Abstract
Inclusive fitness theory predicts that natural selection will favour altruist genes that are more accurate in targeting altruism only to copies of themselves. In this paper, we provide evidence from digital evolution in support of this prediction by competing multiple altruist-targeting mechanisms that vary in their accuracy in determining whether a potential target for altruism carries a copy of the altruist gene. We compete altruism-targeting mechanisms based on (i) kinship (kin targeting), (ii) genetic similarity at a level greater than that expected of kin (similarity targeting), and (iii) perfect knowledge of the presence of an altruist gene (green beard targeting). Natural selection always favoured the most accurate targeting mechanism available. Our investigations also revealed that evolution did not increase the altruism level when all green beard altruists used the same phenotypic marker. The green beard altruism levels stably increased only when mutations that changed the altruism level also changed the marker (e.g. beard colour), such that beard colour reliably indicated the altruism level. For kin- and similarity-targeting mechanisms, we found that evolution was able to stably adjust altruism levels. Our results confirm that natural selection favours altruist genes that are increasingly accurate in targeting altruism to only their copies. Our work also emphasizes that the concept of targeting accuracy must include both the presence of an altruist gene and the level of altruism it produces.
Keywords: kin selection, inclusive fitness, altruism, green beard, digital evolution, Avida
1. Background
Inclusive fitness theory, also known as kin selection theory, describes when a trait will be favoured by natural selection [1]. Applied to altruistic traits, inclusive fitness theory explains that an altruist gene is selected for if it is altruistic (assists another at a cost to itself) towards relatives when the cost of altruism is less than its benefit diluted by the chance that the beneficiary does not have the altruist gene [1]. In its more general form, inclusive fitness theory holds that any gene that directs a net benefit towards other copies of itself will be favoured by selection, even if the altruistic and beneficiary genes do not share common descent [1–7]. Altruist genes can, with varying degrees of reliability, identify carriers of the altruism gene in nature in three ways: (i) by recognizing kin, who are likely to share the altruist gene, (ii) in viscous populations, where surrounding organisms are often related, and (iii) by directly sensing the presence of the altruist gene [8].
Putting the point anthropomorphically, in order to determine whether to target altruism towards an organism (i.e. select it as the beneficiary of altruism), an altruist gene would like to have perfect genetic information as to whether a copy of itself exists in that organism. Given imperfect information, however, altruist genes are forced to settle on less accurate targeting mechanisms. Accuracy in this context is the probability that the recipient of altruism has a copy of the altruistic gene. Targeting altruism based on identifying kinship, which we call kin targeting, is the most commonly used indicator of the presence or absence of an altruist gene [9,10]. Altruism via kin recognition can be evolutionarily stable, although only in certain situations ([11–13]; reviewed in [14]). However, if more accurate indicators of the presence of altruistic genes were available, inclusive fitness theory predicts that natural selection should use them in addition to, or in lieu of, kinship indicators [1,4,5,7,15,16]. This pressure to be as accurate as possible is strongest when donations are costly or otherwise limited, which is the case we focus on in this paper.
Aside from kinship, one possible mechanism for determining whether an organism has a copy of an altruistic gene is to sense the genetic similarity between the potential donor and the recipient, which we will call similarity targeting. One may envision mechanisms based on sensing biochemical signals that would allow the inference of genetic similarity irrespective of kinship. For instance, mice, fishes and humans use scent to preferentially choose mates with genetically dissimilar major histocompatibility complexes, suggesting that genetic information can be acquired and exploited by evolution [17]. Additionally, social amoebas are more likely to form cooperative relationships with genetically similar organisms [18]. If it were possible for organisms to target altruism based on high genetic similarity (greater than expected for kin), inclusive fitness theory predicts that this mechanism would be selected for over kin targeting because it is more accurate.
It is also possible to have altruism-targeting mechanisms that would be even more accurate than those based on high genetic similarity. The most accurate targeting mechanism would be a gene that can identify with certainty whether other organisms possess (and express) its copy, irrespective of kinship or overall genetic similarity. This is the idea behind green beard genes, which were proposed by Hamilton [1], named by Dawkins [4] and subsequently found in nature [19–27]. Green beard genes do two things: (i) display a marker (e.g. a green beard) and (ii) target altruism towards entities bearing that marker and no others. Green beard genes are the ideal implementation of inclusive fitness theory—they direct altruism only towards organisms that contain and express their copies. We refer to this strategy as green beard targeting.
A prediction, then, of inclusive fitness theory is that if more accurate altruism-targeting methods become available, selection will favour their use over less accurate targeting methods. Specifically, inclusive fitness theory predicts that organisms will use kin targeting if it is the only type of altruism targeting available. If genes for both kin targeting and similarity targeting exist in a population, and if the similarity targeting is more accurate, then similarity targeting should have a selective advantage over kin targeting. Finally, if the genes for kin targeting, similarity targeting and green beard targeting all exist in a population, and are mutually exclusive, selection will favour green beard targeting because it is the most accurate. We confirm all of these predictions in this paper. Interestingly, however, we had to implement a novel instantiation of green beard targeting before natural selection favoured it over similarity targeting. Natural selection did not favour green beard targeting over similarity targeting in most situations because green beard targeting by itself cannot evolve the amount of altruism conferred from the altruist to the recipient. Except in rare, contrived situations, natural selection switched away from similarity targeting only when different beard colours existed that each reliably indicated different levels of altruism (a mechanism we call identical beard colour targeting).
To our knowledge, a test of these predictions has not been conducted either in computer models or in natural systems. While it would be ideal to confirm these predictions in natural systems, such tests would be difficult, if not impossible, to perform. As such, the closest we may come to empirically testing these predictions of inclusive fitness theory is in digital evolution systems.
We conduct such tests in Avida, a digital evolution software platform that instantiates evolutionary processes in a computer [28,29]. Avida has repeatedly served as a tractable system to investigate the general properties of evolving systems [30–37].
