Abstract
Anthropogenic alteration of biotic distributions and disturbance regimes has dramatically changed the evolutionary context for the differentiation of species traits. Some of the most striking examples in recent centuries have been on islands where flightless birds, which evolved in the absence of mammalian carnivores, have been decimated following the widespread introduction of exotic predators. Until now, no equivalent case has been reported for plants. Here, we make use of robust analytical tools and an exceptionally well-sampled molecular phylogeny to show that a majority of New Zealand danthonioid grasses (Poaceae) may have adapted to the relaxed vertebrate herbivore pressure during the late Cenozoic through the development of a distinctive and unusual habit: abscission of old leaves. This feature occurs in only about 3 per cent of the world's roughly 11 000 grass species and has been empirically shown to increase plant productivity but to reduce protection against mammal grazing. This result suggests that release from a selective pressure can lead to species radiations. This seemingly anachronistic adaptation may represent an overlooked factor contributing to the severe decline in the geographical extent and species diversity of New Zealand's indigenous grasslands following the introduction of herbivorous terrestrial mammals in the 19th century.
Keywords: evolution, plant–animal interactions, species radiations
1. Introduction
Plant species show many adaptations caused by specific interactions with past and present herbivorous animals, including leaf toughness and spines. However, distinguishing current and historical pressures for different plant traits is hampered by the Late Pleistocene and Holocene extinctions of herbivores associated with human colonization of continents and islands [1–5]. A possible solution to this is to use natural evolutionary experiments by comparing plants in regions with different herbivore histories [6].
Diversification and expansion of grasslands during the late Cenozoic has been linked to the evolution of large herbivorous mammals [4]. Various common features of grasses (family Poaceae), such as phytoliths and rhizomatous growth form, may decrease vulnerability to grazers. New Zealand constitutes an ideal system for investigating the evolution of herbivores and grasses because the animals that lived in the archipelago prior to human colonization around 750 years ago are well documented. During prehistoric times, no terrestrial mammals existed—except for a mouse-sized species that went extinct [7]—and birds were the dominant herbivores, including moas, diverse waterfowl and rails [8]. Mammals and birds feed differently on grasses. Mammals remove and ingest entire leaves by manipulating forage with their lips and tongue, and cutting material with their teeth; birds graze tussock grasses by pulling and cutting foliage, and several (notably rails) use clamping and tugging to remove tillers and access basal meristematic tissue [9]. The historical absence of mammals could thus have resulted in different grass adaptations in New Zealand when compared with the mammal-dominated savannahs of Africa and pampas of South America.
One possible candidate for such an adaptation is the ability to shed dead leaves. Accumulation of dead leaves in grasses is known to reduce light availability and CO2 uptake, convert immediately usable inorganic nitrogen in rainwater to less readily available organic nitrogen in microbial biomass, inhibit nitrogen fixation, decrease soil temperatures and reduce root productivity [10]. Despite the deleterious effects of retaining dead leaves, only about 3 per cent of the world's approximately 11 000 grass species are able to abscise old leaves ([11]; table 1). Leaf abscission typically occurs at a fracture zone at the base of the leaf blade (figure 1). Experimental evidence has led to a recent possible explanation of this paradox. Leaf abscission (mimicked by manual removal of dead leaves) increases biomass production, but it also makes grasses more palatable [12]. Under pressure by grazing mammals, leaf loss to herbivory becomes so severe that it outweighs the benefits of increased biomass production [12].
Table 1.
Number of grass species capable of abscising leaves in different regions; numbers within brackets denote species in subfamily Danthonioideae [11,17].
| Africa | Asia | Australia | Europe | New Zealand | North America | South America | worldwide | |
|---|---|---|---|---|---|---|---|---|
| persistent leavesa | 2537 (120) | 4002 (5) | 1297 (38) | 911 (2) | 385 (24) | 1760 (7) | 3113 (37) | 10 771 (234) |
| abscising leavesa | 45 (8) | 133 (2) | 32 (8) | 19 (0) | 38 (27) | 36 (1) | 111 (5) | 343 (56) |
| total | 2579 (127) | 4134 (5) | 1327 (45) | 930 (2) | 414 (49) | 1790 (8) | 3207 (41) | 11 089 (282) |
aPolymorphic and widespread species were tabulated in each relevant category. Species counts include endemic, indigenous and (except for the Danthonioideae) naturalized species.
