Abstract
Protein scaffolds coordinate the assembly of many multicomponent signaling complexes. Bodemann et al. (2011) now show that the exocyst, a protein complex involved in tethering transport vesicles to the plasma membrane, provides an assembly and activation platform for components of the autophagy machinery via a process requiring the GTPase RalB.
Macroautophagy (henceforth referred to as autophagy) is a cellular degradation pathway for the clearance of damaged or superfluous proteins and organelles (Yang and Klionsky, 2010). Despite its importance in cellular homeostasis and in immunity against pathogens, and despite accumulating knowledge regarding the autophagy machinery itself, the early regulatory signals that activate autophagy are unknown. Furthermore several gaps remain in our understanding of how distinct autophagy complexes assemble and collaborate (Mehrpour et al., 2010). Bodemann et al. (2011) now identify some of the early regulatory and assembly steps, revealing a role for the exocyst, a complex that regulates post-Golgi protein traffic. The authors demonstrate that nutrient deprivation, a condition that promotes autophagy, activates the Ras-like small GTPase, RalB, which then engages the effect or protein and exocyst component, Exo84. This interaction promotes the assembly and activation of the autophagy complex using the exocyst as an assembly scaffold.
During metabolic stress, including starvation, autophagy promotes the degradation of cytoplasmic components by the lysosome, and the recycling of their constituents promotes cell survival (Mehrpour et al., 2010; Yang and Klionsky, 2010). Autophagy involves formation of an isolation membrane, which elongates and fuses to form a double- membrane vesicle called an autophagosome. The autophagosome encloses cytoplasmic cargoes for delivery by fusion to the endosome or lysosome, eventually forming an autolysosome.
Autophagosome formation consists of three steps: nucleation, expansion, and fusion of the isolation membrane. Each step involves a specific set of protein complexes. The ULK (Unc-51 like kinase) and PI3K (phosphatidylinositol-3 kinase) complexes are most important for nucleation, whereas the ubiquitin-like (Ubl) conjugation system and the mAtg9 (mammalian autophagy-related gene 9) cycling complex, which is involved in transit of mAtg9 to and from the isolation membrane, facilitate expansion and closure of the isolation membrane (Mehrpour et al., 2010).
The exocyst, a hetero-octameric complex containing the proteins Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84 (recently renamed EXOC1-EXOC8), is involved in the post-Golgi trafficking and tethering of vesicles to the plasma membrane (He and Guo, 2009; Munson and Novick, 2006). New evidence implicating a role for the exocyst complex in signaling during pathogen infection (Chien et al., 2006), led the authors to screen for proteins interacting with the exocyst subunit, Sec3. Using a high-throughput yeast two-hybrid screen, the authors find that both negative and positive regulators of autophagy interact with Sec3. The interactors include Rubicon (RUN domain and cysteine-rich domain containing), an inhibitor of autophagy, as well as Atg14L, a component of the PI3K complex, and FIP200, part of the ULK complex. In fact, the authors find that several exocyst subunits (Sec3, Sec5 and Exo84) co-immunoprecipitate with Rubicon and Atg14L. Additionally, the core exocyst subunit, Sec8, associates with Atg5 and Atg12 autophagy proteins involved in the ubiquitin-like system, cementing the association between the autophagy machinery and the exocyst.
Given the association between autophagy and exocyst components, and the fact that the small GTPases RalA and RalB mobilize exocyst assembly (Moskalenko et al., 2002; Moskalenko et al., 2003), the authors next inquire whether RalA and RalB also play a role in autophagy. Indeed, Bodemann et al. show convincingly that activation of RalB, but not RalA, in cervical cancer and epithelial cell lines, is necessary for autophagy. They find that competitive inhibitors of RalB inhibit the induction of autophagy during starvation, whereas constitutively activated forms of RalB stimulate autophagy even under nutrientrich conditions. Thus RalB is both necessary and sufficient for activation of autophagy.
RalB, and its related partner, RalA, cooperate in mitogen-induced signaling during oncogenic transformation by Ras. RalA is required to bypass normal restraints on cell proliferation, while RalB bypasses normal restraints on cell survival (Chien et al., 2006). Tumor cells have higher levels of RalB and cells depleted of RalB exhibit survival defects (Bodemann and White, 2008). These observations may be explained, in part, by the finding that RalB promotes cell survival during starvation by inducing autophagy.