2. Methods
In Avida, self-replicating computer programs (i.e. digital organisms) evolve through random mutations and selective pressures. To self-replicate, an organism must copy its own genome, which is a sequence of computer instructions. The copy process is imperfect, however, such that a genomic instruction has a probability of mutating to another instruction when copied. These mutations can alter the execution of the genome and change its behaviour. When an organism self-replicates, a copy of it is placed at random either in one of its parents' cells or in a neighbouring cell of a parent (replacing the resident organism if extant). There are a limited number of cells, creating a competition for space. In the experiments described here, digital organisms obtain extra metabolic units through altruistic donations (explained subsequently), allowing them to execute their genomes more rapidly, which increases their ability to compete for space and thus their fitness.
As Dennett [38] has noted, evolution will occur in any system that has heritable variation and differential fitness. Avida exhibits these traits and can therefore be used to study the general principles of evolving systems [33]. The remainder of §2 describes how Avida was configured for the experiments described in this paper. A general, more thorough description of Avida can be found in [28]. The Avida software is free and can be obtained from http://devolab.cse.msu.edu/software/avida.
Each experiment featured 50 trials that differed only in the seed for the random number generator, which affected stochastic aspects of the trial, such as mutations. Trials began by filling 3600 cells of a virtual toroidal grid with identical copies of an organism that could self-replicate, but exhibited no other behaviours. Each cell within the toroidal grid was adjacent to eight neighbouring cells. When an organism successfully executed a divide command, it reproduced with another organism that had successfully divided. Specifically, offspring resulted from the sexual recombination of two copied genomes using two-point crossover, where both points were randomly chosen from each organism's circular genome and the sections of the genome between each point were swapped [36]. During replication, each instruction in the genome had a 0.75 per cent chance of mutating to any other instruction in the instruction set. Lower mutation rates of 10−3, 10−4 and 10−5 (following Rousset & Roze [13]) did not qualitatively change our results, except evolution was slower and similarity targeting was less competitive versus kin targeting because organisms were more similar, making similarity less informative. The standard Avida instruction set [28] includes instructions that allow organisms to manipulate numbers, copy their instructions, and modify execution flow (e.g. jump to, or skip over, instructions in their genomes). All genomes were fixed at a length of 100 instructions. Organisms died of ‘old age’ and were removed from the population if they executed 2000 instructions prior to completing replication. Organisms started their lives with 100 metabolic units.
For these experiments, we extended Avida to include altruistic instructions that allowed organisms to donate their metabolic units to neighbouring organisms. Organisms lost five metabolic units per donation made and gained 50 per donation received. This asymmetry created the possibility of non-zero sum gains, which are necessary for the evolution of altruism. Altering the ratio or magnitude of cost and benefit did not qualitatively change the results of the experiments, provided the benefit was at least approximately three times the cost. For all experiments, the number of donations of any type that an organism could perform was capped at 100 (by ignoring subsequent attempts), which makes it easier to determine which donation strategies are the most effective in any given setup. If an organism executed a donation instruction but there was not a suitable recipient in the surrounding eight cells, the organism was credited with performing the donation, but no energy transfer or deduction took place.
3. Results
To test the prediction that natural selection would favour the most accurate altruism-targeting mechanism available, we ran a series of experiments with different instructions that enable organisms to altruistically donate metabolic units using different targeting mechanisms.
(a). Kin targeting
We initially wanted to confirm that kin targeting would evolve in Avida. For this first experiment, we added three instructions to the standard Avida instruction set [33]. When an organism executed donate-kin, it made a donation to a neighbouring parent or offspring. We excluded full siblings for simplicity (they are extremely rare because mates are chosen randomly from the population, meaning that it is unlikely that two organisms will have the same two parents). The other two additional instructions were controls: executing donate-random donated to a random neighbour, and executing neutral had no altruistic effect. The latter provided a baseline of how often instructions with no selective advantage or disadvantage were executed. These three instructions were not present in the genome of the starting organism. Because individual organisms can execute multiple donation instructions, we estimate the evolutionary success of a donation instruction by looking at how many times it is executed by the organisms in the final population of each experiment, which is analogous to the expression level for a gene. Expression levels significantly above those for neutral are evidence that an instruction has been selected for. We also report on the frequency of the various donation instructions (alleles) in the populations at the end of evolutionary trials. Those alleles that are significantly more frequent in final populations than the neutral control have probably been selected for.
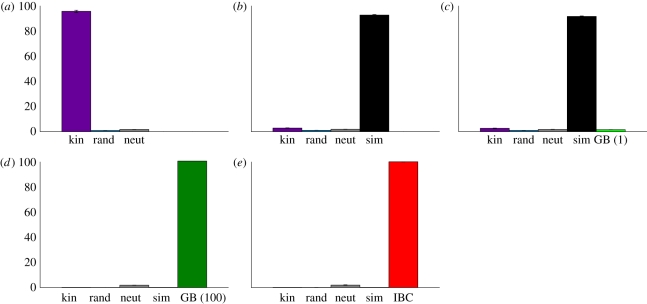
The results show that organisms that donated to their kin were selected for (figure 1a, p < 0.001 comparing the average number of donate-kin executions versus donate-random and neutral in final populations; all p-values in this paper are generated using Matlab's Mann–Whitney test). The average number of donations per organism is nearly the maximum number of donations allowed (100). The average number of donate-kin instructions per genome in the final populations was 7.95 (±0.58 s.d.), which was significantly higher than both the donate-random (2.13 ± 0.22 s.d.) and neutral (2.72 ± 0.25 s.d.) controls (p < 0.001). These results confirm that kin targeting is selected for in Avida, as predicted by inclusive fitness theory. Changing which relatives were considered kin (e.g. including cousins) did not qualitatively change the results (data not shown). By repeating this experiment without the donate-kin instruction, we also found that indiscriminate altruism is selected against in our experimental setup (donate-random expression is significantly lower than neutral, p < 0.001), although this might not have been expected because the population is viscous [39]. The increased accuracy of kin targeting over indiscriminate altruism thus enables the elevated levels of altruism observed with donate-kin.
