Abstract
The insulin/insulin-like signalling (IIS) network is conserved among animals and is central to growth and development. In eusocial honeybees (Apis mellifera), IIS is hypothesized to shape female caste fate. We tested this hypothesis via RNA interference (RNAi) knockdown of the insulin receptor substrate (IRS) homologue, a key adaptor protein in IIS. Female larvae naturally develop into queens (reproductives) or workers (helpers) after being fed rich versus limited diets, respectively. Feeding larvae a rich diet mixed with dsRNA (double stranded RNA) targeting irs gene transcript decreased irs mRNA abundance and caused development of worker morphology. Controls receiving rich larval diet and control dsRNA developed queen morphology. Whole-body mass spectrometry profiling of larvae collected 72, 96 and 120 h after dsRNA treatments revealed proteomic differences between irs gene knockdowns and controls, including levels of hexamerin 110, a storage protein connected to natural caste differences.
Keywords: insulin signalling, Apis mellifera, queen bee, worker bee, RNA interference, proteomics
1. Introduction
Female honeybees develop into two castes, queens and workers, which differ in morphology, physiology, social function and longevity. The reproductive queens lay up to 2000 eggs daily, develop into adults faster, are bigger, heavier, have larger reproductive organs and survive longer (more than 1.5–2 years) than workers [1]. Workers live for two weeks to several months and are mostly sterile helpers that nourish larvae and collect food. The queen–worker diphenism is believed to be central to the ecological success of honeybees, and is mediated socially by larval feeding: high quantity and quality of larval provisions result in queens while more limited nourishment yields workers, independent of genetic background [1]. On the molecular level, juvenile hormone, DNA methylation and TOR (target of rapamycin), a protein kinase involved in nutrient-sensing and growth control, have been independently shown to influence caste development [2,3].
Insulin-like signalling (IIS), which can be coupled to TOR activation through downstream integration of Tsc-1 and 2 (tuberous sclerosis complex genes 1 and 2; e.g. [4]), is similarly crucial to nutrient-sensing and growth regulation in animals. IIS influences development and final body size, food-related behaviour, reproduction and longevity across many taxa ([5] and references therein). Thus, this pathway is also likely to influence honeybee caste development [6,7]. However, functional evidence for this hypothesis has been limited.
Most of what has been revealed about IIS in animal model systems relies on impaired insulin signalling caused by either knockout (disruptive mutation) or knockdown (downregulation via RNA interference, RNAi) of key insulin signalling proteins like the insulin receptor substrate (IRS) protein. Downregulation of IRS confers a substantial reduction of IIS [8]. Recently, we established RNAi-mediated irs gene knockdown in adult honeybee workers to show that the IRS product influences food-choice behaviour towards pollen (amino acids) and nectar (carbohydrate) [9].
Downregulation of IIS via disruptive mutation of irs reduces body size and fertility in Drosophila [10]. Thus, we reasoned that the course of larval development in response to rich queen diet could be blocked by decreasing IIS through irs repression. This should yield the small, essentially sterile worker phenotype, as previously shown, following downregulation of TOR gene transcript [3]. To test this hypothesis, we suppressed irs expression by RNAi in larvae reared on laboratory diet that elicits queen development. This study provides functional evidence for IIS roles in honeybee caste development.
2. Material and methods
(a). irs dsRNA synthesis, bees, feeding treatments, sample collections
Synthesis of irs dsRNA was conducted as before [9]. Larvae were produced from two wild-type (and multiply mated) queens and genotype was a random factor in the experiment. In vitro feeding methods were essentially identical to those of [3]. We produced two treatment groups: larvae-fed queen diet mixed with irs dsRNA, and larvae-fed queen diet mixed with a standard control of gfp (green fluorescent protein) dsRNA.
Two replicate studies with 100 larvae per treatment group were performed. A random subset of larvae was collected into liquid nitrogen 72, 96 and 120 h after the first dsRNA feeding (see the electronic supplementary material).
(b). RNAi validation, morphological phenotyping
irs RNA abundance was tested by quantitative real-time PCR 72 h after dsRNA feeding began (see the electronic supplementary material). Developmental time (larva to adult) was assessed (n = 50), and a random subset (n = 20) measured at adult emergence for fresh weight. The first 12 individuals (only queens in the control group as intercastes typically emerge later) were dissected to determine ovary size by counting ovarian filaments (ovarioles) as before [3].
