature. Berlin, Germany: Springer-Verlag [Google Scholar]
- 34.Cristini V., Lowengrub J. 2010. Mutliscale modeling of cancer. Cambridge, UK: Cambridge University Press [Google Scholar]
- 35.Cristini V., Lowengrub J., Nie Q. 2003. Nonlinear simulation of tumor growth. J. Math. Biol. 46, 191–224 10.1007/s00285-002-0174-6 (doi:10.1007/s00285-002-0174-6) [DOI] [PubMed] [Google Scholar]
- 36.Checa S., Prendergast P. J. 2008. Capillary network formation during tissue differentiation. A mechano-biological model. In Proc. 4th European Conf. of the International Federation for Medical and Biological Engineering, pp. 2195–2199 Berlin, Germany: Springer-Verlag [Google Scholar]
- 37.Owen M. R., Alarcon T., Maini P. K., Byrne H. M. 2009. Angiogenesis and vascular remodelling in normal and cancerous tissue. J. Math. Biol. 58, 689–721 10.1007/s00285-008-0213-z (doi:10.1007/s00285-008-0213-z) [DOI] [PubMed] [Google Scholar]
- 38.Druckenbrod N. R., Epstein M. L. 2005. The pattern of neural crest advance in the cecum and colon. Dev. Biol. 287, 125–133 10.1016/j.ydbio.2005.08.040 (doi:10.1016/j.ydbio.2005.08.040) [DOI] [PubMed] [Google Scholar]