Abstract
Aphids possess several facultative bacterial symbionts that have important effects on their hosts' biology. These have been most closely studied in the pea aphid (Acyrthosiphon pisum), a species that feeds on multiple host plants. Whether secondary symbionts influence host plant utilization is unclear. We report the fitness consequences of introducing different strains of the symbiont Hamiltonella defensa into three aphid clones collected on Lathyrus pratensis that naturally lack symbionts, and of removing symbionts from 20 natural aphid–bacterial associations. Infection decreased fitness on Lathyrus but not on Vicia faba, a plant on which most pea aphids readily feed. This may explain the unusually low prevalence of symbionts in aphids collected on Lathyrus. There was no effect of presence of symbiont on performance of the aphids on the host plants of the clones from which the H. defensa strains were isolated. Removing the symbiont from natural aphid–bacterial associations led to an average approximate 20 per cent reduction in fecundity, both on the natural host plant and on V. faba, suggesting general rather than plant-species-specific effects of the symbiont. Throughout, we find significant genetic variation among aphid clones. The results provide no evidence that secondary symbionts have a major direct role in facilitating aphid utilization of particular host plant species.
Keywords: aphid, secondary symbiont, mutualism, Acyrthosiphon pisum, host specialization
1. Introduction
Although insects have long been known to form symbioses with bacteria, it is only in the last few decades that the extent to which this occurs has become apparent. Many insects have obligate mutualistic relationships with micro-organisms that provide essential substances missing in their diet [1]. Examples of these primary symbionts include Buchnera (which is present in nearly all species of aphid and compensates for the deficiency of amino acids and other compounds in their phloem diet [2]) and Wigglesworthia (which performs an analogous function for tsetse flies (Glossinia) feeding on vertebrate blood [3]). In other insect–bacterial associations, the symbionts are not essential for normal host growth and development, which raises the questions of how and why such facultative or secondary symbioses are maintained. In some cases, the bacterium manipulates host reproduction in ways that allow it to spread through the population, such as in the very widespread Wolbachia [4]. In other associations, the symbiont appears to increase host fitness, but only under certain ecological conditions.
Secondary symbionts that confer conditional adaptive advantages to their hosts have been particularly well studied in aphids. In the pea aphid (Acyrthosiphon pisum; Hemiptera: Aphididae), the best-characterized secondary symbionts are three species of γ-Proteobacteria: Regiella insecticola, Hamiltonella defensa and Serratia symbiotica [5]. A large majority of aphid clones contain one and sometimes two of these symbionts [6,7], which are transmitted with high fidelity from mother to offspring and can also be transmitted paternally in the sexual generation [8]. Experiments that have either removed or introduced symbionts have shown that the bacteria can increase their hosts' ability to defend themselves from parasitoids [9] and entomopathogenic fungi [10], as well as to withstand heat shock [11–13]. Aphid secondary symbionts have also been shown to influence aspects of aphid life history such as the frequency of production of winged morphs [14], though the benefits, if any, of these effects are less clear.
Pea aphids feed on plant species belonging to the family Fabaceae and the pea aphid taxon consists of a series of genetically differentiated host-associated populations connected to differing degrees by gene flow [15–17]. However, a curious feature of pea aphid biology is that while most clones collected in the wild are specialized on a particular host plant species, they nearly all perform well on certain species of vetch (Vicia) [18,19]; gene flow between aphid populations might therefore occur on these species. Pea aphids have been frequently used to study the evolution of specialization and ecological speciation [17,18,20–24], and hence it is natural to ask if secondary symbionts have a role in the adaptation of their hosts to different food plants. Surveys of secondary symbionts clearly show that particular species are strongly associated with aphids feeding on certain food plants—most pea aphid clones on clover (Trifolium), for example, harbour R. insecticola, while those on alfalfa (Medicago) usually carry H. defensa [7,25–28]. While these patterns may reflect a role of secondary symbionts in host plant use, they may also arise because of factors correlated with host plant use (risk of exposure to natural enemies for example) or simple historical contingency.
Studies designed to distinguish between these explanations have given somewhat contradictory results. Tsuchida et al. [29] used antibiotics to remove R. insecticola from a clover-associated pea aphid clone and found that performance on Trifolium, but not Vicia, was negatively affected. However, in a similar experiment, Leonardo [30] found no fitness effects of removing R. insecticola from two clones of aphid specialized on Trifolium. A third study [31] found that the artificial introduction of R. insecticola into five symbiont-free clones not previously associated with clover could have positive, negative or no effect on performance of Trifolium (there was no overall effect of the introduction but a significant clone by treatment interaction). These results suggest that interactions involving the genotype of either the host or symbiont (or both) can influence host plant use, as has been found in the study of other traits [13,32,33], and also emphasizes the importance of using replicate genotypes in investigations of symbiont biology.