Figure 1.
Leaf abscission in grasses. (a) Herbarium specimen of the New Zealand grass Chionochloa rigida, a leaf-abscising species. (b) Detail of fracture zone (inset in (a)) where old leaves are shed.
New Zealand grasslands are dominated by long-lived tussock species, a major component of which are the snow grasses (Chionochloa) and allied short tussock species (Rytidosperma). Both genera belong to the grass subfamily Danthonioideae (‘pampas grasses’ and allies), a clade of around 280 species of temperate grasses distributed on all continents except Antarctica ([13]; figure 2), comprising both leaf-abscising species and species in which old leaves are retained. If New Zealand grasses have experienced different selection pressures to their African and American relatives, we would expect these differences to have left a detectable morphological signature in the extant species. We explored this hypothesis by analysing the occurrence of leaf abscission in danthonioid grasses present in New Zealand compared with those occurring elsewhere.
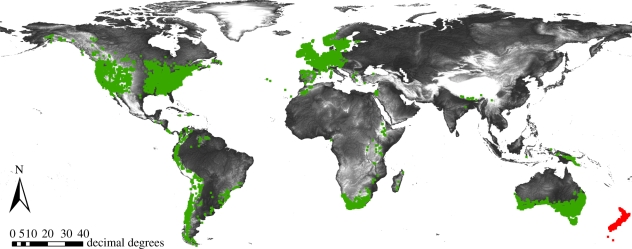
Figure 2.
Distribution of grass subfamily Danthonioideae. GIS-based map based on 22 025 occurrences from the Global Biodiversity Information Facility (GBIF; www.gbif.org) and 19 372 from various sources. Occurrences coded as New Zealand are marked in red, all others in green.
2. Material and methods
(a). Phylogenetic and dating analyses
Recent studies have investigated relationships [14,15] and hybridization [16] within the grass subfamily Danthonioideae, leading to a new taxonomic revision of its genera [17]. To take into account phylogenetic and branch-length uncertainty in our analyses, we generated here a set of optimal trees by combining two recently published datasets [14,15]. The final matrix comprised 299 accessions (representing approx. 81% of the 280 described species), including eight genera outside the Danthonioideae (Amphipogon, Andropogon, Aristida, Arundo, Hordeum, Micraira, Setaria and Stipagrostis), and contained 14 425 aligned nucleotide positions from eight plastid and two nuclear sequence regions: trnL-trnF, rpl16, rbcL, ndhF, matK, atpB-rbcL, trnT-trnL, trnC-trnD, ITS and 26S rDNA.
We inferred phylogenetic relationships in MrBayes v. 3.12 [18,19] by performing 10 topologically unconstrained runs of 1.2 × 106 generations each, under the GTR + Γ + I model as selected by the Akaike Information Criterion in ModelTest v. 3.7 [20], sampling every 500th generation, with one cold and three heated chains. All runs started from the tree with maximum-likelihood (ML) score obtained during 50 independent ML analyses in GARLI v. 0.96b [21]. We estimated absolute divergence times in BEAST v.1.5.2 [22] by calibrating the root of the Danthonioideae with a normally distributed prior (mean 26.1 Ma, s.d. 0.5) following the results of a large analysis of Poaceae based on fossil constraints [23], assuming a Yule model of speciation under an uncorrelated lognormal clock, and following other standard settings. We performed two independent runs of 107 generations each, sampling every 5000th generation. We assessed convergence of runs and effective sample sizes for all Markov chain Monte Carlo (MCMC) parameters in Tracer v. 1.5 [22] and AWTY [24]. An initial burn-in of 500 trees was excluded from each independent MrBayes run, leaving 11 406 trees for calculating a 50 per cent majority-rule consensus. For the BEAST runs, 1500 trees were excluded, leaving 8232 time-calibrated trees (see also [25]).