Figure 1.
Evolved altruism levels for different targeting mechanisms. Plotted is the average number of donation instructions per type executed by organisms in the final populations of 50 trials (±1 s.e., often too small to distinguish). The maximum number of donations was capped at 100. (a) Targeting altruism based on kinship was selected for over two controls. (b) Targeting altruism based on high genetic similarity was favoured over targeting based on kinship. (c) Selection did not favour targeting altruism via a green beard mechanism (with an implicit threshold of 1, see text) over kin and similarity targeting. (d) Selection favoured a green beard mechanism with a threshold of 100 (the maximum number of donations allowed) over kin and similarity targeting. (e) Selection favoured identical beard colour targeting over kin and similarity targeting. Purple, kin; blue, random (rand); grey, neutral (neut); black, similarity (85%); light green, green beard (GB) (1); dark green, green beard (100); red, identical beard colour (IBC).
(b). Similarity targeting
We next investigated whether selection favours altruism-targeting mechanisms that are more accurate than those based on kinship. To test this prediction, we added a donate-similar instruction to the instruction set. When executed, the donate-similar instruction donated to a neighbour that had an edit distance of fewer than 15 (i.e. 85% genetically similar), where edit distance is the number of point, insert and/or delete mutations needed to transform one genome into another [40]. We selected this edit distance of 15 so that donate-similar more accurately targets altruism than donate-kin (otherwise there would be no expected selection for the former over the latter). Specifically, at equilibrium in the previous experiment, 34.69 per cent of donations using donate-kin went to organisms that had an edit distance greater than 15 (±0.0033% s.d.). The average edit distance between the donor and the recipient for donate-kin was 14.44 (±0.12 s.d.), whereas it was 7.48 (±0.05 s.d.) for donate-similar, which is significantly different (p < 0.001).
In accordance with the prediction of inclusive fitness theory, selection did favour this greater accuracy in altruism targeting (figure 1b, p < 0.001 comparing the average number of donate-similar executions versus other donation types in final populations). On average, donate-similar was executed 90 times per organism, whereas donate-kin was executed fewer than 10. The genomic instruction frequencies are consistent with the expression levels: The average number of donate-similar instructions per genome in the final populations was 5.1 (±0.45 s.d.), which was significantly higher (p < 0.001) than donate-kin (3.16 ± 0.32 s.d.), donate-random (2.15 ± 0.26 s.d.) and neutral (2.80 ± 0.24 s.d.). Varying the similarity (edit distance) threshold did not qualitatively change the result as long as it remained below approximately 50, i.e. above about 50 per cent genetic similarity (data not shown).
(c). Green beard targeting
In the next experiment, we provided direct knowledge of the presence of expressed altruist genes by adding a donate-greenbeard instruction. When executed, it caused a donation to a neighbouring organism that (i) had donate-greenbeard in its genome and (ii) executed the donate-greenbeard instruction at least once. The latter condition was added to prevent an organism from having a green beard phenotypic marker (i.e. having donate-greenbeard in its genome), but not being altruistic because the instruction was included in a ‘junk’ section of the genome that was never executed. We determined whether an organism executed the donate-greenbeard instruction by testing each new organism in a separate test environment prior to placing it in the population.
At first pass, it seems that inclusive fitness theory predicts that natural selection should favour the use of donate-greenbeard over both donate-similar and donate-kin because donate-greenbeard is perfectly accurate: it donates only to other green beard donors. In contrast, donate-similar and donate-kin will sometimes donate to non-donors (a false-positive error), as well as fail to donate to others that share their altruism genes (a false-negative error). For example, an organism may donate to kin that did not receive the altruism gene, or an organism may fail to recognize and donate to a cousin that shares the altruism gene. To test the prediction that altruism using the green beard-targeting mechanism will be selected for over kin and similarity altruism targeting, we repeated the previous experiment with the addition of donate-greenbeard to the instruction set.
Contrary to our initial expectations, donate-greenbeard was not competitive with donate-similar (figure 1c). The donate-similar instruction was selected for over all other donation types (p < 0.001 comparing the average number of donate-similar executions versus other donation types in final populations). The genomic instruction frequency data also show that selection favoured similarity targeting over all of the alternatives, including green beard targeting: The average number of donate-similar instructions per genome in the final populations was 4.90 (±0.46 s.d.), which was significantly higher (p < 0.001) than donate-greenbeard (2.60 ± 0.25 s.d.), donate-kin (3.11 ± 0.33 s.d.), donate-random (2.03 ± 0.22 s.d.) and neutral (2.63 ± 0.30 s.d.).
Later in the paper, we demonstrate a variant of the green beard concept that does outcompete donate-similar, but it is first instructive to learn why donate-greenbeard was not selected for. A possible explanation for this result is the disincentive an organism has for performing more than the minimum number of green beard donations necessary to receive green beard donations, which was only 1 in this case. For instance, in a population where all organisms perform two green beard donations, an organism that donates only once would receive more than it donates and thus have a competitive advantage. Such an organism is a variant of a ‘falsebeard’ cheater because it has the altruism-signifying marker, but is not altruistic to the same degree [19]. We hypothesized that green beard organisms are under selective pressure to be as selfish as possible while being just altruistic enough to qualify to receive donations from other green beard donors.
We tested this hypothesis by creating a threshold, which is the number of green beard donations an organism needs to make in order to qualify to receive green beard donations. The original donate-greenbeard instruction has an implicit threshold of 1, but this requirement can be set to any value. If our hypothesis is correct, the amount of green beard altruism should rise as a function of the threshold (T), but should not rise far above T; values slightly above T are expected owing to a pressure for robustness under mutation–selection balance [32].
