Abstract
Microsatellite loci have high mutation rates and high levels of allelic variation, but the factors influencing their mutation rate are not well understood. The proposal that heterozygosity may increase mutation rates has profound implications for understanding the evolution of microsatellite loci, but currently has limited empirical support. We examined 20 microsatellite mutations identified in an analysis of 12 260 meiotic events across three loci in two populations of a songbird, the house wren (Troglodytes aedon). We found that for an allele of a given length, mutation was significantly more likely when there was a relatively large difference in size between the allele and its homologue (i.e. a large ‘allele span’). Our results support the proposal of heterozygote instability at microsatellite loci.
Keywords: microsatellite, mutation, heterozygote instability
1. Introduction
Microsatellites are short tandem repeats (STRs) with high levels of allelic variation and high mutation rates. Mutation rates in microsatellites appear to be influenced by a number of features, including repeat length and composition, as well as flanking sequences outside the microsatellite locus [1]. It has been proposed that heterozygosity increases mutation rates at microsatellite loci [2], a process termed heterozygote instability at STRs (HISTR). If true, this has several interesting implications [3]. Because heterozygosity tends to increase with population size, HISTR would link microsatellite mutation rates to effective population size and demographic history, with mutation rates rising and falling with population size. Furthermore, HISTR could create feedback loops between natural selection and mutation rates. For genetic loci where heterozygosity was tolerated by natural selection, mutation rates would increase, while at loci conserved by natural selection, mutation rates would be reduced. Thus, HISTR provides a mechanism for creating interactions among mutation, drift and natural selection, making elucidation of its significance crucial to an understanding of microsatellite evolution.
Support for HISTR is available from analysis of population data [3,4], and more directly, from pedigree data. Analysis of more than 200 mutations in humans indicated that the probability of mutation for an allele of any length was higher when its homologue was unusually different in length, i.e. when the intra-genomic ‘allele span’ was relatively large [4]. However, while this study controlled for effects of allele length on mutation rate, it made comparisons among different loci and thus could not control for other factors (such as flanking sequences), which could also influence mutation rates. In two pedigree studies involving avian microsatellites, mutation rate was unrelated to allele span [5,6]. In a third avian study, mutation rate correlated with allele span at one of two loci, but it was not possible to disentangle the effects of allele length and span, which are strongly correlated [7]. Here we test HISTR in a set of 20 mutations in three microsatellite loci in the house wren.
2. Material and methods
Pedigree data were obtained during paternity studies in populations in Illinois and Wyoming. We typed males, females and nestlings at three loci (Ltmr6, Mcyµ4 and TA-C3(B)2) as described in detail elsewhere [8,9]. Attendant males that mismatched a nestling at two or more loci were excluded from paternity. Males that mismatched a nestling at one locus were typed at up to four additional loci—TA-A5-15, TA-B4-2 [10], Pocc2 [11] and Fhu2 [12]. The probability that a randomly chosen male would match the nestling at these additional loci was <0.001. We assumed attendant females were mothers of nestlings; in more than 2500 instances, females always matched nestlings at all loci except the handful of cases where mutation provided a more parsimonious explanation than non-maternity. When an attendant, putative parent mismatched a nestling at one locus, we re-typed at that locus to rule out typing error. Mismatches that we could not explain by non-paternity, typing error or the presence of null alleles were assumed to be mutations. In each case, we assigned the mutation to the parental allele that required the smallest change to explain the observed offspring genotype.
If the heterozygote instability hypothesis is correct, then allele spans associated with mutations should be larger than would be expected by chance alone when allele length is controlled. Therefore, we sought to determine if this was the case in our set of mutations. We determined the allele span and allele length associated with each meiotic event in our study. Because allele spans vary substantially by locus, we converted raw allele span values to percentile values so that comparisons among loci would not be disproportionately skewed by the values from a single locus. We then compared the allele span associated with each mutation with the allele spans for all meiotic events in our dataset that matched that mutational event with regard to locus and allele length.