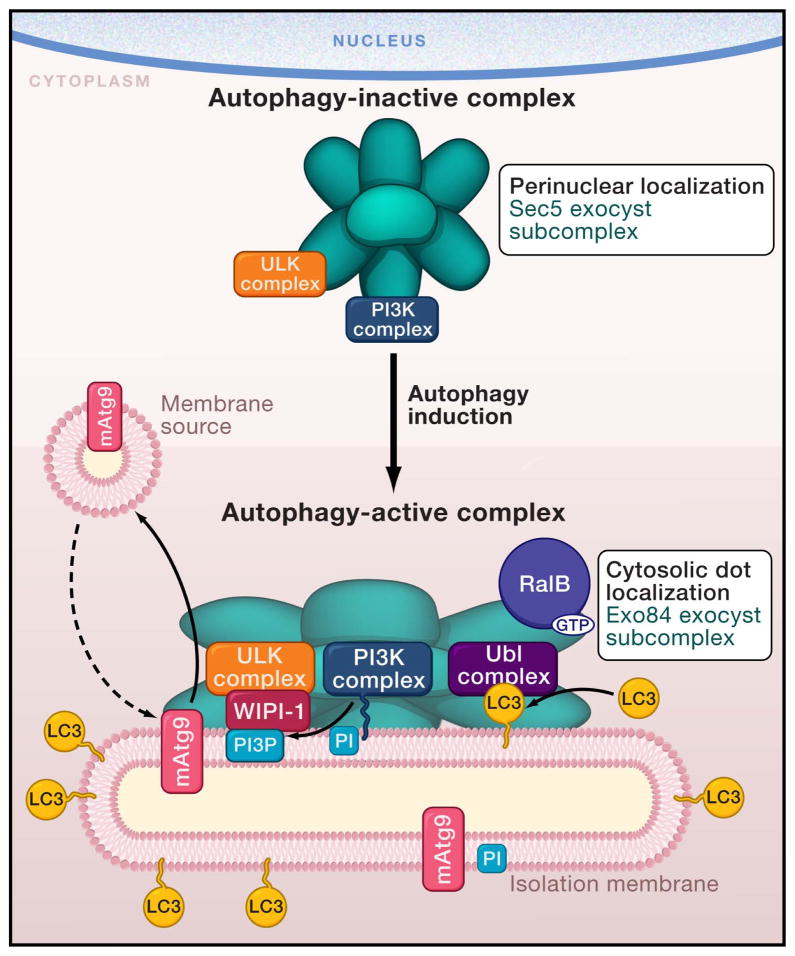
Interestingly, the authors characterize two complexes containing both exocyst and autophagy components: an autophagy-active and autophagy-inactive complex. The RalB-Exo84 complex contains active ULK and PI3K complexes, suggesting that this complex is active during starvation-induced autophagy. In contrast, upon inhibition of RalB signaling, Rubicon, an inhibitor of autophagy, associates with Exo84. The Sec5-ULK-PI3K complex correlates with the inactive autophagy state and is more abundant under nutrient-rich conditions. These observations lead the authors to propose a model for the activation of autophagy (Figure 1). They suggest that assembly of the ULK and PI3K complexes on Exo84 triggers autophagy by generating an autophagy-active complex. In contrast, interaction between these complexes and Sec5 creates an autophagy-inactive complex that is either a pre-initiation complex unable to trigger autophagy, or is a signal termination complex for the process. Consistent with this model, endogenous mTORC1 (mammalian target of rapamycin complex 1), which inhibits autophagy through inactivation of the ULK complex (Mehrpour et al., 2010), is present only in the autophagy-inactive complex.
Figure 1.