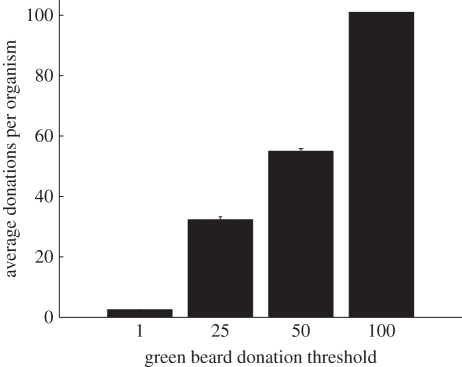
We tested this hypothesis by repeating the previous experiment, but using only the default instruction set and a new donate-threshold-gb instruction, which is identical to donate-greenbeard, but with a threshold that can be set to any integer. We tested four threshold values (T = 1, 25, 50, 100) and the hypothesis was confirmed: the level of altruism rose to the threshold, but did not substantially exceed it (figure 2). These results indicate that green beard targeting is unable to evolve persistent levels of altruism above whatever fixed and arbitrary threshold of altruism is required to qualify for green beard donations. The inability to evolve sustained altruism levels above level T provides a plausible explanation for why green beard targeting did not outcompete similarity targeting in the experiment plotted in figure 1c. In that experiment, the threshold of 1 resulted in a low level of green beard altruism, which prevented selection from taking advantage of the greater accuracy of the green beard-targeting mechanism in all but a few donation opportunities. Natural selection took advantage of the remaining non-zero sum opportunities with similarity targeting.
Figure 2.
Evolved altruism levels for different green beard thresholds. Plotted is the average number of donations executed per organism for different threshold values (T) of the donate-threshold-gb instruction (averaged from the final populations of 50 trials per treatment ±1 s.e., often too small to distinguish). Organisms evolved to perform enough donations to surpass the threshold and thus qualify to receive altruism, but did not perform substantially more than T donations.
These results suggest that in order for an altruist gene to accurately identify another gene as its copy, the level of altruism must also be taken into account. This is because two altruist genes that are altruistic to different degrees are not copies of one another. Previously in this paper, we have discussed altruism-targeting mechanisms solely based on a targeting method, such as targeting based on kinship, shared genetic similarity or the mutual possession of a green beard. In addition to targeting methods, however, targeting mechanisms can also have a discrimination level, which discriminates based on the level of altruism (electronic supplementary material, figure S1). Targeting methods and discrimination levels can work in conjunction to filter out the subset of organisms that will receive altruism. In our experiments, donate-kin and donate-similar had no discrimination level. Green beard targeting has a discrimination level equal to its threshold: the instruction donate-greenbeard had an implicit discrimination level of 1 and donate-threshold-gb had a discrimination level equal to its threshold T. The reason inclusive fitness theory, at first pass, seemed to predict that donate-greenbeard would be favoured over donate-kin and donate-similar is because the altruism level was not being considered when green beard genes were labelled as perfectly accurate. Recognizing the importance of altruism levels reveals that donate-greenbeard is not perfectly accurate because it frequently donates to organisms with more selfish versions of its altruist gene.
In our original green beard experiment (figure 1c), even though kin- and similarity-targeting mechanisms were less accurate, they were employed to take advantage of the remaining donation opportunities. We hypothesized that if the green beard threshold were set to the maximum number of donations allowed, then selection would indeed use the green beard-targeting mechanism instead of similarity targeting owing to its greater accuracy. To test this idea, we used a green beard threshold of 100, which is approximately the level of altruism generated by kin and similarity targeting when they are dominant (figure 1a,b). The experiment reported in figure 1c was repeated, but using donate-threshold-gb (T = 100) instead of donate-greenbeard.
Our prediction was confirmed: selection employed the green beard-targeting mechanism significantly more than kin and similarity targeting (figure 1d, p < 0.001 comparing the average number of donate-greenbeard executions versus other donation types in final populations, and comparing the average number of donate-threshold-gb instructions per genome in the final populations (5.61 ± 0.54 s.d.) versus donate-similar (1.46 ± 0.20 s.d.), donate-kin 1.47 (±0.19 s.d.), donate-random (1.40 ± 0.19 s.d.) and neutral (2.94 ± 0.30 s.d.)). Our results demonstrate that inclusive fitness theory is right that selection will favour the most accurate altruism-targeting mechanism available, but only if the altruism level is controlled for.
The green beard-targeting mechanism can evolve an altruism level near the maximum only if its discrimination level (threshold) is arbitrarily set to be near the maximum. It cannot evolve an altruism level near the maximum if its discrimination level happens to be much lower (figure 1c). By contrast, the altruism level for kin and similarity targeting automatically increased from zero to close to the maximum, exploiting nearly all of the non-zero-sum donation opportunities (figure 1a,b). These results reveal that kin and similarity targeting can evolve ever-higher altruism levels without explicit discrimination levels. One reason a discrimination level is unnecessary is that the organisms they cooperate with (i.e. kin or genetically similar organisms) have similar genomes and thus are likely to have similar altruism levels. Furthermore, organisms using kin and similarity targeting cooperate only with a small group of other organisms, limiting the success of ‘kin-cheaters’, which are organisms within a kin group that mutate to be less altruistic (figure 3a–c) [35,41]. Additionally, if a new kin group is created with a higher altruism level, it needs to survive via drift for only a few generations before some of its members are no longer close enough kin to donate to their less altruistic ancestors, and they can thus successfully evade exploitation (figure 3c,d). These attributes of kin and similarity targeting make possible the evolution of persistent increases in altruism. In other words, kin and similarity targeting do not have an explicit discrimination level, but instead possess an implicit discrimination level that occurs as a side effect of the genetic similarity inherent in these targeting methods.