To minimize sampling biases, where multiple DNA accessions were available for a species and the species was shown to be monophyletic, we subsequently pruned all but one of the sequences from the MrBayes and BEAST post-burn-in tree samples while keeping the remaining branch lengths unaltered. In a few cases where plastid and nuclear DNA partitions for the same species appeared in conflict during pilot runs, we duplicated these taxa in the matrix such that each duplicate was represented by one partition only, following the approach and rationale described by Pirie et al. [16].
(b). Ancestral state optimization
We used a carefully verified DELTA database [17] to code for species distributions (absent in New Zealand = 0; present in New Zealand = 1) and leaf type (leaves persistent = 0, leaves abscising = 1). We used both Fitch parsimony and ML (under two models: the Markov k-state 1 parameter and the asymmetrical Markov k-state 2 parameters) implemented in Mesquite v. 2.72 [26] to reconstruct ancestral states for all nodes in a sample of 5000 Bayesian trees, randomly selected from all independent runs (burn-in excluded). For each node of the 50 per cent majority-rule consensus tree from the Bayesian analysis, we computed the relative frequency of uniquely best states across the tree sample. These results were used to identify clades where shifts to abscission most probably occurred and compute the number of species descending from these shifts.
(c). Directionality and conservativeness of shifts
We tested whether shifts to leaf abscission occurred more times than shifts into the other direction by comparing the distribution of shifts into each direction across our sample of optimal trees. We tested whether shifts in leaf type were phylogenetically conservative by calculating the distribution of observed shifts with the number of shifts across a sample of trees in which terminals had been randomly shuffled (10 × 1000 randomly selected Bayesian trees). To evaluate whether any pattern of conservativeness depended on presence in New Zealand, we performed this test for trees containing all sampled accessions of Danthonioideae and trees in which New Zealand terminals had been pruned while retaining branch lengths of unpruned lineages. These analyses were performed in Mesquite [26].
(d). Correlation analysis
To test whether leaf retention and leaf abscission have evolved depending on geographical distribution, we compared the fit of models of dependent and independent evolution to our data. Correlation analyses were carried out using the Discrete (ML) and BayesDiscrete (MCMC) commands [27,28] in BayesTraits (available from www.evolution.rdg.ac.uk/BayesTraits.html). Eight rate parameters constitute the dependent model, which assumes that each character evolves (forward and backward shifts) at different rates depending on the state of the second character. In the independent model, forward and backward shifts in one character occur at the same rate regardless of the state of the second parameter (coefficients q12 = q34, q13 = q24, q21 = q43 and q31 = q42; electronic supplementary material, table S1). Hence, a model of independent evolution has four parameters. Definitions of rate coefficients and the results for the correlation analyses are provided in electronic supplementary material, table S1. Since the number of reversals inferred seemed unrealistically large, we did not use these values for testing directionality of shifts. Inflated rate coefficients are to be expected when shifts are inferred to have occurred along short branches [29], therefore we only used information on reversals from the parsimony analysis, as this test ignores differences in branch lengths.
Fit of dependent and independent models using ML was compared using a likelihood ratio test over a sample of 5000 randomly selected Bayesian trees (burn-in excluded) and with 10 likelihood iterations per tree. Fit of dependent and independent models in a Bayesian framework were compared using Bayes factors, calculated as twice the difference in log-harmonic mean of the worst-fitting model and the better-fitting model [30]. Multiple long Markov runs were performed to ensure that the harmonic mean remained stable within and among runs. We used the reversible jump MCMC method [27,31], which allows sampling of the various possible models of evolution in proportion to their posterior probabilities [27], as opposed to only the rate parameters being sampled in this way, as in conventional MCMC [29]. We used an exponentially distributed hyperprior [29] with its mean value seeded from a uniform distribution in the interval specified, to ensure that posterior values were contained within, but not determined by, the prior range. We varied the amount by which the rate parameters are allowed to change between iterations of the Markov chain, by varying the ‘ratedev’ value, so that acceptance rates averaged 20 to 40 per cent. This should avoid autocorrelation while ensuring adequate exploration of parameter space. To improve initially low acceptance rates, we used a modified version of the code that accepts either a move to a new model or a move to a different tree (courtesy A. Meade). We ran 1.5 × 108 generations, sampling every 1000 generations, yielding a sample of 145 000 iterations after 5 × 106 iterations were removed as burn-in. In addition, we ran a separate series of analyses restricted to sampling only independent models.
