Abstract
Mitochondria produce up to 95 per cent of the eukaryotic cell's energy. The coding genes of the mitochondrial DNA may therefore evolve under selection owing to metabolic requirements. The killer whale, Orcinus orca, is polymorphic, has a global distribution and occupies a range of ecological niches. It is therefore a suitable organism for testing this hypothesis. We compared a global dataset of the complete mitochondrial genomes of 139 individuals for amino acid changes that were associated with radical physico-chemical property changes and were influenced by positive selection. Two such selected non-synonymous amino acid changes were found; one in each of two ecotypes that inhabit the Antarctic pack ice. Both substitutions were associated with changes in local polarity, increased steric constraints and α-helical tendencies that could influence overall metabolic performance, suggesting a functional change.
Keywords: mitochondria, genome, selection
1. Introduction
Mitochondrial DNA (mtDNA) sequences are used extensively to infer the evolutionary and demographic history of taxa, and many such studies assume that the mitogenome evolves under neutrality [1]. However, recent studies have questioned this assumption [2,3]. Differences in the metabolic requirements of organisms may exert varying selective pressures on the mitochondrion [4,5], which has a key role in the oxidative phosphorylation process [6]. Selection acting on the mitochondrial genome can therefore shape the pattern of mutational variation in the protein-coding sequences in response to metabolic conditions. Although evidence for purifying selection on the mitogenome has been reported [7–9], only a few empirical studies have found evidence for positive selection on the mammalian mitogenome (e.g. [5,7,10,11]), suggesting that adaptive change may be rare.
The killer whale is a useful organism for testing hypotheses of selection on the mammalian mitogenome. It is distributed from the tropics to the polar regions, and has diversified into distinct ecotypes, which differ in diet, body size and latitudinal (and therefore thermal) range [12] (electronic supplementary material). All these factors could select for changes in metabolic requirements and therefore potentially act selectively on the regions of the mitochondrial genome responsible for controlling oxidative phosphorylation [4,5]. Here, we assess the mode of evolution of the mitogenome in the killer whale by comparing a global dataset of mitogenome sequences.
2. Material and methods
We compared the sequences of the 13 protein-coding genes (ATP6, ATP8, COX1, COX2, COX3, CYTB, ND1, ND2, ND3, ND4, ND4L, ND5, ND6) from the complete mitochondrial genomes (16 386–16 392 bp) of 134 individual killer whales and partial mitogenomes (7450–16 355 bp) from an additional five individuals, constituting 67 distinct haplotypes, generated by a recent phylogeography study [12]; see Morin et al. [12] for sequencing details; GenBank accession numbers are provided in the electronic supplementary material. Mitogenome haplotypes are unique to each ecotype [12]. Ecotype was determined before genetic analyses based on morphological or behavioural data (electronic supplementary material) [12]. All eight previously described killer whale ecotypes (electronic supplementary material) and 21 individuals of an unknown ecotype, which included seven sampled from the tropics, were included in this analysis [12]. The samples were therefore taken from across the complete latitudinal range of this species.
A number of criteria for detecting positive selection have been proposed. Some methods, such as the non-synonymous to synonymous rate ratio model (dN/dS), are biased against detecting positive selection in conservative gene sequences where even single amino acid changes can be adaptive. Therefore, we applied the modified MM01 model [13] using the algorithm implemented in TreeSAAP [14]. TreeSAAP categorizes the physico-chemical changes owing to amino acid replacements into eight magnitude categories ranging from 1 to 8, with 1 being the most conservative and 8 the most radical, and then determines whether the observed magnitude of amino acid changes deviates significantly from neutral expectations. The analyses were conducted independently on each gene. Significant positive z-scores indicate that higher magnitude non-synonymous substitutions are more frequent than expected under neutrality, implying change owing to positive selection [11,13]. The phylogenetic tree from which a chronology of divergences was inferred was constructed from the mtDNA control region sequences (electronic supplementary material). The most likely model of evolution for each gene (electronic supplementary material) was selected using jModelTest 1.1 [15]. A sliding window of 15 codons was used, and 31 amino acid properties were included for the selection analysis [11,14]. We only considered amino acid replacements with magnitude categories 6–8 and significant (p < 0.01), positive z-scores as being under positive selection following McClellan et al. [11]. Sites under selection were displayed on the bovine cytochrome b structure using the software VMD [16] to aid with the qualitative assessment of their functional relevance.