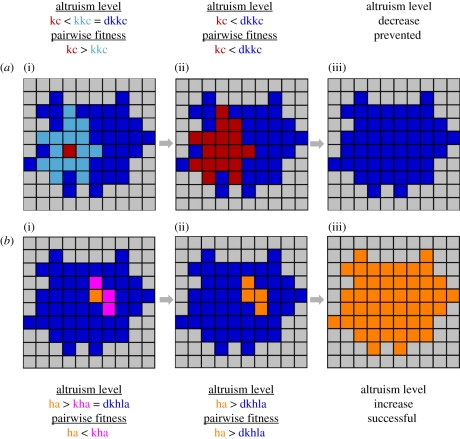
Figure 3.
How kin and similarity targeting can evolve persistently high altruism levels. A thought experiment illustration showing how (a) kin-based altruism naturally thwarts kin-cheaters (kc) and (b) enables enduring increases in altruism levels. (a(i)) Consider a group of related organisms that are altruistic to each other (blue and light blue). One organism may mutate to be less altruistic, becoming a kin-cheater (red), but since only its closest relatives (light blue) will consider it kin, only they will be altruistic towards it. (a(ii)) The kin-cheater will tend to supplant its kin because it receives more donations from them than it gives. (a(iii)) Once the kin-cheater has replaced those that considered it kin, the kin-cheater is left receiving donations only from other kin-cheaters. This group (red) will have a lower altruism level than their distant kin (blue) and will come to be replaced by them. (b(i)) Now consider an organism (orange) that mutates to have a higher level of altruism (ha) than its ancestors (blue). Initially, it will be selected against because it gives more donations to those that it considers kin (pink) than it receives from them. (b(ii)) If the less-altruistic kin of the higher level altruist are killed off by drift, then the higher level altruist and its offspring (orange) will have a competitive advantage over their distant ancestors (blue). (b(iii)) While chance is required to start the process, once it has occurred, there will be selection for the higher level of altruism. There are additional factors that complicate all of these fitness comparisons, but for clarity, we have sketched these scenarios only in broad strokes. kc, kin cheater; kkc, kin of kin cheater; dkkc, distant kin of kin cheater; ha, higher-level altruist; kha, kin of higher-level altruist; dkhla, distant kin of higher-level altruist.
The previous experiment demonstrates that if a green beard-targeting mechanism happens to have a discrimination level near the optimum (here, the maximum number of opportunities to exploit non-zero sum gains), then the green beard mechanism will be favoured by natural selection over kin and similarity targeting (figure 1d). However, it is unlikely that random mutations would produce a green beard gene with a near-optimal discrimination level de novo. This small probability is compounded by the already unlikely prospect of random mutations producing a gene that both creates a phenotypic marker and targets altruism towards bearers of the marker [4]. These improbable requirements diminish the expectations of finding such a gene in natural settings. Furthermore, if the optimal altruism level changed over time, the descendents of a bearer of a green beard gene that happened to be near the optimum would no longer be optimal, and would probably be replaced by a kin- or similarity-targeting mechanism. An unchangeable discrimination level, therefore, appears to be an evolutionary disadvantage.
(d). Identical beard colour targeting
If a green beard-targeting mechanism were able to automatically optimize its altruism level across generations, then such adaptability could make the green beard-targeting mechanism more competitive with kin- and similarity-targeting mechanisms. This type of adaptability is possible with a slight variation on the green beard idea wherein mutations to the altruism gene simultaneously change the level of altruism, the phenotypic marker (e.g. beard colour) and the level of discrimination. We call this targeting mechanism identical beard colour targeting, because organisms carrying the gene would be altruistic only towards others bearing the same beard colour. There would thus be many beard colours in a population, and the beard colour would perfectly indicate the altruism level of its bearer. This proposal is different from previous work with multiple beard colours in a population where the beard colour did not reliably indicate the altruism level [42]. We tested the efficacy of such an identical beard colour targeting gene by creating a donate-identical-beard-colour instruction. This instruction, when executed, donated to another organism that both: (i) had the donate-identical-beard-colour instruction and (ii) donated the same number of times as the donor. Thus, the phenotypic marker accurately signifies both the altruism and discrimination levels.
We repeated the previous experiment, but substituted the donate-identical-beard-colour instruction for the donate-greenbeard-threshold instruction, and found that identical beard colour targeting was selected for over kin and similarity targeting (figure 1e, p < 0.001 comparing the average number of donate-identical-beard-colour executions versus other donation types in final populations), which confirms our prediction. The genomic instruction frequency data also confirmed that selection favoured identical beard colour targeting over all of the alternative targeting mechanisms. The average number of donate-identical-beard-colour instructions per genome in the final populations was 3.61 (±1.60 s.d.), which was significantly higher (p < 0.001) than donate-similar (1.50 ± 0.22 s.d.), donate-kin (1.46 ± 0.21 s.d.) and donate-random (1.44 ± 0.21 s.d.). In this experiment, the difference in genomic instruction frequency between donate-identical-beard-colour and neutral (3.01 ± 0.21 s.d.) was not significant (p > 0.05), but the significant difference in the level of execution of those instructions (figure 1e, p < 0.001) clearly shows that selection favoured a much higher expression of donate-identical-beard-colour via regulatory instructions. We also performed this experiment with a population structure where offspring are placed at random in the population and found that the level of donate-identical-beard-colour expression was significantly higher than all other donation types (p < 0.001).