To clarify the role of secondary symbionts in host plant use by pea aphids, we manipulated symbiont composition and then assessed aphid fitness on different species of plant using a much broader range of aphid clones (from different host plant-associated populations and with different symbionts) than hitherto studied. We did this in two ways. Aphids specialized on Lathyrus pratensis have unusually low rates of infection with secondary symbionts; first, we took such clones without secondary symbionts and established novel infections by injecting strains of the secondary symbiont H. defensa that had been harvested from aphids associated with different host plants. The fitness of the same clone of Lathyrus aphid with and without the symbiont was measured on (i) Lathyrus, (ii) the plant species from which the symbiont's host was collected (where possible) and (iii) the widely acceptable host, Vicia faba. Second, we took 20 clones of aphids drawn from five different host-associated populations, each of which contained the secondary symbiont most commonly associated with their host plant species. By oral administration of antibiotics that are known not to affect the primary symbiont [34], aphid lineages that did not contain secondary symbionts were created. The fitness of aphid clones with and without secondary symbionts was compared on the host plant from which they were collected and on Vicia. This allows us to investigate whether or not any observed fitness effects of secondary symbionts are specific to the host plants with which they are found associated in nature, and thus assess the likelihood that the symbionts are influencing aphid specialization.
2. Material and methods
(a). Experimental organisms
All pea aphid clones were derived from single individuals originally collected in Berkshire and Oxfordshire (southern England) during June and July 2003, and July and August 2008. It was confirmed that these aphid clones were specialized on the plants from which they were collected by assaying their survival and fecundity on the collection plant species. The aphid clones were confirmed as being genetically distinct from one another through microsatellite typing [18].
The facultative symbiont complement of the different aphid clones was assessed by repeated amplification (or attempted amplification) of the bacterial 16S ribosomal RNA gene using a ‘universal’ bacterial primer pair (10F, 35R) with partial sequencing of the fragment [11,35]. These primers span the 16S–23S rRNA genes and therefore detect a wide range of Eubacteria, with the primary symbionts Buchnera being a notable exception. This was followed by diagnostic PCRs using primers specific to the 16S ribosomal RNA genes of four known pea aphid facultative symbionts (Hamiltonella, Regiella, Serratia and a bacterium currently referred to as ‘X-type’) to check for multiple infections. ‘X-type’ is a less well-characterized γ-Proteobacteria secondary symbiont [36]. Aphid clones were screened using diagnostic PCR for the presence of Rickettsia and Spiroplasma infections, both of which have been recorded as facultative associates of pea aphids [37–39]. Appropriate negative and positive controls were used in each case, and details of all primers used are given in the electronic supplementary material, table S1. The secondary symbionts found in the different aphid clones are shown in table 1.
Table 1.
The aphid clones used in the symbiont-removal experiments, the host plants on which they are specialized and the secondary symbionts they contained.
| host plant | symbiont | clones | clone codes |
|---|---|---|---|
| Medicago sativa | Hamiltonella defensa | 5 clones; one (341) also contained the ‘X-type’ symbiont, one (222) also contained R. insecticola, one (161) also contained Spiroplasma | 161, 222, 328, 340, 341 |
| Trifolium repens | Regiella insecticola | 5 clones | 102, 126, 313, 317, 319 |
| Lotus pedunculatus | Hamiltonella defensa | 5 clones; three (141, 184 and 208) also contained Rickettsia | 132, 141, 184, 208, 224 |
| Pisum sativum | Serratia symbiotica | 2 clones | 256, 316 |
| Ononis spinosa | Hamiltonella defensa | 3 clones | 101, 123, 133 |
(b). Creating artificial secondary symbiont infections
Novel associations between pea aphids and the facultative symbiont H. defensa were created by injecting naturally symbiont-free aphids (the ‘recipient’ clones; shown to be symbiont-free by the absence of PCR product after attempted amplification using the universal 10F/35R bacterial primer pair and specific primers for Rickettsia and Spiroplasma) with haemolymph extracted from four different naturally infected pea aphid clones (the ‘donor’ clones). Four different donor clones were used: two from Lotus pedunculatus, one from Medicago sativa and one from Ononis spinosa. All were infected with H. defensa, the facultative symbiont most commonly found in aphids on each of these host plants. One clone (208) also contained Rickettsia and one (161) Spiroplasma. Following injection, recipient aphids were maintained at 14°C in Petri dishes containing healthy leaves of V. faba (cv. The Sutton), which were kept fresh by inserting the petiole into 2 per cent agar gel. Vicia faba is a host plant species on which almost all pea aphid clones have been found to perform well [18,20,40]. Each artificially infected clone was derived from a single injected individual and was screened regularly to confirm the continued presence of H. defensa (we observed no loss of symbionts after the first few generations; in the case of the two co-infections, both symbionts were established). Experiments were carried out at least six generations after the initial injection.