In total, five sampled species are polymorphic for leaf type, and three for distribution. All analyses above were repeated coding polymorphic species with (0,1) followed by coding only for presence (1), with the rationale of retaining information about all those species able to abscise their leaves and being native to New Zealand even if they show polymorphism in one or both traits (except ML optimizations in Mesquite, unable to handle polymorphisms). All statistical tests yielded the same level of significance in both cases.
3. Results and discussion
The 50 per cent majority-rule consensus tree of the Bayesian analysis in MrBayes, together with posterior probabilities of all resolved clades, is provided in electronic supplementary material, figure S1. Figure 3 shows a simplified version of that tree, with duplicated species pruned and indicating the presence of leaf abscission and distribution in New Zealand. Relationships obtained are congruent with previous estimates of the danthonioid phylogeny [14,15]. The time-calibrated tree of the subfamily, estimated in BEAST and showing 95 per cent highest posterior densities of node ages, is provided in electronic supplementary material, figure S2.
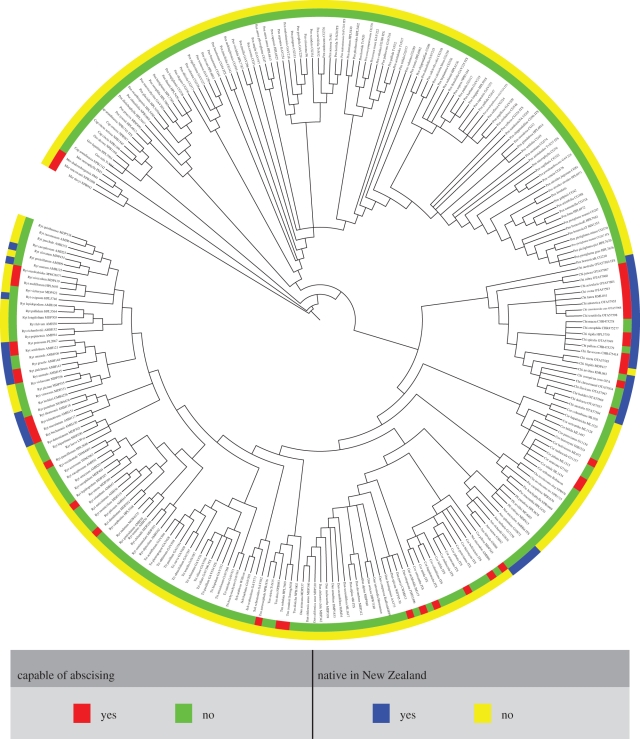
Figure 3.
Phylogeny of Danthonioideae. Bayesian consensus cladogram showing leaf type and distribution for 270 accessions (representing approx. 81% of all described species; only presence is coded). Relationships were derived from the analysis of 14 425 aligned nucleotide positions from eight plastid and two nuclear DNA sequence regions. See electronic supplementary material, figure S1 for the fully annotated MrBayes consensus tree reporting posterior probabilities for all clades.
Results from the parsimony and ML optimizations for leaf abscission and distribution, coding only for presence, produced nearly identical results with respect to ancestral states (the main difference being a higher level of ambiguity for the reconstruction of early diverging nodes under the 2-rate ML model, when compared with parsimony and the 1-rate ML model). Given these similarities, and since ML in Mesquite cannot handle polymorphic taxa and thus requires an arbitrary simplification of reality, we report here only the parsimony results for leaf type and distribution (electronic supplementary material, figures S3 and S4, respectively). The optimizations over 5000 trees resulted in a mean of 29 shifts in leaf abscission and 12 shifts in distribution.
Leaf abscission occurs in the majority of danthonioid species native to New Zealand, a proportion that is markedly higher than that found in other regions (table 1; Fisher's exact test, one-tailed p < 0.001). This striking imbalance could be the result of differences in the rates of shifts between leaf abscission and leaf retention, differences in the retention of one state over the other, differences in speciation rates between lineages with abscising and persistent leaves, or a combination of two or more of these factors.