Because allele spans for a given locus and allele length were not distributed normally, we used bootstrap analysis as follows: for each mutational event, we randomly selected a meiotic event from our dataset with the same allele length at the same microsatellite locus as our mutational event. For example, in one instance a mutation occurred at the Ltmr6 locus at an allele 191 bp long, while the length of the homologous allele was 197 bp. Microsatellites in this study consist of dinucleotide repeats; therefore, a difference of 6 bp = 3 repeat units = an allele span of 3. The percentile value for an allele span of 3 for a 191 bp allele of Ltmr6 is 77, since 77 per cent of all meiotic events for alleles of that length had allele spans of less than 3 (i.e. 0, 1 or 2). The mean allele span for all meiotic events involving a 191 bp allele of Ltmr6 (n = 988) was 1.58, and the mean percentile value for these meiotic events was 38.4. In this instance, our analysis randomly selected, with replacement, a meiotic event from among all 988 meiotic events in our dataset with a parental allele length of 191 bp at the Ltmr6 locus. Similarly, for each of the 20 observed mutations, we randomly selected a meiotic event that matched the locus and allele length (but without regard to allele span) of the actual meiotic event associated with the mutation. We reiterated this process 10 000 times and determined the number of times out of 10 000 that the mean allele span in the randomly selected meiotic events was equal to or greater than the mean allele span in the actual observed mutations. In each of the two cases where it was not possible to determine in which parent the mutation arose, we used the lesser of the two allele spans for our observed mutation, making our analysis conservative.
3. Results
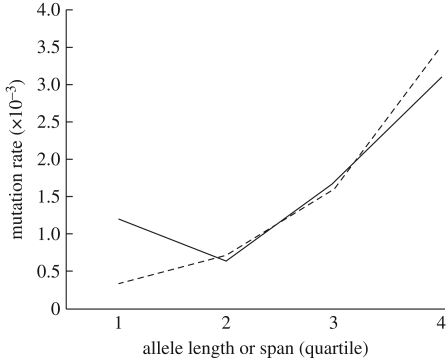
There were a total of 20 mutations in 12 260 meiotic events across three loci (table 1), for an overall mutation rate of 1.6 × 10−3. By locus, rates were 2.1 × 10−3 (8 mutations/3878 meiotic events) for Ltmr6, 2.1 × 10−3 (9/4197) for TA-C3(B)2 and 7.2 × 10−4 (3/4185) for Mcyµ4. Mutation rates did not differ significantly between sexes (11 mutations in males, seven mutations in females, two undetermined; χ2 = 1.1, p > 0.2). Downward mutations outnumbered upward mutations across all three loci, and half of all mutations involved an insertion or deletion of more than one repeat length, including eight that involved an insertion or deletion of three or more repeats. Mutation rate tended to increase as both allele length and span increased (figure 1).
Table 1.
Data for 20 mutational events at three loci in the house wren. (Shown first are male, female and nestling genotypes (specifically, allele length as measured by number of base pairs). Alleles associated with mutations appear in bold. In two cases it was not possible to identify in which parent the mutation occurred and both possible mutated alleles are in bold. Also shown is the change in number of tandem repeats associated with the mutation, the allele span of the parental genotype associated with the mutation followed by its percentile value in parentheses, the mean allele span for all meiotic events in our study that included an allele of the same length as the mutated allele followed by the mean percentile value for the allele spans for all such meiotic events in parentheses, and the number of meiotic events in our dataset for that locus and allele length.)
| locus | male genotype | female genotype | nestling genotype | mutation | allele span (percentile) | mean allele span (mean percentile) | meiotic events |
|---|---|---|---|---|---|---|---|
| TA-C3(B)2 | 204/218 | 192/218 | 216/218 | −1 | 7 (28) | 9.1 (43.7) | 153 |
| TA-C3(B)2 | 198/270 | 196/204 | 194/196 | −2 | 36 (98) | 6.3 (41.8) | 454 |
| TA-C3(B)2 | 196/200 | 218/254 | 196/200 | −9 | 18 (88) | 9.1 (43.7) | 153 |
| TA-C3(B)2 | 196/200 | 200/236 | 200/226 | −5 | 18 (38) | 16.2 (39.7) | 42 |
| TA-C3(B)2 | 200/204 | 194/208 | 194/208 | 2 | 2 (19) | 4.6 (44.0) | 260 |
| TA-C3(B)2 | 210/220 | 190/196 | 190/200 | −5 | 5 (33) | 5.4 (44.5) | 153 |
| TA-C3(B)2 | 198/200 | 198/232 | 198/226 | −3 | 17 (57) | 14.6 (41.4) | 60 |
| TA-C3(B)2 | 198/260 | 200/200 | 200/200 | 1 | 31 (96) | 6.3 (41.8) | 454 |
| TA-C3(B)2 | 198/224 | 210/232 | 198/226 | −3 | 11 (13) | 14.6 (41.4) | 60 |
| Ltmr6 | 195/199 | 191/193 | 191/193 | −1 | 2 (40) | 1.92 (38.9) | 601 |
| Ltmr6 | 193/207 | 189/197 | 205/207 | 4 | 4 (67) | 2.94 (42.7) | 400 |
| Ltmr6 | 193/197 | 187/193 | 191/193 | −1 | 2 (57) | 1.8 (38.8) | 430 |