(c). Curing natural secondary symbiont infections
Natural γ-Proteobacteria secondary symbiont infections were eliminated from 20 aphid clones using oral administration of antibiotics. The clones were collected on L. pedunculatus, M. sativa, O. spinosa, Pisum sativum and Trifolium pratense, and all possessed the secondary symbiont most commonly associated with that host plant (table 1). Cut leaves of V. faba were placed in 1.5 ml Eppendorf tubes containing 100 µg ml−1 Ampicillin, 50 µg ml−1 Cefotaxime and 50 µg ml−1 Gentomicin [34,41], and second instar aphids were allowed to feed on them for 3–4 days at 14°C. Surviving aphids were then transferred to a Vicia leaf with its petiole inserted into agar gel. Once adult, the aphids were separated and the late offspring (over tenth in birth order) retained. After they had begun reproducing, second generation aphids were tested for the presence of secondary endosymbionts using symbiont-specific 16S rRNA primers as described above. Offspring of adults found to lack γ-Proteobacteria were retained to found an infection-free clonal line, which was considered cured if there was a complete lack of amplification at least six generations following antibiotic treatment. The absence of γ-Proteobacteria was reconfirmed after the experiments had been conducted.
Our antibiotic curing protocol did not affect Spiroplasma or Rickettsia. We found the former in one clone and the latter in three clones (table 1) and confirmed that the infection status was the same in both the original lines and in the lines cured of the γ-proteobacterial secondary symbionts. Differences between the two sets of lines were thus owing to changes in the γ-Proteobacteria symbiont infection.
(d). Effects of artificial infection with Hamiltonella defensa on aphid fitness
Aphids were reared on pre-flowering plants of V. faba for two generations prior to the experiments in cultures maintained at 20°C, 60 per cent relative humidity and a 16 L : 8 D light : dark cycle. Experiments were carried out under the same environmental conditions (further details of the plants used in the experiments can be found in the electronic supplementary material, table S2). The fitness of aphids with and without the symbiont H. defensa was assessed on Lathyrus, Vicia and on the plant species from which the donor aphid was collected (except that for logistic reasons this was not possible for O. spinosa). Aphids were allowed to reproduce on fresh leaves of Vicia for up to 24 h, and 8–10 of the resultant offspring (in a very small number of cases fewer) were placed on similarly sized, fresh and healthy plants of the appropriate species. The total number of offspring produced by the aphids between days 7 and 12, controlling for the number of aphids alive at day 7, was used as our measure of fitness. This composite measure incorporates differences in development time, age at first reproduction and number of offspring produced in early adulthood. Five replicates (stratified across five temporal blocks) were carried out for aphid clones tested on Lathyrus, Lotus and Medicago and six replicates (in three temporal blocks) for tests on Vicia.
(e). Effects of secondary symbiont removal upon aphid fitness
Aphid pre-treatment and experiments were conducted under the same conditions as described in §2d above. The fitness of aphids with and without their natural facultative symbiont infections was assessed on the plant species from which they were collected and on Vicia. Adult aphids were allowed to reproduce on fresh leaves of either Vicia or their natural host for 24 h and the offspring produced (typically 6–10) transferred to plants of the same species. After 7 days, any second-generation offspring and all except three of the adult aphids were removed, and the total number of further offspring produced over the next 8 days was recorded as a measure of fitness (in the case of Vicia, the three adult aphids were transferred to a fresh plant as the quality of the seedling plant declines rapidly when fed upon). On average, six replicates were carried out for each aphid clone on each plant species, distributed across nine (Lotus, Medicago and Trifolium), three (Ononis) or four (Pisum) temporal blocks.
(f). Statistical analysis
The data were analysed using generalized linear modelling techniques. The data on fecundity are counts and hence were analysed using log-linear techniques with the assumption of quasi-Poisson error variance to allow for overdispersion. All analyses were performed using R (v. 2.6.1; http://www.r-project.org). In six of the 31 replicates used to estimate the fecundity of artificially infected aphids feeding on Medicago, all aphids died in the first 7 days and hence the replicates were omitted from the analysis.
3. Results
(a). Symbiont injection
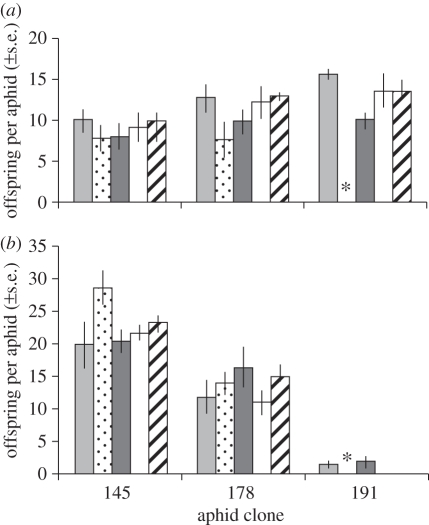
If secondary symbionts improve the ability of their aphid hosts to feed on a particular plant species, then we would expect improved performance of the recipient clone on the plant to which the donor strain's host is specialized. We were able to test this for Lotus and Medicago. The uninfected recipient clones performed very poorly on Lotus, producing no offspring on this species. This poor performance was not affected by the injection of H. defensa derived from Lotus aphids; infected aphids also produced no offspring. Uninfected recipient clones were able to survive and reproduce on Medicago, though their fecundity on this host plant was considerably less than that on Lathyrus (F1,100 = 251.4, p < 0.001). However, contrary to the hypothesis, lines that had been injected with H. defensa derived from aphids collected on Medicago produced fewer offspring on Medicago than the uninfected controls (F1,22 = 5.53, p = 0.029). We also observed significant differences among recipient clones in their response to the introduction of H. defensa when feeding on Medicago (F2,20 = 7.01, p = 0.005).