(a). Evolution and retention of leaf abscission
Shifts in leaf type (persistent ↔ abscising) are strongly correlated with geographical distribution (present in New Zealand ↔ absent in New Zealand; figures 3 and 4a,b; average likelihood ratio 16.1, average log Bayes factor 14.6; in both cases 4 d.f.). The rate of transition to leaf abscission is higher in New Zealand than elsewhere (Wilcoxon rank-sum test based on Bayesian posterior rate coefficients, n = 1.8 × 108, one-tailed p < 0.001). On all continents, shifts from persistent to abscising leaves have been more common than shifts in the opposite direction (electronic supplementary material, figure S5; parsimony ancestral state reconstruction, t-test for independent samples, n = 5000, one-tailed p < 0.001). However, the ultimate consequences of evolving abscission have been very different in New Zealand when compared with other regions: lineages that evolved this ability outside New Zealand have rarely diversified, producing at most six leaf-abscising species in genus Merxmuellera in the southern African Drakensberg—a region that, interestingly, and in accordance with the patterns obtained for New Zealand, historically has only sustained a very low density of small-sized mammal herbivores [32,33]. Within New Zealand, on the other hand, lineages that evolved leaf abscission have become significantly more species-rich, as indicated by a Wilcoxon rank-sum test (n1 = 16, n2 = 3, one-tailed p = 0.001; electronic supplementary material, figure S3 shows which clades were included in the test). The same test remains significant even after correcting for incomplete species sampling in Merxmuellera (four out of seven species sampled) and the potential effect of time in species accumulation. Time was taken into account by calculating speciation rates λ under a pure birth (Yule) model for each leaf-abscising clade, based on the number of descendent species S and the median stem age t obtained in the BEAST analysis, such that λ = ln[S]/t ([34]; p = 0.004 in both cases). Retention of leaf abscission over more diversification events in New Zealand is also evident by the fact that in all mammal-dominated regions, the phylogenetic distribution of leaf-abscising species is not distinguishable from a random distribution, whereas in New Zealand leaf-abscising species show a phylogenetically conserved pattern (electronic supplementary material, figures S6 and S7; t-test for independent samples, n = 5000, one-tailed p < 0.001).
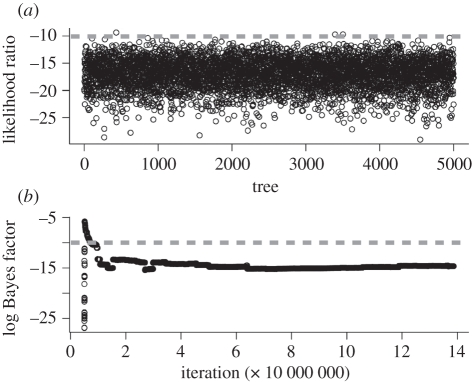
Figure 4.
Results from the phylogeny-based analyses of correlation between distribution and leaf type. (a) Likelihood ratio between dependent and independent models, based on ML analyses implemented in BayesTraits [27]; (b) Bayes factor (calculated as twice the difference of the log-harmonic mean between dependent and independent analyses) plotted against iteration in the reversible-jump MCMC analysis in BayesTraits [27]. The analysis stabilizes after an initial burn-in phase. The dashed lines indicate significance levels, under which there is ‘very strong evidence’ for correlation [30].
These results show that the higher proportion of leaf-abscising danthonioid grass species native to New Zealand compared with species absent from New Zealand is owing to two factors: a higher frequency of evolution of leaf abscission in New Zealand lineages and greater net diversification rates of these lineages.
(b). A ‘ghost’ adaptation?
There has been a long debate on the factors shaping the evolution of New Zealand plants. New Zealand enjoys a rich endemic flora with many peculiar adaptations, such as the ‘divaricate’ life form that characterizes roughly 20 per cent of the endemic woody species [9]. Juveniles of these species produce densely intertangled branches no taller than 2–3 m, forming cage-like barriers to predation. In adult individuals, branches are acute-angled, often with larger leaves, and reach above 3 m in height. Although the divaricate habit has long been interpreted as a response to climatic conditions [35], recent studies have provided overwhelming evidence that it instead served as an adaptation against browsing by the now-extinct moas [9,36]. However, it is unlikely that leaf-abscission evolved as a response to evolutionary pressures exerted by browsing birds. From detailed analyses of fossilized dung and gizzards, moas appear to have eaten tussock grasses only rarely [36]. Similarly, there is no indication that the extant native grassland rail (Porphyrio hochstetteri) would have influenced the evolution of leaf-abscising grasses, since it has been documented to access leaf bases of certain tussock species irrespective of dehiscent features [37]. The absence of mammals in New Zealand therefore appears to have been the major factor conferring a strong evolutionary advantage to leaf-abscising grasses in New Zealand in the past. The results presented here support this hypothesis, and even if they do not provide evidence against other possible causative factors (e.g. soil nutrient differences) they do provide insight into a possibly important factor in the shaping of the New Zealand grass flora.