| Ltmr6 | 189/203 | 185/189 | 185/191 | 1 | 7 (95) | 2.11 (40.6) | 753 |
| Ltmr6 | 195/201 | 189/191 | 187/191 | −4 | 3 (69) | 1.92 (38.9) | 601 |
| Ltmr6 | 191/197 | 181/189 | 189/189 | −1 | 3 (77) | 1.58 (38.4) | 988 |
| Ltmr6 | 189/203 | 181/197 | 181/197 | −3 | 7 (57) | 5.77 (38.7) | 58 |
| Ltmr6 | 173/199 | 191/195 | 195/197 | −1 | 13 (99) | 4.15 (42.4) | 69 |
| Mcyµ4 | 153/161 | 153/159 | 155/161 | 1 | 3 (34) | 3.9 (44.0) | 366 |
| Mcyµ4 | 153/157 | 151/169 | 153/167 | −1 | 9 (96) | 4.1 (42.7) | 403 |
| Mcyµ4 | 157/157 | 151/159 | 151/155 | −1 | 0 (1) | 3 (44.3) | 451 |
Figure 1.
Data for each locus were grouped into four quartiles by allele length or allele span. These data were then combined by quartile to calculate the overall mutation rate as a function of either allele length or allele span across all three loci. Dashed line, span; continuous line, length.
Out of the 10 000 sets of randomly selected meiotic events that were generated without regard to allele span, only in 76 was the mean allele span greater than or equal to the observed mean allele span for the 20 actual mutations. Therefore, if increased allele span at a given allele length had no effect on mutation rate, the probability of obtaining a mean allele span equal to or greater than the one we observed at our 20 actual mutations was 0.0076 (i.e. p ≤ 0.0076).
4. Discussion
The mutation rate that we detected is an order of magnitude lower that reported in several avian species [6,7,13,14], including at one of the same loci (Mcyµ4; [7]). However, the rate is similar to that found in humans [15] and across four loci in one bird species, the lesser kestrel (Falco naumanni; [5]). Although microsatellite mutations often involve insertion or deletion of one repeat length, larger insertions and deletions were common in the loci analysed in lesser kestrels and house wrens. Mutation types are known to vary considerably by locus [13], and by species for a given locus, possibly because multiple mechanisms cause mutation in microsatellites including replication slippage, mismatch repair and recombination [16].
Our data support the hypothesis that heterozygosity can increase the mutation rate at microsatellite loci (HISTR). In our study, mutation rate increased with either allele length or allele span. Because allele span tends to increase with allele length, the effect of the two measures is difficult to disentangle. However, our comparisons allowed us to examine allele span directly while controlling for allele length. Furthermore, unlike the only previous study to demonstrate HISTR from pedigree data [4], we directly compared meiotic events at the same loci within the same populations, so our observations cannot be explained by other potential factors influencing mutation rate, such as flanking sequences, that might arise when comparing among loci. Therefore, the analysis we describe here indicates an association between mutation and increased allele span that cannot be explained by an effect of allele length or flanking sequences.
The association between mutation and increased allele span we observed was highly significant when data from all three loci were combined. The effect seemed particularly strong for Ltmr6, and we saw a trend in the appropriate direction for TA-C3(B)2; however, Mcyµ4 showed no such trend. Because we observed only three mutations at this locus, the lack of a trend may reflect sample size. However, pedigree analysis in another species also found no association between mutation rate and allele length or allele span for Mcyµ4 [7]. It would not necessarily be surprising if HISTR varied by locus. The frequency, size and direction of mutations vary substantially among loci, and it is possible that mutational mechanisms do as well, with some favouring HISTR and others not.
Our study does not examine the association between allele length and mutation, the association of either allele span or allele length with different types of mutations, or the possibility of an interaction between allele length and allele span that affects either mutation rate or mutation type. Analyses of more extensive datasets are likely to be valuable in this regard. However, our results clearly indicate the potential for a significant role for HISTR in microsatellite mutation and evolution.
Acknowledgements
All experiments performed for this paper were approved by the Towson University Institutional Biosafety Committee. The collection of all samples used in this paper was done using methods approved by the Institutional Animal Care and Use Committees of Towson University and Illinois State University.