If carriage of secondary symbionts incurs costs for their hosts, then we might expect reduced fitness on the natural host of the recipients (Lathyrus) or on Vicia, the host on which most aphid clones perform well. We found that carrying H. defensa reduced the fecundity of recipient clones on their natural host (figure 1 and table 2), and also observed that the strength of this effect was significantly influenced by donor strain. Interestingly, the co-infections (one Spiroplasma, one Rickettsia) appear to have, if anything, a slightly less severe impact on their hosts than the infections with Hamiltonella alone. On Vicia, there was a large and significant difference in the performance of the aphid genotype (recipient clones), with one performing very poorly irrespective of infection status. For the remaining recipient clones, we found no effect of infection status on fecundity on Vicia (figure 1 and table 2).
Figure 1.
The effect of symbiont presence on aphid fecundity when feeding on (a) Lathyrus and (b) Vicia. The figure compares three recipient clones (codes 145, 178 and 191) when they carry no symbiont or after the injection of symbiont from four donor clones (codes 101, 132, 208 and 161). In one case, indicated by an asterisk, we failed to establish an infection. Symbiont status: Light grey bars, none; dotted bars, 101; dark grey bars, 132; white bars, 208; striped bars, 161.
Table 2.
Effects of artificial symbiont infection on the cumulative number of offspring produced by groups of aphids over the first 12 days of adult life, measured on either Lathyrus or Vicia. Terms were added in the order shown in the table, and the change in degrees of freedom and the deviance explained (the raw figure and as a percentage of the total deviance) are given along with the F-statistic and its associated probability (in bold when <0.05).
| factor(s) | d.f. | deviance | % deviation | F | p |
|---|---|---|---|---|---|
| Lathyrus | |||||
| no. of adults | 1 | 70.9 | 7.3 | 7.71 | 0.007 |
| block | 4 | 56.2 | 5.8 | 1.53 | 0.207 |
| recipient clone | 2 | 92.1 | 9.5 | 5.01 | 0.010 |
| infection status | 1 | 57.0 | 5.9 | 6.20 | 0.016 |
| donor strain | 3 | 101.6 | 10.5 | 3.69 | 0.017 |
| recipient × infection | 2 | 14.5 | 1.5 | 0.79 | 0.459 |
| recipient × donor | 5 | 11.2 | 1.2 | 0.24 | 0.941 |
| remaining deviance | 57 | 593.6 | |||
| Vicia | |||||
| no. of adults | 1 | 4879.2 | 22.79 | 35.79 | <0.001 |
| block | 2 | 13.6 | 6.21 | 4.88 | 0.011 |
| recipient clone | 1 | 2380.3 | 27.73 | 43.55 | <0.001 |
| infection status | 1 | 39.0 | 2.34 | 3.67 | 0.061 |
| donor strain | 3 | 90.3 | 4.23 | 2.21 | 0.097 |
| recipient × infection | 1 | 6.6 | 0.03 | 0.05 | 0.830 |
| recipient × donor | 3 | 229.9 | 2.71 | 1.42 | 0.247 |
| remaining deviance | 76 | 905.7 | |||
(b). Symbiont removal
Secondary symbionts were removed from 20 clones of aphids collected on five species of host plant (details in table 1). We initially analysed the complete dataset together, fitting a ‘minimal model’ that controlled for all the factors that were not of immediate interest, before adding terms representing infection status and its interactions. The minimal model included terms for collection plant species, the test plant species on which the aphids' fitness was assessed (this was either the collection plant species or Vicia), aphid clone, experimental block, the aphid clone × test plant interaction and the proportion of alate adults among the progeny produced in the experiment. An analysis of the components of the minimal model is given in the electronic supplementary material, table S3.
Loss of the symbiont on average reduced aphid fecundity. Adding a term for infection status to the minimal model significantly increased the explanatory power of the model (table 2) and for 1 d.f. explained 11.1 per cent of the remaining variation (the residual deviance after fitting the minimal model) in the data. On average, the loss of symbionts leads to a 21 per cent reduction (s.e. 19–24%) in offspring numbers. However, there was significant variation in the response of aphids collected on different host plants, as well as among the different aphid clones themselves (table 3). We explored whether symbiont loss affected aphid performance differently on their collection plant or on Vicia by adding the interaction of test plant and infectious status to the statistical model containing all the above terms. A significant difference was found, with fecundity on average being more affected on Vicia, but the effect was not large (it explained only 0.6% of the variation in the data for 1 d.f.) and a significant three-way interaction revealed that its strength varied among aphids from different collection plants (table 3).