(c). A current disadvantage?
If leaf abscission in New Zealand grasses evolved as a response to the absence of grazing mammals, native grasses would be expected to be poorly adapted to the introduction of sheep, cattle and rabbits during European settlement after 1850. There is strong evidence that endemic grasses are inherently susceptible to mammalian grazers [38–40], but grassland deterioration at large scales is complex and usually involves multiple causes, including burning, stocking rates, feral animals and invasive weeds [41]. To date, no studies have experimentally tested whether adult leaf-abscising grasses native to New Zealand are more susceptible to grazing than grasses with persistent leaves, because leaf abscission has never been considered a relevant parameter in ecological studies. There is, however, evidence that dominance (m2 ha−1) of a leaf-abscising species (Chionochloa flavescens) increased significantly during the 21 years that followed the removal of mammal grazers in north-facing plots of New Zealand's South Island [39]. A comparable significant increase in dominance was not observed in the same plots for Chionochloa macra, a closely related species with persistent leaves. Although further studies are clearly needed to test this prediction, these differences reinforce the idea that leaf abscission confers higher productivity, and thus a competitive advantage in the absence of mammal grazers [12]—an advantage that may have been turned into a disadvantage in many areas of New Zealand today.
Acknowledgements
We thank J. Wood, C. Carbutt and T. O'Connor for discussion, R. Wüest for help with GIS, and the Computational Biology Service Unit hosted by Cornell University, USA (http://cbsuapps.tc.cornell.edu), where the MrBayes and BEAST analyses were performed. We are very thankful for the constructive comments by the associated editor Susanne Renner and two anonymous reviewers. This project was financed by a grant from the Swiss Science Foundation to H.P.L.
References
- 1.Temple S. A. 1979. The dodo and the tambalacoque tree. Science 203, 1364. 10.1126/science.424762 (doi:10.1126/science.424762) [DOI] [PubMed] [Google Scholar]
- 2.Guimarães P. R., Jr, Galetti M., Jordano P. 2008. Seed dispersal anachronisms: rethinking the fruits extinct megafauna ate. PLoS ONE 3, (doi:10.1371/journal.pone.0001745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janzen D. H., Martin P. S. 1982. Neotropical anachronisms: the fruits the gomphotheres ate. Science 215, 19–27 10.1126/science.215.4528.19 (doi:10.1126/science.215.4528.19) [DOI] [PubMed] [Google Scholar]
- 4.Johnson C. N. 2009. Ecological consequences of Late Quaternary extinctions of megafauna. Proc. R. Soc. B 276, 2509–2519 10.1098/rspb.2008.1921 (doi:10.1098/rspb.2008.1921) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bond W. J., Silander J. A. 2007. Springs and wire plants: anachronistic defences against Madagascar's extinct elephant birds. Proc. R. Soc. B 274, 1985–1992 10.1098/rspb.2007.0414 (doi:10.1098/rspb.2007.0414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mack R. N., Thompson J. N. 1982. Evolution in steppe with few large hooved mammals. Am. Nat. 119, 757–773 [Google Scholar]
- 7.Worthy T. H., Tennyson A. J. D., Archer M., Musser A. M., Hand S. J., Jones C., Douglas B. J., McNamara J. A., Beck R. M. D. 2006. Miocene mammal reveals a Mesozoic ghost lineage on insular New Zealand, southwest Pacific. Proc. Natl Acad. Sci. USA 103, 19 419–19 423 10.1073/pnas.0605684103 (doi:10.1073/pnas.0605684103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee W. G., Wood J. R., Rogers G. M. 2010. Legacy of avian-dominated plant–herbivore systems in New Zealand. N. Z. J. Ecol. 34, 28–47 [Google Scholar]