(c). Protein extraction, tryptic digestion and proteomics analysis
These steps (see the electronic supplementary material) were essentially carried out as before [11]. Proteomics analysis was conducted using liquid chromatography coupled to tandem mass spectrometry (LC-MS2).
(d). Statistical analysis
Non-parametric Mann Whitney U-tests were used to individually compare irs expression, fresh weight, developmental time and ovary size between groups. Proteomic data were analysed by an established combination of Mann Whitney U-tests with correction for type 1 error inflation (see the electronic supplementary material; [12]).
3. Results
(a). Verification of irs RNAi
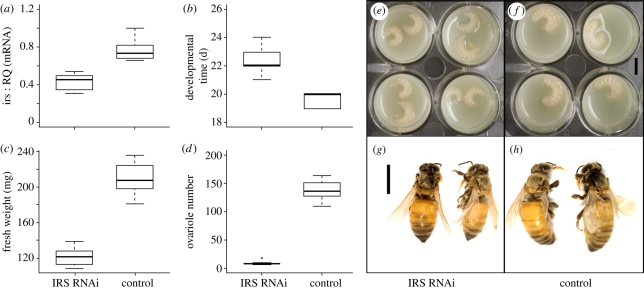
Larvae treated with irs dsRNA in queen diet showed reduced whole-body irs expression compared with control larvae-fed queen diet with gfp dsRNA (p < 7.4E − 07, n = 12, figure 1a). RNA quantification to actin or tubulin housekeeper genes gave corresponding results (see the electronic supplementary material).
Figure 1.
Physiological characters differ between irs knockdowns and control honeybees. (a) irs expression, (b) developmental time, (c) fresh weight and (d) ovariole number. Boxplots show medians and 25–75 percentiles. All traits diverged between groups (Mann Whitney U-tests, p < 7.4E − 07). (e,f) Images of representative individuals in early 5th larval instar and (g,h) as newly emerged adults visualize the phenotypic differences (scale bar, 0.5 cm).
(b). Morphological phenotyping
Compared with controls, irs knockdowns took longer to develop (p < 2.2E − 16, n = 50, figure 1b), had lower fresh weight (p < 1.5E − 11, n = 20, figure 1c), fewer ovarioles (p < 7.4E − 07, n = 12, figure 1d), and were smaller in size throughout development (figure 1e–h). As adults (n = 50), more than 80 per cent irs knockdowns exhibited a worker-specific structure on their hind leg (corbicula, used to collect pollen) and lacked the mandibular notch typical of queens. The residual individuals (less than 20%) had one intercaste character; either lack of corbicula or presence of mandibular notch. As previously noted [3], more than 50 per cent of controls had queen morphology, while the remaining individuals (less than 50%) displayed one intercaste trait (presence of corbicula or lack of mandibular notch, total n = 30). Queen versus worker caste frequencies were significantly different between groups (Chi-square = 55.0, d.f. = 1, p < 1.0E − 5).
(c). Proteotyping
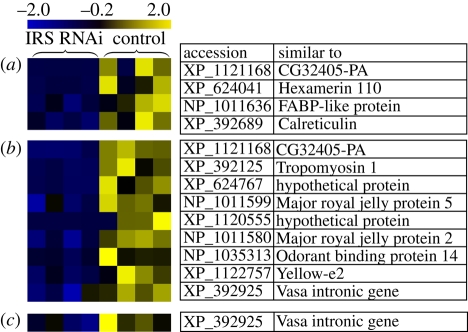
LC-MS/MS-based proteomics was conducted on larvae collected 72, 96 and 120 h after the first feeding of dsRNA. A conservative data analysis corrected for an over-abundance of royal jelly proteins in the sample material suggested diverse metabolic differences between knockdown larvae and the control group (Mann Whitney U-tests, p < 0.05, n = 4, figure 2) including hexamerin 110, a fatty-acid binding protein and the product of vasa intronic gene (VIG). All proteomics data have been deposited in NCBI's Peptidome database (http://www.ncbi.nlm.nih.gov/peptidome/repository/PSE129).
Figure 2.
Proteomic profiling reveals larval proteins affected by the irs knockdown. Proteomic analysis detected significant differences (a) 72, (b) 96 and (c) 120 h after treatments with dsRNA began (Mann Whitney U-tests, p < 0.05); visualized in a heatmap. Columns represent replicates and rows individual proteins. Values range from low (blue) to high (yellow). Only proteins with higher abundance in controls are shown to curtail type 1 error (see §4 for details).