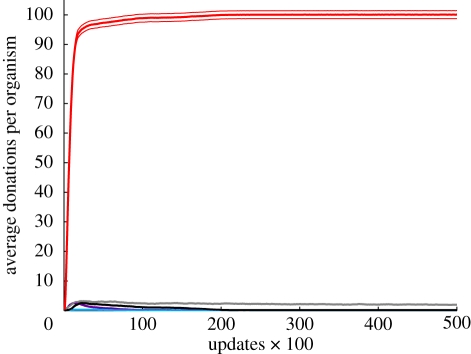
Interestingly, the average edit distance between the donor and the recipient for donate-identical-beard-colour was 52.78 (±0.15 s.d.), which was significantly greater than the edit distances for either donate-kin (14.44 ± 0.12 s.d.) or donate-similar (7.48 ± 0.05 s.d.), indicating that the identical beard colour targeting mechanism did indeed find altruism recipients that kin- and similarity-targeting mechanisms would not have identified. The average number of identical beard colour donations per organism in the final population was near the maximum amount allowed, as was the case in previous experiments for the kin-, similarity- and (threshold) green beard-targeting mechanisms when they were dominant. This high average means that most of the organisms in the population donated nearly 100 times, which was the maximum allowed. A look at altruism levels across evolutionary time reveals that this high level of altruism was evolutionarily stable in the sense that it was maintained for thousands of generations (figure 4). Plots of altruism levels across evolutionary time look qualitatively similar from the previous experiments when other targeting mechanisms were dominant (data not shown). These time plots reveal that the altruism via the targeting mechanisms discussed in this paper was maintained at high levels across thousands of generations.
Figure 4.
Identical beard colour altruism levels over evolutionary time. The data from the experiment plotted in figure 1e are shown here across evolutionary time. Plotted is the average number of donation instructions per type executed per organism for each update. The plot shows averages over 50 evolutionary trials (±1 s.e.). The altruism level of identical beard colour targeting rises early and remains at a high level for most of the experiment, which lasted thousands of generations. Purple, kin; light blue, random; grey, neutral; black, similarity (85%); red, identical beard colour.
Another alternate way altruism levels could be adjusted via a green beard mechanism is with multiple, fixed-threshold green beard genes with different markers (e.g. a blue hand, a red foot etc.). While each has a fixed threshold, the overall organismal level of altruism could be adjusted by having as many of these genes as necessary. We implemented this concept, and it worked qualitatively the same as identical beard colour targeting in the previous experiments, and evolution probabilistically chose one or the other when both options were available (electronic supplementary material). This alternate implementation further shows that being able to adjust altruism levels is a key fitness component of the green beard mechanism.
4. Discussion
It is commonly assumed that kin-based altruism involves poor, but adequate estimations of the presence of an altruist gene and that such altruism would be more effective if it were based on true knowledge of the presence of that gene [1,4,5,15]. In this paper, we provided organisms with such perfect knowledge via green beard targeting with a fixed and low discrimination level, but it was not selected for over altruism targeting based on ‘imperfect’ kin information. We discovered that this unexpected outcome is because kin targeting can naturally adjust its level of altruism, whereas green beard targeting using a single phenotypic marker (e.g. a single beard colour) cannot. This important feature of kin-based altruism may help explain its widespread occurrence in nature [9,10] versus the rare examples of green beard altruism [15,20–26]. It is interesting to note that the documented cases of green beard targeting in nature do involve binary decisions, for example, to kill or not [21] or to bind to or not [23,26].
Our results support that green beard targeting will be the method of choice only if the decision is binary, consisting of whether to be altruistic or not, or if the discrimination level of a green beard gene happens to be near the optimal altruism level. In such cases, green beard targeting is indeed more accurate than kin or similarity targeting. However, when adjusting the level of altruism is advantageous, selection will favour kin targeting (and would favour similarity targeting were it possible) over green beard targeting. Green beard targeting is not favoured by selection in such situations because a gene with a single beard colour cannot evolve its discrimination level threshold. However, as we demonstrated with the identical beard colour targeting mechanism, if mutations can simultaneously change the phenotypic marker, the altruism level and the discrimination level, then the amount of altruism can be continuously optimized by evolution. While kin and similarity targeting have an advantage over green beard targeting in adaptability, they do not have this advantage over identical beard colour targeting. It is not surprising, therefore, that identical beard colour targeting, which is more accurate and as adaptive as kin and similarity targeting, was selected for over these alternatives in our experiments.
Jansen & van Baalen [42] found that non-zero altruism levels could be maintained with multiple beard colours, but only when altruism levels were averaged across a population. In their model, beard colours and altruism traits were unlinked, allowing cheaters to easily appear and replace any subpopulation of altruists that temporarily emerged. Frequent altruist evolution and extinction meant that at any given time it was likely there were some altruist organisms in the population. Jansen & van Baalen conclude that altruism levels become increasingly unstable as the linkage increases between the phenotypic marker (in their case, beard colour) and altruism level. By contrast, such linkage is perfect in our identical beard colour mechanism, but we find that altruism levels quickly evolve to be maximally high and persist at that high level across evolutionary time (figure 4). We believe the phenomena Jansen & van Baalen describe are not due to green beard dynamics, but instead resemble the dynamics of kin selection, wherein altruism is maintained by the constant formation of groups that have not yet been invaded by cheaters (figure 3). In this situation, altruism can be maintained at a population level only if new cheater-free groups are constantly being created. Such kin-based altruism could not be maintained in a well-mixed population structure, yet such a population structure should not preclude high levels of green beard altruism (and did not in our experiments with the identical beard colour mechanism). The stability of altruism levels reported by Jansen & van Baalen is thus a property of a constantly changing subset of the population, but no altruist genotype can persist across evolutionary time because it can be invaded once a cheater emerges. In contrast, the identical beard colour mechanism can produce genotypes that exhibit high levels of altruism and resist invasion indefinitely (figure 4).
Although we have shown that identical beard colour targeting can maintain high levels of altruism, it is unlikely that natural, biological organisms use it as an altruism-targeting mechanism. We consider it improbable because identical beard colour targeting requires an even more unlikely phenomenon to be caused by a single gene than is the case for green beard genes. In addition to the requirements of green beard targeting, the gene must respond to mutations in such a way that new beard colours are created simultaneously with changes in the bearer's altruism level and discrimination level. That said, green beard genes were not thought to exist when they were invented as thought experiments [4]. Another possible green beard mechanism involves a collection of different green beard genes, each with its own marker, that independently contribute a fixed level of altruism. The costs, benefits and existence in nature of this strategy remain interesting open areas of research.