Table 3.
Effects of artificial symbiont removal on the cumulative number of offspring produced by groups of three aphids aged between 8 and 15 days. A minimal model described in the text and analysed further in the electronic supplementary material was fitted to the data. Further terms were added in the order shown in the table and the change in degrees of freedom and the deviance explained (the raw figure and as a percentage of the total deviance) are given along with the F-statistic and its associated probability (in bold when <0.05).
| factor(s) | d.f. | deviance | % deviation | F | p |
|---|---|---|---|---|---|
| minimal model | 50 | 13 125.6 | |||
| infection | 1 | 759.0 | 11.1 | 86.12 | <0.001 |
| infection × collection plant | 4 | 425.1 | 6.2 | 12.06 | <0.001 |
| infection × clone | 15 | 1158.1 | 17.0 | 8.76 | <0.001 |
| infection × test plant | 1 | 44.1 | 0.6 | 5.00 | 0.026 |
| infection × test plant × collection plant | 4 | 129.4 | 1.9 | 3.67 | 0.006 |
| infection × test plant × clone | 15 | 224.4 | 3.3 | 1.70 | 0.049 |
| remaining deviance | 419 | 4113.0 |
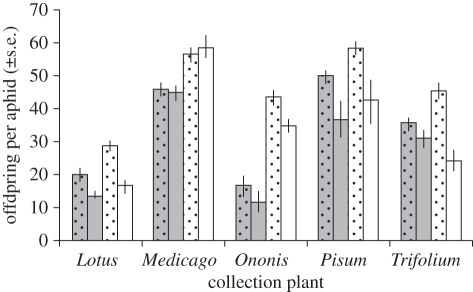
To understand better how aphids from different host plant-associated populations responded to the loss of their symbionts we conducted separate analyses for the clones collected from each of the five host plants (figure 2; further details in the electronic supplementary material, table S4). Aphids from Lotus and Trifolium on average experienced reduced fecundity after symbiont removal, and this effect was stronger on Vicia than on their collection plant. There were also significant differences in response to symbionts across clones for aphids from Trifolium. Clones from Pisum and Ononis again on average showed reduced fecundity in the absence of the symbiont, though this effect was not consistently stronger when the aphids were tested on Vicia. With the Ononis clones, there was a significant three-way interaction; this group contained the only clone to be disadvantaged on its collection plant, but not on Vicia, by the loss of the symbiont. Finally, the response of Medicago clones was variable: different clones had unchanged or lower fecundity when their symbiont was removed (and hence there was no clear overall response), though the response of infected and uninfected aphids within a clone was similar when tested on either Medicago or Vicia.
Figure 2.
The effects of eliminating the natural secondary symbiont infections from aphid clones collected on Lotus, Medicago, Ononis, Pisum and Trifolium. The figure shows fecundity of infected (dotted) and uninfected (undotted) aphids when feeding on the plant species on which they were collected (grey bars) or Vicia (white bars).
4. Discussion
We found no evidence that secondary symbionts have a major effect on host-plant specialization in the pea aphid. Introducing symbionts to aphid lineages that naturally had no bacteria did not improve aphid performance on the plant species with which the symbionts were originally associated. In fact, while aphid performance on Vicia was little affected by introduced symbionts, performance on their natural host plant, Lathyrus, was reduced. Removing symbionts from a variety of natural aphid–bacteria associations on average reduced fecundity by approximately 20 per cent, but the drop in fitness was greater on Vicia than on the original host plant, suggesting that this was a general rather than a plant-species-specific cost of symbiont removal. Throughout, we found that the effects on aphid fitness of removing secondary symbionts tended to vary among aphid genotypes.
Strong correlative associations have been reported between certain symbiont species and aphid populations adapted to feeding on particular plant species; for example, Trifolium-feeding clones normally carry R. insecticola. This combination has previously been investigated experimentally by Tsuchida et al. [29], Leonardo [30] and Ferrari et al. [31]; our study included five such clones. We found that curing aphids of their natural R. insecticola infections had a negative impact on host fitness, in agreement with Tsuchida et al. [29]; however, the results on Vicia confirm the conclusions of Leonardo [30] and Ferrari et al. [31] that the presence of R. insecticola does not provide specific fitness advantages on Trifolium. We obtained similar results for the four other host plants we investigated. Overall, we conclude that no symbiont studied provides any specific advantage to feeding on the host plant with which it is associated, though the presence of often substantial interactions between host genotype and infection status may mean that a particular clone enjoys this benefit, as observed by Tsuchida et al. [29]. We cannot of course rule out the possibility that there is geographical variation in the influence of secondary symbionts on host plant use.