- 9.Bond W. J., Lee W. G., Craine J. M. 2004. Plant structural defences against browsing birds: a legacy of New Zealand's extinct moas. Oikos 104, 500–508 10.1111/j.0030-1299.2004.12720.x (doi:10.1111/j.0030-1299.2004.12720.x) [DOI] [Google Scholar]
- 10.Knapp A., Seastedt T. 1986. Detritus accumulation limits productivity of tallgrass prairie. Bioscience 36, 662–668 10.2307/1310387 (doi:10.2307/1310387) [DOI] [Google Scholar]
- 11.Clayton W. D., Harman K. T., Williamson H. 2006. GrassBase—The Online World Grass Flora. http://www.kew.org/data/grasses-db.html [accessed December 2009]. [Google Scholar]
- 12.Mingo A., Oesterheld M. 2009. Retention of dead leaves by grasses as a defense against herbivores. A test on the palatable grass Paspalum dilatatum. Oikos 118, 753–757 10.1111/j.1600-0706.2008.17293.x (doi:10.1111/j.1600-0706.2008.17293.x) [DOI] [Google Scholar]
- 13.Linder H. P., Barker N. P. 2000. Biogeography of the Danthonieae. In Grass systematics and evolution. Vol. 2 of Proc. Second Int. Conf. on the Comparative Biology of the Monocots, Sydney, September 1998 (eds Jacobs S. W. L., Everett J.). Melbourne, Australia: CSIRO [Google Scholar]
- 14.Pirie M. D., et al. 2008. A novel supermatrix approach improves resolution of phylogenetic relationships in a comprehensive sample of danthonioid grasses. Mol. Phylogenet. Evol. 48, 1106–1119 10.1016/j.ympev.2008.05.030 (doi:10.1016/j.ympev.2008.05.030) [DOI] [PubMed] [Google Scholar]
- 15.Humphreys A. M., Pirie M. D., Linder H. P. 2010. A plastid tree can bring order to the chaotic generic taxonomy of Rytidosperma Steud. s.l. (Poaceae). Mol. Phylogenet. Evol. 55, 911–928 10.1016/j.ympev.2009.12.010 (doi:10.1016/j.ympev.2009.12.010) [DOI] [PubMed] [Google Scholar]
- 16.Pirie M. D., Humphreys A. M., Barker N. P., Linder H. P. 2009. Reticulation, data combination, and inferring evolutionary history: an example from Danthonioideae (Poaceae). Syst. Biol. 58, 612–628 10.1093/sysbio/syp068 (doi:10.1093/sysbio/syp068) [DOI] [PubMed] [Google Scholar]
- 17.Linder H. P., et al. In press A classification of the Danthonioideae (Poaceae). Ann. Missouri Botanic Garden. [Google Scholar]
- 18.Huelsenbeck J. P., Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 10.1093/bioinformatics/17.8.754 (doi:10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 19.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 20.Posada D., Crandall K. A. 1998. ModelTest: testing the model of DNA substitution. Bioinformatics 14, 817–818 10.1093/bioinformatics/14.9.817 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 21.Zwickl D. J. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD thesis, University of Texas, Austin, TX: Software available at www.bio.utexas.edu/faculty/antisense/garli/Garli.html [Google Scholar]
- 22.Drummond A. J., Ho S. Y., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christin P. A., Besnard G., Samaritani E., Duvall M. R., Hodkinson T. R., Savolainen V., Salamin N. 2008. Oligocene CO2 decline promoted C4 photosynthesis in grasses. Curr. Biol. 18, 37–43 10.1016/j.cub.2007.11.058 (doi:10.1016/j.cub.2007.11.058) [DOI] [PubMed] [Google Scholar]
- 24.Nylander J. A. A., Wilgenbusch J. C., Warren D. L., Swofford D. L. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583 10.1093/bioinformatics/btm388 (doi:10.1093/bioinformatics/btm388) [DOI] [PubMed] [Google Scholar]