Ancestral sequences were reconstructed with Datamonkey [17] for taxa and genes showing adaptive divergence as inferred by TreeSAAP, using joint and marginal maximum likelihood as well as sampled reconstruction, using the evolutionary model selected as above.
3. Results and discussion
We found 62 amino acid changes in the coding genes of the mitogenome (electronic supplementary material); however, we only identified two putatively adaptive changes, both in the cytochrome b gene: one shared by all 15 sequenced Antarctic type B individuals, and another shared by 32 out of the 36 Antarctic type C individuals sequenced. Specifically, all 15 type B individuals had a threonine substituted for alanine at site 279, and the 32 type C individuals had an alanine substituted for threonine at site 193. The z-scores for these substitutions were positive and significant (>2.326, p < 0.01) for all pairwise comparisons with the other killer whale sequences, implying positive selection on the physico-chemical amino acid properties.
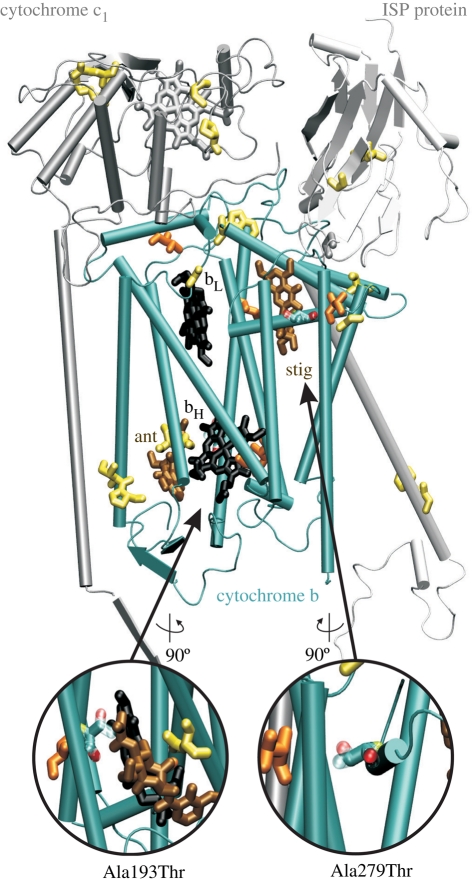
Cytochrome b catalyses the reversible electron transfer from ubiquinol to cytochrome c coupled to proton translocation against the gradient (Q-cycle). The proton gradient is used by ATP synthase to produce adenosine triphosphate (ATP) from adenosine diphosphate [6]. Both amino acid changes imply a variation in local polarity (alanine is neutral, threonine is polar) and increased steric constraints. Changes in the protein environment in the vicinity of electron transfer moieties can have an impact on the redox properties of these molecules [18–20]. We therefore expect that the substitution in site 193, which is adjacent to bH haem (figure 1), will be more significant and potentially interfere with the electron transfer that occurs at the bH haem during the Q-cycle. Although the biochemical complexity of the oxidative phosphorylation processes prevents us from predicting the exact functional implications of the substitution, we suggest that it will have an impact on the overall ATP production by the respiratory chain, and consequently on the overall metabolic performance of type C. The physico-chemical property determined by TreeSAAP to be influenced by positive selection in both changes was α-helical tendencies, and it is known that alterations in the protein conformation can also influence the redox properties of the prosthetic group [18]. McClellan et al. [11] also found α-helical tendencies to be influenced by positive selection in an interspecific comparison of cetacean and artiodactyl cytochrome b sequences.
Figure 1.
Structure of the bovine cytochrome b sequences (pdb code: 1PPJ; ISP, iron–sulphur protein) [23]. The amino acids corresponding to the non-synonymous sites in the killer whales are displayed in orange and the positively selected sites (193 and 279) are coloured by atom type (red, oxygen; blue, carbon; yellow, nitrogen; the alanine and threonine side chains are represented as solid and transparent cylinders, respectively). The prosthetic groups are represented in black, and bound inhibitors in brown (ant, antimycin; stig, stigmatellin). Exercise intolerance in humans is associated with genetic variants at the sites shown in yellow [5].