There remains the question of why secondary symbiont distribution is correlated strongly with particular host-plant-associated aphid populations. It seems unlikely that the associations are merely founder effects, an echo of the symbiont flora that was coincidentally associated with the aphids that first colonized a new host plant. Some associations, such as that between R. insecticola and Trifolium-feeding aphids, are found throughout the world [7,25–28]. It is therefore much more likely that the symbiont–host-plant correlation occurs because different secondary symbionts are selectively advantageous on different host plants for reasons other than direct nutrition-related fitness benefits. It would be difficult but very interesting to see if, for example, aphids on Trifolium are more often subject to infection by entomopathogenic fungi, a natural enemy against which R. insecticola provides protection [10].
A further explanation may be that the presence of a symbiont species is costly on certain plant species and this outweighs the benefits of its carriage. Cases where novel aphid–symbiont associations experience reduced overall fitness have been noted before [42], but here we find that the injection of H. defensa into Lathyrus-adapted clones reduced fitness on this host plant but made little difference to aphid fitness on Vicia; indeed, this was the only plant-specific fitness effect observed in our experiments that was consistent across aphid clones. There are two distinct genetic populations of pea aphid found on Lathyrus and both are unusual in having low or very low frequencies of secondary symbionts (unpublished data). A cost to carrying symbionts on Lathyrus might explain these findings and similar interactions might influence the distribution of symbionts across other host-plant-adapted populations.
Given that secondary symbionts influence many other aspects of pea aphid biology, why do they not affect host plant use? The answer may lie in the widespread distribution of the symbionts outside the pea aphid: the three major secondary symbionts are known also to occur in a large number of different aphid species, and in other related homopterans [35,43]. We understand neither how secondary symbionts move among aphid lineages nor what determines the long-term persistence of the interaction at the aphid community level, but it probably involves their ability to transfer horizontally between species and to provide conditional benefits to their hosts [44]. If this is the case, secondary symbionts may need to provide advantages that may be useful to a wide range of aphid hosts. Most aphids are attacked by parasitoids and fungi, often the same or closely related species, and conferring the ability to withstand this challenge may be valuable to many host species. Similarly, all aphids are potentially vulnerable to heat shock. By contrast, the vast majority of aphid species are highly host-specific, often restricted to one or a set of closely related plants. Assisting an aphid host to feed on a specific food plant may not be a transferable benefit beyond one or a few aphid species, and hence is less likely to evolve in a symbiont moving among a range of different aphids.
Were secondary symbionts to have had a consistent effect on host plant use, it would have complicated the interpretation of the many studies of the evolution of specialization and ecological speciation that have made use of the pea aphid system [17,18,20–24]. Our results suggest that direct effects of symbiont presence on host plant species use are not pervasive, although they do not rule out indirect effects mediated by other aspects of the aphid's biology that might be correlated with host plant species.
Acknowledgements
The work was partially supported by the UK Biotechnology and Biology Research Council award BB/E010857/1 and by a studentship from the UK Natural Environment Research Council to A.H.C.M. We are grateful to two anonymous reviewers for their constructive comments on an earlier version of this manuscript.
References
- 1.Moran N. A. 2001. The coevolution of bacterial endosymbionts and phloem-feeding insects. Ann. Mo. Bot. Gard. 88, 35–44 10.2307/2666130 (doi:10.2307/2666130) [DOI] [Google Scholar]
- 2.Douglas A. E. 2006. Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot. 57, 747–754 10.1093/jxb/erj067 (doi:10.1093/jxb/erj067) [DOI] [PubMed] [Google Scholar]
- 3.Aksoy S. 1995. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45, 848–851 10.1099/00207713-45-4-848 (doi:10.1099/00207713-45-4-848) [DOI] [PubMed] [Google Scholar]
- 4.Werren J. H. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42, 587–609 10.1146/annurev.ento.42.1.587 (doi:10.1146/annurev.ento.42.1.587) [DOI] [PubMed] [Google Scholar]