- 25.Humphreys A. M., Antonelli A., Pirie M. D., Linder H. P. Submitted Ecology and evolution of the diaspore ‘burial syndrome’. [DOI] [PubMed] [Google Scholar]
- 26.Maddison W. P., Maddison D. R. 2009. Mesquite: a modular system for evolutionary analysis. v. 2.71. http://mesquiteproject.org
- 27.Pagel M., Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 167, 808–825 [DOI] [PubMed] [Google Scholar]
- 28.Pagel M. 1994. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc. R. Soc. Lond. B 255, 37–45 10.1098/rspb.1994.0006 (doi:10.1098/rspb.1994.0006) [DOI] [Google Scholar]
- 29.Pagel M., Meade A., Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 10.1080/10635150490522232 (doi:10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- 30.Raftery A. E. 1996. Hypothesis testing and model selection. In Markov chain Monte Carlo in practice (eds Gilks W. R., Richardson S., Spiegelhalter D. J.). London, UK: Chapman & Hall [Google Scholar]
- 31.Green P. J. 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82, 711–732 10.1093/biomet/82.4.711 (doi:10.1093/biomet/82.4.711) [DOI] [Google Scholar]
- 32.O'Connor T. G. 2005. Influence of land use on plant community composition and diversity in Highland Sourveld grassland in the southern Drakensberg, South Africa. J. Appl. Ecol. 42, 975–988 10.1111/j.1365-2664.2005.01065.x (doi:10.1111/j.1365-2664.2005.01065.x) [DOI] [Google Scholar]
- 33.Rowe-Rowe D., Scotcher J. 1986. Ecological carrying capacity of the Natal Drakensberg for wild ungulates. S. Afr. J. Wildl. Res. 16, 12–16 [Google Scholar]
- 34.Nee S., Mooers A. O., Harvey P. H. 1992. Tempo and mode of evolution revealed from molecular phylogenies. Proc. Natl Acad. Sci. USA 89, 8322–8326 10.1073/pnas.89.17.8322 (doi:10.1073/pnas.89.17.8322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howell C. J., Kelly D., Turnbull M. H. 2002. Moa ghosts exorcised? New Zealand's divaricate shrubs avoid photoinhibition. Funct. Ecol. 16, 232–240 10.1046/j.1365-2435.2002.00613.x (doi:10.1046/j.1365-2435.2002.00613.x) [DOI] [Google Scholar]
- 36.Wood J. R., Rawlence N. J., Rogers G. M., Austin J. J., Worthy T. H., Cooper A. 2008. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quart. Sci. Rev. 27, 2593–2602 10.1016/j.quascirev.2008.09.019 (doi:10.1016/j.quascirev.2008.09.019) [DOI] [Google Scholar]
- 37.Mills J. A., Lavers R. B., Lee W. G., Mara M. K. 1991. Food selection by Takahe Notornis mantelli in relation to chemical composition. Ornis Scand. 22, 111–128 10.2307/3676542 (doi:10.2307/3676542) [DOI] [Google Scholar]
- 38.Lee W. G., Fenner M., Loughnan A., Lloyd K. M. 2000. Long-term effects of defoliation: incomplete recovery of a New Zealand alpine tussock grass, Chionochloa pallens, after 20 years. J. Appl. Ecol. 37, 348–355 10.1046/j.1365-2664.2000.00498.x (doi:10.1046/j.1365-2664.2000.00498.x) [DOI] [Google Scholar]
- 39.Rose A. B., Platt K. H. 1992. Snow tussock (Chionochloa) population responses to removal of sheep and European hares, Canterbury, New Zealand. NZ J. Bot. 30, 373–382 [Google Scholar]
- 40.Mark A. F., McLennan B. 2005. The conservation status of New Zealand's indigenous grasslands. NZ J. Bot. 43, 245–270 10.1080/0028825X.2005.9512953 (doi:10.1080/0028825X.2005.9512953) [DOI] [Google Scholar]
- 41.Mark A. F. 1994. Effects of burning and grazing on sustainable utilisation of upland snow tussock (Chionochloa spp.) rangelands for pastoralism in South Island, New Zealand. Aust. J. Bot. 42, 149–161 10.1071/BT9940149 (doi:10.1071/BT9940149) [DOI] [Google Scholar]