Given the conserved nature of the cytochrome b, any deleterious non-synonymous changes would be expected to be subject to purifying selection (e.g. [8]) and not to rise to fixation. Neutral changes may occur and become fixed owing to demographic influences such as a population bottleneck. As an advantageous mutation approaches fixation in a non-recombining locus such as the mitogenome, non-linked neutral variation is swept to low frequency in a population and linked variation is swept to high frequency, reducing variation [2]. Eight out of the 18 nucleotide polymorphisms found throughout the mitogenome in the 36 type C individuals were private within the four individuals without the putatively advantageous mutation (electronic supplementary material). There was no variation in 18 of the 32 type C individuals with the mutation. Further comparison with non-linked loci is needed to discriminate between the removal of variation owing to a selective sweep and demographic influences.
Based on morphological differences [21] and reciprocal monophyly of the mitogenome sequences [12], it has been suggested that type B and type C are distinct species. Positive selection on the cytochrome b could therefore be caused by adaptive divergence relating to a combination of variables that influence metabolic requirements, such as body size or diet; type C is a fish-eating dwarf form of killer whale, whereas type B is one of the largest forms of killer whale and primarily feeds upon seals [21,22] (J. W. Durban & R. L. Pitman 2010, unpublished data). However, the amino acid changes in both ecotypes could be the result of parallel evolution owing to environmental conditions such as oxygen concentration or sea temperature. Both type B and type C at least seasonally inhabit Antarctic pack ice, and both have been sighted over-wintering in the pack ice [21]. The third Antarctic ecotype, for which we found no evidence of positive selection, inhabits the offshore ice-free waters during the austral summer and over-winters at lower latitudes [21]. However, the mutations are in the opposite direction for each ecotype, suggesting that divergent evolution may be more likely. The two changes were private alleles within type B and type C, respectively, and neither substitution was found in the reconstructed ancestral sequence (electronic supplementary material), suggesting that each mutation has occurred and become fixed and almost fixed, respectively, since type B and type C diverged from their most recent common ancestor, approximately 0.15 Ma [12]. Therefore, the ancestral form may not have been subject to the same selective pressures.
References
- 1.Avise J. C. 1994. Molecular markers, natural history, and evolution. New York, NY: Chapman & Hall [Google Scholar]
- 2.Bazin E., Glemin S., Galtier N. 2006. Population size does not influence mitochondrial genetic diversity in animals. Science 312, 570–572 10.1126/science.1122033 (doi:10.1126/science.1122033) [DOI] [PubMed] [Google Scholar]
- 3.Galtier N., Nabholz B., Glemin S., Hurst G. D. D. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550 10.1111/j.1365-294X.2009.04380.x (doi:10.1111/j.1365-294X.2009.04380.x) [DOI] [PubMed] [Google Scholar]
- 4.Ballard J. W. O., Melvin R. G. 2010. Linking the mitochondrial genotype to the organismal phenotype. Mol. Ecol. 19, 1523–1539 10.1111/j.1365-294X.2010.04594.x (doi:10.1111/j.1365-294X.2010.04594.x) [DOI] [PubMed] [Google Scholar]
- 5.da Fonseca R., Johnson W., O'Brien S., Ramos M., Antunes A. 2008. The adaptive evolution of the mammalian mitochondrial genome. BMC Genomics 9, 119. 10.1186/1471-2164-9-119 (doi:10.1186/1471-2164-9-119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saraste M. 1999. Oxidative phosphorylation at the fin de siécle. Science 283, 1488–1493 10.1126/science.283.5407.1488 (doi:10.1126/science.283.5407.1488) [DOI] [PubMed] [Google Scholar]
- 7.Ruiz-Pesini E., Mishmar D., Brandon M., Procaccio V., Wallace D. C. 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303, 223–226 10.1126/science.1088434 (doi:10.1126/science.1088434) [DOI] [PubMed] [Google Scholar]