- 5.Moran N. A., Russell J. A., Koga R., Fukatsu T. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71, 3302–3310 10.1128/AEM.71.6.3302-3310.2005 (doi:10.1128/AEM.71.6.3302-3310.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver K. M., Moran N. A., Hunter M. S. 2006. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc. R. Soc. B 273, 1273–1280 10.1098/rspb.2005.3436 (doi:10.1098/rspb.2005.3436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchida T., Koga R., Shibao H., Matsumoto T., Fukatsu T. 2002. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 11, 2123–2135 10.1046/j.1365-294X.2002.01606.x (doi:10.1046/j.1365-294X.2002.01606.x) [DOI] [PubMed] [Google Scholar]
- 8.Moran N. A., Dunbar H. E. 2006. Sexual acquisition of beneficial symbionts in aphids. Proc. Natl Acad. Sci. USA 103, 12 803–12 806 10.1073/pnas.0605772103 (doi:10.1073/pnas.0605772103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oliver K. M., Degnan P. H., Hunter M. S., Moran N. A. 2009. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science 325, 992–994 10.1126/science.1174463 (doi:10.1126/science.1174463) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scarborough C. L., Ferrari J., Godfray H. C. J. 2005. Aphid protected from pathogen by endosymbiont. Science 310, 1781–1781 10.1126/science.1120180 (doi:10.1126/science.1120180) [DOI] [PubMed] [Google Scholar]
- 11.Russell J. A., Moran N. A. 2005. Horizontal transfer of bacterial symbionts: heritability and fitness effects in a novel aphid host. Appl. Environ. Microbiol. 71, 7987–7994 10.1128/AEM.71.12.7987-7994.2005 (doi:10.1128/AEM.71.12.7987-7994.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montllor C. B., Maxmen A., Purcell A. H. 2002. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 27, 189–195 10.1046/j.1365-2311.2002.00393.x (doi:10.1046/j.1365-2311.2002.00393.x) [DOI] [Google Scholar]
- 13.Chen D. Q., Montllor C. B., Purcell A. H. 2000. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid A. kondoi. Entomol. Exp. Appl. 95, 315–323 10.1023/A:1004083324807 (doi:10.1023/A:1004083324807) [DOI] [Google Scholar]
- 14.Leonardo T. E., Mondor E. B. 2006. Symbiont modifies host life-history traits that affect gene flow. Proc. R. Soc. B 273, 1079–1084 10.1098/rspb.2005.3408 (doi:10.1098/rspb.2005.3408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peccoud J., Ollivier A., Plantegenest M., Simon J. C. 2009. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl Acad. Sci. USA 106, 7495–7500 10.1073/pnas.0811117106 (doi:10.1073/pnas.0811117106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peccoud J., Simon J. C., McLaughlin H. J., Moran N. A. 2009. Post-Pleistocene radiation of the pea aphid complex revealed by rapidly evolving endosymbionts. Proc. Natl Acad. Sci. USA 106, 16 315–16 320 10.1073/pnas.0905129106 (doi:10.1073/pnas.0905129106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Via S. 1991. The genetic structure of host plant adaptation in a spatial patchwork–demographic variability among reciprocally transplanted pea aphid clones. Evolution 45, 827–852 10.2307/2409692 (doi:10.2307/2409692) [DOI] [PubMed] [Google Scholar]
- 18.Ferrari J., Via S., Godfray H. C. J. 2008. Population differentiation and genetic variation in performance on eight hosts in the pea aphid complex. Evolution 62, 2508–2524 10.1111/j.1558-5646.2008.00468.x (doi:10.1111/j.1558-5646.2008.00468.x) [DOI] [PubMed] [Google Scholar]
- 19.Sandström J. 1996. Temporal changes in host adaptation in the pea aphid, Acyrthosiphon pisum. Ecol. Entomol. 21, 56–62 10.1111/j.1365-2311.1996.tb00266.x (doi:10.1111/j.1365-2311.1996.tb00266.x) [DOI] [Google Scholar]
- 20.Ferrari J., Godfray H. C. J., Faulconbridge A. S., Prior K., Via S. 2006. Population differentiation and genetic variation in host choice among pea aphids from eight host plant genera. Evolution 60, 1574–1584 [PubMed] [Google Scholar]
- 21.Hawthorne D. J., Via S. 2001. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904–907 10.1038/35091062 (doi:10.1038/35091062) [DOI] [PubMed] [Google Scholar]
- 22.Via S. 1991. Specialized host plant performance of pea aphid clones is not altered by experience. Ecology 72, 1420–1427 10.2307/1941114 (doi:10.2307/1941114) [DOI] [Google Scholar]
- 23.Via S., Bouck A. C., Skillman S. 2000. Reproductive isolation between divergent races of pea aphids on two hosts. II. Selection against migrants and hybrids in the parental environments. Evolution 54, 1626–1637 [DOI] [PubMed] [Google Scholar]
- 24.Caillaud M. C., Via S. 2000. Specialized feeding behavior influences both ecological specialization and assortative mating in sympatric host races of pea aphids. Am. Nat. 156, 606–621 10.1086/316991 (doi:10.1086/316991) [DOI] [PubMed] [Google Scholar]