4. Discussion
It was hypothesized that IIS plays a central role during honeybee caste development, but functional evidence for the idea was largely missing. We fill this void by establishing that honeybee queen development is blocked by RNAi-mediated repression of irs, encoding a central player in IIS. We observed longer developmental times, reduced fresh weights, smaller ovaries and presence of corbicula but absence of mandibular notch in irs knockdowns. These findings are in good agreement with characteristic differences between queen and worker bees reared in natural nests [1].
Our study complements that of Patel et al. [3], who reported the same developmental effect of reducing nutrient sensing by TOR RNAi. Taken together, these results show that honeybee queen development cannot proceed if IIS or TOR transduction is reduced.
We profiled the proteome of larvae to observe the molecular response to irs RNAi. As a conservative measure, we opted to exclude proteins with reduced abundance in controls because royal jelly proteins were more abundant in this group. Honeybee larvae are submerged in jelly protein, and differences in amounts of jelly carried over to the proteomic analysis (e.g. owing to different larval size or gut content of food), could lead to underestimation of some relative protein levels in controls. Our analysis excluded these (putative) false positives.
Hexamerin 110 was reduced in irs knockdown (figure 2). Hexamerin protein levels were previously reported to be lower in worker-destined than in queen-destined larvae of Apis mellifera and other social Hymenoptera, e.g. Polistes metricus [13,14]. Furthermore, a link between hexamerin and caste is found in termites, where hexamerin RNAi biases development toward soldiers and away from reproductives [15]. The hexamerin protein family, therefore, can be central to caste differentiation in social insects, perhaps by representing a caste-specific nutrient storage and supply [16].
Higher levels of a fatty acid binding protein (FABP-like) homologue in controls suggested that fatty acid metabolism was altered by irs RNAi. In vertebrates, FABPs may have indirect effects on IIS with lower levels conferring increased insulin sensitivity [17]. A putative connection of IIS to the metabolism of fatty acids can now be explored in honeybees.
The difference in CG32 405-PA (annotated as cuticular protein 65Acv) levels suggests diverging cuticular profiles in larvae of the two castes, which could be connected to caste recognition.
VIG (the product of the vasa introgenic gene) associates with microRNAs and the RNA-induced silencing complex (RISC) of the Drosophila RNAi machinery [18]. These features make VIG a possible master regulator of gene expression. Our finding that a VIG encoding gene homologue was differently expressed between irs knockdowns and controls supports the proposition that differential microRNA regulation plays an important role in honeybee caste differentiation [19].
In summary, we show here that honeybee queen development requires IIS transduction. Our protein profiling confirms physiological changes within hours following irs downregulation, lending support to the hypothesis that honeybee caste development relies on IRS-dependent mechanisms. Our findings bring us closer to solving the puzzle of honeybee caste development and exemplify how ancient gene networks can be co-opted during caste evolution in social insects.
Acknowledgements
This work was supported by the PEW Foundation, the Alexander-von-Humboldt Foundation, the Norwegian Research Council (180504, 185306, 191699), National Institute of Ageing (PO1AG22500), and the Binational Science Foundation (2007465). Additional acknowledgements in the electronic supplementary material.