- 8.Stewart J. B., Freyer C., Elson J. L., Wredenberg A., Cansu Z., Trifunovic A., Larsson G. 2008. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 6, e10. 10.1371/journal.pbio.0060010 (doi:10.1371/journal.pbio.0060010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun C., Kong Q.-P., Zhang P. 2007. The role of climate in human mitochondrial DNA evolution: a reappraisal. Genomics 89, 338–342 10.1016/j.ygeno.2006.11.005 (doi:10.1016/j.ygeno.2006.11.005) [DOI] [PubMed] [Google Scholar]
- 10.Balloux F., Lawson Handley L.-J., Jombart T., Liu H., Manica A. 2009. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc. R. Soc. B 276, 3447–3455 10.1098/rspb.2009.0752 (doi:10.1098/rspb.2009.0752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClellan D. A., Palfreyman E. J., Smith M. J., Moss J. L., Christensen R. G., Sailsbery J. K. 2005. Physicochemical evolution and molecular adaptation of the cetacean and artiodactyl cytochrome b proteins. Mol. Biol. Evol. 22, 437–455 10.1093/molbev/msi028 (doi:10.1093/molbev/msi028) [DOI] [PubMed] [Google Scholar]
- 12.Morin P. A., et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 20, 908–916 10.1101/gr.102954.109 (doi:10.1101/gr.102954.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClellan D. A., McCracken K. G. 2001. Estimating the influence of selection on the variable amino acid sites of the cytochrome b protein functional domains. Mol. Biol. Evol. 18, 917–925 [DOI] [PubMed] [Google Scholar]
- 14.Woolley S., Johnson J., Smith M. J., Crandall K. A., McClellan D. A. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19, 671–672 10.1093/bioinformatics/btg043 (doi:10.1093/bioinformatics/btg043) [DOI] [PubMed] [Google Scholar]
- 15.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 10.1093/molbev/msn083 (doi:10.1093/molbev/msn083) [DOI] [PubMed] [Google Scholar]
- 16.Humphrey W., Dalke A., Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 10.1016/0263-7855(96)00018-5 (doi:10.1016/0263-7855(96)00018-5) [DOI] [PubMed] [Google Scholar]
- 17.Pond S. L. K., Frost S. D. W. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 10.1093/bioinformatics/bti320 (doi:10.1093/bioinformatics/bti320) [DOI] [PubMed] [Google Scholar]
- 18.Dolla A., Blanchard L., Guerlesquin F., Bruschi M. 1994. The protein moiety modulates the redox potential in cytochromes c. Biochimie 76, 471–479 10.1016/0300-9084(94)90171-6 (doi:10.1016/0300-9084(94)90171-6) [DOI] [PubMed] [Google Scholar]
- 19.Krishnan N., Becker D. F. 2005. Characterization of a bifunctional Puta homologue from Bradyrhizobium japonicum and identification of an active site residue that modulates proline reduction of the flavin adenine dinucleotide cofactor. Biochemistry 44, 9130–9139 10.1021/bi050629k (doi:10.1021/bi050629k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J.-S., Ohmura T., Katayama A., Sagara M., Niki K., Cusanovich M. A., Akutsu H. 1997. A comparison of the redox potentials of cytochrome c3 from Desulfovibrio vulgaris Hildenborough with those from Desulfovibrio vulgaris Miyazaki F effects of amino acid substitutions on the redox potentials. J. Electroanal. Chem. 438, 231–236 10.1016/S0022-0728(97)00053-3 (doi:10.1016/S0022-0728(97)00053-3) [DOI] [Google Scholar]
- 21.Pitman R. L., Ensor P. 2003. Three forms of killer whale (Orcinus orca) in Antarctic waters. J. Cetacean Res. Manage. 5, 131–139 [Google Scholar]
- 22.Pitman R. L., Perryman W. L., LeRoi D., Eilers E. 2007. A dwarf form of killer whale in Antarctica. J. Mammal. 88, 43–48 10.1644/06-MAMM-A-118R1.1 (doi:10.1644/06-MAMM-A-118R1.1) [DOI] [Google Scholar]
- 23.Huang L. S., Cobessi D., Tung E. Y., Berry E. A. 2005. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc(1) complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J. Mol. Biol. 351, 573–597 10.1016/j.jmb.2005.05.053 (doi:10.1016/j.jmb.2005.05.053) [DOI] [PMC free article] [PubMed] [Google Scholar]