- 25.Darby A. C., Tosh C. R., Walters K. F. A., Douglas A. E. 2003. The significance of a facultative bacterium to natural populations of the pea aphid Acyrthosiphon pisum. Ecol. Entomol. 28, 145–150 10.1046/j.1365-2311.2003.00492.x (doi:10.1046/j.1365-2311.2003.00492.x) [DOI] [Google Scholar]
- 26.Ferrari J., Darby A. C., Daniell T. J., Godfray H. C. J., Douglas A. E. 2004. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 29, 60–65 10.1111/j.1365-2311.2004.00574.x (doi:10.1111/j.1365-2311.2004.00574.x) [DOI] [Google Scholar]
- 27.Leonardo T. E., Muiru G. T. 2003. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc. R. Soc. Lond. B 270, S209–S212 10.1098/rsbl.2003.0064 (doi:10.1098/rsbl.2003.0064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon J. C., Carre S., Boutin M., Prunier-Leterme N., Sabater-Munoz B., Latorre A., Bournoville R. 2003. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. R. Soc. Lond. B 270, 1703–1712 10.1098/rspb.2003.2430 (doi:10.1098/rspb.2003.2430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuchida T., Koga R., Fukatsu T. 2004. Host plant specialization governed by facultative symbiont. Science 303, 1989–1989 10.1126/science.1094611 (doi:10.1126/science.1094611) [DOI] [PubMed] [Google Scholar]
- 30.Leonardo T. E. 2004. Removal of a specialization-associated symbiont does not affect aphid fitness. Ecol. Lett. 7, 461–468 10.1111/j.1461-0248.2004.00602.x (doi:10.1111/j.1461-0248.2004.00602.x) [DOI] [Google Scholar]
- 31.Ferrari J., Scarborough C. L., Godfray H. C. J. 2007. Genetic variation in the effect of a facultative symbiont on host-plant use by pea aphids. Oecologia 153, 323–329 10.1007/s00442-007-0730-2 (doi:10.1007/s00442-007-0730-2) [DOI] [PubMed] [Google Scholar]
- 32.Koga R., Tsuchida T., Sakurai M., Fukatsu T. 2007. Selective elimination of aphid endosymbionts: effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol. Ecol. 60, 229–239 [DOI] [PubMed] [Google Scholar]
- 33.Oliver K. M., Moran N. A., Hunter M. S. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl Acad. Sci. USA 102, 12 795–12 800 10.1073/pnas.0506131102 (doi:10.1073/pnas.0506131102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandler S. M., Wilkinson T. L., Douglas A. E. 2008. Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proc. R. Soc. B 275, 565–570 10.1098/rspb.2007.1478 (doi:10.1098/rspb.2007.1478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandström J. P., Russell J. A., White J. P., Moran N. A. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol. Ecol. 10, 217–228 10.1046/j.1365-294X.2001.01189.x (doi:10.1046/j.1365-294X.2001.01189.x) [DOI] [PubMed] [Google Scholar]
- 36.Guay J.-F., Boudreault S., Michaud D., Cloutier C. 2009. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J. Insect Physiol. 55, 919–926 10.1016/j.jinsphys.2009.06.006 (doi:10.1016/j.jinsphys.2009.06.006) [DOI] [PubMed] [Google Scholar]
- 37.Chen D. Q., Campbell B. C., Purcell A. H. 1996. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris). Curr. Microbiol. 33, 123–128 10.1007/s002849900086 (doi:10.1007/s002849900086) [DOI] [PubMed] [Google Scholar]
- 38.Fukatsu T., Tsuchida T., Nikoh N., Koga R. 2001. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera). Appl. Environ. Microbiol. 67, 1284–1291 10.1128/AEM.67.3.1284-1291.2001 (doi:10.1128/AEM.67.3.1284-1291.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakurai M., Koga R., Tsuchida T., Meng X. Y., Fukatsu T. 2005. (Rickettsia) symbiont in the pea aphid (Acyrthosiphon pisum): novel cellular tropism, effect on host fitness, and interaction with the essential symbiont (Buchnera). Appl. Environ. Microbiol. 71, 4069–4075 10.1128/AEM.71.7.4069-4075.2005 (doi:10.1128/AEM.71.7.4069-4075.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandström J., Pettersson J. 1994. Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 40, 947–955 10.1016/0022-1910(94)90133-3 (doi:10.1016/0022-1910(94)90133-3) [DOI] [Google Scholar]
- 41.Douglas A. E., Prosser W. A. 1992. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J. Insect Physiol. 38, 565–568 10.1016/0022-1910(92)90107-O (doi:10.1016/0022-1910(92)90107-O) [DOI] [Google Scholar]
- 42.Koga R., Tsuchida T., Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. R. Soc. Lond. B 270, 2543–2550 10.1098/rspb.2003.2537 (doi:10.1098/rspb.2003.2537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes S., Darby A. C., Daniell T. J., Webster G., van Veen F. J. F., Godfray H. C. J., Prosser J. I., Douglas A. E. 2003. Diversity of bacteria associated with natural aphid populations. Appl. Environ. Microbiol. 69, 7216–7223 10.1128/AEM.69.12.7216-7223.2003 (doi:10.1128/AEM.69.12.7216-7223.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver K. M., Degnan P. H., Burke G. R., Moran N. A. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266 10.1146/annurev-ento-112408-085305 (doi:10.1146/annurev-ento-112408-085305) [DOI] [PubMed] [Google Scholar]