References
- 1.Winston M. L. 1987. The biology of the honey bee. Cambridge, MA: Harvard University Press [Google Scholar]
- 2.Maleszka R. 2008. Epigenetic integration of environmental and genomic signals in honey bees: the critical interplay of nutritional, brain and reproductive networks. Epigenetics 3, 188–192 10.4161/epi.3.4.6697 (doi:10.4161/epi.3.4.6697) [DOI] [PubMed] [Google Scholar]
- 3.Patel A., Fondrk M. K., Kaftanoglu O., Emore C., Hunt G., Frederick K., Amdam G. V. 2007. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE 2, e509. 10.1371/journal.pone.0000509 (doi:10.1371/journal.pone.0000509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. 2004. Regulation of lifespan in Drosophila by modulation of genes in the tor signaling pathway. Curr. Biol. 14, 885–890 10.1016/j.cub.2004.03.059 (doi:10.1016/j.cub.2004.03.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldham S., Hafen E. 2003. Insulin/igf and target of rapamycin signaling: a TOR de force in growth control. Trends Cell. Biol. 13, 79–85 10.1016/S0962-8924(02)00042-9 (doi:10.1016/S0962-8924(02)00042-9) [DOI] [PubMed] [Google Scholar]
- 6.de Azevedo S. V., Hartfelder K. 2008. The insulin signaling pathway in honey bee (Apis mellifera) caste development—differential expression of insulin-like peptides and insulin receptors in queen and worker larvae. J. Insect Physiol. 54, 1064–1071 10.1016/j.jinsphys.2008.04.009 (doi:10.1016/j.jinsphys.2008.04.009) [DOI] [PubMed] [Google Scholar]
- 7.Wheeler D. E., Buck N., Evans J. D. 2006. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol. Biol. 15, 597–602 10.1111/j.1365-2583.2006.00681.x (doi:10.1111/j.1365-2583.2006.00681.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaburagi Y., et al. 1997. Role of insulin receptor substrate-1 and pp60 in the regulation of insulin-induced glucose transport and glut4 translocation in primary adipocytes. J. Biol. Chem. 272, 25 839–25 844 10.1074/jbc.272.41.25839 (doi:10.1074/jbc.272.41.25839) [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Mutti N. S., Ihle K. E., Siegel A., Dolezal A. G., Kaftanoglu O., Amdam G. V. 2010. Down-regulation of honey bee irs gene biases behavior toward food rich in protein. PLoS Genet. 6, e1000896. 10.1371/journal.pgen.1000896 (doi:10.1371/journal.pgen.1000896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhni R., Riesgo-Escovar J., Oldham S., Brogiolo W., Stocker H., Andruss B. F., Beckingham K., Hafen E. 1999. Autonomous control of cell and organ size by chico, a Drosophila homolog of vertebrate irs1–4. Cell 97, 865–875 10.1016/S0092-8674(00)80799-0 (doi:10.1016/S0092-8674(00)80799-0) [DOI] [PubMed] [Google Scholar]
- 11.Wolschin F., Munch D., Amdam G. V. 2009. Structural and proteomic analyses reveal regional brain differences during honeybee aging. J. Exp. Biol. 212, 4027–4032 10.1242/jeb.033845 (doi:10.1242/jeb.033845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolschin F., Amdam G. 2007. Plasticity and robustness of protein patterns during reversible development in the honey bee (Apis mellifera). Anal. Bioanal. Chem. 389, 1095–1100 10.1007/s00216-007-1523-5 (doi:10.1007/s00216-007-1523-5) [DOI] [PubMed] [Google Scholar]
- 13.Evans J. D., Wheeler D. E. 1999. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc. Natl Acad. Sci. USA 96, 5575–5580 10.1073/pnas.96.10.5575 (doi:10.1073/pnas.96.10.5575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt J. H., Kensinger B. J., Kossuth J. A., Henshaw M. T., Norberg K., Wolschin F., Amdam G. V. 2007. A diapause pathway underlies the gyne phenotype in polistes wasps, revealing an evolutionary route to caste-containing insect societies. Proc. Natl Acad. Sci. USA 104, 14 020–14 025 10.1073/pnas.0705660104 (doi:10.1073/pnas.0705660104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X., Oi F. M., Scharf M. E. 2006. Social exploitation of hexamerin: RNAi reveals a major caste-regulatory factor in termites. Proc. Natl Acad. Sci. USA 103, 4499–4504 10.1073/pnas.0508866103 (doi:10.1073/pnas.0508866103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bitondi M. M., Nascimento A. M., Cunha A. D., Guidugli K. R., Nunes F. M., Simoes Z. L. 2006. Characterization and expression of the Hex 110 gene encoding a glutamine-rich hexamerin in the honey bee, Apis mellifera. Arch. Insect Biochem. Physiol. 63, 57–72 10.1002/arch.20142 (doi:10.1002/arch.20142) [DOI] [PubMed] [Google Scholar]
- 17.Makowski L., Hotamisligil G. S. 2004. Fatty acid binding proteins—the evolutionary crossroads of inflammatory and metabolic responses. J. Nutr. 134, 2464S–2468S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caudy A. A., Myers M., Hannon G. J., Hammond S. M. 2002. Fragile x-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16, 2491–2496 10.1101/gad.1025202 (doi:10.1101/gad.1025202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weaver D. B., et al. 2007. Computational and transcriptional evidence for micrornas in the honey bee genome. Genome Biol. 8, R97. 10.1186/gb-2007-8-6-r97 (doi:10.1186/gb-2007-8-6-r97) [DOI] [PMC free article] [PubMed] [Google Scholar]