A model for exocyst function in autophagy. Bodemann et al. (2011) provide evidence that the exocyst, a protein complex involved in post-Golgi protein traffic, may function as a scaffold for the assembly of autophagy complexes. The authors suggest the following model for activation of autophagy. Under nutrient-rich conditions, an exocyst subcomplex containing the Sec5 protein associates with the ULK (Unc-51 like kinase) and phosphatidylinositol-3 kinase (PI3K) complexes at the perinuclear region forming an autophagy-inactive complex. Induction of autophagy (e.g. in response to starvation) leads to the activation of the Ras-like small GTPase, RalB. The activated RalB interacts with the exocyst, promoting the replacement of Sec5 by another exocyst component, Exo84, and formation of an active autophagy complex that includes the ubiquitin-like (Ubl) conjugation system and the ubiquitin-like molecule LC3. This autophagy-active complex localizes in cytosolic dots that could correspond to the isolation membrane. The Exo84 exocyst subcomplex may bring together complexes of the core autophagic machinery or facilitate their concerted action. The exact subunit compositions of the autophagy-inactive and autophagy-active exocyst subcomplexes remain unknown. PI3P: phosphatidylinositol 3- phosphate, PI: phosphatidylinositol, WIPI-1 is a WD40 repeat autophagy protein that interacts with phosphoinositides such as PI3P.
The cellular localization of complex components under different conditions suggests that the transition from an autophagy-inactive to an autophagy-active complex may involve a change in localization. Bodemann et al. find that under nutrient-rich conditions RalB associates with an exocyst subcomplex containing Sec5, and components of the PI3K autophagy complex colocalize with RalB in perinuclear regions of the cell. Upon starvation and activation of RalB, Exo84 replaces Sec5. Under these conditions (starvation), the authors observe the Exo84 exocyst subcomplex at cytosolic dots, along with the PI3K autophagy complex, components of the Ubl complex and phosphatidylinositol 3-phosphate (PI3P, the product of an active PI3K complex). These results suggest that activation of RalB and recruitment of Exo84 may trigger translocation of the autophagy complex to sites of autophagosome membrane formation.
Epistasis analysis also supports an early role for RalB in triggering autophagy upstream of the initial nucleation step mediated by the ULK complex. Furthermore depletion of several components of the active exocyst complex (Sec3, Sec8, Exo70, Exo84, but not Sec5) suppresses both general autophagy and the selective autophagic degradation of bacteria. Together the data are consistent with the idea that the exocyst functions as a scaffold for the core autophagy machinery in mammalian cells.
Earlier work showed that RalB competes with phosphatidylinositol 3,4,5- trisphosphate (PIP3) for binding to the pleckstrin homology (PH) domain of Exo84 (Moskalenko et al., 2003). It is therefore exciting to hypothesize that the binding of RalB to Exo84 may trigger the movement of the complex from a PIP3-enriched environment, such as the plasma membrane or recycling endosomes, to the autophagosome assembly site.
The precise mechanism linking the activities of the ULK and PI3K complexes to the elongation and completion of the isolation membrane by the Ubl and mAtg9 cycling complexes has been unclear (Mehrpour et al., 2010). The direct interaction of these complexes with the exocyst complex may provide this missing link. The work of Bodemann and colleagues shows that all these complexes assemble on a common scaffold, a recurring theme exploited by other signaling systems (Shaw and Filbert, 2009), suggesting that the exocyst may coordinate molecular events in autophagy.
The primary known function of the exocyst is the targeting and tethering of post- Golgi vesicles to plasma membrane domains. By analogy, the exocyst may also provide a 7 targeting site for the autophagy machinery, perhaps tethering this machinery to the isolation membrane for autophagosome expansion.
Acknowledgments
Our autophagy expertise is made possible by NIH grant GM069373 to SS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou Y-H, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. Cell. 2011 doi: 10.1016/j.cell.2010.12.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodemann BO, White MA. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M, Jr, et al. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrpour M, Esclatine A, Beau I, Codogno P. Cell Res. 2010;20:748–762. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Henry DO, Rosse C, Mirey G, Camonis JH, White MA. Nat Cell Biol. 2002;4:66–72. doi: 10.1038/ncb728. [DOI] [PubMed] [Google Scholar]
- Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, Camonis J, White MA. J Biol Chem. 2003;278:51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- Munson M, Novick P. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- Shaw AS, Filbert EL. Nat Rev Immunol. 2009;9:47–56. doi: 10.1038/nri2473. [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]