5. Conclusion
The main contribution of this paper is to provide empirical verification of the prediction of inclusive fitness theory that natural selection will favour altruism-targeting strategies that are increasingly accurate in targeting copies of the altruist gene. Additionally, our experimental results underscore that the altruism level of a gene is an important aspect of an altruist gene. It has not been common in the literature to discuss how altruistic a green beard gene is, but this level of altruism is a key attribute when green beard-targeting mechanisms compete with kin- and similarity-targeting mechanisms. A further issue highlighted in the paper is the importance of the adaptability, or evolvability, of the altruism levels of different targeting mechanisms. The evolvability of altruism-targeting mechanisms plays a significant role in determining which ones will succeed in evolving populations. Finally, in this paper, we demonstrate that a variant on the green beard-targeting concept, identical beard colour targeting, provides both perfect accuracy and adaptability. When identical beard colour targeting is available, natural selection favours it over its less accurate rivals. However, the unlikelihood of an identical beard colour targeting mechanism arising by chance in nature is sufficiently high that it is likely to remain confined to the realms of thought experiments and in silico evolution.
Acknowledgements
Kevin Foster and Andy Gardner provided helpful reviews, including Andy Gardner's idea of multiple, fixed-threshold green beard genes with different markers. We also thank Elizabeth Ostrowski, Phil McKinley and the members of the digital evolution laboratory at Michigan State University. This work was supported by NSF grants CCF-0643952, CNS-0751155 and CNS-0915885, by the Cambridge Templeton Consortium Emerging Intelligence grant, by a Michigan State University Dean's Recruitment Fellowship to J.C. and by a fellowship to J.C. from the Quantitative Biology and Modelling Initiative at Michigan State University.
References
- 1.Hamilton W. D. 1964. The genetical evolution of social behaviour. I-II. J. Theor. Biol. 7, 1–52 10.1016/0022-5193(64)90038-4 (doi:10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 2.Hamilton W. D. 1963. The evolution of altruistic behavior. Am. Nat. 97, 354–356 10.1086/497114 (doi:10.1086/497114) [DOI] [Google Scholar]
- 3.Maynard Smith J. 1964. Group selection and kin selection. Nature 201, 1145–1147 10.1038/201145a0 (doi:10.1038/201145a0) [DOI] [Google Scholar]
- 4.Dawkins R. 1976. The selfish gene. Oxford, UK: Oxford University Press [Google Scholar]
- 5.Dawkins R. 1982. The extended phenotype. Oxford, UK: Oxford University Press [Google Scholar]
- 6.Foster K. R., Wenseleers T., Ratnieks F. L. W. 2006. Kin selection is the key to altruism. Trends Ecol. Evol. 21, 57–60 10.1016/j.tree.2005.11.020 (doi:10.1016/j.tree.2005.11.020) [DOI] [PubMed] [Google Scholar]
- 7.Lehmann L., Keller L. 2006. The evolution of cooperation and altruism—a general framework and a classification of models. J. Evol. Biol. 19, 1365–1376 10.1111/j.1420-9101.2006.01119.x (doi:10.1111/j.1420-9101.2006.01119.x) [DOI] [PubMed] [Google Scholar]
- 8.West S. A., Griffin A. S., Gardner A. 2007. Evolutionary explanations for cooperation. Curr. Biol. 17, R661–R672 10.1016/j.cub.2007.06.004 (doi:10.1016/j.cub.2007.06.004) [DOI] [PubMed] [Google Scholar]
- 9.Hepper P. G. 1991. Kin recognition. New York, NY: Cambridge University Press [Google Scholar]
- 10.Holmes W. 2004. The early history of Hamiltonian-based kin recognition research theory. Past and future. Ann. Zool. Fennici 41, 691–711 [Google Scholar]
- 11.Crozier R. H. 1986. Genetic clonal recognition abilities in marine invertebrates must be maintained by selection for something else. Evolution 40, 1100–1101 10.2307/2408769 (doi:10.2307/2408769) [DOI] [PubMed] [Google Scholar]
- 12.Grafen A. 1990. Do animals really recognize kin? Anim. Behav. 39, 42–54 10.1016/S0003-3472(05)80724-9 (doi:10.1016/S0003-3472(05)80724-9) [DOI] [Google Scholar]
- 13.Rousset F., Roze D. 2007. Constraints on the origin and maintenance of genetic kin recognition. Evolution 61, 2320–2330 10.1111/j.1558-5646.2007.00191.x (doi:10.1111/j.1558-5646.2007.00191.x) [DOI] [PubMed] [Google Scholar]
- 14.Gardner A., West S. A. 2007. Social evolution: the decline and fall of genetic kin recognition. Curr. Biol. 17, R810–R812 10.1016/j.cub.2007.07.030 (doi:10.1016/j.cub.2007.07.030) [DOI] [PubMed] [Google Scholar]
- 15.Crespi B., Springer S. 2003. Ecology: social slime molds meet their match. Science 299, 56–57 10.1126/science.1080776 (doi:10.1126/science.1080776) [DOI] [PubMed] [Google Scholar]
- 16.West S. A., Griffin A. S., Gardner A., Diggle S. P. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4, 597–607 10.1038/nrmicro1461 (doi:10.1038/nrmicro1461) [DOI] [PubMed] [Google Scholar]
- 17.Penn D. J. 2002. The scent of genetic compatibility: sexual selection and the major histocompatibility complex . Ethology 108, 1–21 10.1046/j.1439-0310.2002.00768.x (doi:10.1046/j.1439-0310.2002.00768.x) [DOI] [Google Scholar]