Our research was supported financially by NSF grants IBN-0316541 and IBN-0316580. We are grateful for the valuable insights of Joel Snodgrass, Susan Gresens, the editor and four anonymous reviewers.
References
- 1.Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 10.1038/nrg1348 (doi:10.1038/nrg1348) [DOI] [PubMed] [Google Scholar]
- 2.Amos W., Sawcer S. J., Feakes R., Rubinsztein D. C. 1996. Microsatellites show mutational bias and heterozygote instability. Nat. Genet. 13, 390–391 10.1038/ng0896-390 (doi:10.1038/ng0896-390) [DOI] [PubMed] [Google Scholar]
- 3.Amos W. 2010. Heterozygosity and mutation rate: evidence for an interaction and its implications. BioEssays 32, 82–90 10.1002/bies.200900108 (doi:10.1002/bies.200900108) [DOI] [PubMed] [Google Scholar]
- 4.Amos W., Flint J., Xu X. 2008. Heterozygosity increases microsatellite mutation rate, linking it to demographic history. BMC Genet. 9, 72. 10.1186/1471-2156-9-72 (doi:10.1186/1471-2156-9-72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortego J., Aparicio J. M., Cordero P. J., Calabuig G. 2008. Characteristics of loci and individuals are associated with germline microsatellite mutation rates in lesser kestrels (Falco naumanni). Mutation Res. 648, 82–86 [DOI] [PubMed] [Google Scholar]
- 6.Primmer C. R., Saino N., Møller A. P., Ellegren H. 1998. Unraveling the processes of microsatellite evolution through analysis of germ line mutations in barn swallows Hirundo rustica. Mol. Biol. Evol. 15, 1047–1054 [Google Scholar]
- 7.Beck N. R., Double M. C., Cockburn A. 2003. Microsatellite evolution at two hypervariable loci revealed by extensive avian pedigrees. Mol. Biol. Evol. 20, 54–61 10.1093/molbev/msg005 (doi:10.1093/molbev/msg005) [DOI] [PubMed] [Google Scholar]
- 8.Forsman A. M., Vogel L. A., Sakaluk S. K., Johnson B. G., Masters B. S., Johnson L. S., Thompson C. F. 2008. Female house wrens (Troglodytes aedon) increase the size, but not immunocompetence, of their offspring through extra-pair mating. Mol. Ecol. 17, 3697–3706 10.1111/j.1365-294X.2008.03860.x (doi:10.1111/j.1365-294X.2008.03860.x) [DOI] [PubMed] [Google Scholar]
- 9.Johnson L. S., Brubaker J. L., Johnson B. G. P., Masters B. S. 2009. Evidence for a maternal effect benefiting extra-pair offspring in a songbird, the house wren, Troglodytes aedon. J. Avian Biol. 40, 1–6 [Google Scholar]
- 10.Cabe P. R., Marshall K. E. 2001. Microsatellite loci from the house wren (Troglodytes aedon). Mol. Ecol. Notes 1, 155–156 10.1046/j.1471-8278.2001.00057.x (doi:10.1046/j.1471-8278.2001.00057.x) [DOI] [Google Scholar]
- 11.Bensch S., Price T., Kohn J. 1996. Isolation and characterization of microsatellite loci in Phylloscopus warbler. Mol. Ecol. 5, 150–151 [DOI] [PubMed] [Google Scholar]
- 12.Ellegren H. 1992. Polymerase chain reaction (PCR) analysis of microsatellites: a new approach to studies of genetic relationships in birds. Auk 109, 86–895 [Google Scholar]
- 13.Brohede J., Primmer C. R., Møller A., Ellegren H. 2002. Heterogeneity in the rate and pattern of germline mutation at individual microsatellite loci. Nucleic Acids Res. 30, 1997–2003 10.1093/nar/30.9.1997 (doi:10.1093/nar/30.9.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brohede J. C. R., Møller A., Ellegren H. 2004. Individual variation in microsatellite mutation rate in barn swallows. Mutation Res. 545, 73–80 [DOI] [PubMed] [Google Scholar]
- 15.Ellegren H. 2000. Heterogeneous mutation processes in human microsatellites. Nat. Genet. 24, 400–402 10.1038/74249 (doi:10.1038/74249) [DOI] [PubMed] [Google Scholar]
- 16.Bhargava A., Fuentes F. F. 2010. Mutational dynamics of microsatellites. Mol. Biotechnol. 44, 250–266 10.1007/s12033-009-9230-4 (doi:10.1007/s12033-009-9230-4) [DOI] [PubMed] [Google Scholar]