- 18.Ostrowski E. A., Katoh M., Shaulsky G., Queller D. C., Strassmann J. E. 2008. Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 6, 2376–2382 10.1371/journal.pbio.0060287 (doi:10.1371/journal.pbio.0060287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner A., West S. A. 2010. Greenbeards. Evolution. 64, 25–38 10.1111/j.1558-5646.2009.00842.x (doi:10.1111/j.1558-5646.2009.00842.x) [DOI] [PubMed] [Google Scholar]
- 20.Haig D. 1997. Behavioural ecology: an evolutionary approach (eds Krebs J. R., Davies N. B.), pp. 284–306 Cambridge, UK: Cambridge University Press [Google Scholar]
- 21.Keller L., Ross K. G. 1998. Selfish genes: a green beard in the red fire ant. Nature 394, 573–575 10.1038/29064 (doi:10.1038/29064) [DOI] [Google Scholar]
- 22.Lizé A., Carval D., Cortesero A. M., Fournet S., Poinsot D. 2006. Kin discrimination and altruism in the larvae of a solitary insect. Proc. R. Soc. B 273, 2381–2386 10.1098/rspb.2006.3598 (doi:10.1098/rspb.2006.3598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queller D. C., Ponte E., Bozzaro S., Strassmann J. E. 2003. Single-gene greenbeard effects in the social amoeba Dictyostelium discoideum. Science 299, 105–106 10.1126/science.1077742 (doi:10.1126/science.1077742) [DOI] [PubMed] [Google Scholar]
- 24.Sinervo B., Clobert J. 2003. Morphs, dispersal behavior, genetic similarity and the evolution of cooperation. Science 300, 1949–1951 10.1126/science.1083109 (doi:10.1126/science.1083109) [DOI] [PubMed] [Google Scholar]
- 25.Smukalla S., et al. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell 135, 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Summers K., Crespi B. 2005. Cadherins in maternal–foetal interactions: red queen with a green beard? Proc. R. Soc. B 272, 643–649 10.1098/rspb.2004.2890 (doi:10.1098/rspb.2004.2890) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.West S. A., Gardner A. 2010. Altruism, spite, and greenbeards. Science 327, 1341–1344 10.1126/science.1178332 (doi:10.1126/science.1178332) [DOI] [PubMed] [Google Scholar]
- 28.Ofria C., Wilke C. O. 2004. Avida: a software platform for research in computational evolutionary biology. Artif. Life 10, 191–229 10.1162/106454604773563612 (doi:10.1162/106454604773563612) [DOI] [PubMed] [Google Scholar]
- 29.Pennock R. T. 2007. Models, simulations, instantiations and evidence: the case of digital evolution. J. Exp. Theor. Artif. Intell. 19, 29–42 10.1080/09528130601116113 (doi:10.1080/09528130601116113) [DOI] [Google Scholar]
- 30.Lenski R. E., Ofria C., Collier T. C., Adami C. 1999. Genome complexity, robustness and genetic interactions in digital organisms. Nature 400, 661–664 10.1038/23245 (doi:10.1038/23245) [DOI] [PubMed] [Google Scholar]
- 31.Adami C., Ofria C., Collier T. C. 2000. Evolution of biological complexity. Proc. Natl Acad. Sci. USA 97, 4463–4468 10.1073/pnas.97.9.4463 (doi:10.1073/pnas.97.9.4463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilke C. O., Wang J., Ofria C., Adami C., Lenski R. E. 2001. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412, 331–333 10.1038/35085569 (doi:10.1038/35085569) [DOI] [PubMed] [Google Scholar]
- 33.Lenski R. E., Ofria C., Pennock R. T., Adami C. 2003. The evolutionary origin of complex features. Nature 423, 139–144 10.1038/nature01568 (doi:10.1038/nature01568) [DOI] [PubMed] [Google Scholar]
- 34.Chow S. S., Wilke C. O., Ofria C., Lenski R. E., Adami C. 2004. Adaptive radiation from resource competition in digital organisms. Science 305, 84–86 10.1126/science.1096307 (doi:10.1126/science.1096307) [DOI] [PubMed] [Google Scholar]
- 35.Goings S., Clune J., Ofria C., Pennock R. T. 2004. Kin-selection: the rise and fall of kin-cheaters. Proc. Artif. Life 9, 303–308 [Google Scholar]
- 36.Misevic D., Ofria C., Lenski R. E. 2006. Sexual reproduction reshapes the genetic architecture of digital organisms. Proc. R. Soc. B 273, 457–464 10.1098/rspb.2005.3338 (doi:10.1098/rspb.2005.3338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clune J., Misevic D., Ofria C., Lenski R. E., Elena S., Sanjuán R. 2008. Natural selection fails to optimize mutation rates for long-term adaptation on rugged fitness landscapes. PLoS Comput. Biol. 4, e1000187. 10.1371/journal.pcbi.1000187 (doi:10.1371/journal.pcbi.1000187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dennett D. 2002. The new replicators. In Encyclopedia of evolution (ed. Pagel M.), pp. E83–E92 New York, NY: Oxford University Press [Google Scholar]
- 39.Rousset F. 2004. Genetic structure and selection in subdivided populations. Princeton, NJ: Princeton University Press [Google Scholar]
- 40.Levenshtein V. I. 1965. Binary codes capable of correcting deletions, insertions and reversals. Dokl. Akad. Nauk SSSR 163, 845–848 [Google Scholar]
- 41.Sober E., Wilson D. S. 1998. Unto others. Cambridge, MA: Harvard University Press [Google Scholar]
- 42.Jansen V. A. A., van Baalen A. 2006. Altruism through beard chromodynamics. Nature 440, 663–666 10.1038/nature04387 (doi:10.1038/nature04387) [DOI] [PubMed] [Google Scholar]